Modulation of the Tissue Expression Pattern of Zebrafish CRP-Like Molecules Suggests a Relevant Antiviral Role in Fish Skin
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Lines and Virus
2.2. Animals
2.3. Ethical Statement of Zebrafish Handling
2.4. In Vivo Viral Infection
2.5. RNA Isolation, cDNA Synthesis, and qPCR
2.6. Statistical Analysis and Graphics
3. Results and Discussion
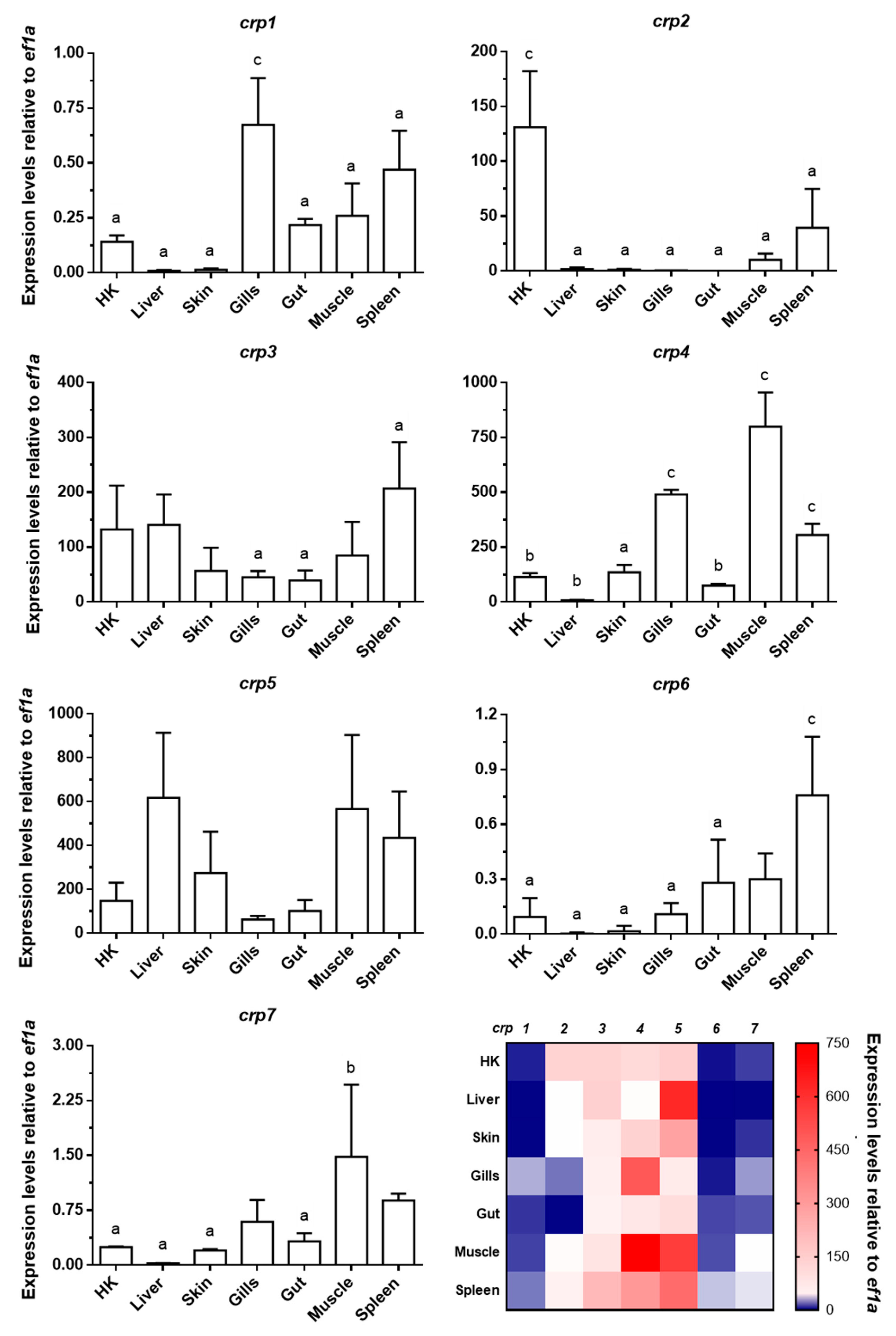
3.1. Broad Tissue Distribution of Zebrafish crp1–7 Expression
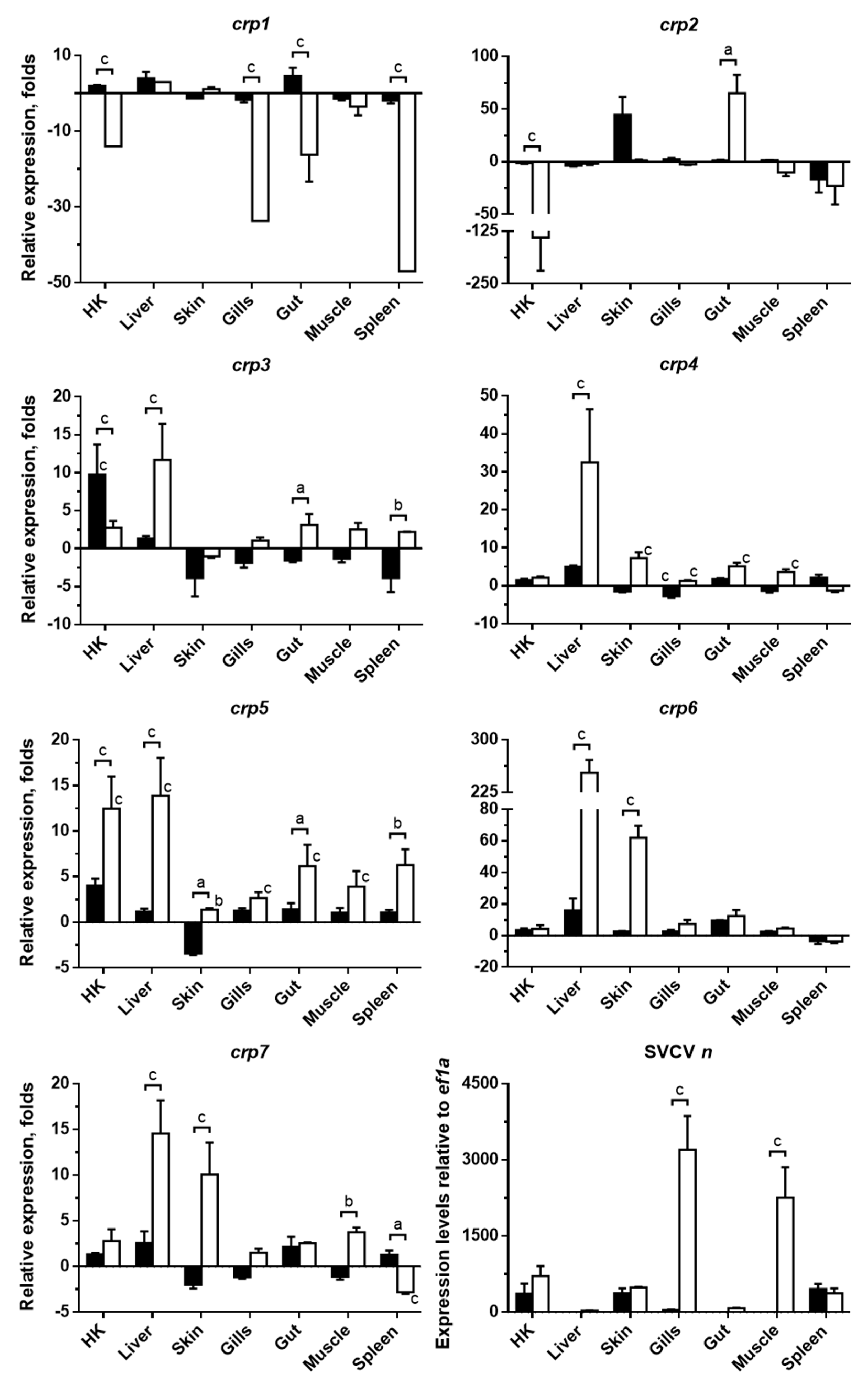
3.2. Systemic Modulation of Zebrafish crp1–7 Expression Levels in Response to SVCV Infection
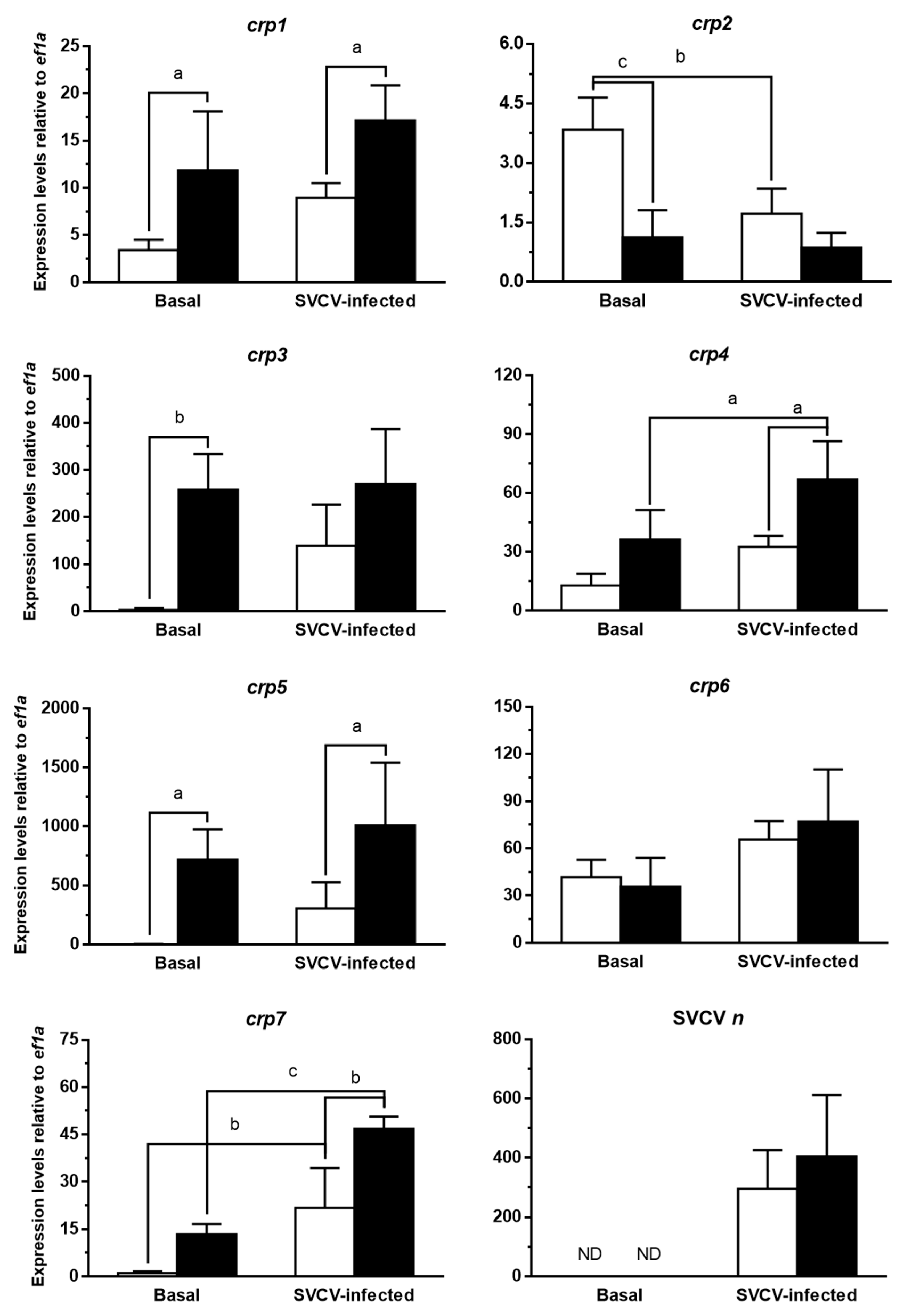
3.3. CRPs Are Constitutively Overexpressed in Skin in the Absence of Adaptive Immunity
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Pepys, M.B.; Hirschfield, G.M. C-reactive protein: A critical update. J. Clin. Investig. 2003, 111, 1805–1812. [Google Scholar] [CrossRef] [PubMed]
- Garlanda, C.; Bottazzi, B.; Bastone, A.; Mantovani, A. Pentraxins at the crossroads between innate immunity, inflammation, matrix deposition, and female fertility. Annu. Rev. Immunol. 2005, 23, 337–366. [Google Scholar] [CrossRef] [PubMed]
- Armstrong, P.B. Comparative biology of the pentraxin protein family: Evolutionarily conserved component of innate immune system. Int. Rev. Cell Mol. Biol. 2015, 316, 1–47. [Google Scholar] [PubMed]
- Pathak, A.; Agrawal, A. Evolution of c-reactive protein. Front. Immunol. 2019, 10. [Google Scholar] [CrossRef]
- Shrive, A.K.; Cheetham, G.M.; Holden, D.; Myles, D.A.; Turnell, W.G.; Volanakis, J.E.; Pepys, M.B.; Bloomer, A.C.; Greenhough, T.J. Three dimensional structure of human c-reactive protein. Nat. Struct. Biol. 1996, 3, 346–354. [Google Scholar] [CrossRef]
- Nguyen, N.Y.; Suzuki, A.; Boykins, R.A.; Liu, T.Y. The amino acid sequence of limulus c-reactive protein. Evidence of polymorphism. J. Biol. Chem. 1986, 261, 10456–10465. [Google Scholar] [CrossRef]
- Tharia, H.A.; Shrive, A.K.; Mills, J.D.; Arme, C.; Williams, G.T.; Greenhough, T.J. Complete cdna sequence of sap-like pentraxin from limulus polyphemus: Implications for pentraxin evolution. J. Mol. Biol. 2002, 316, 583–597. [Google Scholar] [CrossRef]
- Introna, M.; Breviario, F.; d’Aniello, E.M.; Golay, J.; Dejana, E.; Mantovani, A. Il-1 inducible genes in human umbilical vein endothelial cells. Eur. Heart J. 1993, 14 (Suppl. K), 78–81. [Google Scholar]
