Abstract
Background/Objectives: Clostridioides difficile is a “One Health” pathogen and a cause of antibiotics-associated diarrhea and pseudomembranous colitis. Mobile genetic elements (MGEs) have been documented in the genomes of clinical C. difficile strains; however, the presence of MGEs in environmental strains remains poorly characterized. Thus, the present study was conducted with the objective of identifying the prevalence of MGEs, including mobilizable transposons (MTns), conjugative transposons (CTns), plasmids, and insertion sequences, in whole genome sequences (WGSs) of environmental C. difficile isolates. Methods: The analysis of MGEs was conducted using 166 WGSs obtained from C. difficile strains isolated from various environmental sources contaminated with feces. The MGEs were identified using bioinformatic tools. Results: A total of 48.2% (80/166) of the studied genomes were identified to harbor nine transposons, including Tn916, Tn6194-like, Tn5397, Tn6215, Tn4001, Tn6073, Tn6110, Tn6107, or Tn5801-like. The majority of MTns and CTns could be found within C. difficile sequence types ST11, ST3, and ST35. The results demonstrated close genetic relatedness among the studied genomes, the array of antimicrobial resistance (AMR) genes, such as tetM, ermB, and aac(6′)-aph(2″), and the presence of CTns. Furthermore, the analysis revealed that 24.7% (41/166) of the genome sequences of isolates were associated with various predominant plasmid groups, including pCD6, pCD-ECE4-6, pCD-WTSI1-4, pCDBI1, and pCd1_3, which belonged to 16 different sequence types. Furthermore, several plasmids were identified as harboring the prophage phiCDHM19. Conclusions: The results of the current study suggest that the identified plasmids are abundant and may encode functions that are relevant to C. difficile physiology. The genomes of C. difficile strains examined contain closely related CTns, suggesting that horizontal transfer of AMR is important in this species or other bacterial species. Further research is required to ascertain the effect of these genetic elements and their transferability on the biology of C. difficile.
1. Introduction
Clostridioides difficile is a Gram-positive, anaerobic, spore-forming bacterium that is a significant cause of enteric disease in humans and animals [1]. C. difficile infection (CDI) depends on the production of toxins A and B, which induce symptoms ranging from mild to severe diarrhea and can result in potentially fatal pseudomembranous colitis [2]. In the past two decades, PCR and sequence-based techniques—effectively equivalent to whole-genome sequencing (WGS)—have advanced our understanding of the genetic diversity, epidemiology, and pathogenicity of C. difficile [3]. The organism’s evolutionary plasticity, in response to environmental and anthropogenic pressures, has been shown to play a significant role in the rapid emergence and global dissemination of virulent and resistant clonal lineages [3]. Mobile genetic elements (MGEs) have been shown to profoundly influence the ecology and evolution of organisms, including bacteria. The initial WGS of C. difficile, derived from strain CD630, revealed a chromosome of 4.4 Mb and a plasmid, pCD630, of 7881 bp. A high proportion of MGEs, approximately 11% in strain CD630, contributes to the remarkably dynamic and mosaic nature of the C. difficile genome. These elements include conjugative and mobilizable transposons, insertion sequences (ISs), IStrons (group I introns), sigK intervening elements, CRISPR-Cas systems, genomic islands, the ~7 kb plasmid pCD630, and bacteriophages [4,5]. In our previous study, the genome sequences of environmental C. difficile isolates utilized in the current study were found to contain several putative prophages, including 372 intact temperate prophages from the Myoviridae and Siphoviridae families [6].
It is evident that conjugation, transduction, and transformation of MGEs, most notably transposons, among C. difficile strains and/or between C. difficile and other bacterial species, represent pivotal mechanisms through which C. difficile acquires antimicrobial resistance (AMR) genes [7,8]. For instance, it has been demonstrated that Tn916 can be transferred from Bacillus subtilis to C. difficile strains [9]. In addition, the transferability of Tn5397 between C. difficile stains and B. subtilis [10,11] and Enterococcus faecalis [12] was proven. The presence of several conjugative transposons (CTns) and mobilizable transposons (MTns), a significant proportion of which are associated with AMR genes, has been identified in C. difficile [3,4,13]. A putative CTn belonging to the Tn916-like family of transposons and carrying the erm(B) gene has recently been identified in C. difficile 2007855, a strain belonging to the hypervirulent PCR-ribotype 027, and which has been designated as Tn6194 [14,15]. Another CTn, the Tn6194 element, which is associated with the ermB gene, was identified in C. difficile PCR-ribotype 001. This element was subsequently transferred into C. difficile PCR-ribotypes 009 and 027, as well as into a strain of Enterococcus faecalis [16]. This finding could provide further evidence of the transmission risk of AMR between pathogenic bacteria occupying the same human intestinal niche.
Extrachromosomal elements, such as plasmids, have been found to be significant in most bacterial species. C. difficile has been found to possess a significant number of plasmids that encode functions which could be relevant to the pathogenesis of CDI. The majority of previously identified plasmids in C. difficile strains were cryptic. These plasmids may encode functions that are relevant to C. difficile physiology, including putative plasmid replication and partitioning locus [17,18]. To our knowledge, only a limited number of these plasmids have been associated with AMR or virulence genes. For instance, Ramirez-Vargas and Rodrigues reported the presence of conjugative plasmids in association with tcdB and cdtAB in clinical C. difficile isolates belonging to multilocus sequence typing (MLST) clades C-I, 2, and 4 [19,20]. In recent research, a 7 kb plasmid, pCD-METRO, associated with metronidazole resistance, has been identified in both toxigenic and non-toxigenic human and animal C. difficile strains [21]. However, in C. difficile, AMR genes are mostly found on transposons rather than on plasmids [13].
Although a few studies have investigated MGEs—including transposons, plasmids, and prophages—in clinical isolates of C. difficile, the detection of MGEs in environmental C. difficile isolates from various environmental sources remains limited. Therefore, in the present study, the WGSs were collected from 166 C. difficile isolates obtained from fecally contaminated environmental sources to assess the prevalence of MGEs, including MTns, CTns, ISs, and plasmids. Furthermore, the present study focused on MGEs carrying genes potentially associated with AMR. The association between identified MGEs and sequence typing (ST) was also examined. It was hypothesized that the genetic characterization of environmental C. difficile strains would provide further insight into the role of MGEs in the evolution, pathogenicity, and AMR of C. difficile.
2. Results
2.1. Genetic Diversity of MGEs in Environmental C. difficile Isolates
2.1.1. Mobilizable and Conjugative Transposons
The presence of MTns and CTns was screened in the sequenced genomes of 166 environmental C. difficile isolates from diverse fecally contaminated environmental sources, including cattle feces, soil, digested-sludge-amended soils, mixed storage cattle manure, horse feces, thermophilic digesters of biowaste or sewage sludge, biogas plants, anaerobic lab-scale bioreactors for thermophilic digestion of sewage sludge, and samples from a wastewater treatment plant (WWTP), located in northwestern Germany [22,23]. Several MTns and CTns were identified through comparison with reference genomes (Tn5397 (AF333235.1), Tn916 (KC414929.1), Tn6194 (HG475346.1), Tn5801-like (EU918655.2)), including Tn916, Tn6194-like, Tn5397, Tn6215, Tn4001, Tn6073, Tn6110, Tn6107, and Tn5801-like. MTns and CTns were detected in 48.2% (80/166) of C. difficile isolates (Table 1 and Table S1).

Table 1.
MGEs associated with AMR genes in environmental C. difficile strains.
