Screening and Molecular Docking of Bioactive Metabolites of the Red Sea Sponge Callyspongia siphonella as Potential Antimicrobial Agents
Abstract
1. Introduction
2. Results and Discussion
2.1. Metabolomic Profiling
2.2. Assessment of Antimicrobial Activity
2.3. Structure Characterization of the Purified Metabolites
2.4. Biofilm Inhibitory Activity
2.5. Docking Study
3. Materials and Methods
3.1. Sponge Material
3.2. Extraction and Fractionation
3.3. Assessment of Antimicrobial Activity
3.4. Metabolomics Analysis
3.5. Isolation and Purification
3.6. Biofilm Inhibitory Activity
3.7. Docking Study
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Perdicaris, S.; Vlachogianni, T.; Valavanidis, A. Bioactive natural substances from marine sponges: New developments and prospects for future pharmaceuticals. Nat. Prod. Chem. Res 2013, 1, 1–8. [Google Scholar] [CrossRef]
- Hertiani, T.; Edrada-Ebel, R.; Ortlepp, S.; van Soest, R.W.M.; de Voogd, N.J.; Wray, V.; Hentschel, U.; Kozytska, S.; Müller, W.E.G.; Proksch, P. From anti-fouling to biofilm inhibition: New cytotoxic secondary metabolites from two Indonesian Agelas sponges. Bioorg. Med. Chem. 2010, 18, 1297–1311. [Google Scholar] [CrossRef] [PubMed]
- Amade, P.; Charroin, C.; Baby, C.; Vacelet, J. Antimicrobial activities of marine sponges from the Mediterranean Sea. Mar. Biol. 1987, 94, 271–275. [Google Scholar] [CrossRef]
- Ilan, M.; Gugel, J.; Van Soest, R. Taxonomy, reproduction and ecology of new and known Red Sea sponges. Sarsia N. Atl. Mar. Sci. 2004, 89, 388–410. [Google Scholar] [CrossRef]
- Kashman, Y.; Yosief, T.; Carmeli, S. New Triterpenoids from the Red Sea Sponge Siphonochalina siphonella. J. Nat. Prod. 2001, 64, 175–180. [Google Scholar] [CrossRef]
- Tsukamoto, S.; Kato, H.; Hirota, H.; Fusetani, N. Seven new polyacetylene derivatives, showing both potent metamorphosis-inducing activity in ascidian larvae and antifouling activity against barnacle larvae, from the marine sponge Callyspongia truncata. J. Nat. Prod. 1997, 60, 126–130. [Google Scholar] [CrossRef]
- Sera, Y.; Adachi, K.; Shizuri, Y. A New Epidioxy Sterol as an Antifouling Substance from a Palauan Marine Sponge, Lendenfeldia c hondrodes. J. Nat. Prod. 1999, 62, 152–154. [Google Scholar] [CrossRef]
- El-Naggar, H.A.; Bashar, M.A.E.; Rady, I.; El-Wetidy, M.S.; Suleiman, W.B.; Al-Otibi, F.O.; Al-Rashed, S.A.; Abd El-Maoula, L.M.; Salem, E.-S.S.; Attia, E.M.H.; et al. Two red sea sponge extracts (Negombata magnifica and Callyspongia siphonella) induced anticancer and antimicrobial activity. Appl. Sci. 2022, 12, 1400. [Google Scholar] [CrossRef]
