Abstract
Fluorescent molecular probes are very powerful tools that have been generally applied in cell imaging in the research fields of biology, pathology, pharmacology, biochemistry, and medical science. In the last couple of decades, numerous molecular probes endowed with high specificity to particular organelles have been designed to illustrate intracellular images in more detail at the subcellular level. Nowadays, the development of cell biology has enabled the investigation process to go deeply into cells, even at the molecular level. Therefore, probes that can sketch a particular organelle’s location while responding to certain parameters to evaluate intracellular bioprocesses are under urgent demand. It is significant to understand the basic ideas of organelle properties, as well as the vital substances related to each unique organelle, for the design of probes with high specificity and efficiency. In this review, we summarize representative multifunctional fluorescent molecular probes developed in the last decade. We focus on probes that can specially target nuclei, mitochondria, endoplasmic reticulums, and lysosomes. In each section, we first briefly introduce the significance and properties of different organelles. We then discuss how probes are designed to make them highly organelle-specific. Finally, we also consider how probes are constructed to endow them with additional functions to recognize particular physical/chemical signals of targeted organelles. Moreover, a perspective on the challenges in future applications of highly specific molecular probes in cell imaging is also proposed. We hope that this review can provide researchers with additional conceptual information about developing probes for cell imaging, assisting scientists interested in molecular biology, cell biology, and biochemistry to accelerate their scientific studies.
1. Introduction
The cell is the fundamental constituent of living bodies and one of the basic units for biological investigation. At the subcellular level, various organelles, such as nuclei, mitochondria, endoplasmic reticulums (ERs), lysosomes, etc., are distributed inside cells and function synergistically. Thus far, the statuses of different organelles during distinct bioprocesses have attracted intense interest. It is important to provide visualized organelle signals in biological studies because the relative localizations between different organelles or between organelles and particular molecules significantly influence their activations. For example, the release of cytochrome C from mitochondria into the cytoplasm is a special marker of apoptosis [1,2,3]. A large number of proteins, especially transcription factors (TFs), may transfer from other locations into the nucleus to initiate downstream gene expression under different cell stress conditions, such as heat shock [4] and ER stress [5,6,7]. Mitochondria–ER contact (MERC) is important for intracellular calcium homeostasis and other metabolic processes [8,9]. Immunofluorescence (IF) is a widely used method for staining organelles with high specificity [10]. Usually, a primary antibody of a unique biomarker molecule of the special organelle is first incubated with cells to recognize and bind to the target. A secondary antibody labeled with a fluorophore will then be applied to bind to the primary antibody and give the signal. This IF method is highly specific but has some limitations. The recognition between the primary and secondary antibodies relies on the resource species of the primary antibody. For reasons of costs, limited resource species, such as mice, rabbits, and goats, have been chosen, such that the IF method can provide only a few different signals at one time. More importantly, the cells used for IF staining must be fixed and permeabilized to allow the large antibody molecules to enter the targeted localization, such that it cannot be applied to the real-time monitoring of the statuses of organelles in living systems. Due to the development of molecular biology and the discovery of fluorescence proteins (FPs), researchers in the field of biology can construct artificial proteins that fuse FPs with special signal peptides to endow FPs with the capacity of organelle targeting [11]. This method lightens particular organelles and can work in living cells with great specificity; however, the fused proteins have to be expressed intracellularly through elaborate experimental operations involving toxic cell transfection reagents. In addition, the exogenous protein may cause some unexpected bioprocess, such as immune responses [12].
Molecular probes with high specificity toward particular organelles are promising tools for biological studies at the subcellular level [13]. Through the contemporary development of molecular biology, more knowledge can be gained about the particular characteristics of organelles, such as pH and potential. Some proteins have also been found to have unique localizations and may be marked by unique molecules. Hence, scientists can design probes based on these special senses. Some commercial tracker probes have already been employed to stain special organelles, such as nuclei [14], mitochondria [15], ERs [16], and lysosomes [17]. With a particular design, small fluorescent molecules can penetrate the membranes of living cells and target particular organelles with high efficiency.
Up until now, the understanding of cells has extended to the molecular level, where the focus is on more detailed parameters inside cells and even inside unique organelles [18]. For example, scientists have indicated that the pH inside the lysosome is much lower than the normal physiological pH of 7.4 [19], that the temperature of the mitochondrial matrix may be as high as 50 °C [20], that the calcium transient on ER surfaces initiates autophagy [21], and that increased concentrations of Fe2+ ions may cause a new kind of programmable cell death process called ferroptosis [22]. These detailed molecular features have attracted more and more attention because they have been proven to be closely related to numerous bioprocesses and diseases. Therefore, a clear understanding of molecular characteristics at the subcellular level will help explain molecular mechanisms in biology and pathology, thus accelerating studies on the diagnosis and therapeutics of diseases, such as cancers, cardiovascular diseases, and even the recent SARS-CoV-2. To achieve a precise evaluation of the intracellular molecular substances at the organelle level, it is important to develop novel molecular probes that can simultaneously target particular organelles and respond to distinct parameters. The most popular method for designing specific probes is linking different functional molecules as a whole to construct a new molecule with multiple functions. The key to this method is to find and develop appropriate functional groups and linkers to ensure that they can keep their original properties. In this review, we will introduce the developed molecular probes that have emerged in the last decade, where the probes have been applied in the imaging of unique parameters at the organelle level. We will discuss the crucial organelles, including nuclei, mitochondria, ERs, and lysosomes, and introduce the general strategy in designing molecules for specific targeting. Moreover, we will also discuss factors related to different organelles, such as the concentrations of ions as well as bioactive molecules, and the physical properties of the special organelles. We will discuss the meaning of each parameter and the strategies used to monitor them. We hope that this general review can provide some new opinions for people interested in both the chemical and biological fields, to dig out promising molecular probes to better explain the mechanism of different bioprocesses to provide advanced approaches for disease diagnosis and treatment.
2. Design of Molecular Probes for Organelle-Targeted Cell Imaging
2.1. Nucleus-Targeted Molecular Probes
2.1.1. Properties of the Nucleus
The cellular nucleus is the most important organelle inside a cell. The nucleus is the habitat of chromosomes—the fundamental components that store genetic information [23]. Chromosomes constitute DNA molecules that encode the gene information in their sequences, thus controlling the expression of proteins in cells. It is well-known that gene mutations will induce abnormal activity of proteins and influence the health of cells. Plenty of diseases have been proven to be related to the nucleus [24,25,26]. In addition, the status of the nucleus has been recognized as a biomarker of some cell-fate-related bioprocesses, such as apoptosis and necrosis [27,28].
The structure of the nucleus contains double-layer membranes enclosing the contents, which include the nucleolus, DNA, and other genetic materials. The outer nucleus membrane contains many structures termed nuclear pores, which regulate the exchange of molecules. Some small hydrophilic molecules can freely transfer across nuclear pores, whereas the macromolecules, such as nucleic acids and proteins, must pass the membrane with the facility of other transporters, called importins [29].
The abundance of DNA is the most distinguishing factor of the nucleus. Hence, many molecular probes have been designed to interact with DNA molecules for nucleus targeting. The most commonly utilized interaction methods between DNA and small molecules include electrostatic interaction, intercalation, and groove binding [30,31]. The phosphoric backbone of the long-chain DNA structure makes the whole molecule negatively charged, and thus can bind with cationic dyes through electrostatic force. Some molecules, such as anthracyclines and acridines, can intercalate in the double-stranded structure through interaction with the bases. Major and minor grooves are featured sites of the double-stranded DNA. Small molecules can bind along grooves through hydrogen binding interaction, especially narrower minor grooves. Moreover, the importin system was also utilized for nucleus targeting.
2.1.2. Nucleus-Targeted Probe Design
As mentioned above, molecules that can accumulate around DNA are potential specific probes for nucleus targeting. At present, commercial dyes, such as DAPI, PicoGreen, and Hoechst series, have generally been applied to nucleus staining because they can bind with double-stranded DNA structures and display special fluorescent signals [32,33]. Hoechst is a typical molecule that can bind to DNA minor grooves at A–T-rich regions [34]. The basic structure of Hoechst has been used as the guide functional group with which to drive other fluorophores to nuclei. Hamachi’s group has introduced a new concept called self-localizing ligands (SLLs), which can bind with target proteins and tether them to a new position inside cells [35]. They demonstrated that a trimethoprim (TMP) links with Hoechst to form a new ligand (hoeTMP, 1, Scheme 1) that can guide a special protein, dihydrofolate reductase (eDHFR), to nuclei with high efficiency. Inspired by this discovery, the same group developed a Hoechst tagging strategy that can simply guide non-specific molecular fluorophores towards the position of nuclei by linking them with Hoechst [36]. Recently, Zhang et al. reported a new derivative called HoeSR (2, Scheme 1), which was constructed by conjugating sulforhodamine (SR) and Hoechst, for the super-resolution imaging of nuclei in living cells through direct stochastic optical reconstruction microscopy (dSTORM) [37]. This HoeSR probe has been applied to chromosome morphology imaging at a super-resolution level, making the probe itself not only an indicator of nuclei but also a potential dye of DNA structures. In addition to the Hoechst moiety, some other unique chemical structures have also been reported to recognize DNA structures through groove binding. In 2015, Govindaraju and co-workers reported a DNA-specific probe, basing its design on the quinone cyanine-dithiazole (QCy-DT, 3, Scheme 1) structures [38]. They demonstrated that QCy-DT could recognize duplex AT-rich DNA through the minor groove binding mechanism, which is the first switch-on NIR fluorescence probe for sequence-specific DNA staining. They also showed that the probe could be applied to specific parasite staining and treatment. Recently, they have explained the cis/trans isomerization of QCy-DT upon binding to DNA targets and promoted the application of the probe [39]. Pang’s group developed a novel NIR-emitting probe based on phenol-containing m-phenylene building blocks for DNA-targeted staining through a groove binding mechanism (4, Scheme 1). They showed that the probe modification on the m-phenylene bridge group might endow the probe with the ability to target other organelles, such as lysosomes in cells [40]. Intercalation is another general mode of interaction between small molecules and DNA [41,42], which led to the development of numerous molecular probes for DNA targeting and nucleus imaging based on the intercalation method. Acridine orange (AO) is a well-known commercial DNA-intercalating dye. The fluorescence property of AO has been explored further recently [43]. Kandinska et al. prepared a series of asymmetric monomeric monomethine cyanine dyes through a simple and reliable synthetic procedure (5, Scheme 1). They found that the designed probes can bind with DNA through either groove binding or intercalation processes. These probes showed excellent imaging properties of DNA and RNA, completely suitable for application as DNA and RNA fluorescent molecular probes [44]. Polycyclic aromatic hydrocarbons (PAHs), such as naphthalene, anthracene, pyrene, and perylene, are excellent rigid molecular skeletons that are useful for developing highly efficient two-photon excitable fluorescent probes. Ali et al. designed an innovative fluorescent dye by impregnating perylene diimide into the functionalized surface of magnetic core–shell silica nanoparticles [45]. These nanoparticles exhibit features such as low toxicity, being environmentally friendly, and high sensitivity; they also show high DNA binding capacity, which makes the developed particle promising for DNA extraction, delivery, and fluorescent labeling. Pang’s group synthesized a pyrene-based fluorescent probe (6, Scheme 1), which possesses remarkable selectivity to the intracellular nucleus and is also useful for two-photon fluorescence microscopy [42]. They proposed a possible mechanism: due to the rigid planar pyrene structure of it, a possible interaction mode is the intercalation of the molecules into adjacent base pairs in the DNA duplex. The nucleic RNA can also be a target for nucleus imaging. Zhao et al. developed a probe called BEB-A (7, Scheme 1) [46]. With the help of cysteine, the probe could bind with RNA molecules through the hydrogen bonds forming between the probe and the uracil in the RNA molecules, thus achieving accumulation in the nucleolus position. They showed that this BEB-A could image cysteine in mitochondria and RNA in the nucleus simultaneously.