The majority of MTns and CTns identified were Tn916, Tn6107, Tn4001, Tn6194-like, and Tn6073, followed by Tn5397 and Tn6110, Tn5801-like, and Tn6215. These were found in 56 (33.7%), 12 (7.2%), 8 (4.8%), 6 (3.6%), 5 (3%), 3 (1.8%), 2 (1.2%), and 1 (0.6%) isolates, respectively (Figure 1A). To establish a genomic context for the analysis of AMR genes, a screening procedure was employed in which draft genomes were examined for the presence of MTns and CTns. The most prevalent transposons carrying tetM were Tn916 (33.7%, 56/166), Tn5397 (1.8%, 3/166), and Tn5801-like (1.2%, 2/166), which were widely distributed among strains from diverse sources (Table 1 and Table S1). Transposons carrying the ermB gene included Tn6194-like (3.6%, 6/166) and Tn6215 (0.6%, 1/166). Tn6215 was identified exclusively in the RSS11/RT010/ST15 strain (Table 1 and Table S1). Tn4001 was identified as containing IS256 elements flanking the aac(6′)-aph(2″) gene, which confers resistance to gentamicin, tobramycin, and kanamycin. These elements were verified in eight isolates (4.8%) (Table 1 and Table S1).
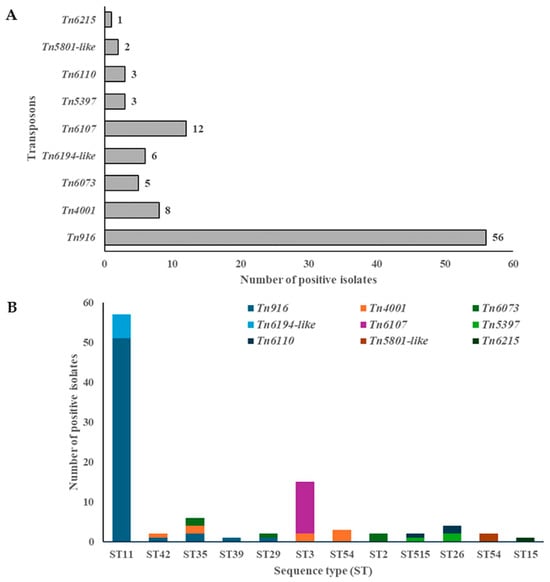
Figure 1.
The presence of MTns and CTns (A) and their association with sequence types (STs) (B) in environmental C. difficile isolates.
Most MTns and CTns were found within ST11 (51 (60.6%)), ST3 (15 (16%)), ST35 (6 (6.4%)), ST26 (4 (4.3%)), and ST54 (3 (3.2%)) (Figure 1B). Among these STs, ST35 demonstrated the highest level of diversity, with three different MTns/CTns identified. The following STs, ST11, ST42, ST3, ST515, and ST26, each exhibited two different MTns/CTns. A single instance of MTns/CTns was identified among the remaining STs (Figure 1B). Tn5397, also known as CTn3, was found in 3 out of 166 isolates (1.8%) (TDS118/RT140/ST515, RSS12/RT140/ST26, and RSS52/RT140/ST26). The tndX gene, which encodes a large serine recombinase, was identified in all Tn5397 transposons within the genomes of three strains that have been isolated from a thermophilic digester treating sewage sludge and raw sewage sludge. This recombinase is essential for the insertion and excision of Tn5397. All Tn5397 transposons were found to carry the tetracycline resistance gene tetM. Tn5397 was identified in only two different STs: ST515 and ST26 (Figure 1B).
Tn916 is a prominent transposon family that has been documented in C. difficile strains and is known to carry the tetM gene. In the present study, Tn916 was identified in the majority (33.7%) of environmental C. difficile isolates (Table 1), which were recovered from calf feces, raw sewage sludge, digested-sludge-amended soils, and biogas plants (Table 1 and Table S1). The tetM and int genes, which is responsible for insertion, were observed in all Tn916 transposons. Tn916 was predominantly identified in hypervirulent RTs, including RT078, RT126, and RT127, which belong to the ST11 group. Notably, a considerable proportion of environmental C. difficile isolates exhibited the presence of multiple Tn916 transposons, with 1–4 copies per isolate, as documented in Table S1.
It was found that a significant proportion of environmental C. difficile strains are characterized by the presence of multiple MTns and/or CTns. For instance, Tn916 and Tn6194-like were identified within the same chromosomal genome of the strain DS158/RT126/ST11, which carried tetM and ermB genes, encoding resistance to tetracycline and aminoglycosides, respectively (Figure 2A). Furthermore, the strain RS35/RT012/ST54 was found to carry the transposons Tn5801-like and Tn4001, which are associated with tetM and aac(6′)-aph(2″) resistance genes, respectively (Figure 2B). In addition, strains TDS118/RT140/ST515 and RSS37/RT031/ST29 carried two different transposons (Tn5397 and Tn6110, Tn916 and Tn6073), respectively (Figure 2C,D).
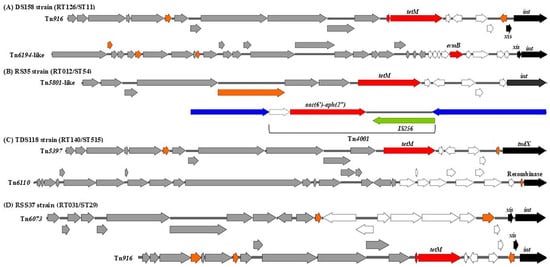
Figure 2.
Schematic view of representative transposons in a selection of C. difficile strains. Black: recombination genes, white: transcriptional regulation, red: accessory genes (AMR genes), grey: conjugation modules, orange: hypothetical proteins or proteins of unknown function. The schematic of transposons was generated using SnapGene (version 8.0.1).
2.1.2. Plasmids
It is hypothesized that this study is the first to demonstrate the presence of a substantial number of plasmids in the genomes of C. difficile isolates obtained from diverse fecally contaminated environmental sources. Plasmids exhibiting nearly 80–100% coverage were identified in 41 out of 166 isolates, corresponding to 24.7%, and ranged in size from 801 bp to 120,098 bp across 16 distinct ST patterns (Figure 3 and Table S2). Most isolates carried between one and sixteen plasmids in their genomes. The most common plasmid counts per isolate were 2, 1, 3, and 5, found in 19, 10, 5, and 4 isolates, respectively. In contrast, two isolates carried four plasmids, and one isolate was found to harbor sixteen plasmids (Figure 3).
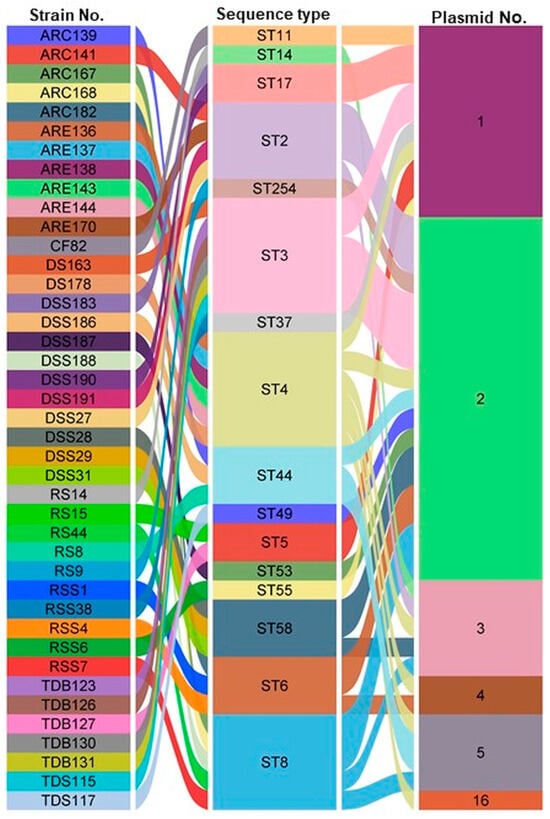
Figure 3.
An alluvial diagram representing, from right to left, the number of identified plasmids and their association with STs and C. difficile strains isolated from different environmental sources. The width of each connection is proportional to the number of positive isolates. RSS: raw sewage sludge, RS: raw sewage, DSS: digested sewage sludge, CF: calf feces, ARC: anaerobic lab-scale bioreactors treating sewage sludge/control, ARE: anaerobic lab-scale bioreactors treating sewage sludge/experiment, DS: digested-sewage-sludge-amended soils, TDS: thermophilic digester treating sewage sludge, TDB: thermophilic digester treating biowaste. The alluvial diagram was generated using SRplot [24].