- Pepys, M.B.; Baltz, M.L. Acute phase proteins with special reference to c-reactive protein and related proteins (pentaxins) and serum amyloid a protein. Adv. Immunol. 1983, 34, 141–212. [Google Scholar]
- Amatayakul-Chantler, S.; Dwek, R.A.; Tennent, G.A.; Pepys, M.B.; Rademacher, T.W. Molecular characterization of limulus polyphemus c-reactive protein. Ii. Asparagine-linked oligosaccharides. Eur. J. Biochem. 1993, 214, 99–110. [Google Scholar] [CrossRef]
- Pepys, M.B.; Baltz, M.; Gomer, K.; Davies, A.J.; Doenhoff, M. Serum amyloid p-component is an acute-phase reactant in the mouse. Nature 1979, 278, 259–261. [Google Scholar] [CrossRef] [PubMed]
- Schwalbe, R.A.; Dahlback, B.; Coe, J.E.; Nelsestuen, G.L. Pentraxin family of proteins interact specifically with phosphorylcholine and/or phosphorylethanolamine. Biochemistry 1992, 31, 4907–4915. [Google Scholar] [CrossRef] [PubMed]
- Coe, J.E.; Ross, M.J. Hamster female protein, a sex-limited pentraxin, is a constituent of syrian hamster amyloid. J. Clin. Investig. 1985, 76, 66–74. [Google Scholar] [CrossRef] [PubMed]
- Srinivasan, N.; White, H.E.; Emsley, J.; Wood, S.P.; Pepys, M.B.; Blundell, T.L. Comparative analyses of pentraxins: Implications for protomer assembly and ligand binding. Structure 1994, 2, 1017–1027. [Google Scholar] [CrossRef]
- Bello-Perez, M.; Falco, A.; Medina, R.; Encinar, J.A.; Novoa, B.; Perez, L.; Estepa, A.; Coll, J. Structure and functionalities of the human c-reactive protein compared to the zebrafish multigene family of c-reactive-like proteins. Dev. Comp. Immunol. 2017, 69, 33–40. [Google Scholar] [CrossRef]
- Shrive, A.K.; Metcalfe, A.M.; Cartwright, J.R.; Greenhough, T.J. C-reactive protein and sap-like pentraxin are both present in limulus polyphemus haemolymph: Crystal structure of limulus sap. J. Mol. Biol. 1999, 290, 997–1008. [Google Scholar] [CrossRef]
- Chen, R.; Qi, J.; Yuan, H.; Wu, Y.; Hu, W.; Xia, C. Crystal structures for short-chain pentraxin from zebrafish demonstrate a cyclic trimer with new recognition and effector faces. J. Struct. Biol. 2015, 189, 259–268. [Google Scholar] [CrossRef]
- Cray, C. Acute phase proteins in animals. Prog. Mol. Biol. Transl. Sci. 2012, 105, 113–150. [Google Scholar]
- Ansar, W.; Ghosh, S. C-reactive protein and the biology of disease. Immunol. Res. 2013, 56, 131–142. [Google Scholar] [CrossRef]
