Proposal of Potent Inhibitors for a Bacterial Cell Division Protein FtsZ: Molecular Simulations Based on Molecular Docking and ab Initio Molecular Orbital Calculations
Abstract
1. Introduction
2. Details of Molecular Simulations
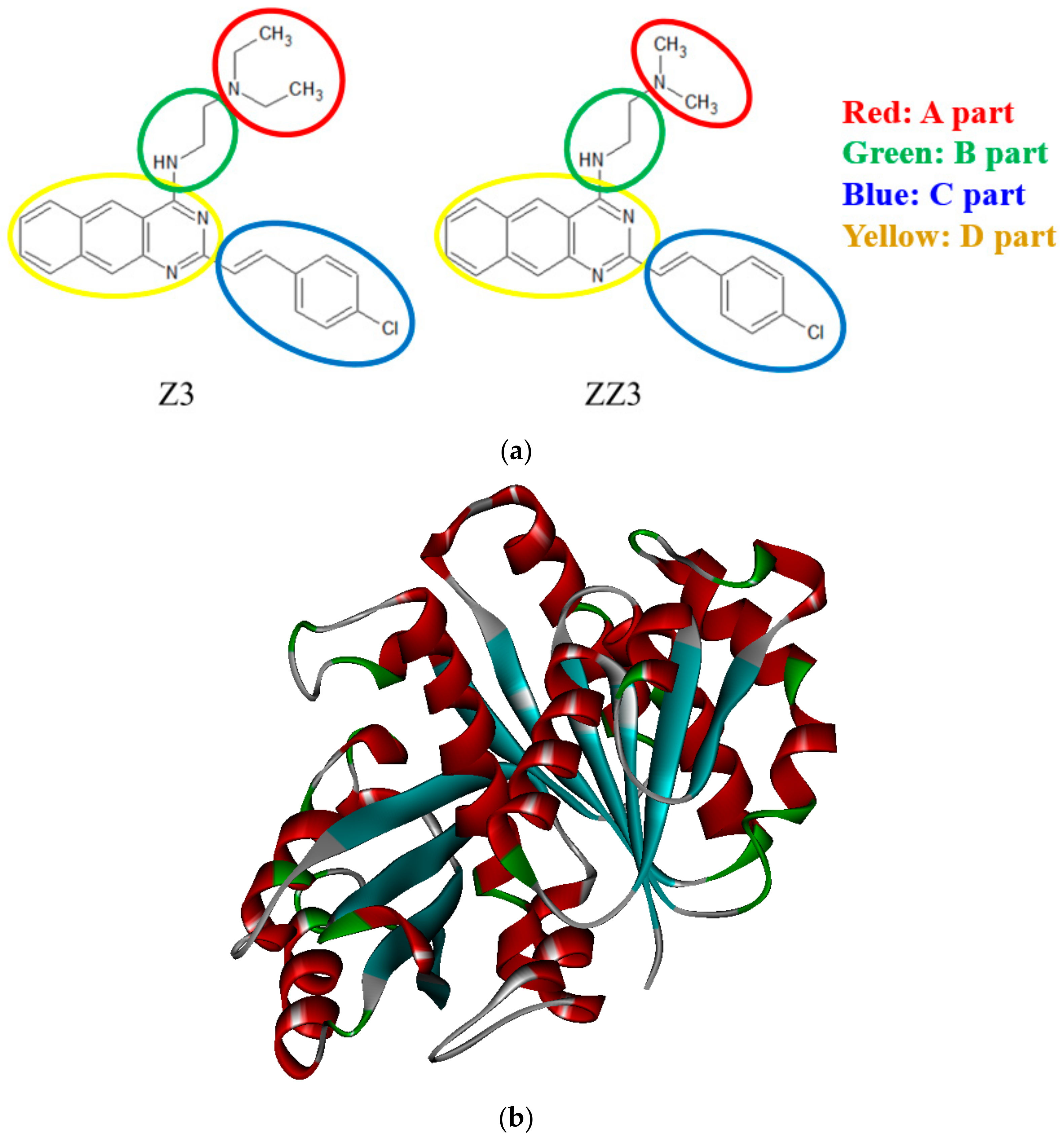
2.1. Proposal of Novel ZZ3 Derivatives as Potent Inhibitors of FtsZ
2.2. Constructions and Optimizations of the FtsZ + Derivative Complexes
2.3. FMO Calculations for the FtsZ + Derivative Complexes
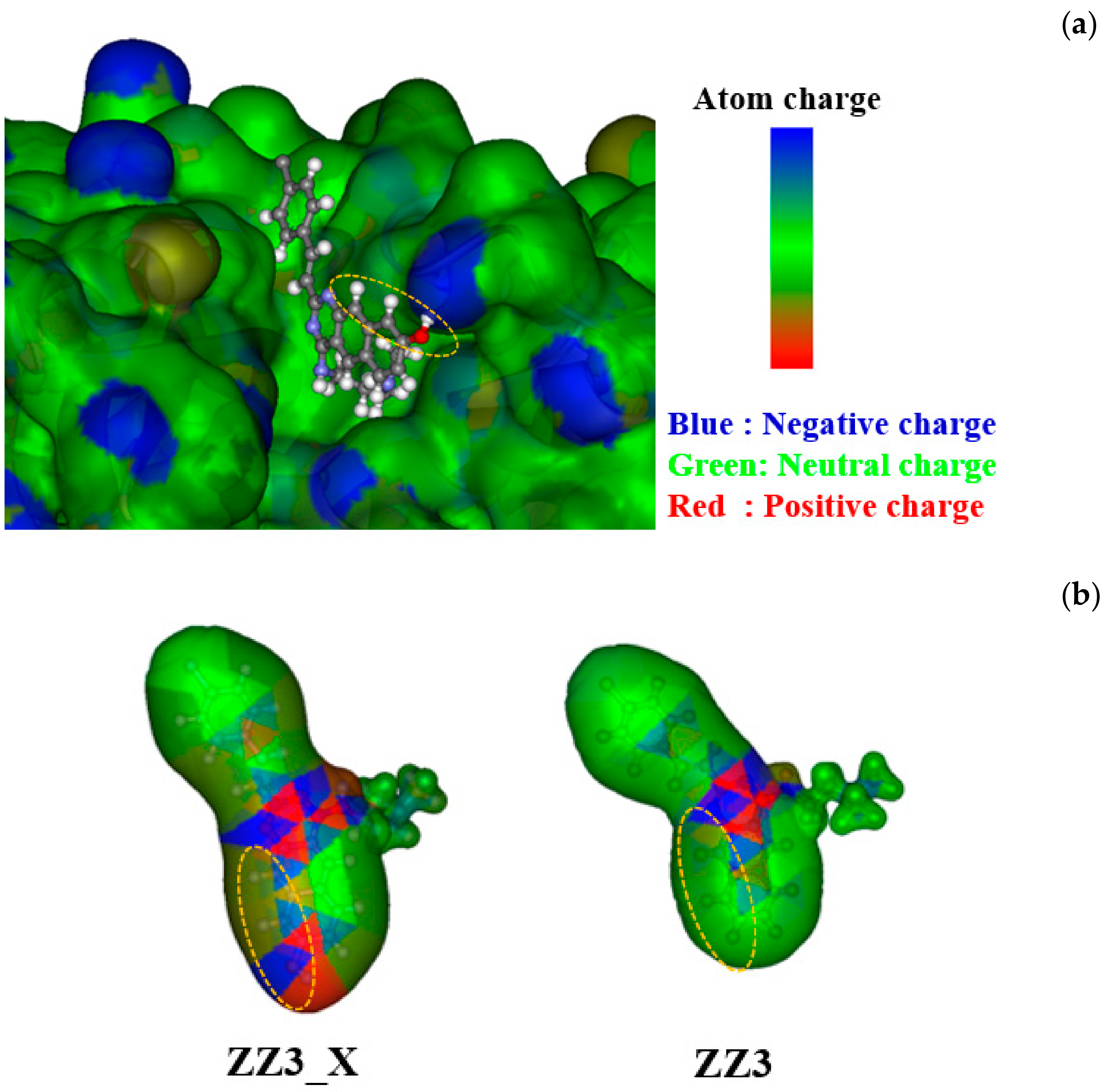
3. Results and Discussion
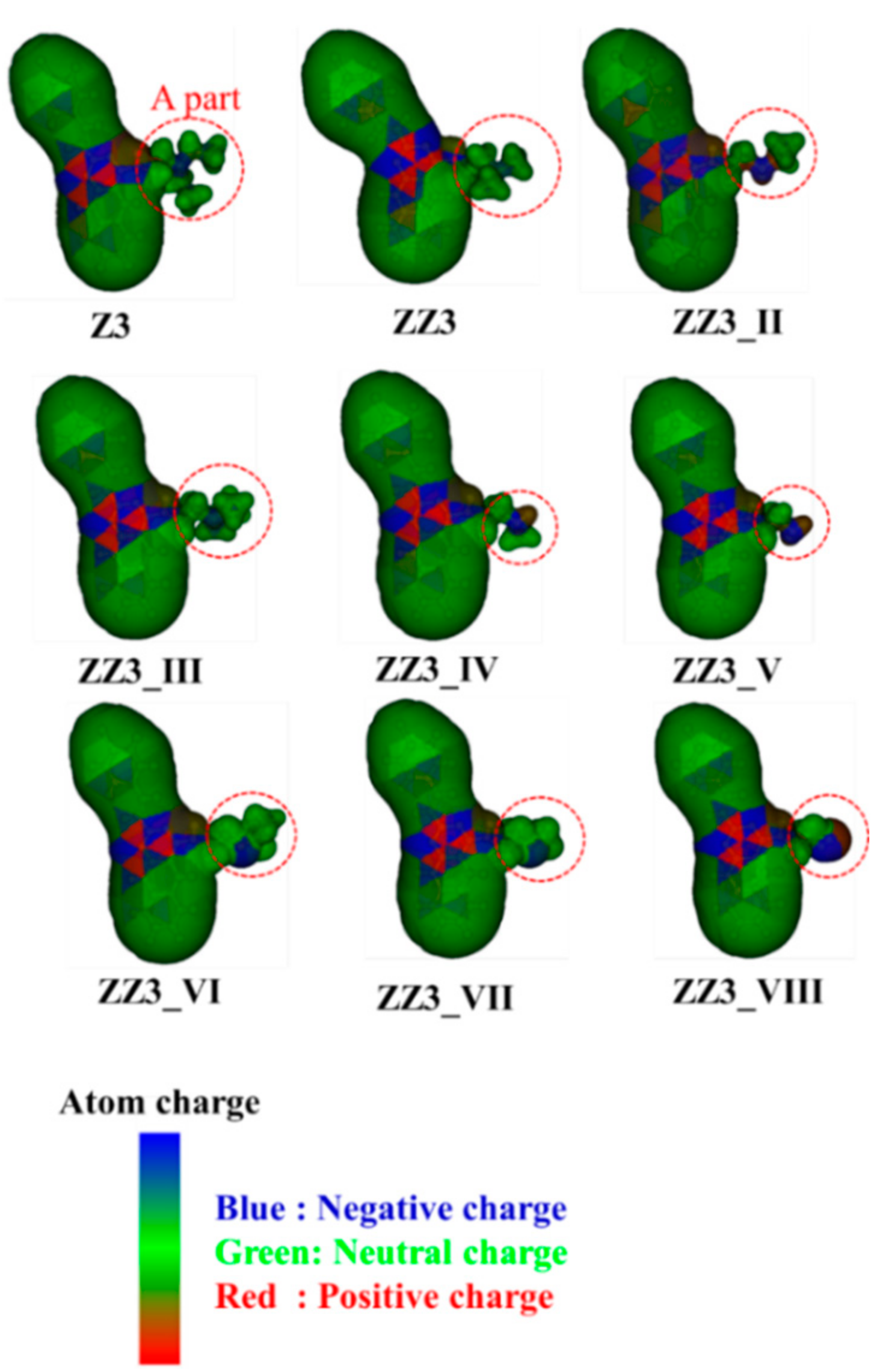
3.1. Binding Properties between FtsZ and the ZZ3 Derivatives by Replacing A-Part
3.2. Binding Properties between FtsZ and the ZZ3 Derivatives by Replacing the B- or D-Part
4. Conclusions
- (1)
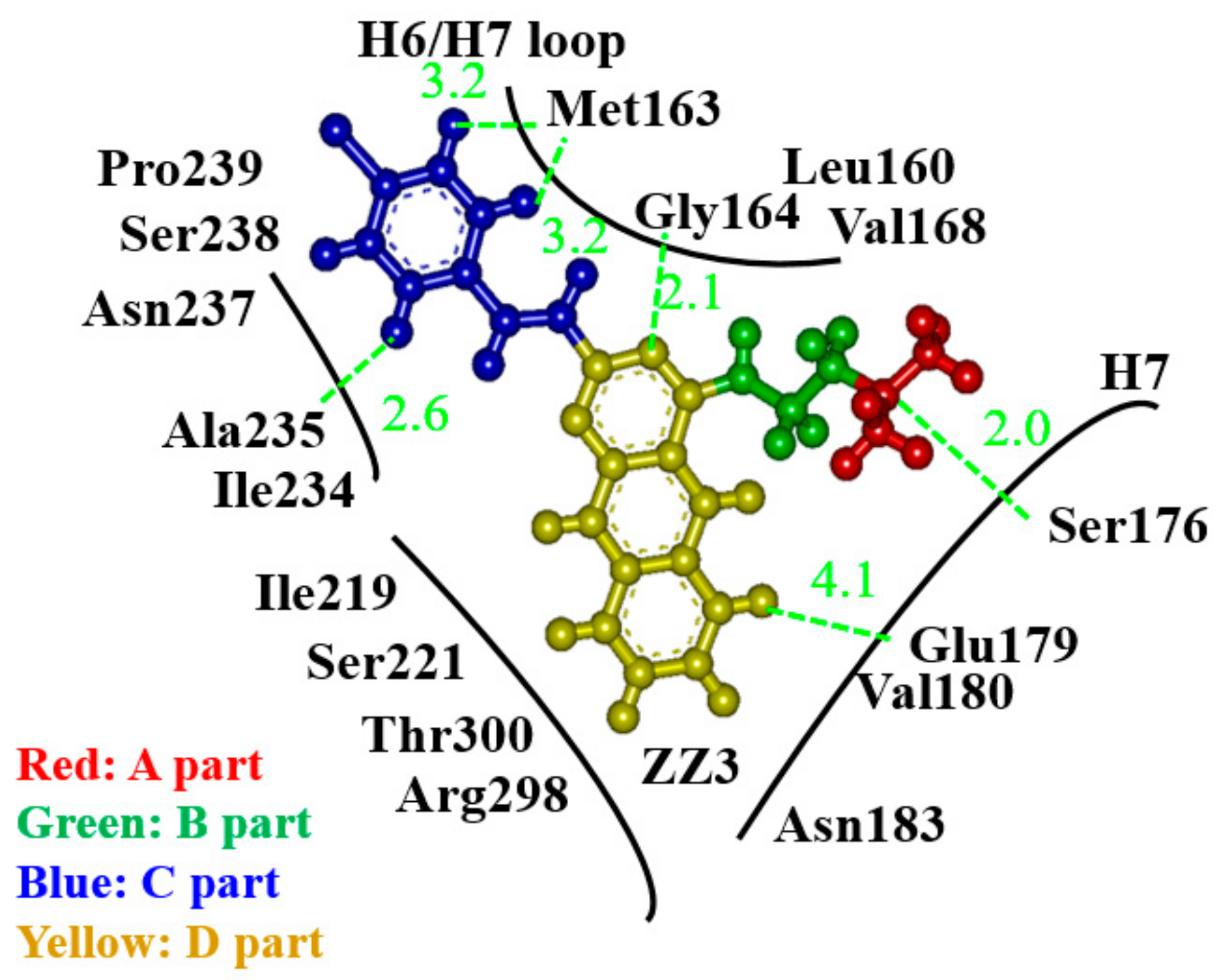
- The derivative, ZZ3_X, in which an OH group was introduced in the D-part of ZZ3, possessed the largest BE to FtsZ due to the strong H-bond between the OH group and Asp165 side chain.
- (2)
- Since Asp165 was included in the H6/H7 loop, which was beneficial for the aggregation of FtsZ, ZZ3_X was expected to change the conformation of the loop to inhibit the aggregations.
- (3)
- The replacement of the A- and B-parts of ZZ3 did not exert any positive effect on the enhancement of the interactions between ZZ3 and FtsZ.
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- World Health Organization. Global Tuberculosis Report 2017; World Health Organization: Geneva, Switzerland, 2017. [Google Scholar]
- World Health Organization Media Centre. WHO. Fact Sheet: Tuberculosis; WHO: Geneva, Switzerland, 2017. [Google Scholar]