The majority of plasmids were identified in the genome sequences of strains belonging to ST4, ST3, and ST8, with seven, six, and five isolates, respectively. Meanwhile, plasmids of the specified types were observed in three isolates each from ST44, ST2, ST58, and ST6, while the other STs occurred at lower frequencies, being represented by only one or two isolates each (Figure 3). This study also demonstrated that 7.2% (12/166) of environmental C. difficile strains carry a plasmid belonging to the pCD6 family (Table S2). The majority of pCD6 plasmids were found in isolates belonging to ST4, ST2, and ST8, which were obtained from DSS, ARC and ARE, and TDB and TDS (Figure 3 and Table S2).
Plasmids belonging to the pCD-WTSI subfamilies were identified in 12.04% (20/166) of environmental C. difficile isolates (Figure 3 and Table S2). The pCD-WTSI subfamilies were distributed across a range of STs, including ST58, ST44, ST3, ST6, ST49, ST14, and ST8 (Figure 3 and Table S2). Within the pCD-ECE families, four plasmids were identified in environmental C. difficile strains belonging to the pCD-ECE4 family (2.4%), whereas the pCD-ECE5 and pCD-ECE6 families were each identified in a single isolate. The pCD-ECE1, pCD-ECE2, and pCD-ECE3 plasmids were not detected. The maximum number of ECEs documented in a single isolate was two. The majority of ECEs were observed in isolates belonging to ST58, ST14, and ST8 (Table S2). The present study has revealed that plasmids of the identified types are present in the genomes of C. difficile isolates recovered from various fecally contaminated environmental sources, with the exception of samples derived from activated sewage sludge (ASS) and biogas plants (BPs) (Figure 3 and Table S2).
2.2. Phylogenetic Analysis of Identified Plasmids in Environmental C. difficile Genome Sequences
A phylogenetic tree was constructed using the complete sequences of both the identified plasmids and reference plasmids. Potential plasmids extracted from the genomes of environmental strains were clustered based on their average nucleotide identity (ANI) with available reference sequences. Among the plasmids identified in this study, 23 fall within a distinct pCD6 cluster (Figure 4). Based on phylogenetic analysis, a significant proportion (4.8%, 8/166) of environmental strains carry cryptic 45–48 kb plasmids belonging to the pDLL3026 family. Additionally, four plasmids were found to cluster distinctly within the pCDBI1 and pDSM1296 plasmids (Figure 4).
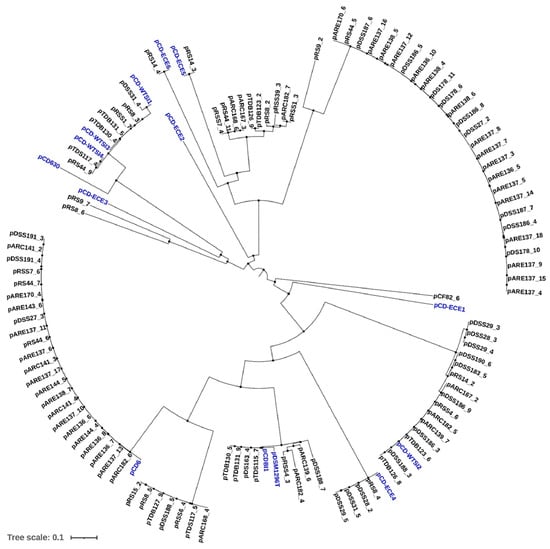
Figure 4.
Phylogenetic analysis of identified plasmids in environmental C. difficile strains. Reference plasmids (blue), pCD-ECE6 (LR594546.1), pCD-ECE4 (LR594545.1), pCD-WTSI2 (NZ_JADKQL030000003.1), pCD6 (NC_005326.1), pCD-WTSI4 (NZ_MG019962.1), pCD-WTSI3 (NZ_MG019961.1), pCD-ECE5 (LR594541.1), pCD-WTSI1 (NZ_MG019959.1), pCDI1 (NC_017176.1), pDSM1296 (NZ_CP011969.1), pCD-ECE1 (NZ_LR594544.1), pCD-ECE2 (NZ_LR594542.1), pCD-ECE3 (NZ_LR594543.1), and pCD630 (NC_008226.2).
It is noteworthy that the identified plasmids carrying phage-related functions were distributed into two distinct clusters. One of these clusters exhibited 100% sequence identity with the reference plasmids pCDBI1 and pDSM1296 and included plasmids, such as pTDB130_5, pTDB131_9, pDS163_4, and pTDS115_7 (Figure 4). Moreover, these plasmids, which belong to the pDLL3026 family, were found exclusively in ST3 isolates (Table S2). The second cluster comprises several plasmids identified in the current study, including pRSS7_4, pRS44_11, pARC167_3, pARC168_6, pTDB123_2, pTDB126_9, pRSS39_3, pRS8_2, pARC182_7, and pRSS1_3 (Figure 4). These plasmids are hypothesized to have arisen through recombination with bacteriophages, despite retaining key plasmid features, such as a replication initiation protein (RepB).
The potential plasmids identified in this study were also found to be similar to particular clusters of pCD-ECE4, pCD-ECE5, or pCD-ECE6 plasmids, with four corresponding to pCD-ECE4 and one each to pCD-ECE5 and pCD-ECE6 (Figure 4). Plasmids pRS9_7, pRS9_2, and pRS8_6 were found to be closely related, exhibiting 100% identity and over 98% coverage with Enterococcus faecium plasmids (Figure 4 and Table S2). Furthermore, an additional 22 plasmids demonstrating 99–100% sequence identity and coverage with the pCD-WTSI subfamilies were also identified (Figure 4 and Table S2).
A total of four identified plasmids, pRS15_2, pRS8_5, pDSS188_5, and pARC168_4, were closely related, showing 100% sequence identity and coverage with the pCD-ECE6 plasmid (Table S2). However, these plasmids did not cluster with the reference plasmid (Figure 4). Instead, they formed a cluster with three other previously identified plasmids (pTDB127_9, pRSS6_4, and pTDS117_5), which were identical to the reference plasmid pAR1090_2 (Table S2). Finally, 27 plasmids—ranging from pARE170_6 to pARE137_4 (Figure 4)—were found to be identical to the reference plasmids pCd1_3, pCd11_5, pCd8, or pCd5_4 (Table S2). Notably, these reference plasmids were all isolated from the same source.
2.3. Comparison of the Identified Plasmids with Reference Plasmids
The assignment of the representative identified plasmids to the reference plasmids is preliminary and based on regions of similarity to the reference plasmid sequences. To gain further insight into the relatedness of the plasmids, nucleotide alignments were visualized using clinker [25]. Several observations emerged from these analyses. Firstly, the pTDS117_4 plasmid appears to contain a 17-gene insertion compared to the pCD-WTSI4 reference sequence. The ORF1 and ORF17 regions of pTDS117_4 appear to be non-conserved in the reference plasmid pCD-WTSI4 (Figure 5E). ORF1 of pTDS117_4 encodes a DEAD/DEAH box helicase, while ORF17 encodes a non-functional (hypothetical) protein. Secondly, the ORF4, ORF6, and ORF8 regions of the reference pCD-ECE5 plasmid appear to be non-conserved in the pRS14_3 plasmid (Figure 5G). These three ORFs in the pCD-ECE5 reference sequence encode non-functional proteins. Thirdly, the pDSS31_4 plasmid contains a conserved region but also includes one insertion compared to the reference plasmid pCD-WTSI4 (Figure 5H). ORF1 of pDSS31_4 is hypothesized to encode an additional DEAD/DEAH box helicase. Finally, the plasmid pARE137_10 contains a 7-gene insertion compared to the pCD6 reference sequence and the pARE136_8 plasmid sequence. ORF1 of pARE137_10 encodes a hypothetical protein of unknown function, while ORF7 is hypothesized to encode an additional replication protein region, including the repA gene, which encodes a 545-amino acid protein (Figure 5D).
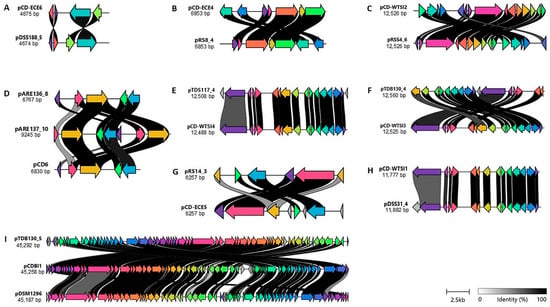
Figure 5.