- Rubio, N.; Sharp, P.M.; Rits, M.; Zahedi, K.; Whitehead, A.S. Structure, expression, and evolution of guinea pig serum amyloid p component and c-reactive protein. J. Biochem. 1993, 113, 277–284. [Google Scholar] [CrossRef]
- MacCarthy, E.M.; Burns, I.; Irnazarow, I.; Polwart, A.; Greenhough, T.J.; Shrive, A.K.; Hoole, D. Serum crp-like protein profile in common carp cyprinus carpio challenged with aeromonas hydrophila and escherichia coli lipopolysaccharide. Dev. Comp. Immunol. 2008, 32, 1281–1289. [Google Scholar] [CrossRef] [PubMed]
- Pionnier, N.; Falco, A.; Miest, J.J.; Shrive, A.K.; Hoole, D. Feeding common carp cyprinus carpio with beta-glucan supplemented diet stimulates c-reactive protein and complement immune acute phase responses following pamps injection. Fish Shellfish Immunol. 2014, 39, 285–295. [Google Scholar] [CrossRef] [PubMed]
- Bello-Perez, M.; Pereiro, P.; Coll, J.; Novoa, B.; Perez, L.; Falco, A. Zebrafish c-reactive protein isoforms inhibit svcv replication by blocking autophagy through interactions with cell membrane cholesterol. Sci. Rep. 2020, 10, 1–18. [Google Scholar] [CrossRef]
- Bello-Perez, M.; Falco, A.; Novoa, B.; Perez, L.; Coll, J. Hydroxycholesterol binds and enhances the anti-viral activities of zebrafish monomeric c-reactive protein isoforms. PLoS ONE 2019, 14, e0201509. [Google Scholar] [CrossRef]
- Bello-Perez, M.; Falco, A.; Medina-Gali, R.; Pereiro, P.; Encinar, J.A.; Novoa, B.; Perez, L.; Coll, J. Neutralization of viral infectivity by zebrafish c-reactive protein isoforms. Mol. Immunol. 2017, 91, 145–155. [Google Scholar] [CrossRef] [PubMed]
- Westerfield, M. The Zebrafish Book: A Guide for the Laboratory Use of Zebrafish (Brachydanio rerio); University of Oregon Press: Eugene, OR, USA, 1995. [Google Scholar]
- MacRae, C.A.; Peterson, R.T. Zebrafish as tools for drug discovery. Nat. Rev. Drug Discov. 2015, 14, 721–731. [Google Scholar] [CrossRef] [PubMed]
- Valenzuela-Muñoz, V.; Pereiro, P.; Álvarez-Rodríguez, M.; Gallardo-Escárate, C.; Figueras, A.; Novoa, B. Comparative modulation of lncrnas in wild-type and rag1-heterozygous mutant zebrafish exposed to immune challenge with spring viraemia of carp virus (svcv). Sci. Rep. 2019, 9, 1–13. [Google Scholar] [CrossRef]
- Petrie-Hanson, L.; Hohn, C.; Hanson, L. Characterization of rag 1 mutant zebrafish leukocytes. BMC Immunol. 2009, 10, 1–8. [Google Scholar] [CrossRef]
