α-Amylase-Mediated Antibiotic Degradation and Sequestration in Pseudomonas aeruginosa Biofilm Therapy
Abstract
1. Introduction
2. Results
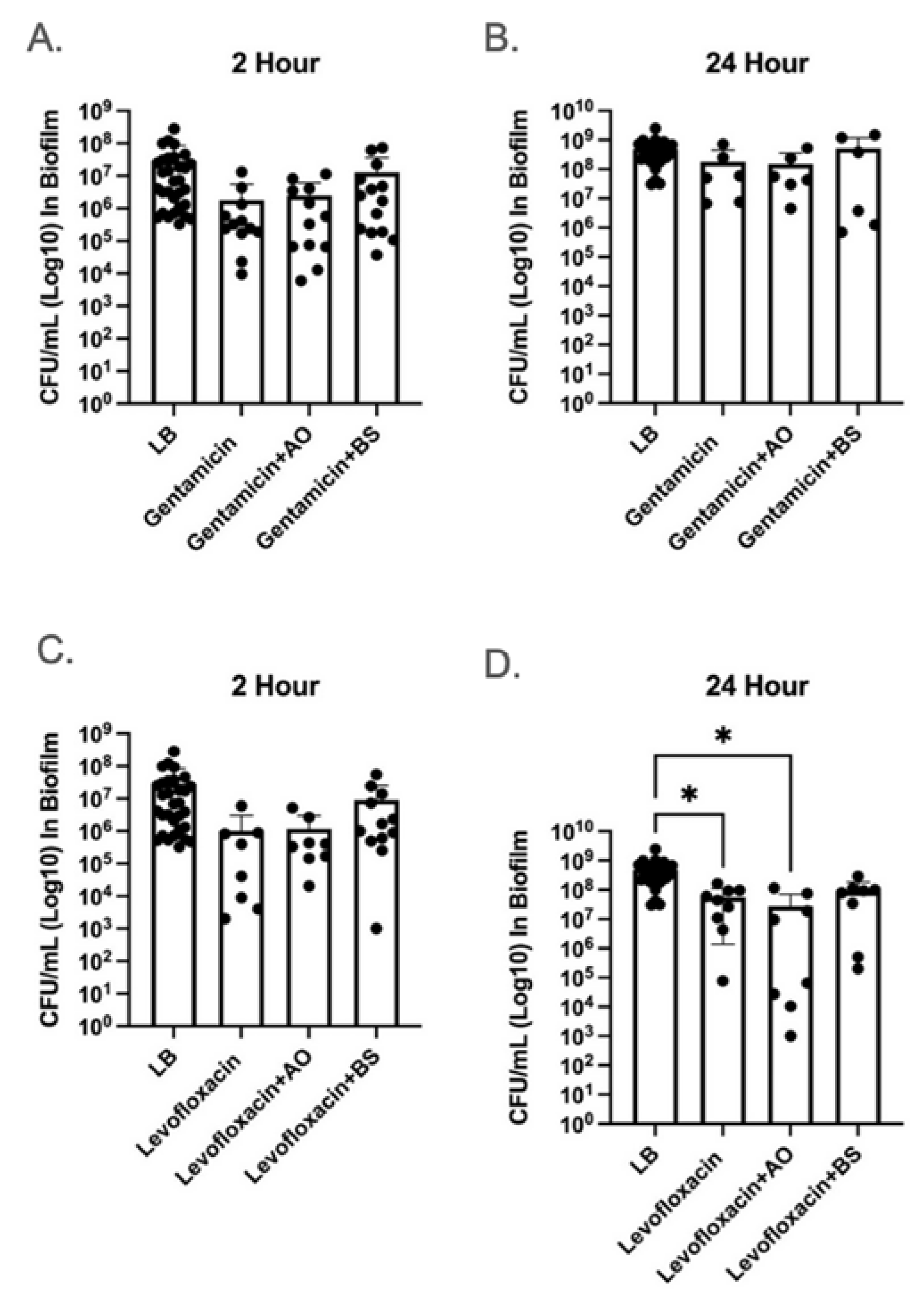
2.1. α-Amylase AO and BS Did Not Improve Antibiotic Clearance
2.2. Exposure to α-Amylase AO and BS Resulted in Altered MIC Values
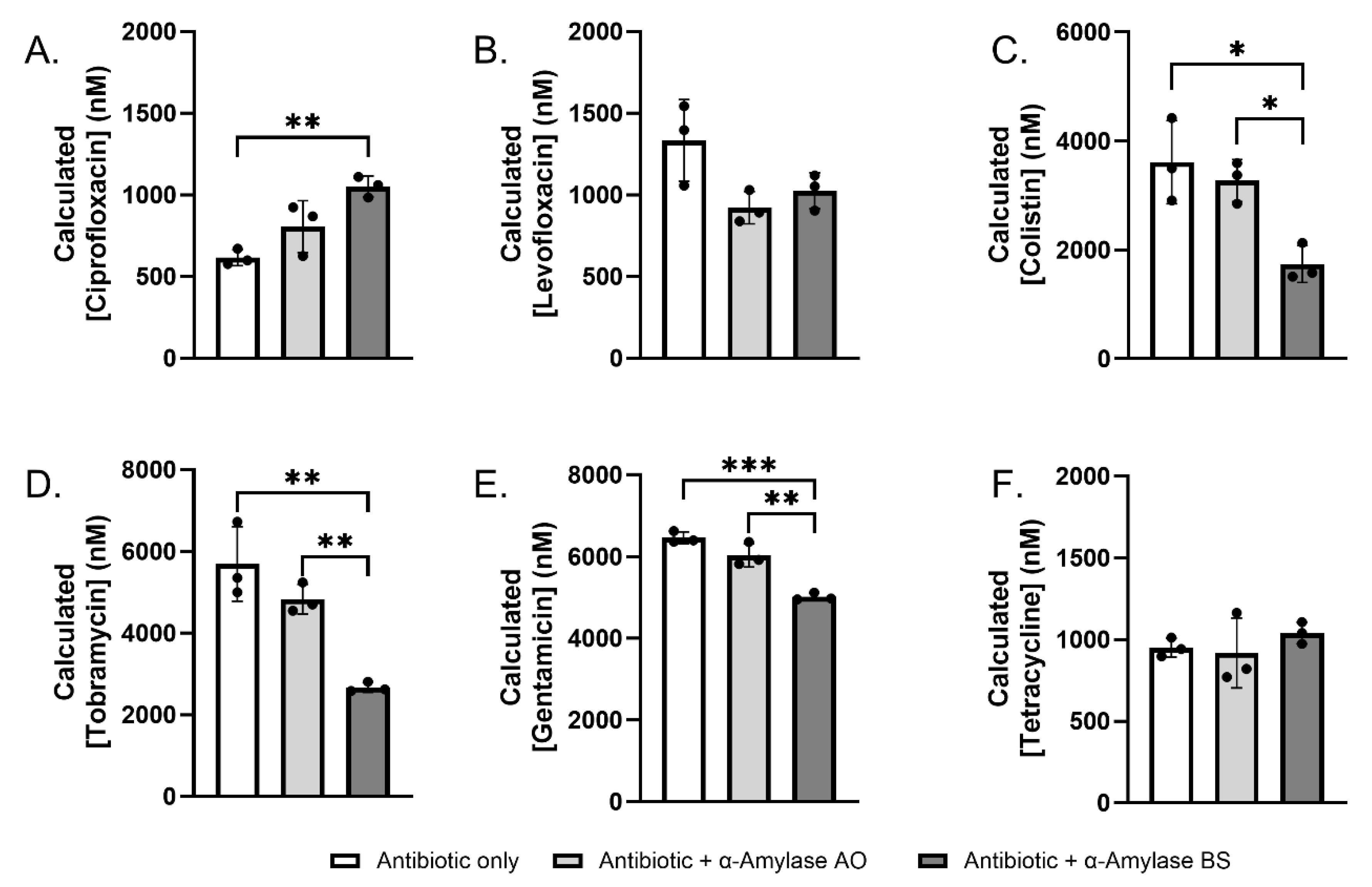
2.3. Sequestration and Degradation of Antibiotics by α-Amylase AO and BS
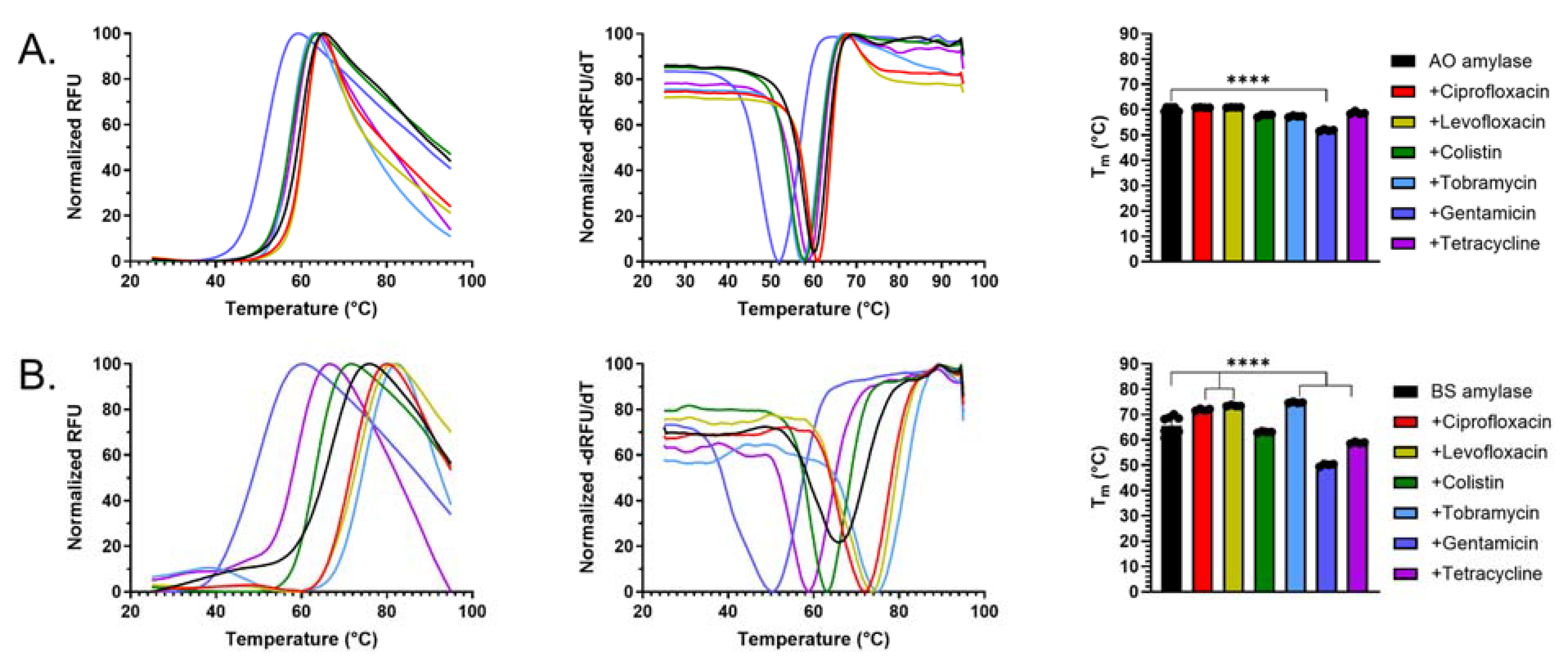
2.4. Antibiotics Interact with α-Amylase AO and BS
3. Discussion
4. Materials and Methods
4.1. Bacterial Strains and Culture Preparation
4.2. α-Amylase Preparation
4.3. Antibiotic Preparation
4.4. Forty-Eight-Hour Well-Plate Biofilm Growth
4.5. Minimum Inhibitory Concentration (MIC)
4.6. α-Amylase Mediated Antibiotic Interactions
4.7. Determination of Multiple Reaction Monitoring (MRM) parameters
4.8. Stability Assays
4.9. Differential Scanning Fluorimetry Assays
4.10. Data Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Antimicrobial Resistance Collaborators. Global burden of bacterial antimicrobial resistance in 2019: A systematic analysis. Lancet 2022, 399, 629–655. [Google Scholar] [CrossRef]
- Lipsitch, M.; Samore, M.H. Antimicrobial use and antimicrobial resistance: A population perspective. Emerg. Infect. Dis. 2002, 8, 347–354. [Google Scholar] [CrossRef]
- Nesme, J.; Cecillon, S.; Delmont, T.O.; Monier, J.M.; Vogel, T.M.; Simonet, P. Large-scale metagenomic-based study of antibiotic resistance in the environment. Curr. Biol. 2014, 24, 1096–1100. [Google Scholar] [CrossRef] [PubMed]
- Baquero, F.; Martinez, J.L.; Canton, R. Antibiotics and antibiotic resistance in water environments. Curr. Opin. Biotechnol. 2008, 19, 260–265. [Google Scholar] [CrossRef]
- Wright, G.D. Antibiotic resistance in the environment: A link to the clinic? Curr. Opin. Microbiol. 2010, 13, 589–594. [Google Scholar] [CrossRef] [PubMed]
- del Pozo, J.L.; Patel, R. The challenge of treating biofilm-associated bacterial infections. Clin. Pharmacol. Ther. 2007, 82, 204–209. [Google Scholar] [CrossRef]