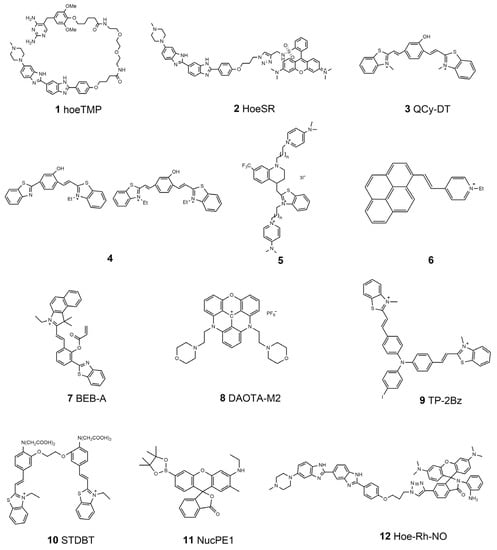
Scheme 1.
Chemical structures of nucleus-targeted fluorescent molecular probes.
The double helix is not the only existing structure of DNA molecules in living cells. For instance, guanine-rich DNA sequences were found to form a tetra-stranded structure called a G-quadruplex (GQ) [47,48]. Many studies have indicated that GQs are implicated in numerous bioprocesses, such as gene expression regulation and telomere replication [49,50]. Therefore, GQs provide a potential marker not only for nucleus-targeted imaging but also for gene activities. Most specific probes for the detection of GQs are based on the interaction between the probe molecules and the unique planar G-quartet units through π–π stacking. Vilar and co-workers designed a small molecule based on trianguleniums, termed DAOTA-M2 (8, Scheme 1) [51]. They demonstrated that DAOTA-M2 could recognize GQs and thus applied this probe to the identification of GQs in the nuclei of living cells using fluorescence lifetime imaging microscopy (FLIM) [52]. Sun et al. synthesized a new probe called TP-2Bz (9, Scheme 1) by linking triphenylamine with N-methylbenzothiazole [53]. TP-2Bz can bind with GQs for specific imaging and reveal the viscosity in the nucleus region because of the unique twistable donor–π–acceptor structure of the probe.
Signal transduction molecules within nuclei always serve important roles in regulating different bioactivities. Zhu et al. linked two benzothiazolium hemicyanine dyes with a special Ca2+-chelating molecule to construct a new probe, called STDBT (10, Scheme 1), for Ca2+ detection [54]. They found that the acetoxymethyl ester of STDBT distributes in both the cytosol and the nucleus with a clear boundary after entering cells; it could be applied to imaging nucleic Ca2+. Meanwhile, endogenous reactive species, such as H2O2 and NO, are general signaling molecules in the nucleus [55,56,57]. In 2011, Dickinson et al. designed the first probe, NucPE1 (11, Scheme 1), for imaging nucleic H2O2 in living cells [58]. They demonstrated that this new probe works in both living cells and in vivo in C. elegans. They found that the nuclear ROS pools in C. elegans were regulated by a longevity-promoting sirtuin protein, demonstrating that NucPE1 is a potential tool for biological investigation. Recently, Zhao et al. developed a probe called Hoe-Rh-NO (12, Scheme 1) for the localized imaging of NO in nuclei [59]. They selected a rhodamine spirolactam as the NO indicator moiety and Hoechst as the nucleus guider. A summary of the properties of the nucleus-targeted molecular probes is listed in Table 1.

Table 1.
Properties of the nucleus-targeted fluorescent molecular probes.
This probe was applied to the detection of both exo-/endogenous NO in a zebrafish inflammation model.
2.2. Mitochondria-Targeted Molecular Probes
2.2.1. Property of the Mitochondria
Mitochondria, known as the “powerhouse” of cells, are important organelles that play a key role in sourcing energy for most creatures [60]. Mitochondria are the primary areas where adenosine triphosphate (ATP) is produced, which fuels most biological processes, such as cell proliferation, biogenesis, biosynthesis, and metabolism. During the development of modern biology, scientists found that the functions of mitochondria are far more complicated than simply that of a powerhouse. Mitochondria modulate the homeostasis of cellular Ca2+ and reactive oxygen species (ROS) [61,62]. They are also responsible for many cellular bioprocesses, such as autophagy and protein degradation [63,64]. In recent studies, mitochondria have been verified to regulate different programmed cell death processes, such as apoptosis, pyroptosis, and ferroptosis [65,66,67,68,69]. Because of the significance of mitochondria, it is not surprising that people find that they are closely related to numerous diseases, such as cancers, cardiovascular diseases (CVDs), and neurodegenerative disorders [70,71,72].
The basic structure of mitochondria contains a double lipid bilayer—the outer membrane and the inner membrane, the matrix coated by the inner membrane, and intermembrane space (IMS) [73]. The outer mitochondrial membrane (OMM) is permeable for molecules smaller than 6 kD, thus keeping the fundamental substance exchange between the cytoplasm and the IMS [74].
The oxidative phosphorylation and tricarboxylic acid cycle processes occur in the matrix to produce ATP through the synergistic function between proteins located in the matrix and the inner mitochondrial membrane (IMM). Notably, a proton transfer process occurs during these bioprocesses. The proton pump on the IMM may drive the protons from the matrix to the IMS, resulting in a membrane potential that is much higher than that of other organelles. In fact, the potential difference between the two sides of the IMM can be as large as 160–180 mV [75]. Therefore, molecules with a positive charge can be accumulated on the matrix side of the IMM spontaneously once they are able to permeate the lipid bilayer; this is the most general physical parameter of mitochondria for the principle of specific probe design.
2.2.2. Mitochondria-Targeted Probe Design
Thus far, the majority of the mitochondria-targeted molecular probes have been lipophilic cations, including intrinsic lipophilic cationic dyes, such as rhodamine (13, Scheme 2) and cyanine (14, Scheme 2). Moreover, triphenylphosphine (TPP, 15, Scheme 2) and pyridinium (16, Scheme 2) are typical building blocks that can be modified as the mitochondria-targeted moieties on natural fluorophores [13,76]. The positive charge of the molecular probes is important for mitochondrial specificity because of the high potential of the IMM, as mentioned above.
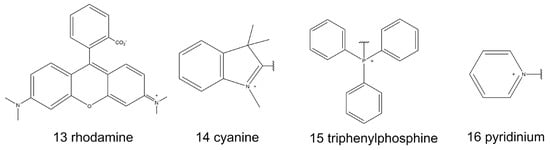
Scheme 2.
Chemical structures of typical mitochondria-targeted building blocks.
The staining of mitochondria provides valuable insights into morphology. The word “mitochondrion” stems from Greek and contains two parts, “mitos” and “khondrion”, which mean thread and small grain, respectively. Natural mitochondria have a highly dynamic morphology, transitioning from dense branches to scattered particles through a fission–fusion process regulated by several proteins. At this time, commercial dyes, such as the MitoTracker series, are available for mitochondria-specific imaging. These powerful tools provide visualized signals of mitochondrial morphology and mitochondrial dynamic information to people for further studies on biology, pathology, and pharmacology [77]. Meanwhile, advanced molecular probes have been developed as structure probes of mitochondria with high specificity and low toxicity. Relying upon these developments, numerous probes have been applied to super-resolution cell imaging to help people in cellular and molecular biology monitor and investigate mitochondria-related bioprocesses [76].
The structures and dynamics of mitochondria provide significant information for intracellular bioactivities. Moreover, mitochondria are the places that carry many important biochemical interactions inside cells. Therefore, probes that can investigate the functional properties of mitochondria are also important and have recently garnered increasing interest. For example, the membrane potential of mitochondria is vital to maintaining respiratory chain reactions, ion transfer, and other bioactivities. Several commercial probes, including TMRM and JC-1, have been widely applied to detect the mitochondria membrane potential [78,79]. Chemical reagents are also important for the normal activity of mitochondria. Mitochondria are the primary areas of ROS production. They produce ROS during the electron transfer mechanism in related bioprocesses, such as oxidative phosphorylation. ROS are important reagents in cells that play multiple roles, acting as signal transducers, stress inducers, and pathogen defenders [80]. A certain concentration of ROS is necessary for cells; however, the accumulation of ROS will cause oxidative damage to biomolecules such as DNA, proteins, and lipids [81]. MitoSOX is a commercial dye used to visualize the broad-spectrum ROS in mitochondria [82]. Thus far, multifunction probes with better performance have been introduced. Li et al. reported a probe, HQPQ-B (17, Scheme 3), for the highly accurate detection of mitochondrial H2O2 [83]. The probe would precipitate and show a signal in situ once it reacted with H2O2, thus avoiding the false signal caused by the dispersion of traditional soluble probes. Recently, Zhou et al. designed a two-photon fluorescence-lifetime-based probe (TFP, 18, Scheme 3) that can monitor the ATP and H2O2 levels of mitochondria in living cells in real time [84]. TFP is a potential tool for the study of the relationship between cellular oxidative state and energy metabolism. Niu et al. developed a two-photon three-channel fluorescence probe, NPClA (19, Scheme 3), which responds to SO2 and ClO− for more detailed mitochondrial oxidative stress imaging [85]. An oxime-containing fluorescent probe, MitoClO (20, Scheme 3), based on a BODIPY scaffold and the C=N isomerization mechanism, was designed by Peng’s group and used for HClO determination with a rapid response, low detection limits, and high selectivity. Triphenylphosphine was introduced at the meso-position to enable the probe to locate mitochondria within living cells [86].
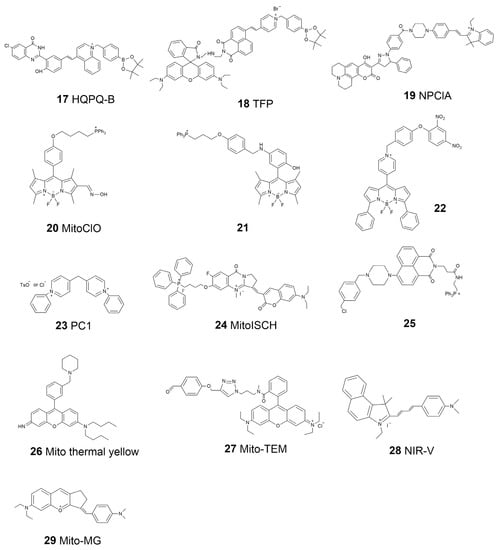
Scheme 3.
Chemical structures of mitochondria-targeted fluorescent molecular probes.
ROS are not the only attractive compounds inside mitochondria. Because of the complex function of mitochondria, several metal ions and other reactive species, such as reactive nitrogen species (RNS) and reactive sulfur species (RSS), are involved in maintaining the activity of mitochondria [87,88]. Nowadays, probes for the mitochondria local imaging of ions such as Ca2+, Zn2+, Fe2+, Hg2+, RNS, such as NO, RSS, such as H2S, and glutathione (GSH) have been introduced; a variety of them are commercially available [89,90,91,92,93,94]. Based on the N-nitrosation of aromatic secondary amines, Miao et al. developed a mitochondrion-targeted NO probe (21, Scheme 3) and successfully visualized the NO levels in macrophages in the resting state or under diverse stimulations [95]. In this BODIPY-based probe, TPP was introduced as the targeting group, where a N-benzyl-4-hydroxyaniline moiety was the reaction site. This probe was applied to detect NO levels in living cells after oxygen–glucose deprivation (OGD) stimulation. Zhang et al. reported a BODIPY-based probe (22, Scheme 3) for monitoring mitochondrial GSH [96]. They successfully used this probe for mitochondrial-targeted GSH imaging in living HeLa cells. The self-immolative group paradinitrophenoxy benzyl pyridinium was introduced at the meso-position of BODIPY, and it could simultaneously act as the targeting ligand and responsive group.