- Youssef, D.T.A.; Ibrahim, A.K.; Khalifa, S.I.; Mesbah, M.K.; Mayer, A.M.S.; van Soest, R.W.M. New antiinflammatory sterols from the Red Sea sponges Scalarispongia aqabaensis and Callyspongia siphonella. Nat. Prod. Commun. 2010, 5, 1934578X1000500107. [Google Scholar] [CrossRef]
- Bjarnsholt, T.; Jensen, P.Ø.; Jakobsen, T.H.; Phipps, R.; Nielsen, A.K.; Rybtke, M.T.; Tolker-Nielsen, T.; Givskov, M.; Høiby, N.; Ciofu, O. Quorum sensing and virulence of Pseudomonas aeruginosa during lung infection of cystic fibrosis patients. PLoS ONE 2010, 5, e10115. [Google Scholar] [CrossRef]
- Li, Y.-H.; Tian, X. Quorum sensing and bacterial social interactions in biofilms. Sensors 2012, 12, 2519–2538. [Google Scholar] [CrossRef] [PubMed]
- Pejin, B.; Talevska, A.; Ciric, A.; Glamoclija, J.; Nikolic, M.; Talevski, T.; Sokovic, M. Anti-quorum sensing activity of selected sponge extracts: A case study of Pseudomonas aeruginosa. Nat. Prod. Res. 2014, 28, 2330–2333. [Google Scholar] [CrossRef] [PubMed]
- Jain, S.; Laphookhieo, S.; Shi, Z.; Fu, L.; Akiyama, S.; Chen, Z.-S.; Youssef, D.T.A.; Van Soest, R.W.M.; El Sayed, K.A. Reversal of P-glycoprotein-mediated multidrug resistance by sipholane triterpenoids. J. Nat. Prod. 2007, 70, 928–931. [Google Scholar] [CrossRef] [PubMed]
- Abraham, I.; Jain, S.; Wu, C.-P.; Khanfar, M.A.; Kuang, Y.; Dai, C.-L.; Shi, Z.; Chen, X.; Fu, L.; Ambudkar, S. V Marine sponge-derived sipholane triterpenoids reverse P-glycoprotein (ABCB1)-mediated multidrug resistance in cancer cells. Biochem. Pharmacol. 2010, 80, 1497–1506. [Google Scholar] [CrossRef]
- Jain, S.; Abraham, I.; Carvalho, P.; Kuang, Y.-H.; Shaala, L.A.; Youssef, D.T.A.; Avery, M.A.; Chen, Z.-S.; El Sayed, K.A. Sipholane triterpenoids: Chemistry, reversal of ABCB1/P-glycoprotein-mediated multidrug resistance, and pharmacophore modeling. J. Nat. Prod. 2009, 72, 1291–1298. [Google Scholar] [CrossRef]
- Chang, H.-S.; Lee, S.-J.; Yang, C.-W.; Chen, I.-S. Cytotoxic sesquiterpenes from Magnolia kachirachirai. Chem. Biodivers. 2010, 7, 2737–2747. [Google Scholar] [CrossRef]
- Delbeke, E.I.P.; Everaert, J.; Uitterhaegen, E.; Verweire, S.; Verlee, A.; Talou, T.; Soetaert, W.; Van Bogaert, I.N.A.; Stevens, C.V. Petroselinic acid purification and its use for the fermentation of new sophorolipids. AMB Express 2016, 6, 28. [Google Scholar] [CrossRef]
- Knothe, G.; Steidley, K.R. Composition of Some Apiaceae Seed Oils Includes Phytochemicals, and Mass Spectrometry of Fatty Acid 2-Methoxyethyl Esters. Eur. J. Lipid Sci. Technol. 2019, 121, 1800386. [Google Scholar] [CrossRef]