- Santos, A.; Galdino, A.C.M.; Mello, T.P.; Ramos, L.S.; Branquinha, M.H.; Bolognese, A.M.; Columbano Neto, J.; Roudbary, M. What are the advantages of living in a community? A microbial biofilm perspective! Mem. Inst. Oswaldo. Cruz. 2018, 113, e180212. [Google Scholar] [CrossRef]
- Vestby, L.K.; Grønseth, T.; Simm, R.; Nesse, L.L. Bacterial Biofilm and its Role in the Pathogenesis of Disease. Antibiotics 2020, 9, 59. [Google Scholar] [CrossRef]
- Ciofu, O.; Tolker-Nielsen, T. Tolerance and Resistance of Pseudomonas aeruginosa Biofilms to Antimicrobial Agents-How P. aeruginosa Can Escape Antibiotics. Front. Microbiol. 2019, 10, 913. [Google Scholar] [CrossRef]
- Campoccia, D.; Mirzaei, R.; Montanaro, L.; Arciola, C.R. Hijacking of immune defences by biofilms: A multifront strategy. Biofouling 2019, 35, 1055–1074. [Google Scholar] [CrossRef]
- Jamal, M.; Ahmad, W.; Andleeb, S.; Jalil, F.; Imran, M.; Nawaz, M.A.; Hussain, T.; Ali, M.; Rafiq, M.; Kamil, M.A. Bacterial biofilm and associated infections. J. Chin. Med. Assoc. 2018, 81, 7–11. [Google Scholar] [CrossRef]
- Darouiche, R.O. Treatment of infections associated with surgical implants. N. Engl. J. Med. 2004, 350, 1422–1429. [Google Scholar] [CrossRef] [PubMed]
- Lamont, R.J.; Jenkinson, H.F. Life below the gum line: Pathogenic mechanisms of Porphyromonas gingivalis. Microbiol. Mol. Biol. Rev. 1998, 62, 1244–1263. [Google Scholar] [CrossRef]
- Pouget, C.; Dunyach-Remy, C.; Pantel, A.; Schuldiner, S.; Sotto, A.; Lavigne, J.-P. Biofilms in Diabetic Foot Ulcers: Significance and Clinical Relevance. Microorganisms 2020, 8, 1580. [Google Scholar] [CrossRef]
- Mirzaei, R.; Mohammadzadeh, R.; Alikhani, M.Y.; Shokri Moghadam, M.; Karampoor, S.; Kazemi, S.; Barfipoursalar, A.; Yousefimashouf, R. The biofilm-associated bacterial infections unrelated to indwelling devices. IUBMB Life 2020, 72, 1271–1285. [Google Scholar] [CrossRef]
- Beloin, C.; Roux, A.; Ghigo, J.M. Escherichia coli biofilms. In Current Topics in Microbiology and Immunology; Springer: Berlin/Heidelberg, Germany, 2008; Volume 322, pp. 249–289. [Google Scholar] [CrossRef]
- Yildiz, F.H.; Visick, K.L. Vibrio biofilms: So much the same yet so different. Trends Microbiol. 2009, 17, 109–118. [Google Scholar] [CrossRef] [PubMed]