Mitochondria are known as semi-autonomous organelles, which means that they have their own genetic system, including mitochondrial DNA (mtDNA) and related enzymes for gene transcription [97]. Human mtDNA is a closed-circle model constructed by double-stranded DNA with a length of 16,569 base pairs. Traditional biology has revealed that mtDNA encodes a few RNAs and peptides that function for mitochondria. Moreover, an increasing number of studies have indicated that mtDNA is also related to different physiological processes, such as aging and inflammation [98]. Therefore, the imaging of mtDNA has become interesting in recent years. The most challenging part of mtDNA staining is the interference from nuclear DNA, which is much more abundant inside cells. Uno et al. presented a new N-aryl pyrido cyanine derivative, PC1 (23, Scheme 3), that shows a dose-dependent mtDNA staining ability [99]. They demonstrated that this probe has a relatively long absorption wavelength and can be applied to the imaging of mtDNA in living cells with the separation of photons via a lifetime tuning method in stimulated emission depletion nanoscopy (SPLIT-STED). Tan’s group reported a probe called MitoISCH (24, Scheme 3) using TPP as the guider moiety and specifically responded to the GQ structure in mtDNA [100]. They further indicated that the GQ in mtDNA would affect the energy metabolism mode inside cells.
The microenvironment is also important for the activity of mitochondria, since most of the enzymes are influenced by pH, temperature, and even the viscosity of the surroundings [101]. Using a piperazine-linked naphthalimide as the pH sensor moiety and TPP as the guider, Lee et al. synthesized a probe (25, Scheme 3) for the real-time monitoring of pH change in mitochondria in living cells [102]. They observed the process of mitophagy in nutrient-starved cells by using the designed probe. In 2015, Chang’s group introduced a probe, Mito thermal yellow (26, Scheme 3), with which to detect the temperature of mitochondria [103]. This probe is a derivative of rosamine with rotatable substituents, demonstrating a proportional fluorescence intensity change according to the change in temperature. Further, scientists found that the temperature of mitochondria can be as high as 50 °C, meaning that some mitochondrial recognition events should be re-evaluated [20]. Xiao’s group introduced a rhodamine-B-based probe to monitor the temperature of mitochondria [104]. This probe, termed Mito-TEM (27, Scheme 3), could anchor on the IMM and thus avoid the signal interference caused by the dispersion due to the dynamics of mitochondria. At present, a 2.0 version of Mito-TEM has been presented by the same group and applied to mitochondrial temperature tracking in vivo in a zebrafish model [105]. Chen et al. designed a probe, NIR-V (28, Scheme 3), by linking 2,3,3-trimethyl-3H-indolenine and 4-(dimethylamino) cinnamaldehyde [106]. This probe was applied to monitor the viscosity changes in the mitochondria in cells as well as in the organs in a mouse model. They demonstrated that NIR-V could be used for the detection of viscosity in different organs in diabetic mice models with insulin treatment, thus providing a new tool for the diagnosis of viscosity-related diseases. Zhang et al. introduced a multifunctional mitochondrial-specific probe that can detect SO2 and viscosity simultaneously [107]. They constructed the Mito-MG (29, Scheme 3) probe by combining benzopyrylium salt and 4-dimethylaminobenzaldehyde. This probe was also applied to imaging in both living cells and in vivo. A summary of the properties of the mitochondria-targeted molecular probes is listed in Table 2.

Table 2.
Properties of the mitochondria-targeted fluorescent molecular probes.
2.3. Endoplasmic-Reticulum-Targeted Molecular Probes
2.3.1. Properties of the Endoplasmic Reticulum
The endoplasmic reticulum (ER) is the center of synthesis and transport for most proteins and lipids inside cells [108]. Besides the key role of biomolecule synthesis, the ER also acts as a regulator of many bioprocesses. The ER is the main storage of intracellular calcium, thus regulating the intracellular Ca2+ signal transfer [109]. ER also regulates the metabolism of proteins and lipids; moreover, the behavior of ER is also closely linked to the activities of cells and the physiological state of life [7]. ER stress is a series of reactions to restore protein homeostasis activated by misfolded or unfolded protein aggregation as well as calcium ion imbalance in the ER lumen [110], related to many diseases such as cancer [111], diabetes [112], and heart diseases [113]. In recent years, ER-phagy has also been shown to be involved in maintaining ER homeostasis and greatly influence ER as well as cell function [114,115].
Proteins are synthesized in the rough ER, whereas lipids are synthesized in the smooth ER [116]. The two categories of the ER are effectively distinguished by their surface morphologies: The rough ER is rich with ribosomes on the surface and thus appears coarse under electron microscopy. The ER is relatively rich in zwitterionic lipids. Thus, amphipathic, lipophilic cations with moderately sized conjugated systems were proven to be suitable for ER probe designs [117]. The cytosolic face of ER membranes is abundant in ATP-sensitive potassium channel sulfonylurea receptors, whereby ER-targeting probes are often designed and synthesized based on molecules with sulfonamide groups [118].
2.3.2. ER-Targeted Probe Design
Special ATP-dependent K+ channels are abundant on the ER membrane; sulfonylurea and sulfonamide groups can be recognized by the special receptors in these channels [119]. Therefore, molecular probes based on sulfonamide (30, Scheme 4) showed great potential for ER specificity. Although commercial probes, such as ER-Tracker Bluewhite DPX, based on the sulfonamide group are now available [16], it is noteworthy that sulfonamide receptors, such as glibenclamide and its derivatives, are still expensive. At this point, scientists have found more affordable molecules with which to design probes that target the ER. For example, pentafluorophenyl (31, Scheme 4) is also a general group in probe design for ER targeting. The ER surface proteins that contain the sulfydryl group may react with the pentafluorophenyl group through a halogen-substituted reaction. Moreover, resorufin (32, Scheme 4), Pennsylvania green (33, Scheme 4), and other groups are also used to design ER-targeting probes [120]. Recently reported hydrophobic resorufamine derivatives, which possess a structural similarity to fluorinated hydrophobic rhodol fluorophores, may accumulate in the ER of mammalian cells and were developed to target the ER [121].
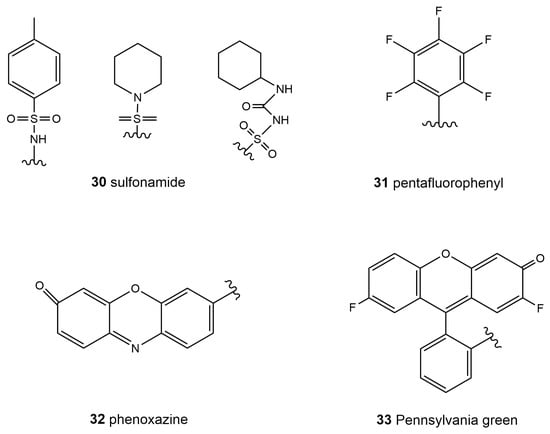
Scheme 4.
Chemical structures of typical ER building blocks.
As in mitochondria, reactive species, including ROS, RNS, and RSS, are critical factors in the ER for signal transduction and bioprocess regulation [122,123]. Reactive-species-induced ER stress and its association with ER-related diseases have drawn significant attention and concerns. Following the discovery that SO2 has the functionality to inhibit ER stress [124], Yue et al. designed a probe, MSO-SO2 (34, Scheme 5), with which to detect SO2 produced during ER stress induced by dithiothreitol (DTT) [125]. Due to the good ER-targeting ability of dimalononitrile isophorone derivatives, the authors selected levulinic acid as the specific recognition site for SO2, as well as a weak electron withdrawing group, and constructed an ER-targeted NIR fluorescent probe based on the dicyanoisophorone framework, realizing directly the visualization of the production as well as consumption of SO2 during DTT-induced ER stress. Zhang et al. reported a novel ER-targetable fluorescent probe, ER-CN (35, Scheme 5), for the visualization of H2S in living cells containing piperazine coumarin as the fluorescence platform, a 7-nitro-1,2,3-benzoxadiazole (NBD) moiety as the H2S-specific recognition site, and a methyl sulfinamide group as the ER-targeting part [126]. Although the mechanism of NO inducing ER stress has not been fully studied, it has become a fact that NO can alter ER functions and then activate ER-stress-mediated apoptosis [127]. Li et al. designed and synthesized an ER-Nap-NO (36, Scheme 5) probe composed of naphthalimide as the two-photon fluorophore, o-phenylenediamine as the NO recognition group, and methyl sulfonamide as the ER-targetable group [128]. This new probe was used for the two-photon imaging of exogenous and endogenous NO in the ER of living cells, exhibiting the advantages of a fast response, high selectivity, and low cytotoxicity. Meanwhile, ER-Nap-NO showed its capability to monitor NO level in a tunicamycin-induced ER stress system, suggesting that this probe is a promising tool for the function investigation of NO in ER-associated vascular diseases. Linking methyl sulphonamide, as an ER-targeting group, to the 1,8-naphthalimide fluorophore, a probe with which to accurately image exogenous and endogenous HNO in ER was obtained recently by Ye’s group [129]. This new probe, R2 (37, Scheme 5), showed high selectivity, a fast response, a low detection limit, and good biocompatibility in ER-specific cell imaging. Lu et al. constructed a probe that employed triflate as the response site for O2•− and p-methylsulfonamide as the ER-specific moiety (ER-Rs, 38, Scheme 5) for the imaging of endogenously produced O2•− in the ER of living cells [130]. They also demonstrated that this probe, ER-Rs, could be applied for O2•− monitoring in vivo in tissues and zebrafish models. In 2018, Pak et al. reported a naphthoimidazolium borane probe that could trace the exo/endogenous HClO in the ER of living cells and tissues (39, Scheme 5) [131].
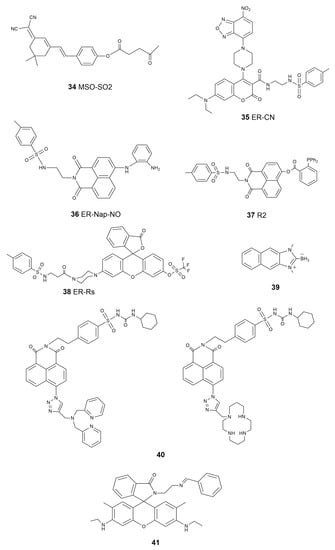
Scheme 5.
Chemical structure of the ER-targeted fluorescent molecular probes.
In addition to calcium, the ER is also the stockroom of zinc, which is necessary for the proper functioning of living organisms [132]. By introducing a simple sulfonylurea-targeting group, Fang et al. developed probes that realized the remarkable ability to image mobile Zn2+ in the ER in different cell lines (40, Scheme 5) [133]. Since the discovery of ferroptosis a decade ago, the level of iron in the ER has become a significant marker. The Fenton reaction, which generates Fe3+ and highly oxidative hydroxyl radicals from H2O2 and Fe2+ reactions, plays a key role in lipid peroxidation inside cells and initiates the process of ferroptosis [22]. Lee et al. prepared a series of probes through a chemical reaction of rhodamine-ethylene diamine and benzaldehyde to detect Fe3+ in the ER [134]. They demonstrated that this particular probe (41, Scheme 5) showed good selectivity to the ER-localized Fe3+ in HepG2 cells. A summary of the properties of the ER-targeted molecular probes is given in Table 3.

Table 3.
Properties of the ER-targeted fluorescent molecular probes.
2.4. Lysosome-Targeted Molecular Probe
2.4.1. Properties of Lysosomes
Lysosomes are spherical catabolic organelles that contain more than 50 kinds of hydrolases, such as protease, phosphatase, lipase, and nuclease, which are used to metabolize and decompose a variety of endogenous and exogenous macromolecular substances, such as sugar, lipids, glycolipids, glycosaminoglycans, nucleic acids, and proteins [135]. As one of the most important digestive organelles in a cell, lysosomes have been proven to be involved in controlling the recycling and signal transduction processes of cell growth, division, differentiation, and cell death; they are vital elements in maintaining metabolic homeostasis, intracellular signal transduction, and plasma membrane repair as well as secretion [136]. Numerous studies have revealed that the dysfunction of lysosomes is related to various diseases, including cancers, atherosclerosis, and neurodegenerative disorders [137]. In the most recent research, a novel therapeutical strategy, termed lysosome-targeting chimaera (LysoTAC), has emerged for disease treatments that rely upon the digestion function of lysosomes [138,139]. Therefore, the imaging of lysosomes and the monitoring of their activities are attractive to people in a wide range of research fields, such as biology, pathology, pharmacology, and biochemistry.