- Nagai, H. Present status of multidrug resistant tuberculosis. Iryo 2004, 58, 595–598. (In Japanese) [Google Scholar]
- Kobayashi, H.; Koyanagi, K.; Kato, O.; Oe, T. Outbreak of extensively drug-resistant pulmonary tuberculosis in a hemodialysis facility. Kekkaku 2013, 88, 477–484. (In Japanese) [Google Scholar] [PubMed]
- Karpov, P.A.; Demchuk, O.M.; Britsun, V.M.; Lytvyn, D.I.; Pydiura, N.O.; Rayevsky, A.V.; Samofalova, D.A.; Spivak, S.I.; Volochnyuk, D.M.; Yemets, A.I.; et al. New imidazole inhibitors of mycobacterial FtsZ: The way from high-throughput molecular screening in grid to in vitro verification. Sci. Innov. 2016, 12, 43–55. [Google Scholar] [CrossRef]
- Tomioka, H. Prospects for the development of new antituberculous drugs putting our hopes on new drug targets. Kekkaku 2010, 85, 815–822. (In Japanese) [Google Scholar] [PubMed]
- Margalit, D.N.; Romberg, L.; Mets, R.B.; Hebert, A.M.; Mitchison, T.J.; Kirschner, M.W.; Raychaudhuri, D. Targeting cell division: Small-molecule inhibitors of FtsZ GTPase perturb cytokinetic ring assembly and induce bacterial lethality. Proc. Natl. Acad. Sci. USA 2004, 101, 11821–11826. [Google Scholar] [CrossRef] [PubMed]
- Osawa, M.; Anderson, D.E.; Erickson, H.P. Reconstitution of Contractile FtsZ Rings in Liposomes. Science 2008, 320, 792–794. [Google Scholar] [CrossRef] [PubMed]
- Haydon, D.J.; Stokes, N.R.; Ure, R.; Galbraith, G.; Bennett, J.M.; Brown, D.R.; Baker, P.J.; Barynin, V.V.; Rice, D.W.; Sedelnikova, S.E.; et al. An inhibitor of FtsZ with potent and selective anti-staphyloccocal aureus activity. Science 2008, 321, 1673–1675. [Google Scholar] [CrossRef] [PubMed]