- Otto, M. Staphylococcal biofilms. In Current Topics in Microbiology and Immunology; Springer: Berlin/Heidelberg, Germany, 2008; Volume 322, pp. 207–228. [Google Scholar] [CrossRef]
- Hoiby, N.; Ciofu, O.; Bjarnsholt, T. Pseudomonas aeruginosa biofilms in cystic fibrosis. Future Microbiol. 2010, 5, 1663–1674. [Google Scholar] [CrossRef] [PubMed]
- Jennings, L.K.; Dreifus, J.E.; Reichhardt, C.; Storek, K.M.; Secor, P.R.; Wozniak, D.J.; Hisert, K.B.; Parsek, M.R. Pseudomonas aeruginosa aggregates in cystic fibrosis sputum produce exopolysaccharides that likely impede current therapies. Cell Rep. 2021, 34, 108782. [Google Scholar] [CrossRef]
- Gawande, P.V.; Clinton, A.P.; LoVetri, K.; Yakandawala, N.; Rumbaugh, K.P.; Madhyastha, S. Antibiofilm Efficacy of DispersinB((R)) Wound Spray Used in Combination with a Silver Wound Dressing. Microbiol. Insights 2014, 7, 9–13. [Google Scholar] [CrossRef]
- Gawande, P.V.; Leung, K.P.; Madhyastha, S. Antibiofilm and antimicrobial efficacy of DispersinB(R)-KSL-W peptide-based wound gel against chronic wound infection associated bacteria. Curr. Microbiol. 2014, 68, 635–641. [Google Scholar] [CrossRef]
- Donelli, G.; Francolini, I.; Romoli, D.; Guaglianone, E.; Piozzi, A.; Ragunath, C.; Kaplan, J.B. Synergistic activity of dispersin B and cefamandole nafate in inhibition of staphylococcal biofilm growth on polyurethanes. Antimicrob. Agents Chemother. 2007, 51, 2733–2740. [Google Scholar] [CrossRef]
- Sauer, K.; Camper, A.K.; Ehrlich, G.D.; Costerton, J.W.; Davies, D.G. Pseudomonas aeruginosa displays multiple phenotypes during development as a biofilm. J. Bacteriol. 2002, 184, 1140–1154. [Google Scholar] [CrossRef] [PubMed]
- Wille, J.; Coenye, T. Biofilm dispersion: The key to biofilm eradication or opening Pandora’s box? Biofilm 2020, 2, 100027. [Google Scholar] [CrossRef] [PubMed]
- Fleming, D.; Rumbaugh, K. The Consequences of Biofilm Dispersal on the Host. Sci. Rep. 2018, 8, 10738. [Google Scholar] [CrossRef] [PubMed]
- Fleming, D.; Rumbaugh, K.P. Approaches to Dispersing Medical Biofilms. Microorganisms 2017, 5, 15. [Google Scholar] [CrossRef]