As the activity of hydrolase is usually the highest under acidic conditions (pH = 4.5~5), lysosomes continuously pump protons into the lysosome through V-type ATPase, i.e., a proton pump, to maintain a stable acidic microenvironment and metabolic capacity [140]. The acidic pH in the lysosome chamber is also an important characteristic, mainly regulating the circulation of proteins and macromolecules. Because of this lower pH of the lysosome chamber compared to other parts of the cell, the pH-sensitive probe has attracted the attention of scientists when designing lysosome-targeted probes.
2.4.2. Lysosome-Targeted Probe Design
Lysosome-targeted molecular probes are usually derived from lipophilic amines, a lipophilic weak base group. Amine-containing molecules can accumulate in lysosomes through passive diffusion or active transport actions, including endocytosis and autophagy; however, compared to the accumulation mechanism mediated by passive diffusion, endocytosis or autophagy contributes less to the number of accumulation probes [141,142]. The majority of probe design is still focused on the accumulation mechanism mediated by passive diffusion. Small molecules below 1 kDa diffuse freely in the cell membrane system [143]. Once the small-molecule probe diffuses into the lysosome, the amine moiety can be protonated. Due to the impermeability of the membrane, a probe that becomes positively charged will be trapped in the lysosome organelle. For example, commercial LysoTracker probes, constructed based on the same lipophilic amino moiety, have been widely applied to label lysosomes with different fluorescences [144,145].
In addition to weakly alkaline anchoring, scientists have also reported fluorescent probes based on an “AND” logic gate strategy. Probes that only release fluorescence signals in an acidic environment and after reacting with analytes are designed for the detection of lysosomal location matters. These kinds of molecules may not accumulate in lysosomes automatically, but would still exhibit an excellent signal-to-noise ratio for lysosome-targeted cell imaging.
Metal ions are involved in the mediation of the activation of lysosomal enzymes, including the proton pump V-type ATPase [140]. On the other hand, lysosomes are the main regulators of the metabolism of ions such as iron [146,147]. The liver is the main organ for storing iron in the body. The concentration of iron ions (especially Fe3+) in the lysosome of liver cells is one of the important biological indicators for diagnosing liver diseases such as cirrhosis, liver injury, and liver cancer [148]. Wang’s group designed three new Fe3+ fluorescence probes, using galactose as the liver-cell-targeting group, imidazole as the lysosome-targeting group, and naphthimide, with good photophysical and chemical properties, as the fluorescence response group to obtain monodentate, bidentate, and tridentate galactose probes—IM-Gal-1, IM-Gal-2, and IM-Gal-3 (42–44, Scheme 6) [149]. A fluorescence enhancement signal is generated in the presence of Fe3+ due to photoinduced electron transfer. Galactosyl probes can specifically recognize the overexpressed asialoglycoprotein receptor (ASGPR) in hepatocytes, which enables the probes to exhibit good liver-targeting ability. Cu2+ in lysosomes is reported to relate to Munk’s syndrome, Wilson’s disease, Alzheimer’s disease, and other diseases [150]. Chen et al. designed and synthesized a lysosome-targeted fluorescence probe, DHUCu-1 (45, Scheme 6), with near-infrared emission using methylene blue as the fluorescence carrier [151]. It can detect and image Cu2+ in lysosomes, in addition to having the advantages of good water solubility and low toxicity. When the nitrogen-containing structural probe coordinates with Cu2+ in lysosomes, a conjugated system is formed to generate a fluorescence-enhanced signal.
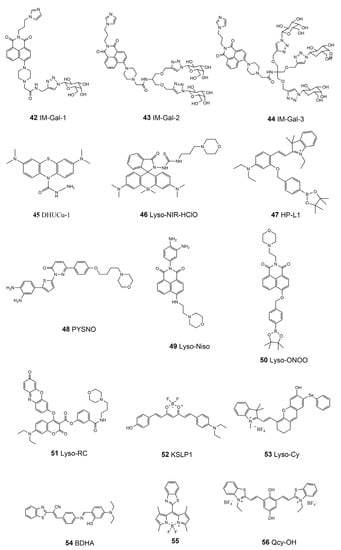
Scheme 6.
Chemical structure of the lysosome-targeted fluorescent molecular probes.
Unsurprisingly, the reactive species are also important in lysosomes. For example, ROS and RNS may induce lysosomal oxidative stress; the abnormal homocysteine (HCy) level may cause or aggravate cardiovascular and Alzheimer’s disease [152,153]. Mao et al. reported a lysosome-targeted near-infrared fluorescence probe, Lyso-NIR-HClO (46, Scheme 6), for highly selective as well as sensitive detection and imaging of endogenous and exogenous HClO in lysosomes [154]. This probe is composed based on Si-rhodamine modified with a morpholine unit as a lysosome-targeting group and a HClO-mediated cyclization reaction site as a response group. This probe was applied for HClO imaging in living cells and mice models. In 2016, Liu et al. introduced a probe (HP-L1, 47 Scheme 6) activated in an acidic environment for detecting H2O2 in lysosomes [155]. The fluorescence probe uses borate as the response sensing unit of H2O2 and spirobenzopyran as the pH-switchable fluorophores. The probe is constructed based on the “AND” logic gate method, meaning that it can only emit strong fluorescence if it reacts with H2O2 in acidic conditions in the lysosome. Recently, Zhou et al. designed a lysosomal-targeted NO fluorescence probe, PYSNO (48, Scheme 6), with high sensitivity and selectivity on the basis of a pyridazinone scaffold and photo-induced electron transfer (PET) mechanism, using morpholine as the acidic lysosomal-targeting group [156]. The fluorescence was turned on through the photo-induced electron transfer mechanism, which was used to explore the relationship between myocardial NO production and myocardial fibrosis in mice models. Based on the principle of PET, Yu et al. introduced the lysosome-targeted N-aminoethyl morpholine into the efficient two-photon fluorescence carrier naphthalimide. Using o-phenylenediamine as the recognition group, they designed a lysosome-targeted two-photon fluorescent free probe, Lyso-Nino (49, Scheme 6), which is highly selective and sensitive to NO [157]. The probe was used to capture NO from the lysosomes of macrophages for the first time and successfully monitored the production of NO in lysosomes under different stimuli. In the most recent study, Wang et al. synthesized a lysosomal-targeted naphthalimide-derived fluorescent probe, Lyso-ONOO (50, Scheme 6), that can monitor the exogenous and endogenous peroxynitrite (ONOO−) of human hepatic stellate cells [158]. The probe uses the phenyl borate pinacol ester group as the recognition group of ONOO− and morpholine as the lysosomal-targeting group. This probe can infer the order of severity in liver injury processes through the change in the level of ONOO−. In 2018, Zhang et al. developed a lysosome-targeted fluorescence probe, Lyso-RC (51, Scheme 6), for the simultaneous detection of Cys/Hcy, GSH, and H2S [159]. The target group of lysosomes is the morpholine group, and the probe contains two fluorophores, resorufin and 7-diethylaminocoumarin. This particular probe exhibits different fluorescences when responding to different RSS. Its emission spectra are generally free of crosstalk, which allows for simultaneous detection.
The lysosomal microenvironment affects the molecular transportation and activity of the enzymes, thus being related to the lysosomal function and status [160]. Li and co-workers proposed a curcumin-based polar near-infrared lysosomal-targeting fluorescence probe, KSLP1 (52, Scheme 6), with which to monitor the changes in lysosomal polarity in the process of cell aging [161]. Using this probe, scientists found that the polarity of lysosomes in aging cells increased for the first time and proposed the potential mechanism of changes in lysosomal polarity in the process of cell aging. The viscosity of lysosomes plays an important role in biological regulation [162]. The real-time tracking and monitoring of the dynamic changes in lysosomal viscosity are of great significance for diagnosing viscosity-related lysosomal diseases. Recently, Zeng’s group constructed a near-infrared lysosome-targeted viscosity probe, Lyso-cy (53, Scheme 6), with which to monitor the change in viscosity of biological systems [163]. The phenyl selenide group was introduced into the oxaanthrene indole dye, making the probe an ideal lysosome-targeting tracer. In a viscous medium, this probe has strong fluorescence emission near 710 nm and has been demonstrated to work well in the monitoring of the change in the lysosomal viscosity of living cells. The benzothiazole group was found to target lysosomes in living cells. Chen et al. synthesized a lysosome-targeted fluorescent probe, BDHA (54, Scheme 6), based on benzothiazole compounds, realizing the dual detection of OCl- and viscosity [164]. BDHA can be used to image the viscosity and OCl- level of HeLa cells and zebrafish. In 2022, Shi et al. designed and synthesized a BODIPY-based fluorescence probe (55, Scheme 6) with a rotatable meso-benzothiazole group, showing both good viscosity responsiveness and AIE properties [165]. This probe can sense lysosomal viscosity changes induced by lipopolysaccharides, nystatin, a low temperature, and dexamethasone in living cells, which can be further applied in autophagy monitoring via tracking viscosity changes. Recently, a new NIR fluorescent probe, Qcy-OH (56, Scheme 6), was developed by Zeng’s group to track lysosomal pH changes [166]. Two benzothiazolium units were introduced in the backbone of the probe to enhance the selectivity for intracellular lysosomes. This sulfur-driving lysosome-targeting ability of the probe affords a guarantee for achieving the long-term monitoring of lysosomal pH biology through the elimination of harmful protonating effects of the probe. A summary of the properties of the lysosome-targeted molecular probes is listed in Table 4.

Table 4.
Properties of the lysosome-targeted fluorescent molecular probes.
2.5. Dual-Targeted Molecular Probe
Different organelles play their roles inside cells as parts; however, as a whole, they must communicate with each other to keep entities running smoothly. Different molecules, such as ions, lipids, and proteins, transfer between different organelles under sophisticated regulation. For example, the unfolded protein response (UPR) of the ER and mitochondria may cause the translocation of some transcription factors toward the nucleus to initiate feedback bioprocesses [167,168]. The fission of mitochondria requires contact between mitochondria and the ER [169,170]. Lysosomes may attend to the autophagy of different organelles, such as mitophagy, through signal-induced recruitment [171]. Therefore, dual-targeted probes that can monitor different organelles simultaneously have attracted more and more interest because they have shown great potential for the investigation of communication and signal transfer processes between different organelles.