- Hohn, C.; Petrie-Hanson, L. Rag1−/− mutant zebrafish demonstrate specific protection following bacterial re-exposure. PLoS ONE 2012, 7, e44451. [Google Scholar] [CrossRef]
- Espin-Palazon, R.; Martinez-Lopez, A.; Roca, F.J.; Lopez-Munoz, A.; Tyrkalska, S.D.; Candel, S.; Garcia-Moreno, D.; Falco, A.; Meseguer, J.; Estepa, A. Tnfα impairs rhabdoviral clearance by inhibiting the host autophagic antiviral response. PLoS Pathog. 2016, 12, e1005699. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative pcr and the 2(-delta delta c(t)) method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Lee, P.; Bird, S.; Zou, J.; Martin, S. Phylogeny and expression analysis of c-reactive protein (crp) and serum amyloid-p (sap) like genes reveal two distinct groups in fish. Fish Shellfish Immunol. 2017, 65, 42–51. [Google Scholar] [CrossRef] [PubMed]
- Kovacevic, N.; Hagen, M.O.; Xie, J.; Belosevic, M. The analysis of the acute phase response during the course of trypanosoma carassii infection in the goldfish (Carassius auratus L.). Dev. Comp. Immunol. 2015, 53, 112–122. [Google Scholar] [CrossRef]
- Falco, A.; Cartwright, J.R.; Wiegertjes, G.F.; Hoole, D. Molecular characterization and expression analysis of two new c-reactive protein genes from common carp (Cyprinus carpio). Dev. Comp. Immunol. 2012, 37, 127–138. [Google Scholar] [CrossRef] [PubMed]
- Choi, K.-M.; Shim, S.H.; An, C.M.; Nam, B.-H.; Jeong, J.-M.; Kim, J.-W.; Park, C.-I. Functional characterisation and expression analysis of recombinant serum amyloid p isoform 1 (rbsap1) from rock bream (Oplegnathus fasciatus). Fish Shellfish Immunol. 2015, 45, 277–285. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.-H.; Chen, K.; Ma, W.-J.; Chen, J. Ayu c-reactive protein/serum amyloid p agglutinates bacteria and inhibits complement-mediated opsonophagocytosis by monocytes/macrophages. Fish Shellfish Immunol. 2018, 76, 58–67. [Google Scholar] [CrossRef]
- Elvitigala, D.A.S.; Wan, Q.; Kim, H.C.; Lee, J. Identification of a c-reactive protein like homologue from black rockfish (Sebastes schlegelii) evidencing its potent anti-microbial properties at molecular level. Dev. Comp. Immunol. 2015, 53, 169–178. [Google Scholar] [CrossRef]