- Redman, W.K.; Welch, G.S.; Williams, A.C.; Damron, A.J.; Northcut, W.O.; Rumbaugh, K.P. Efficacy and safety of biofilm dispersal by glycoside hydrolases in wounds. Biofilm 2021, 3, 100061. [Google Scholar] [CrossRef]
- Morgan, R.; Kohn, S.; Hwang, S.H.; Hassett, D.J.; Sauer, K. BdlA, a chemotaxis regulator essential for biofilm dispersion in Pseudomonas aeruginosa. J. Bacteriol. 2006, 188, 7335–7343. [Google Scholar] [CrossRef]
- Marques, C.N.; Davies, D.G.; Sauer, K. Control of Biofilms with the Fatty Acid Signaling Molecule cis-2-Decenoic Acid. Pharmaceuticals 2015, 8, 816–835. [Google Scholar] [CrossRef]
- Kalia, M.; Amari, D.; Davies, D.G.; Sauer, K. cis-DA-dependent dispersion by Pseudomonas aeruginosa biofilm and identification of cis-DA-sensory protein DspS. mBio 2023, 14, e0257023. [Google Scholar] [CrossRef]
- Nguyen, T.K.; Selvanayagam, R.; Ho, K.K.K.; Chen, R.; Kutty, S.K.; Rice, S.A.; Kumar, N.; Barraud, N.; Duong, H.T.T.; Boyer, C. Co-delivery of nitric oxide and antibiotic using polymeric nanoparticles. Chem. Sci. 2016, 7, 1016–1027. [Google Scholar] [CrossRef]
- Grayton, Q.E.; Nguyen, H.K.; Broberg, C.A.; Ocampo, J.; Nagy, S.G.; Schoenfisch, M.H. Biofilm Dispersal, Reduced Viscoelasticity, and Antibiotic Sensitization via Nitric Oxide-Releasing Biopolymers. ACS Infect. Dis. 2023, 9, 1730–1741. [Google Scholar] [CrossRef]
- Chen, Y.T.; Ma, Y.M.; Peng, X.X.; Li, H. Glutamine potentiates gentamicin to kill lab-evolved gentamicin-resistant and clinically isolated multidrug-resistant Escherichia coli. Front. Microbiol. 2022, 13, 1071278. [Google Scholar] [CrossRef] [PubMed]
- Goodwine, J.; Gil, J.; Doiron, A.; Valdes, J.; Solis, M.; Higa, A.; Davis, S.; Sauer, K. Pyruvate-depleting conditions induce biofilm dispersion and enhance the efficacy of antibiotics in killing biofilms in vitro and in vivo. Sci. Rep. 2019, 9, 3763. [Google Scholar] [CrossRef] [PubMed]
- Rumbaugh, K.P.; Sauer, K. Biofilm dispersion. Nat. Rev. Microbiol. 2020, 18, 571–586. [Google Scholar] [CrossRef] [PubMed]
- Chambers, J.R.; Cherny, K.E.; Sauer, K. Susceptibility of Pseudomonas aeruginosa Dispersed Cells to Antimicrobial Agents Is Dependent on the Dispersion Cue and Class of the Antimicrobial Agent Used. Antimicrob. Agents Chemother. 2017, 61, e00846-17. [Google Scholar] [CrossRef] [PubMed]