In recent years, many functional multi-organelle-targeting probes have been developed. Lin’s group designed and synthesized a fluorescent probe, MNQI (57, Scheme 7), which has a weak electronic donor and a positively charged group to bind DNA, for the visualization of the mitochondria and nucleus in dual channels [172]. Furthermore, they proved that the probe could reversibly detect the changes in MMP levels from both the localization and the emission color; reversible changes in cell viability have been successfully observed with the probe. The same group employed a semi-cyanine unit as the SO2-responsive site, a naphthalimide unit and semi-cyanine unit as the fluorophores, and semi-cyanine as well as morpholine groups to target mitochondria and lysosomes, constructing the first dual-targeting organelle-targeted fluorescent probe, DML-P (58, Scheme 7), capable of tracing the mitochondria and lysosome SO2 simultaneously [173]. Most recently, Ho et al. utilized an electron push–pull π-conjugation skeleton and a rarely reported diethyl phosphonate moiety on the pyridinium ring to develop an AIE-based bioprobe, ASP-PE (59, Scheme 7), for dual plasma membrane–mitochondria targeting through its phosphonate moiety and pyridinium, visualizing the changes in the membrane tension of the plasma membrane as well as mitochondria in response to varied osmotic pressure and substrate stiffness [174]. Therefore, this probe revealed the significance of actin and microtubules for plasma membranes and mitochondria in response to mechanobiological stimulations. A probe named RT-ER (60, Scheme 7), which could image the ER and lysosomes via sequencing under the stimulation of ClO-, was designed by Shan et al. through combining rhodamine B and 1,8-naphthalimide [175]. It was proven that this probe showed fast responses and was highly specific to ClO-, in addition to having an extremely large linear range, allowing it to be used in quantitative analyses in vitro. Duan et al. rationally designed a fluorescent probe, NADH-R (61, Scheme 7), via a simple graft of pyridiniumylbutenenitrile on a 1-methylquinolinium moiety in the 3-position, which could visualize exogenous and endogenous NAD(P)H generation in living cells. People applied NADH-R to monitor the fluctuation in the level of NAD(P)H under metabolic perturbation, and observed decreased NAD(P)H levels in the brains of stroke mice models. These results indicated that the new probe has a certain reference value for the treatment of clinical diseases [176]. Based on the TICT mechanism, a new NIR pH-dependent fluorescent probe, DCIC (62, Scheme 7), with a push–pull electronic moiety was synthesized by Wang et al. to identify the lysosome viscosity. The fluorescence quenching of DCIC did not occur with the environment changing from acidic to neutral or alkaline, providing a basis for multi-organelle localization [177]. DCIC can monitor lysosomal viscosity fluctuations while localized towards lysosomes, mitochondria, the Golgi apparatus, and the ER. Recently, Zhuang et al. developed an esterase-responsive prodrug, TPE-QC (63, Scheme 7), for dual organelle-targeted imaging and synergetic chemo-photodynamic cancer therapy [178]. They demonstrated that activated TPE-QC accumulated in both lysosomes and mitochondria. Benefitting from the overexpressed esterase in cancer cells, TPE-QC could be efficiently activated in cancer cells rather than in normal cells, resulting in enhanced anticancer potency. Lin’s group synthesized a fluorescent probe, LN-2 (64, Scheme 7), to visualize cell death based on its subcellular immigration from lysosomes towards the nucleus by regulating the binding affinity of the probe to lysosomes and DNA [179]. LN-2 was successfully used to display cell death induced by hydrogen peroxide and apoptosis induced by rotenone. It is a potential tool for studying apoptosis-related bioprocesses. Yapici et al. designed and synthesized a broad-range pH probe, RCPP (65, Scheme 7), to detect organellar acidity/alkalinity by integrating an acidic-pH-responsive rhodamine label with an alkaline-pH-responsive coumarin moiety [180]. Depending on the organellar acidity/alkalinity, RCPP can simultaneously move between mitochondria and lysosome subcellular locations, facilitating the simultaneous monitoring of acidity/alkalinity alterations in mitochondria and lysosomes in living cells. Recently, Yin’s group designed a dual-targeting SO2 fluorescent probe, Mito-SO2-Lyso (66, Scheme 7), based on the FRET regulation strategy, which realized the ratio detection of endogenous and exogenous SO2 in vivo [181]. Among them, benzopyranium salt units and morpholine groups were conjugated to target mitochondria and lysosomes. They demonstrated that the Mito-SO2-Lyso could quantitatively detect sulfite concentration in Yuba and crystal sugar, showing potential applications in cell imaging and food analyses. A summary of the properties of the dual-targeted molecular probes is listed in Table 5.
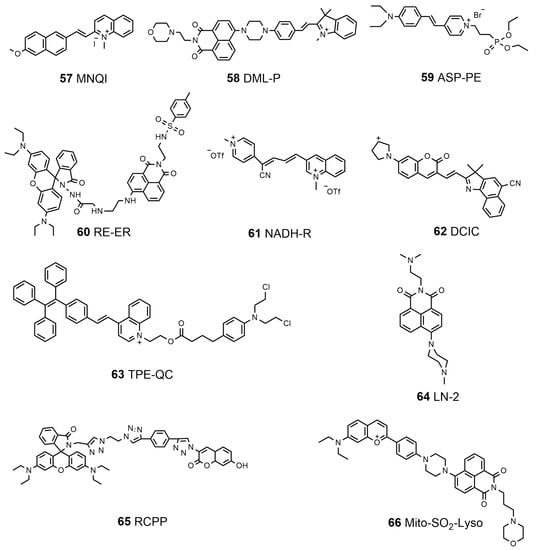
Scheme 7.
Chemical structure of the dual-targeted molecular probes.

Table 5.
Properties of the dual-targeted molecular probes.
3. Summary and Outlook
The development of fluorescent molecular probes provides powerful tools in the fields of cell-related research. At this point, elaborate cell images covering subcellular information, such as the patterns and statuses of organelles, can be obtained by utilizing specially designed probes. From these images, it is possible to investigate the relationship between particular organelles and different bioprocesses, revealing the signal molecules that play a key role in regulating bioactivity, explaining the molecular mechanisms of some diseases and bringing potential therapeutic targets to light. Hence, advanced fluorescent molecular probes facilitate the understanding of biology, pathology, and pharmacology. Here, we review the probes developed in the past decade for targeting organelles, such as nucleus, mitochondria, ERs, and lysosomes. We mostly focused on the probes proposed in the past five years and discussed the types of building blocks that can be selected for achieving organelle specificity according to the unique chemical/physical properties of different organelles. We also introduced the representative probes that can respond to special intracellular chemical/biological reagents at the subcellular level. The novel probes certainly showed advanced performances in comprehensive applications, but some drawbacks and challenges remain. We believe some of the following directions might receive more attention in the future in order to promote the capacity of the probes.
- (1)
- Low cytotoxicity: The cytotoxicity of the fluorescent molecular probes includes two parts. The first is the phototoxicity generated when the probes were excited by irradiation sources. Under the excitation conditions during cell imaging, the generation of ROS is inevitable. The exceedingly exogenous ROS may break the intracellular redox balance and cause oxidative damage to intracellular biomacromolecules, including DNA, proteins, and lipids, thus influencing the statuses of organelles and living cells. This property of the molecules has been applied to photodynamic therapy for disease treatment; however, it is a limitation of probes in the application of cell imaging, especially in cases of super-resolution and longtime imaging, which need higher emission energy. The modification of the fluorophore is still necessary to obtain a probe that works better inside cells. Chen and co-workers have introduced a triplet-state engineering strategy with which to construct mitochondria-targeted probes with reduced phototoxicity, providing a potential strategy for this direction [182]. The other part of the cytotoxicity of the molecular probes is the interruption of cell activity from the molecules themselves. For example, the membrane potential of the mitochondria will be somehow neutralized by the cationic molecules accumulated around the IMM. The underlying mechanism can introduce interferences with the normal activity of the respiratory chain reaction and thus cause the dysfunction of mitochondria. Therefore, a suitable targeted strategy is needed. The mitochondria-penetrating peptides (MPPs) are promising choices [183,184], which were reported to guide exogenous molecules into the mitochondrial location. Similarly, the sulfonamide group that can react with the K+ channel on the ER surface may influence the bioactivity of the ER. Therefore, several ER localization signal sequences that guide intracellular protein distribution, such as ER retention signal sequences, KDEL (—Lys-Asp-Glu-Leu-COOH), and ER insertion signal sequences, Eriss (ER insertion signal sequence), have been conjugated with molecular probes for ER-targeted imaging [185]. Some special short peptides, named nuclear localization signals (NLSs), or other chemical motifs that may recognize the importins in nuclei were applied to construct the nucleus-targeted probes [186].
- (2)
- The capacity of in vivo imaging: Fluorescent imaging in vivo is always challenging but engaging because it provides the most straightforward biological information. Many of the probes developed at this point possess a relatively short wavelength of excitation/emission located in the UV–Vis wavelength region. This region overlaps with the excitation/emission spectra of biomolecules and biosystems. Therefore, the result might be cluttered by the high background signals. In addition, the absorbance of the short-wavelength light by the biological samples will also limit the application of the probes in deep sample penetration imaging. The satisfying wavelength for in vivo imaging is in the near-infrared range (NIR, 650–950 nm), which can avoid interference form the biological samples. A number of NIR fluorophores have been developed, suggesting the importance of considering the link design with particular moieties for in vivo imaging at the organelle level. Another encouraging method is the design of two-photon-excited fluorophores, which can be excited by two lower-energy NIR photons. Therefore, two-photon fluorescent microscopy can be applied for imaging in vivo by using fluorescent probes with an increased penetration depth and other advantages, such as a prolonged observation time.
- (3)
- Biological guidance: Most molecular probes are developed by chemists, but the utilization of probes is most likely carried out by biologists. The gap between these two fields is obvious, implying that communication between scientists is integral to major breakthroughs and beyond. Our standpoint suggests that the development of biology should guide probe design. The studies on ferroptosis created an urgent demand for the detection of cellular iron and lipid peroxidase, as an example [187]. It was recently found that the calcium transients on the ER surface would trigger the process of autophagy [21], meaning that the ER-targeted Ca2+ probes are promising tools for autophagy monitoring. Moreover, the development of novel probes may also encourage biologists to discover unknown bioprocesses. For example, high-resolution methods can assist in distinguishing the different modes of mitochondrial fission; this may explain how other organelles participate in modulating the fission process in cells [188]. Furthermore, a series of novel probes that can monitor the membrane tension on the plasma membrane and other organelles were developed recently and promote the understanding of mechanobiological processes, thus indicating the importance of mechanoforce as an interesting parameter in the regulation of bioactivities [189,190,191,192]. Therefore, it is critical for scientists in different fields to exchange their cutting-edge knowledge to discover the most suitable molecular probes, thus advancing the growth of science.
Overall, the journey of organelle-targeted fluorescent molecular probes is just beginning. Deeper growth of chemical and biological knowledge will certainly lead to new probes as powerful tools for the relative fields.