Comparison of representative identified plasmids with reference plasmids. Within the panels, the presence of colored arrows indicates the presence of similar genes; links are drawn between similar genes on neighboring clusters and shaded based on sequence identity (0% white, 100% black, identity threshold for visualization 0.30). (A). Comparison of pDSS188_5 with pCD-ECE6 (LR594546.1). (B). Comparison of pRS8_4 with pCD-ECE4 (LR594545.1). (C). Comparison of pRSS4_6 with pCD-WTSI2 (NZ_JADKQL030000003.1). (D). Comparison of pARE137_10 and pARE136_8 with pCD6 (NC_005326.1). (E). Comparison of pDTS117_4 with pCD-WTSI4 (NZ_MG019962.1). (F). Comparison of pTDB130_4 with pCD-WTSI3 (NZ_MG019961.1). (G). Comparison of pRS14_3 with pCD-ECE5 (LR594541.1). (H). Comparison of pDSS31_4 with pCD-WTSI1 (NZ_MG019959.1). (I). Comparison of pTDB130_5 with pCDI1 (NC_017176.1) and pDSM1296 (NZ_CP011969.1). The image has been derived from a visualization using clinker [25].
It is probable that the non-conjugative cryptic plasmids belong to the pDLL3026 plasmid family, which includes pCDBI1, pDSM1296, and other plasmids identified in the present study, such as pTDB130_5 (Figure 5I). These plasmids contain a RepB, genes involved in transcriptional regulation, and members of the ParM/StbA family, as well as several phage-related functions. Furthermore, the annotation of the pTDB130_5 plasmid revealed the presence of 69 ORFs, whereas the reference plasmids pCDBI1 and pDSM1296 contain 66 and 67 ORFs, respectively. The predicted functions of these ORFs were categorized into seven groups: plasmid maintenance, DNA replication and repair, restriction endonuclease and DNA modification, transcriptional regulation, recombination, hypothetical proteins and proteins of unknown function, and phage or phage-related functions (Figure 5I).
2.4. The Genome Organization of Identified Representative Cryptic Plasmids of C. difficile
All plasmids identified in the studied genome sequences of different environmental C. difficile strains were found to be cryptic. Therefore, representative identified plasmid genomes are displayed schematically in Figure 6. The plasmids pRS8_5 and pRSS6_4 contained only four annotated genes, while pDSS31_5, pTDS117_4, pDSS190_6, and pARE170_4 contained 7, 17, 17, and 21 genes, respectively. The majority of these genes encode proteins involved in transcriptional regulation, as well as proteins of hypothetical or unknown function. In many bacterial species, plasmids play a significant role in AMR, often mediating the transmission of resistance genes. However, in C. difficile, AMR genes are predominantly located on transposons rather than on plasmids [13]. Additionally, a total of 17 identified plasmids were found to carry phage-related genes. These genes were specifically associated with the intact prophage phiCDHM19. One plasmid, pARC182_4 (RT005/ST6), was found to be linked to the incomplete prophage phiCDHM11 (Table S3). Reference plasmids—including pCDBI1, p830101_1, pCd11_3, pCd10_2, pJ21_1, pDSM1296, and unnamed 1_CD4 that are identical to the identified plasmids—were screened for the presence of prophage phiCDHM19. The results indicated that all reference plasmids harbored the same prophage as those identified in the environmental C. difficile genomes.
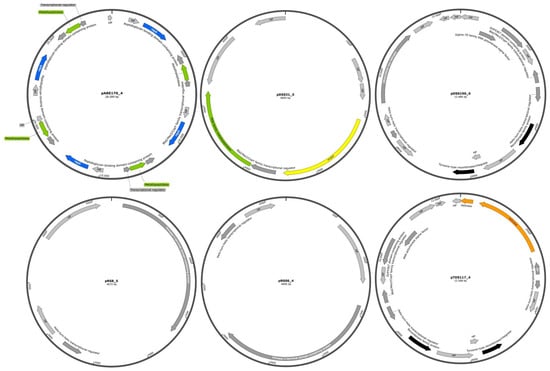
Figure 6.
Genome organization of representative cryptic C. difficile plasmids. Predicted CDSs are marked with arrows and colors that indicate functional assignments: plasmid maintenance (blue), DNA replication and repair (orange), restriction endonuclease and DNA modification (green), transcriptional regulation (dark grey), recombination (black), hypothetical proteins and proteins of unknown function (light grey), and conjugation modules (yellow). The genome maps of representative identified plasmids were generated using SnapGene (version 8.0.1).
3. Discussion
It is evident that essential MGEs and integrative conjugative elements (ICEs) play a crucial role in horizontal gene transfer (HGT), facilitating increased genetic diversity and the acquisition of exogenous genetic material [14]. Transposons, including Tn916, Tn5397, Tn5398, and Tn4453, have been widely distributed across diverse STs of C. difficile isolates [13]. In this study, nine types of MTns or CTns could be identified in the genome sequences of environmental C. difficile isolates, including Tn916, Tn6194-like, Tn5397, Tn6215, Tn4001, Tn6073, Tn6110, Tn6107, and Tn5801-like. These transposons were identified in 11 distinct STs, the majority of which belonged to hypervirulent RTs, including RT126, RT127, and RT78, which were found to be associated with the ST11 group.
The most commonly detected transposons in environmental C. difficile genomes were Tn916, Tn6107, Tn4001, and Tn6194-like, with Tn916 being the most prevalent, present in 33.7% of isolates. Notably, Tn916 was predominantly linked to hypervirulent RTs, such as RT126, RT127, and RT078, which are all part of the ST11 lineage and carry the tetM gene. The acquisition of tetracycline resistance in C. difficile strains has previously been linked to transposons including Tn5397, the Tn916-like family, and Tn6164 [26]. The findings of the present study align with previous research showing Tn916 (53.7%) and Tn5397 (31.48%) as the most prevalent tetM-carrying transposons among environmental, animal, and clinical strains in China [27]. Furthermore, tetM associated with Tn916 has also been identified in both toxigenic and non-toxigenic C. difficile strains from Southeast Asia [28]. Dingle et al. reported that 76.5% of agriculture-associated C. difficile RT078 strains were positive for the tetM gene [29]. Overall, Tn5397 and Tn916-like transposons are the predominant genetic elements mediating tetracycline resistance through the tetM gene [7,30].
In addition to Tn916-like elements, tetM was linked to Tn5397 and Tn5801-like transposons, detected in three and two isolates, respectively. This highlights the potential role of CTns in driving the evolution and widespread dissemination of tetM among both toxigenic and non-toxigenic C. difficile strains and possibly to other enteric pathogens. For instance, it has been demonstrated that Tn916, originally identified in C. difficile, can be transferred to Bacillus subtilis [9]. Tn5397, formerly known as CTn3, was first identified in C. difficile strain 630. This 21 kb element closely resembles the Tn916-like family [4], which is the paradigm of this MGE family. The transferability of Tn5397 has been demonstrated between C. difficile stains and B. subtilis [10,11] and Enterococcus faecalis [12]. Vries et al. reported that Tn5801-like genomic islands (GIs) are widespread in tetracycline-resistant Staphylococcus pseudintermedius, as well as in other Gram-positive species [31]. While Tn5801 shares several ORFs with Tn916, it differs by containing a unique integrase gene (intTn5801) instead of the xisTn916/intTn916 excisionase/integrase system found in Tn916. Tn916 was detected in C. difficile isolates from calf feces and WWTP samples. Agricultural use of tetracyclines likely drives selection pressure and, coupled with AMR spread via the food chain, facilitates the persistence of resistant C. difficile strains in both clinical and environmental contexts. Resistance to macrolide–lincosamide–streptogramin B (MLSB) antimicrobials in C. difficile strains is predominantly mediated by the ermB gene. In the present study, transposons carrying the ermB gene were identified, including Tn6194-like (3.6%) and Tn6215 (0.6%). The Tn6194-like element was detected in the genome sequences of RT126 and RT078 strains belonging to the ST11 group, while Tn6215 was identified in an RT010/ST15 strain. A recent study involving environmental, animal, and clinical isolates reported that ermB-carrying transposons could be classified into four categories: Tn6189 (9.0%), Tn6194 (5.3%), Tn6215 (9.0%), and Tn6218 (64.0%), based on their distribution among different origins [27]. Additionally, Imwattana et al. demonstrated that both toxigenic and non-toxigenic C. difficile strains from Southeast Asia harbor the ermB gene on various transposons, such as Tn6218, Tn6189, Tn6194-like, and Tn6215 [28]. Further analysis of publicly available genomes of ermB-positive C. difficile strains revealed that the most common ermB-positive transposon is Tn6194 (44.4%), followed by Tn6189 (23.9%) and Tn6218 (12.2%), among 1775 strains [32]. Wasels et al. showed that the ermB-carrying Tn6194-like, identified in a C. difficile RT001 strain, was capable of horizontal transfer of AMR not only within C. difficile strains (RT009 and RT027) but also between C. difficile and Enterococcus faecalis [16].