- Kalia, M.; Sauer, K. Distinct transcriptome and traits of freshly dispersed Pseudomonas aeruginosa cells. mSphere 2024, 9, e0088424. [Google Scholar] [CrossRef]
- Wolcott, R.D.; Kennedy, J.P.; Dowd, S.E. Regular debridement is the main tool for maintaining a healthy wound bed in most chronic wounds. J. Wound Care 2009, 18, 54–56. [Google Scholar] [CrossRef]
- Ousey, K.; Ovens, L. Comparing methods of debridement for removing biofilm in hard-to-heal wounds. J. Wound Care 2023, 32, S4–S10. [Google Scholar] [CrossRef]
- Kaplan, J.B. Biofilm dispersal: Mechanisms, clinical implications, and potential therapeutic uses. J. Dent. Res. 2010, 89, 205–218. [Google Scholar] [CrossRef]
- Fleming, D.; Chahin, L.; Rumbaugh, K. Glycoside Hydrolases Degrade Polymicrobial Bacterial Biofilms in Wounds. Antimicrob. Agents Chemother. 2017, 61, e01998-16. [Google Scholar] [CrossRef]
- Aksoy, N.; Vatansever, C.; Adali, C.; Adakli Aksoy, B.; Fisgin, T. The Inhibitory Effects of Amylase and Streptokinase on Minimum Inhibitory Concentration of Antibiotics Used to Treat Gram Negative Bacteria Biofilm Infection on Indwelling Devices. Indian J. Microbiol. 2023, 63, 533–540. [Google Scholar] [CrossRef] [PubMed]
- Yu, S.; Su, T.; Wu, H.; Liu, S.; Wang, D.; Zhao, T.; Jin, Z.; Du, W.; Zhu, M.J.; Chua, S.L.; et al. PslG, a self-produced glycosyl hydrolase, triggers biofilm disassembly by disrupting exopolysaccharide matrix. Cell Res. 2015, 25, 1352–1367. [Google Scholar] [CrossRef] [PubMed]
- Baker, P.; Hill, P.J.; Snarr, B.D.; Alnabelseya, N.; Pestrak, M.J.; Lee, M.J.; Jennings, L.K.; Tam, J.; Melnyk, R.A.; Parsek, M.R.; et al. Exopolysaccharide biosynthetic glycoside hydrolases can be utilized to disrupt and prevent Pseudomonas aeruginosa biofilms. Sci. Adv. 2016, 2, e1501632. [Google Scholar] [CrossRef]
- Ramakrishnan, R.; Singh, A.K.; Singh, S.; Chakravortty, D.; Das, D. Enzymatic dispersion of biofilms: An emerging biocatalytic avenue to combat biofilm-mediated microbial infections. J. Biol. Chem. 2022, 298, 102352. [Google Scholar] [CrossRef] [PubMed]
- Jujjavarapu, S.E.; Dhagat, S. Evolutionary Trends in Industrial Production of alpha-amylase. Recent Pat. Biotechnol. 2019, 13, 4–18. [Google Scholar] [CrossRef]