Author Contributions
S.L., Z.D. and Y.C. contributed to the writing of the original draft; Y.C. and D.-M.K. contributed to the reviewing and editing. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the National Natural Science Foundation of China (21874075, 22074068), the Tianjin Natural Science Foundation (JCQNJC00240), and the Fundamental Research Funds for the Central Universities (JCQNJC00240).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
No new data were created or analyzed in this study. Data sharing is not applicable to this article.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Kluck, R.; Bossy-Wetzel, E.; Green, D.; Newmeyer, D. The release of cytochrome c from mitochondria: A primary site for Bcl-2 regulation of apoptosis. Science 1997, 275, 1132–1136. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Liu, X.; Bhalla, K.; Kim, C.; Ibrado, A.; Cai, J.; Peng, T.-I.; Jones, D.; Wang, X. Prevention of apoptosis by Bcl-2: Release of cytochrome c from mitochondria blocked. Science 1997, 275, 1129–1132. [Google Scholar] [CrossRef] [PubMed]
- Yarman, A.; Neumann, B.; Bosserdt, M.; Gajovic-Eichelmann, N.; Scheller, F. Peroxide-dependent analyte conversion by the heme prosthetic group, the heme Peptide “microperoxidase-11” and cytochrome C on chitosan capped gold nanoparticles modified electrodes. Biosensors 2012, 2, 189–204. [Google Scholar] [CrossRef]
- Gomez-Pastor, R.; Burchfiel, E.; Thiele, D. Regulation of heat shock transcription factors and their roles in physiology and disease. Nat. Rev. Mol. Cell Biol. 2018, 19, 4–19. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, H.; Matsui, T.; Yamamoto, A.; Okada, T.; Mori, K. XBP1 mRNA is induced by ATF6 and spliced by IRE1 in response to ER stress to produce a highly active transcription factor. Cell 2001, 107, 881–891. [Google Scholar] [CrossRef]
- Han, J.; Backa, S.; Hur, J.; Lin, Y.; Gildersleeve, R.; Shan, J.; Yuan, C.; Krokowski, D.; Wang, S.; Hatzoglou, M.; et al. ER-stress-induced transcriptional regulation increases protein synthesis leading to cell death. Nat. Cell Biol. 2013, 15, 481–490. [Google Scholar] [CrossRef]
- Wu, J.; Kaufman, R. From acute ER stress to physiological roles of the Unfolded Protein Response. Cell Death Differ. 2006, 13, 374–384. [Google Scholar] [CrossRef]
- Ziegler, D.; Vindrieux, D.; Goehrig, D.; Jaber, S.; Collin, G.; Griveau, A.; Wiel, C.; Bendridi, N.; Djebali, S.; Farfariello, V.; et al. Calcium channel ITPR2 and mitochondria-ER contacts promote cellular senescence and aging. Nat. Commun. 2021, 12, 720. [Google Scholar] [CrossRef]
- Simoes, I.; Morciano, G.; Lebiedzinska-Arciszewska, M.; Aguiari, G.; Pinton, P.; Potes, Y.; Wieckowski, M. The mystery of mitochondria-ER contact sites in physiology and pathology: A cancer perspective. Biochim. Et Biophys. Acta Mol. Basis Dis. 2020, 1866, 165834. [Google Scholar] [CrossRef]
- Chen, C.; Peng, J.; Xia, H.-S.; Yang, G.-F.; Wu, Q.-S.; Chen, L.-D.; Zeng, L.-B.; Zhang, Z.-L.; Pang, D.-W.; Li, Y. Quantum dots-based immunofluorescence technology for the quantitative determination of HER2 expression in breast cancer. Biomaterials 2009, 30, 2912–2918. [Google Scholar] [CrossRef]
- Wang, Y.; Shyy, J.; Chien, S. Fluorescence proteins, live-cell imaging, and mechanobiology: Seeing is believing. Annu. Rev. Biomed. Eng. 2008, 10, 1–38. [Google Scholar] [CrossRef]
- Bikoff, E. Formation of complexes between self-peptides and MHC class II molecules in cells defective for presentation of exogenous protein antigens. J. Immunol. 1992, 149, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Xu, W.; Zeng, Z.; Jiang, J.-H.; Chang, Y.-T.; Yuan, L. Discerning the Chemistry in Individual Organelles with Small-Molecule Fluorescent Probes. Angew. Chem. Int. Ed. 2016, 55, 13658–13699. [Google Scholar] [CrossRef]
- Kapuscinski, J. DAPI: A DNA-specific fluorescent probe. Biotech. Histochem. Off. Publ. Biol. Stain. Comm. 1995, 70, 220–233. [Google Scholar] [CrossRef]
- Pendergrass, W.; Wolf, N.; Poot, M. Efficacy of MitoTracker Green (TM) and CMXRosamine to measure changes in mitochondrial membrane potentials in living cells and tissues. Cytom. Part A 2004, 61A, 162–169. [Google Scholar] [CrossRef]
- Cole, L.; Davies, D.; Hyde, G.; Ashford, A. ER-Tracker dye and BODIPY-brefeldin A differentiate the endoplasmic reticulum and Golgi bodies from the tubular-vacuole system in living hyphae of Pisolithus tinctorius. J. Microsc. 2000, 197, 239–248. [Google Scholar] [CrossRef]
- Chikte, S.; Panchal, N.; Warnes, G. Use of LysoTracker Dyes: A Flow Cytometric Study of Autophagy. Cytom. Part A 2014, 85, 169–178. [Google Scholar] [CrossRef] [PubMed]
- Dutta, T.; Pal, K.; Koner, A. Intracellular Physical Properties with Small Organic Fluorescent Probes: Recent Advances and Future Perspectives. Chem. Rec. 2022, 22, e202200035. [Google Scholar] [CrossRef] [PubMed]
- Pierzynska-Mach, A.; Janowski, P.; Dobrucki, J. Evaluation of Acridine Orange, LysoTracker Red, and Quinacrine as Fluorescent Probes for Long-Term Tracking of Acidic Vesicles. Cytom. Part A 2014, 85A, 729–737. [Google Scholar] [CrossRef]
- Chretien, D.; Benit, P.; Ha, H.-H.; Keipert, S.; El-Khoury, R.; Chang, Y.-T.; Jastroch, M.; Jacobs, H.; Rustin, P.; Rak, M. Mitochondria are physiologically maintained at close to 50 degrees C. PLoS Biol. 2018, 16, e2003992. [Google Scholar] [CrossRef]
- Zheng, Q.; Chen, Y.; Chen, D.; Zhao, H.; Feng, Y.; Meng, Q.; Zhao, Y.; Zhang, H. Calcium transients on the ER surface trigger liquid-liquid phase separation of FIP200 to specify autophagosome initiation sites. Cell 2022, 185, 4082–4098. [Google Scholar] [CrossRef] [PubMed]
- Stockwell, B.; Angeli, J.; Bayir, H.; Bush, A.; Conrad, M.; Dixon, S.; Fulda, S.; Gascon, S.; Hatzios, S.; Kagan, V.; et al. Ferroptosis: A Regulated Cell Death Nexus Linking Metabolism. Redox Biol. Dis. Cell 2017, 171, 273–285. [Google Scholar]
- Downs, J.; Nussenzweig, M.; Nussenzweig, A. Chromatin dynamics and the preservation of genetic information. Nature 2007, 447, 951–958. [Google Scholar] [CrossRef] [PubMed]
- Abdalla, S.; Letarte, M. Hereditary haemorrhagic telangiectasia: Current views on genetics and mechanisms of disease. J. Med. Genet. 2006, 43, 97–110. [Google Scholar] [CrossRef] [PubMed]
- Rotthier, A.; Baets, J.; Timmerman, V.; Janssens, K. Mechanisms of disease in hereditary sensory and autonomic neuropathies. Nat. Rev. Neurol. 2012, 8, 73–85. [Google Scholar] [CrossRef]
- Solowska, J.; Baas, P. Hereditary spastic paraplegia SPG4: What is known and not known about the disease. Brain 2015, 138, 2471–2484. [Google Scholar] [CrossRef]
- Prokhorova, E.; Zamaraev, A.; Kopeina, G.; Zhivotovsky, B.; Lavrik, I. Role of the nucleus in apoptosis: Signaling and execution. Cell. Mol. Life Sci. 2015, 72, 4593–4612. [Google Scholar] [CrossRef]
- Kang, Y.; He, R.; Yang, L.; Qin, D.; Guggilam, A.; Elks, C.; Yan, N.; Guo, Z.; Francis, J. Brain tumour necrosis factor-alpha modulates neurotransmitters in hypothalamic paraventricular nucleus in heart failure. Cardiovasc. Res. 2009, 83, 737–746. [Google Scholar] [CrossRef]
- Paine, P.; Moore, L.; Horowitz, S. Nuclear envelope permeability. Nature 1975, 254, 109–114. [Google Scholar] [CrossRef]
- Dervan, P. Molecular recognition of DNA by small molecules. Bioorganic Med. Chem. 2001, 9, 2215–2235. [Google Scholar] [CrossRef]
- Sirajuddin, M.; Ali, S.; Badshah, A. Drug-DNA interactions and their study by UV-Visible, fluorescence spectroscopies and cyclic voltametry. J. Photochem. Photobiol. B Biol. 2013, 124, 1–19. [Google Scholar] [CrossRef]
- Kim, Y.; Sah, R.; Doong, J.; Grodzinsky, A. Fluorometric assay of DNA in cartilage explants using Hoechst 33258. Anal. Biochem. 1988, 174, 168–176. [Google Scholar] [CrossRef]
- Ahn, S.; Costa, J.; Emanuel, J. PicoGreen quantitation of DNA: Effective evaluation of samples pre- or post-PCR. Nucleic Acids Res. 1996, 24, 2623–2625. [Google Scholar] [CrossRef]
- Bucevicius, J.; Lukinavicius, G.; Gerasimaite, R. The Use of Hoechst Dyes for DNA Staining and Beyond. Chemosensors 2018, 6, 18. [Google Scholar] [CrossRef]
- Ishida, M.; Watanabe, H.; Takigawa, K.; Kurishita, Y.; Oki, C.; Nakamura, A.; Hamachi, I.; Tsukiji, S. Synthetic Self-Localizing Ligands That Control the Spatial Location of Proteins in Living Cells. J. Am. Chem. Soc. 2013, 135, 12684–12689. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, A.; Takigawa, K.; Kurishita, Y.; Kuwata, K.; Ishida, M.; Shimoda, Y.; Hamachi, I.; Tsukiji, S. Hoechst tagging: A modular strategy to design synthetic fluorescent probes for live-cell nucleus imaging. Chem. Commun. 2014, 50, 6149–6152. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Ye, Z.; Zhang, X.; Man, H.; Huang, Z.; Li, N.; Xiao, Y. A targetable fluorescent probe for dSTORM super-resolution imaging of live cell nucleus DNA. Chem. Commun. 2019, 55, 1951–1954. [Google Scholar] [CrossRef]
- Narayanaswamy, N.; Das, S.; Samanta, P.; Banu, K.; Sharma, G.; Mondal, N.; Dhar, S.; Pati, S.; Govindaraju, T. Sequence-specific recognition of DNA minor groove by an NIR-fluorescence switch-on probe and its potential applications. Nucleic Acids Res. 2015, 43, 8651–8663. [Google Scholar] [CrossRef] [PubMed]
- Ganguly, S.; Murugan, N.; Ghosh, D.; Narayanaswamy, N.; Govindaraju, T.; Basu, G. DNA Minor Groove-Induced cis-trans Isomerization of a Near-Infrared Fluorescent Probe. Biochemistry 2021, 60, 2084–2097. [Google Scholar] [CrossRef] [PubMed]
- Abeywickrama, C.; Bertman, K.; Plescia, C.; Stahelin, R.; Pang, Y. Structural Effect on the Cellular Selectivity of an NIR-Emitting Cyanine Probe: From Lysosome to Simultaneous Nucleus and Mitochondria Selectivity with Potential for Monitoring Mitochondria Dysfunction in Cells. ACS Appl. Bio Mater. 2019, 2, 5174–5181. [Google Scholar] [CrossRef]
- Pratihar, S.; Suseela, Y.; Govindaraju, T. Threading Intercalator-Induced Nanocondensates and Role of Endogenous Metal Ions in Decondensation for DNA Delivery. ACS Appl. Bio Mater. 2020, 3, 6979–6991. [Google Scholar] [CrossRef]