- Bakos, K.; Havass, J.; Fülöp, F.; Gera, L.; Stewart, J.M.; Falkay, G.; Tóth, G.K. Synthesis and receptor binding of oxytocin analogs containing conformationally restricted amino acids. Lett. Pept. Sci. 2001, 8, 35–40. [Google Scholar] [CrossRef]
- Azcuna, M.; Tun, J.O.; Yap, H.T.; Concepcion, G.P. Callyspongia samarensis (Porifera) extracts exhibit anticancer activity and induce bleaching in Porites cylindrica (Scleractinia). Chem. Ecol. 2018, 34, 397–411. [Google Scholar] [CrossRef]
- Shi, Z.; Jain, S.; Kim, I.; Peng, X.; Abraham, I.; Youssef, D.T.A.; Fu, L.; El Sayed, K.; Ambudkar, S.V.; Chen, Z. Sipholenol A, a marine-derived sipholane triterpene, potently reverses P-glycoprotein (ABCB1)-mediated multidrug resistance in cancer cells. Cancer Sci. 2007, 98, 1373–1380. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Wang, T.-T.; Gao, K. A new cytotoxic stigmasterone from Agathis macrophylla. Nat. Prod. Commun. 2017, 12, 1934578X1701200307. [Google Scholar] [CrossRef]
- Sandjo, L.P.; Rincheval, V.; Ngadjui, B.T.; Kirsch, G. Cytotoxic effect of some pentacyclic triterpenes and hemisynthetic derivatives of stigmasterol. Chem. Nat. Compd. 2011, 47, 731–734. [Google Scholar] [CrossRef]
- Hong, R.; Qianqun, G.; Chengbin, C.; Weiming, Z. The cytotoxic constituents from marine-derived streptomyces 3320. J. Ocean Univ. China 2006, 5, 75–81. [Google Scholar] [CrossRef]
- Herraiz, T.; Galisteo, J. Tetrahydro-β-carboline alkaloids that occur in foods and biological systems act as radical scavengers and antioxidants in the ABTS assay. Free Radic. Res. 2002, 36, 923–928. [Google Scholar] [CrossRef] [PubMed]
- Placek, L.L. A review on petroselinic acid and its derivatives. J. Am. Oil Chem. Soc. 1963, 40, 319–329. [Google Scholar] [CrossRef]
- Carmely, S.; Kashman, Y. The sipholanes, a novel group of triterpenes from the marine sponge Siphonochalina siphonella. J. Org. Chem. 1983, 48, 3517–3525. [Google Scholar] [CrossRef]
- Bragina, M.E.; Daina, A.; Perez, M.A.S.; Michielin, O.; Zoete, V. The SwissSimilarity 2021 Web Tool: Novel Chemical Libraries and Additional Methods for an Enhanced Ligand-Based Virtual Screening Experience. Int. J. Mol. Sci. 2022, 23, 811. [Google Scholar] [CrossRef] [PubMed]
- Bodley, J.W.; Zieve, F.J.; Lin, L.; Zieve, S.T. Formation of the ribosome-G factor-GDP complex in the presence of fusidic acid. Biochem. Biophys. Res. Commun. 1969, 37, 437–443. [Google Scholar] [CrossRef]
- Gao, Y.-G.; Selmer, M.; Dunham, C.M.; Weixlbaumer, A.; Kelley, A.C.; Ramakrishnan, V. The structure of the ribosome with elongation factor G trapped in the posttranslocational state. Science 2009, 326, 694–699. [Google Scholar] [CrossRef]