- Colvin, K.M.; Irie, Y.; Tart, C.S.; Urbano, R.; Whitney, J.C.; Ryder, C.; Howell, P.L.; Wozniak, D.J.; Parsek, M.R. The Pel and Psl polysaccharides provide Pseudomonas aeruginosa structural redundancy within the biofilm matrix. Environ. Microbiol. 2012, 14, 1913–1928. [Google Scholar] [CrossRef]
- Redman, W.K.; Welch, G.S.; Rumbaugh, K.P. Differential Efficacy of Glycoside Hydrolases to Disperse Biofilms. Front. Cell. Infect. Microbiol. 2020, 10, 379. [Google Scholar] [CrossRef]
- Abo-Kamar, A.M.; Mustafa, A.A.; Al-Madboly, L.A. Purified alpha-Amylase from Bacillus cereus exhibits antibiofilm and antiquorum sensing activities against uropathogenic Escherichia coli, Downregulating fimH, and papC virulence genes: Implications for urinary tract infections. BMC Microbiol. 2024, 24, 502. [Google Scholar] [CrossRef]
- Dias-Souza, M.V.; Alves, A.L.; Pagnin, S.; Veiga, A.A.; Haq, I.U.; Alonazi, W.B.; Dos Santos, V.L. The activity of hydrolytic enzymes and antibiotics against biofilms of bacteria isolated from industrial-scale cooling towers. Microb. Cell Fact. 2024, 23, 282. [Google Scholar] [CrossRef]
- Schneider, R.E.; Hamdan, J.V.; Rumbaugh, K.P. Biofilm Dispersal and Wound Infection Clearance With Preclinical Debridement Agents. Int. Wound J. 2025, 22, e70145. [Google Scholar] [CrossRef]
- Amiri, B.; Hosseini, N.S.; Taktaz, F.; Amini, K.; Rahmani, M.; Amiri, M.; Sadrjavadi, K.; Jangholi, A.; Esmaeili, S. Inhibitory effects of selected antibiotics on the activities of alpha-amylase and alpha-glucosidase: In-vitro, in-vivo and theoretical studies. Eur. J. Pharm. Sci. 2019, 138, 105040. [Google Scholar] [CrossRef]
- Thatoi, H.; Mohapatra, P.K.D.; Mohapatra, S.; Mondal, K.C. Mondal. Microbial Fermentation and Enzyme Technology; CRC Press: Boca Raton, FL, USA, 2020. [Google Scholar] [CrossRef]
- de Souza, P.M.; de Oliveira Magalhaes, P. Application of microbial alpha-amylase in industry—A review. Braz. J. Microbiol. 2010, 41, 850–861. [Google Scholar] [CrossRef] [PubMed]