- Abeywickrama, C.; Wijesinghe, K.; Plescia, C.; Fisher, L.; Goodson, T.; Stahelin, R.; Pang, Y. A pyrene-based two-photon excitable fluorescent probe to visualize nuclei in live cells. Photochem. Photobiol. Sci. 2020, 19, 1152–1159. [Google Scholar] [CrossRef]
- Sayed, M.; Gubbala, G.; Pal, H. Contrasting interactions of DNA-intercalating dye acridine orange with hydroxypropyl derivatives of -cyclodextrin and -cyclodextrin hosts. New J. Chem. 2019, 43, 724–736. [Google Scholar] [CrossRef]
- Kandinska, M.; Cheshmedzhieva, D.; Kostadinov, A.; Rusinov, K.; Rangelov, M.; Todorova, N.; Ilieva, S.; Ivanov, D.; Videva, V.; Lozanov, V.; et al. Tricationic asymmetric monomeric monomethine cyanine dyes with chlorine and trifluoromethyl functionality—Fluorogenic nucleic acids probes. J. Mol. Liq. 2021, 342, 117501. [Google Scholar] [CrossRef]
- Ali, T.H.; Mandal, A.M.; Alhasan, A.; Dehaen, W. Surface fabrication of magnetic core-shell silica nanoparticles with perylene diimide as a fluorescent dye for nucleic acid visualization. J. Mol. Liq. 2022, 359, 119345. [Google Scholar]
- Zhao, L.; He, X.; Li, D.; Xu, S.; Huang, Y.; Li, X.; Wang, X.; Sun, Y.; Ma, P.; Song, D. A novel fluorescent probe for the localization of nucleoli developed via a chain reaction of endogenous cysteine in cells. J. Mater. Chem. B 2020, 8, 7652–7658. [Google Scholar] [CrossRef]
- Bochman, M.; Paeschke, K.; Zakian, V. DNA secondary structures: Stability and function of G-quadruplex structures. Nat. Rev. Genet. 2012, 13, 770–780. [Google Scholar] [CrossRef]
- Biffi, G.; Tannahill, D.; McCafferty, J.; Balasubramanian, S. Quantitative visualization of DNA G-quadruplex structures in human cells. Nat. Chem. 2013, 5, 182–186. [Google Scholar] [CrossRef]
- Cui, Y.; Kong, D.; Ghimire, C.; Xu, C.; Mao, H. Mutually Exclusive Formation of G-Quadruplex and i-Motif Is a General Phenomenon Governed by Steric Hindrance in Duplex DNA. Biochemistry 2016, 55, 2291–2299. [Google Scholar] [CrossRef]
- Sutherland, C.; Cui, Y.; Mao, H.; Hurley, L. A Mechanosensor Mechanism Controls the G-Quadruplex/i-Motif Molecular Switch in the MYC Promoter NHE III1. J. Am. Chem. Soc. 2016, 138, 14138–14151. [Google Scholar] [CrossRef] [PubMed]
- Kotar, A.; Wang, B.; Shivalingam, A.; Gonzalez-Garcia, J.; Vilar, R.; Plavec, J. NMR Structure of a Triangulenium-Based Long-Lived Fluorescence Probe Bound to a G-Quadruplex. Angew. Chem. Int. Ed. 2016, 55, 12508–12511. [Google Scholar] [CrossRef]
- Summers, P.; Lewis, B.; Gonzalez-Garcia, J.; Porreca, R.; Lim, A.; Cadinu, P.; Martin-Pintado, N.; Mann, D.; Edel, J.; Vannier, J.; et al. Visualising G-quadruplex DNA dynamics in live cells by fluorescence lifetime imaging microscopy. Nat. Commun. 2021, 12, 162. [Google Scholar] [CrossRef]
- Sun, W.; Cui, J.-X.; Ma, L.-L.; Lu, Z.-L.; Gong, B.; He, L.; Wang, R. Imaging nucleus viscosity and G-quadruplex DNA in living cells using a nucleus-targeting two-photon fluorescent probe. Analyst 2018, 143, 5799–5804. [Google Scholar] [CrossRef] [PubMed]
- Zhu, B.; Jia, H.; Zhang, X.; Chen, Y.; Liu, H.; Tan, W. Engineering a subcellular targetable, red-emitting, and ratiometric fluorescent probe for Ca2+ and its bioimaging applications. Anal. Bioanal. Chem. 2010, 397, 1245–1250. [Google Scholar] [CrossRef]
- Exposito-Rodriguez, M.; Laissue, P.; Yvon-Durocher, G.; Smirnoff, N.; Mullineaux, P. Photosynthesis-dependent H2O2 transfer from chloroplasts to nuclei provides a high-light signalling mechanism. Nat. Commun. 2017, 8, 49. [Google Scholar] [CrossRef]
- Maruta, T.; Noshi, M.; Tanouchi, A.; Tamoi, M.; Yabuta, Y.; Yoshimura, K.; Ishikawa, T.; Shigeoka, S. H2O2-triggered Retrograde Signaling from Chloroplasts to Nucleus Plays Specific Role in Response to Stress. J. Biol. Chem. 2012, 287, 11717–11729. [Google Scholar] [CrossRef] [PubMed]
- Nathan, C.; Xie, Q. Nitric oxide synthases: Roles, tolls, and controls. Cell 1994, 78, 915–918. [Google Scholar] [CrossRef] [PubMed]
- Dickinson, B.; Tang, Y.; Chang, Z.; Chang, C. A Nuclear-Localized Fluorescent Hydrogen Peroxide Probe for Monitoring Sirtuin-Mediated Oxidative Stress Responses In Vivo. Chem. Biol. 2011, 18, 943–948. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Huang, Z.; Ma, D.; Yan, Y.; Zhang, X.; Xiao, Y. A nucleus targetable fluorescent probe for ratiometric imaging of endogenous NO in living cells and zebrafishes. Analyst 2021, 146, 4130–4134. [Google Scholar] [CrossRef] [PubMed]
- McBride, H.; Neuspiel, M.; Wasiak, S. Mitochondria: More than just a powerhouse. Curr. Biol. 2006, 16, R551–R560. [Google Scholar] [CrossRef] [PubMed]
- Shadel, G.; Horvath, T. Mitochondrial ROS Signaling in Organismal Homeostasis. Cell 2015, 163, 560–569. [Google Scholar] [CrossRef]
- Orrenius, S.; Zhivotovsky, B.; Nicotera, P. Regulation of cell death: The calcium-apoptosis link. Nat. Rev. Mol. Cell Biol. 2003, 4, 552–565. [Google Scholar] [CrossRef]
- Narendra, D.; Tanaka, A.; Suen, D.-F.; Youle, R. Parkin is recruited selectively to impaired mitochondria and promotes their autophagy. J. Cell Biol. 2008, 183, 795–803. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Xue, Y.; Xu, X.; Wang, G.; Liu, Y.; Wu, H.; Li, W.; Wang, Y.; Chen, Z.; Zhang, W.; et al. A mitochondrial FUNDC1/HSC70 interaction organizes the proteostatic stress response at the risk of cell morbidity. EMBO J. 2019, 38, e98786. [Google Scholar] [CrossRef]
- Green, D.; Reed, J. Mitochondria and apoptosis. Science 1998, 281, 1309–1312. [Google Scholar] [CrossRef] [PubMed]
- Tait, S.; Green, D. Mitochondria and cell death: Outer membrane permeabilization and beyond. Nat. Rev. Mol. Cell Biol. 2010, 11, 621–632. [Google Scholar] [CrossRef] [PubMed]
- Angelova, P.; Esteras, N.; Abramov, A. Mitochondria and lipid peroxidation in the mechanism of neurodegeneration: Finding ways for prevention. Med. Res. Rev. 2021, 41, 770–784. [Google Scholar] [CrossRef] [PubMed]
- Picca, A.; Calvani, R.; Coelho-Junior, H.; Marzetti, E. Cell Death and Inflammation: The Role of Mitochondria in Health and Disease. Cells 2021, 10, 537. [Google Scholar] [CrossRef]
- Carinci, M.; Vezzani, B.; Patergnani, S.; Ludewig, P.; Lessmann, K.; Magnus, T.; Casetta, I.; Pugliatti, M.; Pinton, P.; Giorgi, C. Different Roles of Mitochondria in Cell Death and Inflammation: Focusing on Mitochondrial Quality Control in Ischemic Stroke and Reperfusion. Biomedicines 2021, 9, 169. [Google Scholar] [CrossRef]
- Obrador, E.; Salvador-Palmer, R.; Lopez-Blanch, R.; Jihad-Jebbar, A.; Valles, S.; Estrela, J. The Link between Oxidative Stress, Redox Status, Bioenergetics and Mitochondria in the Pathophysiology of ALS. Int. J. Mol. Sci. 2021, 22, 6352. [Google Scholar] [CrossRef]
- Huang, M.; Myers, C.; Wang, Y.; You, M. Mitochondria as a Novel Target for Cancer Chemoprevention: Emergence of Mitochondrial-targeting Agents. Cancer Prev. Res. 2021, 14, 285–306. [Google Scholar] [CrossRef]
- Zielonka, J.; Joseph, J.; Sikora, A.; Hardy, M.; Ouari, O.; Vasquez-Vivar, J.; Cheng, G.; Lopez, M.; Kalyanaraman, B. Mitochondria-Targeted Triphenylphosphonium-Based Compounds: Syntheses, Mechanisms of Action, and Therapeutic and Diagnostic Applications. Chem. Rev. 2017, 117, 10043–10120. [Google Scholar] [CrossRef]
- Frey, T.; Mannella, C. The internal structure of mitochondria. Trends Biochem. Sci. 2000, 25, 319–324. [Google Scholar] [CrossRef]
- Henze, K.; Martin, W. Evolutionary biology: Essence of mitochondria. Nature 2003, 426, 127–128. [Google Scholar] [CrossRef]
- Liu, Z.; Bushnell, W.; Brambl, R. Pontentiometric cyanine dyes are sensitive probes for mitochondria in intact plant cells: Kinetin enhances mitochondrial fluorescence. Plant Physiol. 1987, 84, 1385–1390. [Google Scholar] [CrossRef] [PubMed]
- Zhai, R.; Fang, B.; Lai, Y.; Peng, B.; Bai, H.; Liu, X.; Li, L.; Huang, W. Small-molecule fluorogenic probes for mitochondrial nanoscale imaging. Chem. Soc. Rev. 2022, 52, 942–972. [Google Scholar] [CrossRef]
- Chazotte, B. Labeling mitochondria with MitoTracker dyes. Cold Spring Harb. Protoc. 2011, 2011, 990–992. [Google Scholar] [CrossRef]
- Kasparov, S. Feasibility of Photodynamic Therapy for Glioblastoma with the Mitochondria-Targeted Photosensitizer Tetramethylrhodamine Methyl Ester (TMRM). Biomedicines 2021, 9, 1453. [Google Scholar]
- Sivandzade, F.; Bhalerao, A.; Cucullo, L. Analysis of the Mitochondrial Membrane Potential Using the Cationic JC-1 Dye as a Sensitive Fluorescent Probe. Bio-Protocol 2019, 9, e3128. [Google Scholar] [CrossRef] [PubMed]
- Winterbourn, C. Reconciling the chemistry and biology of reactive oxygen species. Nat. Chem. Biol. 2008, 4, 278–286. [Google Scholar] [CrossRef] [PubMed]
- Chouchani, E.; Pell, V.; Gaude, E.; Aksentijevic, D.; Sundier, S.; Robb, E.; Logan, A.; Nadtochiy, S.; Ord, E.; Smith, A.; et al. Ischaemic accumulation of succinate controls reperfusion injury through mitochondrial ROS. Nature 2014, 515, 431–435. [Google Scholar] [CrossRef] [PubMed]
- Robinson, K.; Janes, M.; Pehar, M.; Monette, J.; Ross, M.; Hagen, T.; Murphy, M.; Beckman, J. Selective fluorescent imaging of superoxide in vivo using ethidium-based probes. Proc. Natl. Acad. Sci. USA 2006, 103, 15038–15043. [Google Scholar] [CrossRef]
- Li, Z.; Ren, T.-B.; Zhang, X.-X.; Xu, S.; Gong, X.-Y.; Yang, Y.; Ke, G.; Yuan, L.; Zhang, X.-B. Precipitated Fluorophore-Based Probe for Accurate Detection of Mitochondrial Analytes. Anal. Chem. 2021, 93, 2235–2243. [Google Scholar] [CrossRef]
- Wu, Z.; Liu, M.; Liu, Z.; Tian, Y. Real-Time Imaging and Simultaneous Quantification of Mitochondrial H2O2 and ATP in Neurons with a Single Two-Photon Fluorescence-Lifetime-Based Probe. J. Am. Chem. Soc. 2020, 142, 7532–7541. [Google Scholar] [CrossRef]
- Niu, H.; Tang, J.; Zhu, X.; Li, Z.; Zhang, Y.; Ye, Y.; Zhao, Y. A three-channel fluorescent probe to image mitochondrial stress. Chem. Commun. 2020, 56, 7710–7713. [Google Scholar] [CrossRef] [PubMed]
- Cheng, G.; Fan, J.; Sun, W.; Sui, K.; Jin, X.; Wang, J.; Peng, X. A highly specific BODIPY-based probe localized in mitochondria for HClO imaging. Analyst 2013, 138, 6091–6096. [Google Scholar] [CrossRef] [PubMed]