The transposon Tn4001 was found to contain a single copy of the IS256 element, which encodes a bifunctional aminoglycoside-modifying enzyme with both acetyltransferase and phosphotransferase activity—specifically, aac(6′)-aph(2″)—in 4.8% of the 166 C. difficile strains analyzed. As demonstrated by Ramírez-Vargas et al., a sequence copy of the composite Tn4001 could be detected in human C. difficile isolates of the NAPCR1/RT012/ST54 genotype. This sequence was found to be inserted within a novel putative MTn, which also carried the aac(6′)-aph(2″) gene [33]. The composite transposon Tn4001, which consists of two flanking IS256 elements, was originally identified in Staphylococcus aureus [34].
In addition, three other transposons, Tn6073, Tn6107 (CTn5-like element), and Tn6110, were detected in 3.0%, 7.2%, and 1.8% of C. difficile isolates, respectively. Brouwer et al. reported the presence of Tn6073, Tn6107, and Tn6110 in C. difficile strains QCD-23M63, QCD-23M63, and QCD-66C26, respectively [4]. The accessory modules of these transposons encode a diverse set of genes, including those for a predicted N-terminal hydrolase, a sigma factor, putative transcriptional regulators, and ABC transporters, all of which may play roles in facilitating the adaptation and persistence of C. difficile across diverse environmental niches.
Among the 166 C. difficile genomes analyzed in the present study, 41 (24.7%) were found to harbor potential plasmids, distributed across 16 distinct STs. These plasmids were predominantly classified into major groups such as pCD6, pCD-ECE4–6, pCD-WTSI1–4, pCDBI1, and pCd1_3, highlighting the widespread occurrence and diversity of plasmids within the species. Previous studies have demonstrated that C. difficile strains from diverse geographic regions harbor plasmids of varying sizes [35]. For example, Dost et al. reported plasmid presence in 87.7% (471/537) of a C. difficile genome collection belonging to ST8/RT002, with the most prevalent plasmids being pCDBI1, pAK2, pCDT4, pCD116-S, and plasmid 1 from C. difficile strain FDAARGOS_267 [36]. In the present study, it was also observed that individual isolates can carry multiple plasmids from specific families. This observation aligns with prior in silico analyses of publicly available genome data, which have indicated the co-occurrence of multiple plasmids within a single isolate and suggested that certain plasmid families may be compatible and capable of co-existing within the same host strain [18,37]. Roseboom et al. demonstrated that a human clinical isolate carried three plasmids from three different plasmid families, including pCD-ECE1, pCDWTSI1, and pCD630 [38].
The present study investigated the prevalence of a specific plasmid, pCD6. A total of 7.2% of C. difficile strains from different environmental sources were found to carry this plasmid. pCD6 remains the only C. difficile plasmid that has been fully characterized. Its 6.8 kb nucleotide sequence was originally identified during attempts to establish genetic manipulation methods in strain CD6 [39]. Moreover, the copy number of the pCD6 plasmid ranged between 4 and 10 [40]. The pCD6 replicons are characterized by encoding a RepA that shows significant similarity to the RepA protein of the C. perfringens plasmid pIP404 [41]. A comprehensive sequencing study conducted at a major Australian hospital revealed the presence of various plasmids, including pCD6, pCDBI1, pDLL3026, and pCD630, in approximately 27% of C. difficile strains analyzed (19 out of 71). Notably, pCD6 was identified in 12 of these strains, constituting the most prevalent plasmid detected [42].
Approximately 4.8% of environmental C. difficile strains were found to harbor cryptic 45–48 kb plasmids, classified within the pDLL3026 family. This group includes pCDBI1, which carries prophage sequences homologous to the intact temperate phage phiCDHM19, suggesting a potential role in HGT or lysogenic conversion. In our previous study, 17% of intact phiCDHM19 prophages were identified within contigs of environmental C. difficile genome sequences [6]. This finding is consistent with a prior study in which 4.9% of C. difficile isolates were found to carry cryptic plasmids ranging from 42 to 47 kb. These plasmids are believed to have arisen through recombination with bacteriophages but still retain essential plasmid features [17]. Furthermore, the pDLL3026 plasmid was found to contain multiple putative bacteriophages, including temperate phages from the Myoviridae family and one intact temperate phage from the Siphoviridae family [17].
In the present study, pCD-ECE family elements were identified in 3.6% of the 166 C. difficile genome sequences, with 1–2 elements per strain from different subfamilies. In contrast, Hornung et al. reported that approximately 13% of C. difficile sequence data contained ECEs, with 1–6 elements per strain [18]. In this study, most ECEs were affiliated with various STs, such as ST58, ST14, ST44, ST8, and ST5. Conversely, analysis of publicly available C. difficile genomes revealed that the majority of ECEs were associated with ST1, ST11, and ST37 [18].
In addition to the many new findings, the currently existing limitations of this investigation should also be mentioned. First, a comparative genomic analysis of environmental and clinical C. difficile strains from the same geographic region could not be performed, as only a limited number of human isolates were available. This constraint limits the ability to assess potential transmission dynamics or genetic overlap between environmental and clinical populations. Second, it is evident that research in this area is limited, with no studies to date investigating the transferability of AMR-associated MGEs both within C. difficile populations and between C. difficile and other pathogenic bacteria originating from diverse environmental niches.
4. Materials and Methods
4.1. Isolation and Identification of C. difficile
C. difficile isolates utilized in the present study were recovered from 81 fecally contaminated environmental samples collected from March 2021 to June 2022, including WWTP samples, calf feces, cattle-feces-contaminated soil, thermophilic digesters treating biowaste and sewage sludge, and digested-sewage-sludge-amended soils, as previously described [22]. Briefly, environmental samples were inoculated in C. difficile selective (CD) broth supplemented with 12 mg/L norfloxacin (Sigma-Aldrich Chemie GmbH, Munich, Germany) and 32 mg/L moxalactam (Biomol GmbH, Hamburg, Germany) and 0.1% sodium taurocholate (Carl Roth GmbH & Co. KG, Karlsruhe, Germany) for spore germination. All inoculated CD broths were prepared anaerobically in an anaerobic chamber (Coy Laboratory Products, Inc. Los Angeles, CA, USA) and flushed with a gas mixture (80% N2 and 20% CO2). All inoculated CD broths were incubated at 37 °C for 7–10 days. Each incubated CD broth was then mixed with an equal volume of absolute alcohol (1:1) and incubated at room temperature for 50–60 min. The mixtures were then centrifuged at 4000 rpm for 10 min and the supernatant was discarded. The pellet was resuspended in 1 phosphate-buffered saline (PBS) and plated on Clostridium difficile agar (CDA, Fisher Scientific GmbH, Schwerte, Germany) supplemented with 7% defibrinated horse blood (Fisher Scientific GmbH, Schwerte, Germany), 12 mg/mL norfloxacin, 32 mg/mL moxalactam, and 0.1% sodium taurocholate. All plates were incubated anaerobically in anaerobic jars (Schuett-Biotec GmbH, Göttingen, Germany) (80% N2, 10% H2, and 10% CO2) at 37 °C for 2–5 days. Selected colonies were evaluated by morphology and confirmed by the Oxoid C. difficile latex agglutination test (Fisher Scientific GmbH, Schwerte, Germany). The final confirmation was made by analyzing the specific housekeeping gene, triose phosphate isomerase (tpi), as previously described by Leeme et al. [43].