- Fujimori, M.; Sogawa, H.; Ota, S.; Karpov, P.; Shulga, S.; Blume, Y.; Kurita, N. Specific interactions between mycobacterial FtsZ protein and curcumin derivatives: Molecular docking and ab initio molecular simulations. Chem. Phys. Lett. 2018, 692, 166–173. [Google Scholar] [CrossRef]
- Sogawa, H.; Sato, R.; Suzuki, K.; Tomioka, S.; Shinzato, T.; Karpov, P.; Shulga, S.; Blume, Y.; Kurita, N. Binding sites of Zantrin inhibitors to the bacterial cell division protein FtsZ: Molecular docking and ab initio molecular orbital calculations. Chem. Phys. 2020, 530, 110603. [Google Scholar] [CrossRef]
- Aggarwal, B.B.; Ichikawa, H.; Garodia, P.; Weerasinghe, P.; Sethi, G.; Bhatt, I.D.; Pandey, M.K.; Shishodia, S.; Nair, M.G. From traditional Ayurvedic medicine to modern medicine: Identification of therapeutic targets for suppression of inflammation and cancer. Expert Opin. Ther. Targets 2006, 10, 87–118. [Google Scholar] [CrossRef] [PubMed]
- Ray, B.; Bisht, S.; Maitra, A.; Maitra, A.; Lahiri, D.K. Neuroprotective and Neurorescue Effects of a Novel Polymeric Nanoparticle Formulation of Curcumin (NanoCurc™) in the Neuronal Cell Culture and Animal Model: Implications for Alzheimer’s disease. J. Alzheimer’s Dis. 2011, 23, 61–77. [Google Scholar] [CrossRef] [PubMed]
- Rai, D.; Singh, J.K.; Roy, N.; Panda, D. Curcumin inhibits FtsZ assembly: An attractive mechanism for its antibacterial activity. Biochem. J. 2008, 410, 147–155. [Google Scholar] [CrossRef] [PubMed]
- Nepomuceno, G.M.; Chan, K.M.; Huynh, V.; Martin, K.S.; Moore, J.T.; O’Brien, T.E.; Pollo, L.A.E.; Sarabia, F.J.; Tadeus, C.; Yao, Z.; et al. Synthesis and evaluation of quinazolines as inhibitors of the bacterial cell division protein FtsZ. ACS Med. Chem. Lett. 2015, 6, 308–312. [Google Scholar] [CrossRef] [PubMed]