- Rundlet, E.J.; Holm, M.; Schacherl, M.; Natchiar, S.K.; Altman, R.B.; Spahn, C.M.T.; Myasnikov, A.G.; Blanchard, S.C. Structural basis of early translocation events on the ribosome. Nature 2021, 595, 741–745. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Koripella, R.K.; Sanyal, S.; Selmer, M. Staphylococcus aureus elongation factor G–structure and analysis of a target for fusidic acid. FEBS J. 2010, 277, 3789–3803. [Google Scholar]
- El-Ghorab, A.H.; Behery, F.A.; Abdelgawad, M.A.; Alsohaimi, I.H.; Musa, A.; Mostafa, E.M.; Altaleb, H.A.; Althobaiti, I.O.; Hamza, M.; Elkomy, M.H. LC/MS Profiling and Gold Nanoparticle Formulation of Major Metabolites from Origanum majorana as Antibacterial and Antioxidant Potentialities. Plants 2022, 11, 1871. [Google Scholar] [CrossRef] [PubMed]
- Seeliger, D.; de Groot, B.L. Ligand docking and binding site analysis with PyMOL and Autodock/Vina. J. Comput.-Aided Mol. Des. 2010, 24, 417–422. [Google Scholar] [CrossRef] [PubMed]
- Shmueli, U.; Carmely, S.; Groweiss, A.; Kashman, Y. Sipholenol and sipholenone, two new triterpenes from the marine sponge siphonochalina siphonella (levi). Tetrahedron Lett. 1981, 22, 709–712. [Google Scholar] [CrossRef]
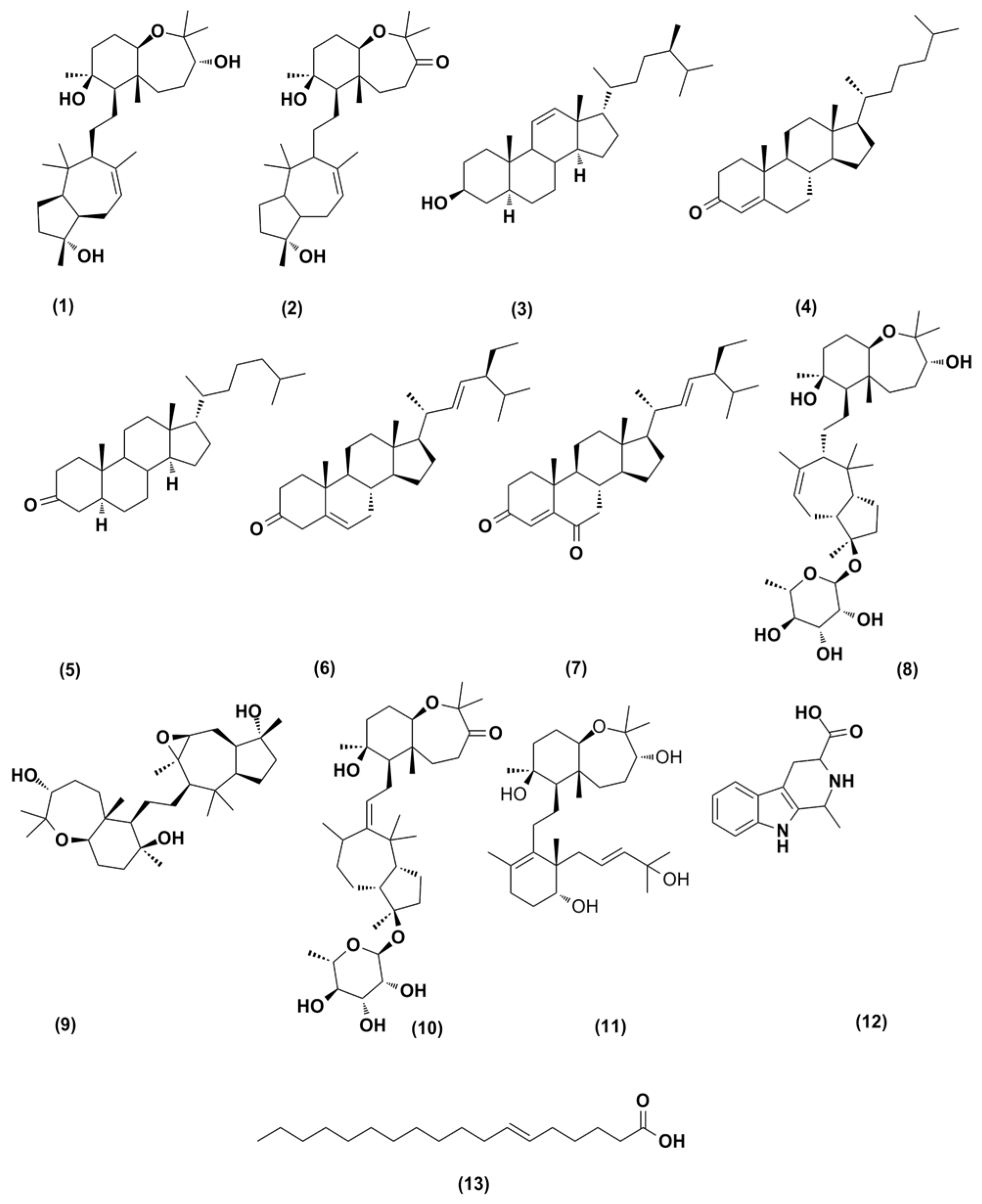


| NP | Tentative Identification b | Quasi-Form | Suggested Formula a | Calculated Accurate m/z | Experimentally Accurate m/z |
|---|---|---|---|---|---|
| 1 | Sipholenol A | [M+H]+ | C30H53O4 | 477.3944 | 477.3941 |
| 2 | Sipholenone A | [M+H]+ | C30H51O4 | 475.3785 | 475.3787 |
| 3 | Callysterol | [M+H]+ | C28H49O | 401.381 | 401.3783 |
| 4 | Cholestenone | [M+H]+ | C27H45O | 385.3472 | 385.347 |
| 5 | 5α-cholestanone | [M+H]+ | C27H47O | 387.3625 | 387.3627 |
| 6 | Stigmasterone | [M+H]+ | C29H47O | 411.3634 | 411.3637 |
| 7 | stigmasta-4,22-dien-3,6-dione | [M+H]+ | C29H45O2 | 425.345 | 425.342 |
| 8 | Sipholenoside B | [M+H]+ | C36H63O8 | 623.452 | 623.4523 |
| 9 | Sipholenol G | [M+H]+ | C30H53O5 | 493.3889 | 493.3893 |
| 10 | Sipholenoside A | [M+H]+ | C36H61O8 | 621.4368 | 621.4366 |
| 11 | Siphonellinol C | [M+H]+ | C30H52O5 | 492.3814 | 492.3820 |
| 12 | 1,2,3,4-tetrahydro-1-methyl-β-carboline-3-carboxylic acid | [M+H]+ | C13H15N2O2 | 231.1133 | 231.1134 |
| 13 | Petroselenic acid | [M+H]+ | C18H35O2 | 283.2634 | 283.2637 |
| NP | Tentative Identification b | Intensity | Suggested Formula a | Calculated Accurate m/z | Experimentally Accurate m/z |
|---|---|---|---|---|---|
| 1 | Sipholenol A | 2.2 × 104 | C30H53O4 | 477.3944 | 477.3941 |
| 2 | Sipholenone A | 1.2 × 107 | C30H51O4 | 475.3785 | 475.3787 |
| 3 | Callysterol | 4.4 × 107 | C28H49O | 401.381 | 401.3783 |
| 11 | Siphonellinol C | 8.8 × 105 | C30H52O5 | 492.3814 | 492.3820 |