4.2. Whole Genome Sequencing and Data Analysis
In total, 166 environmental C. difficile isolates were subjected to WGS using the Pacific Biosciences long-read platform Sequel IIe (Pacific Biosciences Inc., Menlo Park, CA, USA) and were subsequently assembled de novo using the SMRT Link software versions 10 and 11 (Pacific Biosciences Inc.) as described recently [44]. The MLSTs (STs) were determined according to the C. difficile MLST database of the PubMLST website (https://pubmlst.org/organisms/clostridioides-difficile (accessed on 15 November 2022)). All sequenced contigs were annotated using the rapid annotation using subsystem technology (RAST) web server version 2.0 (https://rast.nmpdr.org/ (accessed on 15 November 2022)). AMR genes were identified in the genome sequences of 166 C. difficile strains with the comprehensive antibiotic resistance databases (CARD) version 2 using resistance gene identifier (RGI) (https://card.mcmaster.ca/ (accessed on 11 April 2023)), BacAnt [45], ResFinder 4.1 (https://cge.food.dtu.dk/services/ResFinder/ (accessed on 11 April 2023)) [46], ARG-ANNOT [47], and Vrprofile2 [48]. The previously deposited WGS data, including the complete set of contig sequences, were submitted to the National Center for Biotechnology Information (NCBI) GenBank database under BioProject accession number PRJNA1011814 [23]. The accession numbers for the individual plasmid sequences identified in this study are listed in Table S2.
4.3. Analysis of MGEs
MTns, CTns, and their association with AMR genes were identified in C. difficile genome sequences using web servers, such as BacAnt [45] and Vrprofile2 [48]. These findings were complemented by a BLASTn search of the C. difficile genome sequences or transposon sequences available at NCBI (https://www.ncbi.nlm.nih.gov/ (accessed on 24 October 2024)). The MTns and CTns were defined as having >80% nucleotide identity and coverage [49]. To identify plasmids, contigs were screened for plasmids using BLASTn against the NCBI complete plasmid database. The potential plasmids were defined as having ≥70% coverage and ≥80% identity [49]. Prophages were identified using the PHASTEST web server in the identified plasmids which carry phage-related genes. The intact, questionable, and incomplete prophage sequences were defined by score values of >90, 70 to 90, and <70, respectively [50].
4.4. Phylogenetic Tree of Identified Plasmids in Environmental C. difficile Strains
The construction of the phylogenetic tree was achieved by employing the complete sequences of the identified and reference plasmids in order to ascertain the genetic relatedness between the identified plasmids and well-known reference plasmids, such as pCD-ECE6 (LR594546.1), pCD-ECE4 (LR594545.1), pCD-WTSI2 (NZ_JADKQL030000003.1), pCD6 (NC_005326.1), pCD-WTSI4 (NZ_MG019962.1), pCD-WTSI3 (NZ_MG019961.1), pCD-ECE5 (LR594541.1), pCD-WTSI1 (NZ_MG019959.1), pCDI1 (NC_017176.1), pDSM1296 (NZ_CP011969.1), pCD-ECE1 (NZ_LR594544.1), pCD-ECE2 (NZ_LR594542.1), pCD-ECE3 (NZ_LR594543.1), and pCD630 (NC_008226.2). The Newick tree was made using the NGPhylogeny.fr web server (https://ngphylogeny.fr/, accessed on 15 February 2025) [51]. Subsequently, the phylogenetic analysis was visualized using the integrative Tree of Life (iTOL) web server.
4.5. Comparison of the Identified Plasmids with Reference Plasmids
In the present study, the representatively identified plasmids were compared with the reference plasmids. This is achieved by basing the comparison on regions of similarity with the reference plasmid sequences. In order to gain further insight into the genetic relatedness of the plasmids, nucleotide alignments were visualised using Clinker [25].
5. Conclusions
The present study showed that putative MTns and CTns are present in nearly half of the genomes of environmental C. difficile strains. Despite the variability exhibited by the accessory regions of the CTns, several distinct types of core elements were identified, including Tn916, Tn5397, Tn6194, and Tn6215. These elements were found to be associated with AMR genes that encode tetracycline and aminoglycoside resistance. Furthermore, given that some CTns have the capacity to transfer between genera, there is a possibility that they may enable both toxigenic and non-toxigenic C. difficile strains—as well as other enteric pathogens—to access and incorporate parts of the mitogenome of the intestinal tract. The results also reveal a high abundance of plasmids in C. difficile, suggesting a potential role in modulating key physiological functions. Further investigation is necessary to determine how these genetic elements influence the organism’s biology and its capacity for pathogenicity.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/antibiotics14070678/s1, Table S1: MGEs associated with AMR genes in the genomes of 166 C. difficile isolates in this study; Table S2: Potential identified plasmids in the genomes of environmental C. difficile strains; Table S3: The occurrence of prophages predicted in identified plasmids of environmental C. difficile strains, which carry phage-related genes.
Author Contributions
Conceptualization, K.B. and C.G.; methodology, K.B.; software, K.B.; validation, K.B. and C.G.; formal analysis, K.B.; investigation, K.B.; resources, K.B.; data curation, K.B.; writing—original draft preparation, K.B.; writing—review and editing, K.B. and C.G.; visualization, K.B.; supervision, C.G.; project administration, C.G.; funding acquisition, C.G. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the German Research Foundation (Deutsche Forschungsgemeinschaft, DFG) within the project SUPERsafe “Survival and pathogenicity of Clostridioides difficile in sewage, sewage sludge, surface water, animal, manure, fodder, crops and silage-Treatment requirements to minimize health risks”, grant number (GA 546/13-1).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
All supporting data and protocols have been provided within the article or through Supplementary Data Files.
Acknowledgments
The authors extend thanks to Alexander Mellmann for carrying out the whole genome sequencing of the strains at the Institute of Hygiene, Münster University, Münster, Germany.
Conflicts of Interest
The authors declare no conflicts of interest.