- Wirth, M.; Zoete, V.; Michielin, O.; Sauer, W.H.B. SwissBioisostere: A database of molecular replacements for ligand design. Nucleic Acids Res. 2013, 41, D1137–D1143. [Google Scholar] [CrossRef]
- Daina, A.; Michielin, O.; Zoete, V. SwissADME: A free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci. Rep. 2017, 7, 42717. [Google Scholar] [CrossRef]
- Shah, S.; Patel, B.; Savjani, J.K. Pharmacophore mapping based virtual screening, molecular docking and ADMET studies of ROCK II inhibitors. Mult. Scler. Relat. Disord. 2018, 21, 35–41. [Google Scholar] [CrossRef]
- Lipinski, C.A.; Lombardo, F.; Dominy, B.W.; Feeney, P.J. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv. Drug Deliver. Rev. 1997, 23, 3–25. [Google Scholar] [CrossRef]
- Leung, A.K.; White, E.L.; Ross, L.J.; Reynolds, R.C.; DeVito, J.A.; Borhani, D.W. Structure of Mycobacterium tuberculosis FtsZ Reveals Unexpected, G Protein-like Conformational Switches. J. Mol. Biol. 2004, 342, 953–970. [Google Scholar] [CrossRef]
- Roy, A.; Kucukural, A.; Zhang, Y. I-TASSER: A unified platform for automated protein structure and function prediction. Nat. Protoc. 2010, 5, 725–738. [Google Scholar] [CrossRef]
- Roy, A.; Yang, J.; Zhang, Y. COFACTOR: An accurate comparative algorithm for structure-based protein function annotation. Nucleic Acids Res. 2012, 40, W471–W477. [Google Scholar] [CrossRef]
- Søndergaard, C.R.; Olsson, M.H.M.; Rostkowski, M.; Jensen, J.H. Improved Treatment of Ligands and Coupling Effects in Empirical Calculation and Rationalization of pKa Values. J. Chem. Theory Comput. 2011, 7, 2284–2295. [Google Scholar] [CrossRef] [PubMed]