| 13 | Petroselenic acid | 2.3 × 107 | C18H35O2 | 283.2634 | 283.2637 |
| Tested Extract | S. aureus | B. subtilis | E. coli | P. aeruginosa |
|---|---|---|---|---|
| MeOH Ext | 1.1 ± 0.5 | 1.2 ± 0.2 | - | - |
| Hex Fr | 2.3 ± 0.9 | 1.1 ± 0.4 | - | 1 ± 0.4 |
| DCM Fr | 6.6 ± 0.2 | 5.4 ± 0.3 | 1.5 ± 0.7 | - |
| ButOH Fr | - | 0.5 ± 0.2 | - | - |
| Ampicillin | 13.7 ± 0.9 | 12.3 ± 1.2 | 3.9 ± 0.9 | 3.6 ± 0.3 |
| Gentamicin | 9.8 ± 1.2 | 10.1 ± 1.1 | 15.5 ± 0.1 | 14.8 ± 1.3 |
| Inhibition Ratio (%) | |||||||
|---|---|---|---|---|---|---|---|
| Kle | Sal | Sta | MRSA | Ech | Can | Asp | |
| 11 * | NA | NA | 60.90 | 50.374 | NA | NA | NA |
| 1 * | NA | NA | 35.216 | NA | 45.50 | NA | NA |
| 2 * | NA | NA | 55.92 | 40.30 | 50.0 | NA | NA |
| 3 * | NA | NA | 60.90 | NA | NA | NA | NA |
| 13 * | NA | NA | 30.00 | NA | 70.50 | NA | NA |
| Nys | - | - | - | - | - | 97 | 98 |
| Cip | 98 | - | 96 | - | 98 | - | - |
| MIC (µg/mL) | |||
|---|---|---|---|
| S. aureus | MRSA | E. coli | |
| 11 * | 6.25 | 12.50 | NA |
| 1 * | 12.5 | NA | 25.00 |
| 2 * | 12.5 | 25.00 | NA |
| 3 * | 6.25 | NA | NA |
| 13 * | 25.00 | NA | NA |
| Cip | 1.25 | - | 0.390 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Musa, A.; Abdelgawad, M.A.; Shaker, M.E.; El-Ghorab, A.H.; Parambi, D.G.T.; Hamed, A.A.; Sayed, A.M.; Hassan, H.M.; Aboseada, M.A. Screening and Molecular Docking of Bioactive Metabolites of the Red Sea Sponge Callyspongia siphonella as Potential Antimicrobial Agents. Antibiotics 2022, 11, 1682. https://doi.org/10.3390/antibiotics11121682
Musa A, Abdelgawad MA, Shaker ME, El-Ghorab AH, Parambi DGT, Hamed AA, Sayed AM, Hassan HM, Aboseada MA. Screening and Molecular Docking of Bioactive Metabolites of the Red Sea Sponge Callyspongia siphonella as Potential Antimicrobial Agents. Antibiotics. 2022; 11(12):1682. https://doi.org/10.3390/antibiotics11121682
Chicago/Turabian StyleMusa, Arafa, Mohamed A. Abdelgawad, Mohamed E. Shaker, Ahmed H. El-Ghorab, Della Grace Thomas Parambi, Ahmed A. Hamed, Ahmed M. Sayed, Hossam M. Hassan, and Mahmoud A. Aboseada. 2022. "Screening and Molecular Docking of Bioactive Metabolites of the Red Sea Sponge Callyspongia siphonella as Potential Antimicrobial Agents" Antibiotics 11, no. 12: 1682. https://doi.org/10.3390/antibiotics11121682
APA StyleMusa, A., Abdelgawad, M. A., Shaker, M. E., El-Ghorab, A. H., Parambi, D. G. T., Hamed, A. A., Sayed, A. M., Hassan, H. M., & Aboseada, M. A. (2022). Screening and Molecular Docking of Bioactive Metabolites of the Red Sea Sponge Callyspongia siphonella as Potential Antimicrobial Agents. Antibiotics, 11(12), 1682. https://doi.org/10.3390/antibiotics11121682