- Li, M.-f.; Chen, C.; Li, J.; Sun, L. The c-reactive protein of tongue sole cynoglossus semilaevis is an acute phase protein that interacts with bacterial pathogens and stimulates the antibacterial activity of peripheral blood leukocytes. Fish Shellfish Immunol. 2013, 34, 623–631. [Google Scholar] [CrossRef]
- Wang, T.; Sun, L. Cssap, a teleost serum amyloid p component, interacts with bacteria, promotes phagocytosis, and enhances host resistance against bacterial and viral infection. Dev. Comp. Immunol. 2016, 55, 12–20. [Google Scholar] [CrossRef]
- Pietrzak, E.; Mazurkiewicz, J.; Slawinska, A. Innate immune responses of skin mucosa in common carp (Cyprinus carpio) fed a diet supplemented with galactooligosaccharides. Animals 2020, 10, 438. [Google Scholar] [CrossRef]
- Magnadóttir, B.; Hayes, P.; Gísladóttir, B.; Bragason, B.Þ.; Hristova, M.; Nicholas, A.P.; Guðmundsdóttir, S.; Lange, S. Pentraxins crp-i and crp-ii are post-translationally deiminated and differ in tissue specificity in cod (Gadus morhua L.) ontogeny. Dev. Comp. Immunol. 2018, 87, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Tsutsui, S.; Yamaguchi, M.; Hirasawa, A.; Nakamura, O.; Watanabe, T. Common skate (raja kenojei) secretes pentraxin into the cutaneous secretion: The first skin mucus lectin in cartilaginous fish. J. Biochem. 2009, 146, 295–306. [Google Scholar] [CrossRef] [PubMed]
- Patel, D.M.; Brinchmann, M.F. Skin mucus proteins of lumpsucker (Cyclopterus lumpus). Biochem. Biophys. Rep. 2017, 9, 217–225. [Google Scholar] [CrossRef] [PubMed]
- Ramos, F.; Smith, A.C. The c-reactive protein (crp) test for the detection of early disease in fishes. Aquaculture 1978, 14, 261–266. [Google Scholar] [CrossRef]
- Shao, L.; Xiao, Y.; He, Z.; Gao, L. An n-targeting real-time pcr strategy for the accurate detection of spring viremia of carp virus. J. Virol. Methods 2016, 229, 27–34. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Kuang, M.; Lu, Y.; Lin, L.; Liu, X. Characterization and biological function analysis of the trim47 gene from common carp (Cyprinus carpio). Gene 2017, 627, 188–193. [Google Scholar] [CrossRef] [PubMed]
- Uribe, C.; Folch, H.; Enríquez, R.; Moran, G. Innate and adaptive immunity in teleost fish: A review. Vet. Med. 2011, 56, 486–503. [Google Scholar] [CrossRef]
- Encinas, P.; Garcia-Valtanen, P.; Chinchilla, B.; Gomez-Casado, E.; Estepa, A.; Coll, J. Identification of multipath genes differentially expressed in pathway-targeted microarrays in zebrafish infected and surviving spring viremia carp virus (svcv) suggest preventive drug candidates. PLoS ONE 2013, 8, e73553. [Google Scholar] [CrossRef]