- Olsson, M.H.M.; Søndergaard, C.R.; Rostkowski, M.; Jensen, J.H. PROPKA3: Consistent Treatment of Internal and Surface Residues in Empirical pKa Predictions. J. Chem. Theory Comput. 2011, 7, 525–537. [Google Scholar] [CrossRef] [PubMed]
- Frisch, M.J.; Trucks, G.W.; Schlegel, H.B.; Scuseria, G.E.; Robb, M.A.; Cheeseman, J.R.; Calmani, G.; Barone, V.; Mennucci, B.; Petersson, G.A.; et al. Gaussian 09, Revision A.02; Gaussian, Inc.: Wallingford, CT, USA, 2009. [Google Scholar]
- Bayly, C.I.; Cieplak, P.; Cornell, W.; Kollman, P.A. A well-behaved electrostatic potential based method using charge restraints for deriving atomic charges: The RESP model. J. Phys. Chem. 1993, 97, 10269–10280. [Google Scholar] [CrossRef]
- Morris, G.M.; Ruth, H.; Lindstrom, W.; Sanner, M.F.; Belew, R.K.; Goodsell, D.S.; Olson, A.J. AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J. Comput. Chem. 2009, 30, 2785–2791. [Google Scholar] [CrossRef]
- Case, D.A.; Darden, T.A.; Cheatham, T.E.; Simmerling, C.L., III; Wang, J.; Duke, R.E.; Luo, R.; Walker, R.C.; Zhang, W.; Merz, K.M.; et al. AMBER 12; University of California: San Francisco, CA, USA, 2012. [Google Scholar]
- Lindorff-Larsen, K.; Piana, S.; Palmo, K.; Maragakis, P.; Klepeis, J.L.; Dror, R.O.; Shaw, D.E. Improved side-chain torsion potentials for the Amber ff99SB protein force field. Proteins Struct. Funct. Bioinform. 2010, 78, 1950–1958. [Google Scholar] [CrossRef] [PubMed]
- Jorgensen, W.L.; Chandrasekhar, J.; Madura, J.; Klein, M.L. Compatison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983, 79, 926–935. [Google Scholar] [CrossRef]