- Hui, M.; Kwok, A.K.; Pang, C.P.; Cheung, S.W.; Chan, R.C.; Lam, D.S.; Cheng, A.F. An in vitro study on the compatibility and precipitation of a combination of ciprofloxacin and vancomycin in human vitreous. Br. J. Ophthalmol. 2004, 88, 218–222. [Google Scholar] [CrossRef] [PubMed]
- Niesen, F.H.; Berglund, H.; Vedadi, M. The use of differential scanning fluorimetry to detect ligand interactions that promote protein stability. Nat. Protoc. 2007, 2, 2212–2221. [Google Scholar] [CrossRef]
- Shao, H.; Oltion, K.; Wu, T.; Gestwicki, J.E. Differential scanning fluorimetry (DSF) screen to identify inhibitors of Hsp60 protein-protein interactions. Org. Biomol. Chem. 2020, 18, 4157–4163. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Zhang, C. Using Differential Scanning Fluorimetry (DSF) to Detect Ligand Binding with Purified Protein. Methods Mol. Biol. 2021, 2213, 183–186. [Google Scholar] [CrossRef]
- Vivoli, M.; Novak, H.R.; Littlechild, J.A.; Harmer, N.J. Determination of protein-ligand interactions using differential scanning fluorimetry. J. Vis. Exp. 2014, 91, 51809. [Google Scholar] [CrossRef]
- Taylor, I.R.; Paczkowski, J.E.; Jeffrey, P.D.; Henke, B.R.; Smith, C.D.; Bassler, B.L. Inhibitor Mimetic Mutations in the Pseudomonas aeruginosa PqsE Enzyme Reveal a Protein-Protein Interaction with the Quorum-Sensing Receptor RhlR That Is Vital for Virulence Factor Production. ACS Chem. Biol. 2021, 16, 740–752. [Google Scholar] [CrossRef]
- Mah, T.F. Biofilm-specific antibiotic resistance. Future Microbiol. 2012, 7, 1061–1072. [Google Scholar] [CrossRef]
- Pamp, S.J.; Gjermansen, M.; Johansen, H.K.; Tolker-Nielsen, T. Tolerance to the antimicrobial peptide colistin in Pseudomonas aeruginosa biofilms is linked to metabolically active cells, and depends on the pmr and mexAB-oprM genes. Mol. Microbiol. 2008, 68, 223–240. [Google Scholar] [CrossRef]
- Stewart, P.S.; Zhang, T.; Xu, R.; Pitts, B.; Walters, M.C.; Roe, F.; Kikhney, J.; Moter, A. Reaction-diffusion theory explains hypoxia and heterogeneous growth within microbial biofilms associated with chronic infections. NPJ Biofilms Microbiomes 2016, 2, 16012. [Google Scholar] [CrossRef]
- Fleming, D.; Redman, W.; Welch, G.S.; Mdluli, N.V.; Rouchon, C.N.; Frank, K.L.; Rumbaugh, K.P. Utilizing glycoside hydrolases to improve the quantitation and visualization of biofilm bacteria. Biofilm 2020, 2, 100037. [Google Scholar] [CrossRef]