- Pionnier, N.; Adamek, M.; Miest, J.J.; Harris, S.J.; Matras, M.; Rakus, K.Ł.; Irnazarow, I.; Hoole, D. C-reactive protein and complement as acute phase reactants in common carp cyprinus carpio during cyhv-3 infection. Dis. Aquat. Org. 2014, 109, 187–199. [Google Scholar] [CrossRef]
- Gruys, E.; Toussaint, M.; Niewold, T.; Koopmans, S. Acute phase reaction and acute phase proteins. J. Zhejiang Univ. Sci. B 2005, 6, 1045. [Google Scholar] [CrossRef]
- Mantovani, A.; Garlanda, C.; Doni, A.; Bottazzi, B. Pentraxins in innate immunity: From c-reactive protein to the long pentraxin ptx3. J. Clin. Immunol. 2008, 28, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Kushner, I.; Jiang, S.-L.; Zhang, D.; Lozanski, G.; Samols, D. Do post-transcriptional mechanisms participate in induction of c-reactive protein and serum amyloid a by il-6 and il-1? Ann. N. Y. Acad. Sci. 1995, 762, 102–107. [Google Scholar] [CrossRef] [PubMed]
- Paludan, S.R. Requirements for the induction of interleukin-6 by herpes simplex virus-infected leukocytes. J. Virol. 2001, 75, 8008–8015. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Wang, Q.; Han, T.; Li, Y.-K.; Zhu, S.-L.; Ao, F.; Feng, J.; Jing, M.-Z.; Wang, L.; Ye, L.-B. Soluble interleukin-6 receptor is elevated during influenza a virus infection and mediates the il-6 and il-32 inflammatory cytokine burst. Cell. Mol. Immunol. 2015, 12, 633–644. [Google Scholar] [CrossRef] [PubMed]
- Xia, C.; Liu, Y.; Chen, Z.; Zheng, M. Involvement of interleukin 6 in hepatitis b viral infection. Cell. Physiol. Biochem. 2015, 37, 677–686. [Google Scholar] [CrossRef] [PubMed]
- Øvergård, A.-C.; Nerland, A.H.; Fiksdal, I.U.; Patel, S. Atlantic halibut experimentally infected with nodavirus shows increased levels of t-cell marker and ifnγ transcripts. Dev. Comp. Immunol. 2012, 37, 139–150. [Google Scholar] [CrossRef] [PubMed]
- Wei, X.; Li, X.Z.; Zheng, X.; Jia, P.; Wang, J.; Yang, X.; Yu, L.; Shi, X.; Tong, G.; Liu, H. Toll-like receptors and interferon associated immune factors responses to spring viraemia of carp virus infection in common carp (Cyprinus carpio). Fish Shellfish Immunol. 2016, 55, 568–576. [Google Scholar] [CrossRef]
- Black, S.; Kushner, I.; Samols, D. C-reactive protein. J. Biol. Chem. 2004, 279, 48487–48490. [Google Scholar] [CrossRef]