- Wang, J.; Wolf, R.M.; Caldwell, J.W.; Kollman, P.A.; Case, D.A. Development and testing of a general amber force field. J. Comput. Chem. 2004, 25, 1157–1174. [Google Scholar] [CrossRef]
- Kitaura, K.; Ikeo, E.; Asada, T.; Nakano, T.; Uebayasi, M. Fragment molecular orbital method: An approximate computational method for large molecules. Chem. Phys. Lett. 1999, 313, 701–706. [Google Scholar] [CrossRef]
- Mochizuki, Y.; Nakano, T.; Koikegami, S.; Tanimori, S.; Abe, Y.; Nagashima, U.; Kitaura, K. A parallelized integral-direct second-order Mφller? Plesset perturbation theory method with a fragment molecular orbital scheme. Theor. Chem. Acc. 2004, 112, 442–452. [Google Scholar] [CrossRef]
- Mochizuki, Y.; Koikegami, S.; Nakano, T.; Amari, S.; Kitaura, K. Large scale MP2 calculations with fragment molecular orbital scheme. Chem. Phys. Lett. 2004, 396, 473–479. [Google Scholar] [CrossRef]
- Mochizuki, Y.; Yamashita, K.; Nakano, T.; Okiyama, Y.; Fukuzawa, K.; Taguchi, N.; Tanaka, S. Higher-order correlated calculations based on fragment molecular orbital scheme. Theor. Chem. Acc. 2011, 130, 515–530. [Google Scholar] [CrossRef]
- Fukuzawa, K.; Komeiji, Y.; Mochizuki, Y.; Kato, A.; Nakano, T.; Tanaka, S. Intra- and intermolecular interactions between cyclic-AMP receptor protein and DNA:Ab initio fragment molecular orbital study. J. Comput. Chem. 2006, 27, 948–960. [Google Scholar] [CrossRef] [PubMed]

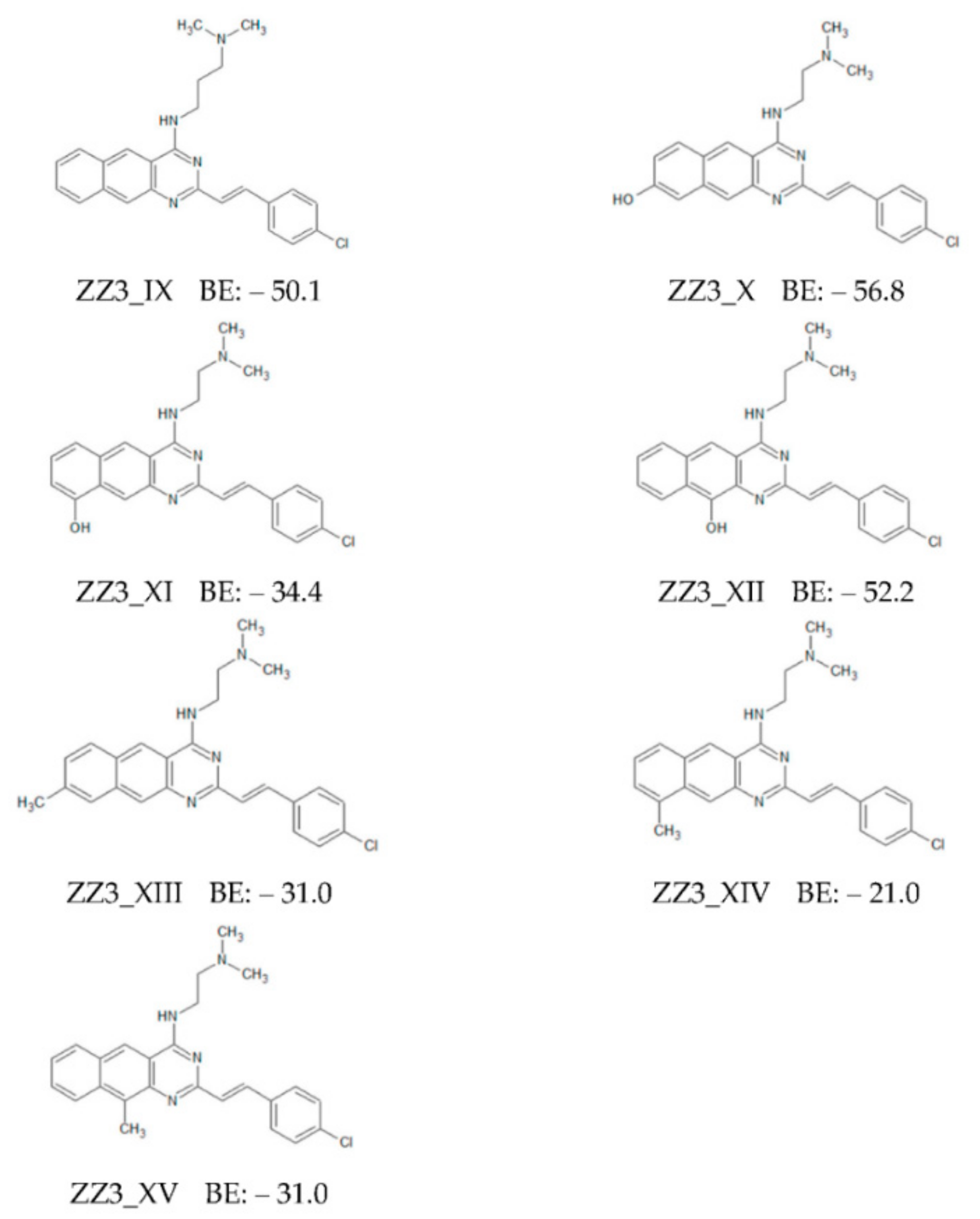

| Ligand | MW | RB | HBA | HBD | LogP | PSA |
|---|---|---|---|---|---|---|
| Z3 | 431.0 | 8 | 3 | 1 | 4.65 | 3.33 |
| ZZ3 | 402.9 | 6 | 3 | 1 | 4.24 | 3.12 |