Abbreviations
The following abbreviations were used in this manuscript:
| CDI | C. difficile infection |
| WGS | Whole genome sequencing |
| CTns | Conjugative transposons |
| MGEs | Mobile genetic elements |
| AMR | Antimicrobial resistance |
| MTns | Mobilizable transposons |
| MLST | Multilocus sequence typing |
| IS | Insertion sequence |
| ICEs | Integrative conjugative elements |
| HGT | Horizontal gene transfer |
| MLSB | Macrolide–lincosamide–streptogramin B |
| GI | Genomic island |
| ST | Sequence type |
| ORF | Open reading frame |
References
- Smits, W.K.; Lyras, D.; Lacy, D.B.; Wilcox, M.H.; Kuijper, E.J. Clostridium difficile infection. Nat. Rev. Dis. Prim. 2016, 2, 16020. [Google Scholar] [CrossRef] [PubMed]
- Aktories, K.; Schwan, C.; Jank, T. Clostridium difficile Toxin Biology. Annu. Rev. Microbiol. 2017, 71, 281–307. [Google Scholar] [CrossRef] [PubMed]
- Knight, D.R.; Elliott, B.; Chang, B.J.; Perkins, T.T.; Riley, T.V. Diversity and Evolution in the Genome of Clostridium difficile. Clin. Microbiol. Rev. 2015, 28, 721–741. [Google Scholar] [CrossRef] [PubMed]
- Brouwer, M.S.M.; Warburton, P.J.; Roberts, A.P.; Mullany, P.; Allan, E. Genetic organisation, mobility and predicted functions of genes on integrated, mobile genetic elements in sequenced strains of Clostridium difficile. PLoS ONE 2011, 6, e23014. [Google Scholar] [CrossRef]
- Sebaihia, M.; Wren, B.W.; Mullany, P.; Fairweather, N.F.; Minton, N.; Stabler, R.; Thomson, N.R.; Roberts, A.P.; Cerdeño-Tárraga, A.M.; Wang, H.; et al. The multidrug-resistant human pathogen Clostridium difficile has a highly mobile, mosaic genome. Nat. Genet. 2006, 38, 779–786. [Google Scholar] [CrossRef]
- Blau, K.; Gallert, C. Prophage Carriage and Genetic Diversity within Environmental Isolates of Clostridioides difficile. Int. J. Mol. Sci. 2023, 25, 2. [Google Scholar] [CrossRef]
- Peng, Z.; Jin, D.; Kim, H.B.; Stratton, C.W.; Wu, B.; Tang, Y.-W.; Sun, X. Update on Antimicrobial Resistance in Clostridium difficile: Resistance Mechanisms and Antimicrobial Susceptibility Testing. J. Clin. Microbiol. 2017, 55, 1998–2008. [Google Scholar] [CrossRef]
- Spigaglia, P. Recent advances in the understanding of antibiotic resistance in Clostridium difficile infection. Ther. Adv. Infect. Dis. 2016, 3, 23–42. [Google Scholar] [CrossRef]
- Mullany, P.; Williams, R.; Langridge, G.C.; Turner, D.J.; Whalan, R.; Clayton, C.; Lawley, T.; Hussain, H.; McCurrie, K.; Morden, N.; et al. Behavior and target site selection of conjugative transposon Tn916 in two different strains of toxigenic Clostridium difficile. Appl. Environ. Microbiol. 2012, 78, 2147–2153. [Google Scholar] [CrossRef]
- Roberts, A.P.; Pratten, J.; Wilson, M.; Mullany, P. Transfer of a conjugative transposon, Tn5397 in a model oral biofilm. FEMS Microbiol. Lett. 1999, 177, 63–66. [Google Scholar] [CrossRef]
- Mullany, P.; Wilks, M.; Lamb, I.; Clayton, C.; Wren, B.; Tabaqchali, S. Genetic analysis of a tetracycline resistance element from Clostridium difficile and its conjugal transfer to and from Bacillus subtilis. J. Gen. Microbiol. 1990, 136, 1343–1349. [Google Scholar] [CrossRef] [PubMed]
- Jasni, A.S.; Mullany, P.; Hussain, H.; Roberts, A.P. Demonstration of conjugative transposon (Tn5397)-mediated horizontal gene transfer between Clostridium difficile and Enterococcus faecalis. Antimicrob. Agents Chemother. 2010, 54, 4924–4926. [Google Scholar] [CrossRef]
- Mullany, P.; Allan, E.; Roberts, A.P. Mobile genetic elements in Clostridium difficile and their role in genome function. Res. Microbiol. 2015, 166, 361–367. [Google Scholar] [CrossRef] [PubMed]
- He, M.; Sebaihia, M.; Lawley, T.D.; Stabler, R.A.; Dawson, L.F.; Martin, M.J.; Holt, K.E.; Seth-Smith, H.M.B.; Quail, M.A.; Rance, R.; et al. Evolutionary dynamics of Clostridium difficile over short and long time scales. Proc. Natl. Acad. Sci. USA 2010, 107, 7527–7532. [Google Scholar] [CrossRef] [PubMed]
- He, M.; Miyajima, F.; Roberts, P.; Ellison, L.; Pickard, D.J.; Martin, M.J.; Connor, T.R.; Harris, S.R.; Fairley, D.; Bamford, K.B.; et al. Emergence and global spread of epidemic healthcare-associated Clostridium difficile. Nat. Genet. 2013, 45, 109–113. [Google Scholar] [CrossRef]
- Wasels, F.; Monot, M.; Spigaglia, P.; Barbanti, F.; Ma, L.; Bouchier, C.; Dupuy, B.; Mastrantonio, P. Inter- and intraspecies transfer of a Clostridium difficile conjugative transposon conferring resistance to MLSB. Microb. Drug Resist. 2014, 20, 555–560. [Google Scholar] [CrossRef]
- Amy, J.; Bulach, D.; Knight, D.; Riley, T.; Johanesen, P.; Lyras, D. Identification of large cryptic plasmids in Clostridioides (Clostridium) difficile. Plasmid 2018, 96–97, 25–38. [Google Scholar] [CrossRef]
- Hornung, B.V.H.; Kuijper, E.J.; Smits, W.K. An in silico survey of Clostridioides difficile extrachromosomal elements. Microb. Genom. 2019, 5, e000296. [Google Scholar] [CrossRef]
- Ramírez-Vargas, G.; López-Ureña, D.; Badilla, A.; Orozco-Aguilar, J.; Murillo, T.; Rojas, P.; Riedel, T.; Overmann, J.; González, G.; Chaves-Olarte, E.; et al. Novel Clade C-I Clostridium difficile strains escape diagnostic tests, differ in pathogenicity potential and carry toxins on extrachromosomal elements. Sci. Rep. 2018, 8, 13951. [Google Scholar] [CrossRef]
- Ramírez-Vargas, G.; Rodríguez, C. Putative Conjugative Plasmids with tcdB and cdtAB Genes in Clostridioides difficile. Emerg. Infect. Dis. 2020, 26, 2287–2290. [Google Scholar] [CrossRef]
- Boekhoud, I.M.; Hornung, B.V.H.; Sevilla, E.; Harmanus, C.; Bos-Sanders, I.M.J.G.; Terveer, E.M.; Bolea, R.; Corver, J.; Kuijper, E.J.; Smits, W.K. Plasmid-mediated metronidazole resistance in Clostridioides difficile. Nat. Commun. 2020, 11, 598. [Google Scholar] [CrossRef] [PubMed]
- Blau, K.; Gallert, C. Prevalence, Antimicrobial Resistance and Toxin-Encoding Genes of Clostridioides difficile from Environmental Sources Contaminated by Feces. Antibiotics 2023, 12, 162. [Google Scholar] [CrossRef] [PubMed]
- Blau, K.; Berger, F.K.; Mellmann, A.; Gallert, C. Clostridioides difficile from Fecally Contaminated Environmental Sources: Resistance and Genetic Relatedness from a Molecular Epidemiological Perspective. Microorganisms 2023, 11, 2497. [Google Scholar] [CrossRef]
- Tang, D.; Chen, M.; Huang, X.; Zhang, G.; Zeng, L.; Zhang, G.; Wu, S.; Wang, Y. SRplot: A free online platform for data visualization and graphing. PLoS ONE 2023, 18, e0294236. [Google Scholar] [CrossRef] [PubMed]
- Gilchrist, C.L.M.; Chooi, Y.-H. clinker & clustermap.js: Automatic generation of gene cluster comparison figures. Bioinformatics 2021, 37, 2473–2475. [Google Scholar] [CrossRef]
- Kartalidis, P.; Skoulakis, A.; Tsilipounidaki, K.; Florou, Z.; Petinaki, E.; Fthenakis, G.C. Clostridioides difficile as a Dynamic Vehicle for the Dissemination of Antimicrobial-Resistance Determinants: Review and In Silico Analysis. Microorganisms 2021, 9, 1383. [Google Scholar] [CrossRef]