- Lazado, C.C.; Caipang, C.M.A. Mucosal immunity and probiotics in fish. Fish Shellfish Immunol. 2014, 39, 78–89. [Google Scholar] [CrossRef]
- Harmache, A.; LeBerre, M.; Droineau, S.; Giovannini, M.; Brémont, M. Bioluminescence imaging of live infected salmonids reveals that the fin bases are the major portal of entry for novirhabdovirus. J. Virol. 2006, 80, 3655–3659. [Google Scholar] [CrossRef]
- Bearzotti, M.; Delmas, B.; Lamoureux, A.; Loustau, A.-M.; Chilmonczyk, S.; Bremont, M. Fish rhabdovirus cell entry is mediated by fibronectin. J. Virol. 1999, 73, 7703–7709. [Google Scholar] [CrossRef] [PubMed]
- Esteban, M.Á.; Cerezuela, R. Fish mucosal immunity: Skin. In Mucosal Health in Aquaculture; Elsevier: Amsterdam, The Netherlands, 2015; pp. 67–92. [Google Scholar]
- Salinas, I. The mucosal immune system of teleost fish. Biology 2015, 4, 525–539. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.; Takizawa, F.; Casadei, E.; Shibasaki, Y.; Ding, Y.; Sauters, T.J.; Yu, Y.; Salinas, I.; Sunyer, J.O. Specialization of mucosal immunoglobulins in pathogen control and microbiota homeostasis occurred early in vertebrate evolution. Sci. Immunol. 2020, 5. [Google Scholar] [CrossRef] [PubMed]
- García-Valtanen, P.; Martínez-López, A.; López-Muñoz, A.; Bello-Perez, M.; Medina-Gali, R.M.; Ortega-Villaizán, M.d.M.; Varela, M.; Figueras, A.; Mulero, V.; Novoa, B. Zebra fish lacking adaptive immunity acquire an antiviral alert state characterized by upregulated gene expression of apoptosis, multigene families, and interferon-related genes. Front. Immunol. 2017, 8, 121. [Google Scholar] [PubMed]
- Agrawal, A.; Singh, P.P.; Bottazzi, B.; Garlanda, C.; Mantovani, A. Pattern recognition by pentraxins. In Target Pattern Recognition in Innate Immunity; Springer: Berlin/Heidelberg, Germany, 2009; pp. 98–116. [Google Scholar]
- Chorny, A.; Casas-Recasens, S.; Sintes, J.; Shan, M.; Polentarutti, N.; García-Escudero, R.; Walland, A.C.; Yeiser, J.R.; Cassis, L.; Carrillo, J. The soluble pattern recognition receptor ptx3 links humoral innate and adaptive immune responses by helping marginal zone b cells. J. Exp. Med. 2016, 213, 2167–2185. [Google Scholar] [CrossRef]
- Bottazzi, B.; Doni, A.; Garlanda, C.; Mantovani, A. An integrated view of humoral innate immunity: Pentraxins as a paradigm. Annu. Rev. Immunol. 2009, 28, 157–183. [Google Scholar] [CrossRef]