| ZZ3_II | 402.9 | 7 | 3 | 2 | 4.24 | 3.11 |
| ZZ3_III | 417.0 | 7 | 3 | 1 | 4.45 | 3.22 |
| ZZ3_IV | 388.9 | 6 | 3 | 2 | 4.04 | 3.01 |
| ZZ3_V | 374.9 | 5 | 3 | 2 | 3.83 | 2.87 |
| ZZ3_VI | 403.9 | 7 | 3 | 1 | 4.24 | 3.09 |
| ZZ3_VII | 389.9 | 6 | 3 | 1 | 4.04 | 2.96 |
| ZZ3_VIII | 375.9 | 5 | 3 | 2 | 3.83 | 2.82 |
| ZZ3_IX | 417.0 | 7 | 3 | 1 | 4.45 | 3.21 |
| ZZ3_X | 418.9 | 6 | 4 | 2 | 3.42 | 3.11 |
| ZZ3_XI | 418.9 | 6 | 4 | 2 | 3.42 | 3.09 |
| ZZ3_XII | 418.9 | 6 | 4 | 2 | 3.42 | 3.17 |
| ZZ3_XIII | 417.0 | 6 | 3 | 1 | 4.45 | 3.25 |
| ZZ3_XIV | 417.0 | 6 | 3 | 1 | 4.45 | 3.24 |
| ZZ3_XV | 417.0 | 6 | 3 | 1 | 4.45 | 3.27 |
| Ligand | BBB | Caco2 | HIA | PPB | Mouse | Rat | hERG |
|---|---|---|---|---|---|---|---|
| Z3 | 3.5 | 55.6 | 97.1 | 86.7 | positive | negative | medium |
| ZZ3 | 1.5 | 54.3 | 97.1 | 84.4 | positive | negative | medium |
| ZZ3_II | 4.2 | 48.2 | 95.9 | 91.9 | positive | negative | medium |
| ZZ3_III | 2.4 | 55.0 | 97.1 | 85.1 | positive | negative | medium |
| ZZ3_IV | 3.1 | 45.8 | 95.9 | 86.6 | positive | negative | medium |
| ZZ3_V | 0.5 | 25.8 | 96.3 | 95.8 | positive | negative | medium |
| ZZ3_VI | 1.0 | 53.5 | 97.1 | 91.8 | positive | negative | medium |
| ZZ3_VII | 0.6 | 52.0 | 97.1 | 91.1 | positive | negative | medium |
| ZZ3_VIII | 2.4 | 29.8 | 95.9 | 92.3 | positive | negative | medium |
| ZZ3_IX | 1.7 | 54.9 | 97.1 | 83.3 | positive | negative | medium |
| ZZ3_X | 2.8 | 37.6 | 96.1 | 84.4 | negative | negative | medium |
| ZZ3_XI | 2.8 | 37.6 | 96.1 | 85.3 | positive | negative | medium |
| ZZ3_XII | 2.8 | 37.6 | 96.1 | 85.0 | positive | negative | medium |