- Zhou, Y.; Zhou, W.; Xiao, T.; Chen, Y.; Lv, T.; Wang, Y.; Zhang, S.; Cai, H.; Chi, X.; Kong, X.; et al. Comparative genomic and transmission analysis of Clostridioides difficile between environmental, animal, and clinical sources in China. Emerg. Microbes Infect. 2021, 10, 2244–2255. [Google Scholar] [CrossRef]
- Imwattana, K.; Kiratisin, P.; Riley, T.V.; Knight, D.R. Genomic basis of antimicrobial resistance in non-toxigenic Clostridium difficile in Southeast Asia. Anaerobe 2020, 66, 102290. [Google Scholar] [CrossRef]
- Dingle, K.E.; Didelot, X.; Quan, T.P.; Eyre, D.W.; Stoesser, N.; Marwick, C.A.; Coia, J.; Brown, D.; Buchanan, S.; Ijaz, U.Z.; et al. A Role for Tetracycline Selection in Recent Evolution of Agriculture-Associated Clostridium difficile PCR Ribotype 078. mBio 2019, 10, e02790-18. [Google Scholar] [CrossRef]
- O’Grady, K.; Knight, D.R.; Riley, T.V. Antimicrobial resistance in Clostridioides difficile. Eur. J. Clin. Microbiol. Infect. Dis. 2021, 40, 2459–2478. [Google Scholar] [CrossRef]
- de Vries, L.E.; Hasman, H.; Jurado Rabadán, S.; Agersø, Y. Sequence-Based Characterization of Tn5801-Like Genomic Islands in Tetracycline-Resistant Staphylococcus pseudintermedius and Other Gram-positive Bacteria from Humans and Animals. Front. Microbiol. 2016, 7, 576. [Google Scholar] [CrossRef]
- Imwattana, K.; Putsathit, P.; Collins, D.A.; Leepattarakit, T.; Kiratisin, P.; Riley, T.V.; Knight, D.R. Global evolutionary dynamics and resistome analysis of Clostridioides difficile ribotype 017. Microb. Genom. 2022, 8, 000792. [Google Scholar] [CrossRef] [PubMed]
- Ramírez-Vargas, G.; Quesada-Gómez, C.; Acuña-Amador, L.; López-Ureña, D.; Murillo, T.; Del Mar Gamboa-Coronado, M.; Chaves-Olarte, E.; Thomson, N.; Rodríguez-Cavallini, E.; Rodríguez, C. A Clostridium difficile Lineage Endemic to Costa Rican Hospitals Is Multidrug Resistant by Acquisition of Chromosomal Mutations and Novel Mobile Genetic Elements. Antimicrob. Agents Chemother. 2017, 61, e02054-16. [Google Scholar] [CrossRef] [PubMed]
- Lyon, B.R.; May, J.W.; Skurray, R.A. Tn4001: A gentamicin and kanamycin resistance transposon in Staphylococcus aureus. Mol. Gen. Genet. 1984, 193, 554–556. [Google Scholar] [CrossRef] [PubMed]
- Amy, J.; Johanesen, P.; Lyras, D. Extrachromosomal and integrated genetic elements in Clostridium difficile. Plasmid 2015, 80, 97–110. [Google Scholar] [CrossRef]
- Dost, I.; Abdel-Glil, M.; Persson, S.; Conza, K.L.; Oleastro, M.; Alves, F.; Maurischat, S.; Scholtzek, A.; Mazuet, C.; Diancourt, L.; et al. Genomic study of European Clostridioides difficile ribotype 002/sequence type 8. Microb. Genom. 2024, 10, 001270. [Google Scholar] [CrossRef]
- Smits, W.K.; Roseboom, A.M.; Corver, J. Plasmids of Clostridioides difficile. Curr. Opin. Microbiol. 2022, 65, 87–94. [Google Scholar] [CrossRef]
- Roseboom, A.M.; Ducarmon, Q.R.; Hornung, B.V.H.; Harmanus, C.; Crobach, M.J.T.; Kuijper, E.J.; Vossen, R.H.A.M.; Kloet, S.L.; Smits, W.K. Carriage of three plasmids in a single human clinical isolate of Clostridioides difficile. Plasmid 2023, 125, 102669. [Google Scholar] [CrossRef]
- Purdy, D.; O’KEeffe, T.A.T.; Elmore, M.; Herbert, M.; McLeod, A.; Bokori-Brown, M.; Ostrowski, A.; Minton, N.P. Conjugative transfer of clostridial shuttle vectors from Escherichia coli to Clostridium difficile through circumvention of the restriction barrier. Mol. Microbiol. 2002, 46, 439–452. [Google Scholar] [CrossRef]
- Ransom, E.M.; Ellermeier, C.D.; Weiss, D.S. Use of mCherry Red fluorescent protein for studies of protein localization and gene expression in Clostridium difficile. Appl. Environ. Microbiol. 2015, 81, 1652–1660. [Google Scholar] [CrossRef]
- Garnier, T.; Cole, S.T. Identification and molecular genetic analysis of replication functions of the bacteriocinogenic plasmid pIP404 from Clostridium perfringens. Plasmid 1988, 19, 151–160. [Google Scholar] [CrossRef]
- Larcombe, S.; Williams, G.C.; Amy, J.; Lim, S.C.; Riley, T.V.; Muleta, A.; Barugahare, A.A.; Powell, D.R.; Johanesen, P.A.; Cheng, A.C.; et al. A genomic survey of Clostridioides difficile isolates from hospitalized patients in Melbourne, Australia. Microbiol. Spectr. 2023, 11, e01352-23. [Google Scholar] [CrossRef]
- Lemee, L.; Dhalluin, A.; Testelin, S.; Mattrat, M.-A.; Maillard, K.; Lemeland, J.-F.; Pons, J.-L. Multiplex PCR Targeting tpi (Triose Phosphate Isomerase), tcdA (Toxin A), and tcdB (Toxin B) Genes for Toxigenic Culture of Clostridium difficile. J. Clin. Microbiol. 2004, 42, 5710–5714. [Google Scholar] [CrossRef] [PubMed]
- van Almsick, V.; Schuler, F.; Mellmann, A.; Schwierzeck, V. The Use of Long-Read Sequencing Technologies in Infection Control: Horizontal Transfer of a blaCTX-M-27 Containing lncFII Plasmid in a Patient Screening Sample. Microorganisms 2022, 10, 491. [Google Scholar] [CrossRef] [PubMed]
- Hua, X.; Liang, Q.; Deng, M.; He, J.; Wang, M.; Hong, W.; Wu, J.; Lu, B.; Leptihn, S.; Yu, Y.; et al. BacAnt: A Combination Annotation Server for Bacterial DNA Sequences to Identify Antibiotic Resistance Genes, Integrons, and Transposable Elements. Front. Microbiol. 2021, 12, 649969. [Google Scholar] [CrossRef] [PubMed]
- Bortolaia, V.; Kaas, R.S.; Ruppe, E.; Roberts, M.C.; Schwarz, S.; Cattoir, V.; Philippon, A.; Allesoe, R.L.; Rebelo, A.R.; Florensa, A.F.; et al. ResFinder 4.0 for predictions of phenotypes from genotypes. J. Antimicrob. Chemother. 2020, 75, 3491–3500. [Google Scholar] [CrossRef]
- Gupta, S.K.; Padmanabhan, B.R.; Diene, S.M.; Lopez-Rojas, R.; Kempf, M.; Landraud, L.; Rolain, J.-M. ARG-ANNOT, a New Bioinformatic Tool To Discover Antibiotic Resistance Genes in Bacterial Genomes. Antimicrob. Agents Chemother. 2014, 58, 212–220. [Google Scholar] [CrossRef]
- Wang, M.; Goh, Y.-X.; Tai, C.; Wang, H.; Deng, Z.; Ou, H.-Y. VRprofile2: Detection of antibiotic resistance-associated mobilome in bacterial pathogens. Nucleic Acids Res. 2022, 50, W768–W773. [Google Scholar] [CrossRef]
- Wu, Y.; Liu, C.; Li, W.-G.; Xu, J.-L.; Zhang, W.-Z.; Dai, Y.-F.; Lu, J.-X. Independent Microevolution Mediated by Mobile Genetic Elements of Individual Clostridium difficile Isolates from Clade 4 Revealed by Whole-Genome Sequencing. mSystems 2019, 4, e00252-18. [Google Scholar] [CrossRef]
- Wishart, D.S.; Han, S.; Saha, S.; Oler, E.; Peters, H.; Grant, J.R.; Stothard, P.; Gautam, V. PHASTEST: Faster than PHASTER, better than PHAST. Nucleic Acids Res. 2023, 51, W443–W450. [Google Scholar] [CrossRef]
- Lemoine, F.; Correia, D.; Lefort, V.; Doppelt-Azeroual, O.; Mareuil, F.; Cohen-Boulakia, S.; Gascuel, O. NGPhylogeny.fr: New generation phylogenetic services for non-specialists. Nucleic Acids Res. 2019, 47, W260–W265. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).