- Novoa, B.; Pereiro, P.; López-Muñoz, A.; Varela, M.; Forn-Cuní, G.; Anchelin, M.; Dios, S.; Romero, A.; Martinez-López, A.; Medina-Gali, R.M. Rag1 immunodeficiency-induced early aging and senescence in zebrafish are dependent on chronic inflammation and oxidative stress. Aging Cell 2019, 18, e13020. [Google Scholar] [CrossRef]
- Netea, M.G.; Joosten, L.A. Master and commander: Epigenetic regulation of macrophages. Cell Res. 2016, 26, 145–146. [Google Scholar] [CrossRef]
- Quintin, J.; Saeed, S.; Martens, J.H.; Giamarellos-Bourboulis, E.J.; Ifrim, D.C.; Logie, C.; Jacobs, L.; Jansen, T.; Kullberg, B.-J.; Wijmenga, C. Candida albicans infection affords protection against reinfection via functional reprogramming of monocytes. Cell Host Microbe 2012, 12, 223–232. [Google Scholar] [CrossRef]
- Medina-Gali, R.; Bello-Perez, M.; Martinez-Lopez, A.; Falco, A.; Ortega-Villaizan, M.M.; Encinar, J.A.; Novoa, B.; Coll, J.; Perez, L. Chromatin immunoprecipitation and high throughput sequencing of svcv-infected zebrafish reveals novel epigenetic histone methylation patterns involved in antiviral immune response. Fish. Shellfish Immunol. 2018, 82, 514–521. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bello-Perez, M.; Adamek, M.; Coll, J.; Figueras, A.; Novoa, B.; Falco, A. Modulation of the Tissue Expression Pattern of Zebrafish CRP-Like Molecules Suggests a Relevant Antiviral Role in Fish Skin. Biology 2021, 10, 78. https://doi.org/10.3390/biology10020078
Bello-Perez M, Adamek M, Coll J, Figueras A, Novoa B, Falco A. Modulation of the Tissue Expression Pattern of Zebrafish CRP-Like Molecules Suggests a Relevant Antiviral Role in Fish Skin. Biology. 2021; 10(2):78. https://doi.org/10.3390/biology10020078
Chicago/Turabian StyleBello-Perez, Melissa, Mikolaj Adamek, Julio Coll, Antonio Figueras, Beatriz Novoa, and Alberto Falco. 2021. "Modulation of the Tissue Expression Pattern of Zebrafish CRP-Like Molecules Suggests a Relevant Antiviral Role in Fish Skin" Biology 10, no. 2: 78. https://doi.org/10.3390/biology10020078
APA StyleBello-Perez, M., Adamek, M., Coll, J., Figueras, A., Novoa, B., & Falco, A. (2021). Modulation of the Tissue Expression Pattern of Zebrafish CRP-Like Molecules Suggests a Relevant Antiviral Role in Fish Skin. Biology, 10(2), 78. https://doi.org/10.3390/biology10020078