| ZZ3_XIII | 2.8 | 54.4 | 97.1 | 84.5 | positive | negative | medium |
| ZZ3_XIV | 3.1 | 54.4 | 97.1 | 84.2 | positive | negative | medium |
| ZZ3_XV | 2.7 | 54.4 | 97.1 | 84.1 | positive | negative | medium |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yamamoto, S.; Saito, R.; Nakamura, S.; Sogawa, H.; Karpov, P.; Shulga, S.; Blume, Y.; Kurita, N. Proposal of Potent Inhibitors for a Bacterial Cell Division Protein FtsZ: Molecular Simulations Based on Molecular Docking and ab Initio Molecular Orbital Calculations. Antibiotics 2020, 9, 846. https://doi.org/10.3390/antibiotics9120846
Yamamoto S, Saito R, Nakamura S, Sogawa H, Karpov P, Shulga S, Blume Y, Kurita N. Proposal of Potent Inhibitors for a Bacterial Cell Division Protein FtsZ: Molecular Simulations Based on Molecular Docking and ab Initio Molecular Orbital Calculations. Antibiotics. 2020; 9(12):846. https://doi.org/10.3390/antibiotics9120846
Chicago/Turabian StyleYamamoto, Shohei, Ryosuke Saito, Shunya Nakamura, Haruki Sogawa, Pavel Karpov, Sergey Shulga, Yaroslav Blume, and Noriyuki Kurita. 2020. "Proposal of Potent Inhibitors for a Bacterial Cell Division Protein FtsZ: Molecular Simulations Based on Molecular Docking and ab Initio Molecular Orbital Calculations" Antibiotics 9, no. 12: 846. https://doi.org/10.3390/antibiotics9120846
APA StyleYamamoto, S., Saito, R., Nakamura, S., Sogawa, H., Karpov, P., Shulga, S., Blume, Y., & Kurita, N. (2020). Proposal of Potent Inhibitors for a Bacterial Cell Division Protein FtsZ: Molecular Simulations Based on Molecular Docking and ab Initio Molecular Orbital Calculations. Antibiotics, 9(12), 846. https://doi.org/10.3390/antibiotics9120846