- Kalita, D.; Holm, D.G.; LaBarbera, D.V.; Petrash, J.M.; Jayanty, S.S. Inhibition of alpha-glucosidase, alpha-amylase, and aldose reductase by potato polyphenolic compounds. PLoS ONE 2018, 13, e0191025. [Google Scholar] [CrossRef]
- Kowalska-Krochmal, B.; Dudek-Wicher, R. The Minimum Inhibitory Concentration of Antibiotics: Methods, Interpretation, Clinical Relevance. Pathogens 2021, 10, 165. [Google Scholar] [CrossRef] [PubMed]
- Wu, T.; Gale-Day, Z.J.; Gestwicki, J.E. DSFworld: A flexible and precise tool to analyze differential scanning fluorimetry data. Protein Sci. 2024, 33, e5022. [Google Scholar] [CrossRef] [PubMed]
| Antibiotic | Original MIC (µg/mL) | α-Amylase AO (µg/mL) | α-Amylase BS (µg/mL) |
|---|---|---|---|
| Ciprofloxacin | 0.4 | 0.78 | 1.563 |
| Levofloxacin | 1.5 | 3.125 | 6.25 |
| Colistin | 31.25 | 31.25 | 15.63 |
| Tobramycin | 6.25 | 6.25 | 25 |
| Gentamicin | 3.125 | 3.125 | 25 |
| Tetracycline | 31.25 | >1000 | 500 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Murray, R.K.; Martin, A.E.; Zipkowitz, S.; Jahan, N.; Davis, T.D.; Redman, W.K. α-Amylase-Mediated Antibiotic Degradation and Sequestration in Pseudomonas aeruginosa Biofilm Therapy. Antibiotics 2025, 14, 941. https://doi.org/10.3390/antibiotics14090941
Murray RK, Martin AE, Zipkowitz S, Jahan N, Davis TD, Redman WK. α-Amylase-Mediated Antibiotic Degradation and Sequestration in Pseudomonas aeruginosa Biofilm Therapy. Antibiotics. 2025; 14(9):941. https://doi.org/10.3390/antibiotics14090941
Chicago/Turabian StyleMurray, Robert K., Allison E. Martin, Sarah Zipkowitz, Nusrat Jahan, Tony D. Davis, and Whitni K. Redman. 2025. "α-Amylase-Mediated Antibiotic Degradation and Sequestration in Pseudomonas aeruginosa Biofilm Therapy" Antibiotics 14, no. 9: 941. https://doi.org/10.3390/antibiotics14090941
APA StyleMurray, R. K., Martin, A. E., Zipkowitz, S., Jahan, N., Davis, T. D., & Redman, W. K. (2025). α-Amylase-Mediated Antibiotic Degradation and Sequestration in Pseudomonas aeruginosa Biofilm Therapy. Antibiotics, 14(9), 941. https://doi.org/10.3390/antibiotics14090941