- Lacza, Z.; Pankotai, E.; Csordas, A.; Geroo, D.; Kiss, L.; Horvath, E.; Kollai, M.; Busija, D.; Szabo, C. Mitochondrial NO and reactive nitrogen species production: Does mtNOS exist? Nitric Oxide Biol. Chem. 2006, 14, 162–168. [Google Scholar] [CrossRef] [PubMed]
- Paulsen, C.; Carroll, K. Cysteine-Mediated Redox Signaling: Chemistry, Biology, and Tools for Discovery. Chem. Rev. 2013, 113, 4633–4679. [Google Scholar] [CrossRef]
- Trollinger, D.; Cascio, W.; Lemasters, J. Selective loading of Rhod 2 into mitochondria shows mitochondrial Ca-2+ transients during the contractile cycle in adult rabbit cardiac myocytes. Biochem. Biophys. Res. Commun. 1997, 236, 738–742. [Google Scholar] [CrossRef] [PubMed]
- Sensi, S.; Ton-That, D.; Weiss, J.; Rothe, A.; Gee, K. A new mitochondrial fluorescent zinc sensor. Cell Calcium 2003, 34, 281–284. [Google Scholar] [CrossRef]
- Taki, M.; Akaoka, K.; Iyoshi, S.; Yamamoto, Y. Rosamine-Based Fluorescent Sensor with Femtomolar Affinity for the Reversible Detection of a Mercury Ion. Inorg. Chem. 2012, 51, 13075–13077. [Google Scholar] [CrossRef]
- Glickstein, H.; El, R.B.; Shvartsman, M.; Cabantchik, Z. Intracellular labile iron pools as direct targets of iron chelators: A fluorescence study of chelator action in living cells. Blood 2005, 106, 3242–3250. [Google Scholar] [CrossRef]
- Gao, P.; Pan, W.; Li, N.; Tang, B. Fluorescent probes for organelle-targeted bioactive species imaging. Chem. Sci. 2019, 10, 6035–6071. [Google Scholar] [CrossRef]
- Zhou, L.; Liu, C.; Zheng, Y.; Huang, Z.; Zhang, X.; Xiao, Y. Bio-orthogonal Toolbox for Monitoring Nitric Oxide in Targeted Organelles of Live Cells and Zebrafishes. Anal. Chem. 2022, 94, 15678–15685. [Google Scholar] [CrossRef] [PubMed]
- Miao, J.; Huo, Y.; Lv, X.; Li, Z.; Cao, H.; Shi, H.; Shi, Y.; Guo, W. Fast-response and highly selective fluorescent probes for biological signaling molecule NO based on N-nitrosation of electron-rich aromatic secondary amines. Biomaterials 2016, 78, 11–19. [Google Scholar] [CrossRef]
- Zhang, J.; Bao, X.; Zhou, J.; Peng, F.; Ren, H.; Dong, X.; Zhao, W. A mitochondria-targeted turn-on fluorescent probe for the detection of glutathione in living cells. Biosens. Bioelectron. 2016, 85, 164–170. [Google Scholar] [CrossRef]
- Michikawa, Y.; Mazzucchelli, F.; Bresolin, N.; Scarlato, G.; Attardi, G. Aging-dependent large accumulation of point mutations in the human mtDNA control region for replication. Science 1999, 286, 774–779. [Google Scholar] [CrossRef] [PubMed]
- Yao, Y.-G.; Kajigaya, S.; Feng, X.; Samsel, L.; McCoy, J.; Torelli, G., Jr.; Young, N. Accumulation of mtDNA variations in human single CD34(+) cells from maternally related individuals: Effects of aging and family genetic background. Stem Cell Res. 2013, 10, 361–370. [Google Scholar] [CrossRef] [PubMed]
- Uno, K.; Sugimoto, N.; Sato, Y. N-aryl pyrido cyanine derivatives are nuclear and organelle DNA markers for two-photon and super-resolution imaging. Nat. Commun. 2021, 12, 2650. [Google Scholar] [CrossRef]
- Chen, X.-C.; Tang, G.-X.; Luo, W.-H.; Shao, W.; Dai, J.; Zeng, S.-T.; Huang, Z.-S.; Chen, S.-B.; Tan, J.-H. Monitoring and Modulating mtDNA G-Quadruplex Dynamics Reveal Its Close Relationship to Cell Glycolysis. J. Am. Chem. Soc. 2021, 143, 20779–20791. [Google Scholar] [CrossRef]
- Burch, S.; Lopez, C. Effects of Cell Density and Microenvironment on Stem Cell Mitochondria Transfer among Human Adipose-Derived Stem Cells and HEK293 Tumorigenic Cells. Int. J. Mol. Sci. 2022, 23, 2003. [Google Scholar] [CrossRef]
- Lee, M.; Park, N.; Yi, C.; Han, J.; Hong, J.; Kim, K.; Kang, D.; Sessler, J.; Kang, C.; Kim, J. Mitochondria-Immobilized pH-Sensitive Off-On Fluorescent Probe. J. Am. Chem. Soc. 2014, 136, 14136–14142. [Google Scholar] [CrossRef]
- Arai, S.; Suzuki, M.; Park, S.-J.; Yoo, J.; Wang, L.; Kang, N.-Y.; Ha, H.-H.; Chang, Y.-T. Mitochondria-targeted fluorescent thermometer monitors intracellular temperature gradient. Chem. Commun. 2015, 51, 8044–8047. [Google Scholar] [CrossRef] [PubMed]
- Huang, Z.; Li, N.; Zhang, X.; Wang, C.; Xiao, Y. Fixable Molecular Thermometer for Real-Time Visualization and Quantification of Mitochondrial Temperature. Anal. Chem. 2018, 90, 13953–13959. [Google Scholar] [CrossRef]
- Huang, Z.; Li, N.; Zhang, X.; Xiao, Y. Mitochondria-Anchored Molecular Thermometer Quantitatively Monitoring Cellular Inflammations. Anal. Chem. 2021, 93, 5081–5088. [Google Scholar] [CrossRef]
- Chen, B.; Mao, S.; Sun, Y.; Sun, L.; Ding, N.; Li, C.; Zhou, J. A mitochondria-targeted near-infrared fluorescent probe for imaging viscosity in living cells and a diabetic mice model. Chem. Commun. 2021, 57, 4376–4379. [Google Scholar] [CrossRef]
- Zhang, G.; Quan, W.; Li, Y.; Song, W.; Lin, W. Near-Infrared Mitochondria-Targetable Single-Molecule probe for Dual-Response of viscosity and sulfur dioxide in vivo. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2022, 270, 120796. [Google Scholar] [CrossRef]
- Nicchitta, C. A platform for compartmentalized protein synthesis: Protein translation and translocation in the ER. Curr. Opin. Cell Biol. 2002, 14, 412–416. [Google Scholar] [CrossRef] [PubMed]
- Mattson, M.; LaFerla, F.; Chan, S.; Leissring, M.; Shepel, P.; Geiger, J. Calcium signaling in the ER: Its role in neuronal plasticity and neurodegenerative disorders. Trends Neurosci. 2000, 23, 222–229. [Google Scholar] [CrossRef] [PubMed]
- Walter, P.; Ron, D. The Unfolded Protein Response: From Stress Pathway to Homeostatic Regulation. Science 2011, 334, 1081–1086. [Google Scholar] [CrossRef]
- Thaxton, J.; Wallace, C.; Riesenberg, B.; Zhang, Y.; Paulos, C.; Beeson, C.; Liu, B.; Li, Z. Modulation of Endoplasmic Reticulum Stress Controls CD4(+) T-cell Activation and Antitumor Function. Cancer Immunol. Res. 2017, 5, 666–675. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, W.; Bone, R.; Sohn, P.; Syed, F.; Reissaus, C.; Mosley, A.; Wijeratne, A.; True, J.; Tong, X.; Kono, T.; et al. Endoplasmic reticulum stress alters ryanodine receptor function in the murine pancreatic cell. J. Biol. Chem. 2019, 294, 168–181. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.; Samidurai, A.; Thompson, J.; Hu, Y.; Das, A.; Willard, B.; Lesnefsky, E. Endoplasmic reticulum stress-mediated mitochondrial dysfunction in aged hearts. Biochim. Et Biophys. Acta Mol. Basis Dis. 2020, 1866, 165899. [Google Scholar] [CrossRef]
- Yang, M.; Luo, S.; Wang, X.; Li, C.; Yang, J.; Zhu, X.; Xiao, L.; Sun, L. ER-Phagy: A New Regulator of ER Homeostasis. Front. Cell Dev. Biol. 2021, 9, 684526. [Google Scholar] [CrossRef]
- Wilkinson, S. ER-phagy: Shaping up and destressing the endoplasmic reticulum. FEBS J. 2019, 286, 2645–2663. [Google Scholar] [CrossRef]
- Markgraf, D.; Klemm, R.; Junker, M.; Hannibal-Bach, H.; Ejsing, C.; Rapoport, T. An ER Protein Functionally Couples Neutral Lipid Metabolism on Lipid Droplets to Membrane Lipid Synthesis in the ER. Cell Rep. 2014, 6, 44–55. [Google Scholar] [CrossRef] [PubMed]
- Colston, J.; Horobin, R.; Rashid-Doubell, F.; Pediani, J.; Johal, K. Why fluorescent probes for endoplasmic reticulum are selective: An experimental and QSAR-modelling study. Biotech. Histochem. 2003, 78, 323–332. [Google Scholar] [CrossRef] [PubMed]
- Zhou, M.; He, H.-J.; Suzuki, R.; Liu, K.-X.; Tanaka, S.; Sekiguchi, M.; Itoh, H.; Kawahara, K.; Abe, H. Localization of sulfonylurea receptor subunits, SUR2A and SUR2B, in rat heart. J. Histochem. Cytochem. 2007, 55, 795–804. [Google Scholar] [CrossRef]
- Yuriev, E.; Kong, D.; Iskander, M. Investigation of structure-activity relationships in a series of glibenclamide analogues. Eur. J. Med. Chem. 2004, 39, 835–847. [Google Scholar] [CrossRef]
- Singh, D.; Rajput, D.; Kanvah, S. Fluorescent probes for targeting endoplasmic reticulum: Design strategies and their applications. Chem. Commun. 2022, 58, 2413–2429. [Google Scholar] [CrossRef]
- Phaniraj, S.; Gao, Z.; Rane, D.; Peterson, B. Hydrophobic resorufamine derivatives: Potent and selective red fluorescent probes of the endoplasmic reticulum of mammalian cells. Dye. Pigment. 2016, 135, 127–133. [Google Scholar] [CrossRef]
- Banerjee, A.; Banerjee, V.; Czinn, S.; Blanchard, T. Increased reactive oxygen species levels cause ER stress and cytotoxicity in andrographolide treated colon cancer cells. Oncotarget 2017, 8, 26142–26153. [Google Scholar] [CrossRef]
- Khandelwal, A.; Hebert, V.; Dugas, T. Essential role of ER-alpha-dependent NO production in resveratrol-mediated inhibition of restenosis. Am. J. Physiol. -Heart Circ. Physiol. 2010, 299, H1451–H1458. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.; Liu, S.; Tan, W.; Tang, F.; Long, J.; Li, Z.; Liang, B.; Chu, C.; Yang, J. Gaseous signalling molecule SO2 via Hippo-MST pathway to improve myocardial fibrosis of diabetic rats. Mol. Med. Rep. 2017, 16, 8953–8963. [Google Scholar] [CrossRef] [PubMed]
- Yue, L.; Huang, H.; Song, W.; Lin, W. A near-infrared endoplasmic reticulum-targeted fluorescent probe to visualize the fluctuation of SO2 during endoplasmic reticulum stress. Chem. Eng. J. 2022, 431, 133468. [Google Scholar] [CrossRef]
- Zhang, H.; Chen, J.; Xiong, H.; Zhang, Y.; Chen, W.; Sheng, J.; Song, X. An endoplasmic reticulum-targetable fluorescent probe for highly selective detection of hydrogen sulfide. Org. Biomol. Chem. 2019, 17, 1436–1441. [Google Scholar] [CrossRef]
- Gotoh, T.; Mori, M. Nitric oxide and endoplasmic reticulum stress. Arterioscler. Thromb. Vasc. Biol. 2006, 26, 1439–1446. [Google Scholar] [CrossRef]
- Li, S.-J.; Zhou, D.-Y.; Li, Y.; Liu, H.-W.; Wu, P.; Ou-Yang, J.; Jiang, W.-L.; Li, C.-Y. Efficient Two-Photon Fluorescent Probe for Imaging of Nitric Oxide during Endoplasmic Reticulum Stress. ACS Sens. 2018, 3, 2311–2319. [Google Scholar] [CrossRef]
- Peng, S.; Li, Z.; Zhang, Y.; Cao, W.; Liu, J.; Zhu, W.; Ye, Y. A two-photon fluorescent probe for HNO rapid visualization in endoplasmic reticulum. Sens. Actuator B Chem. 2020, 317, 128211. [Google Scholar] [CrossRef]
- Lu, Y.; Wang, R.; Sun, Y.; Tian, M.; Dong, B. Endoplasmic reticulum-specific fluorescent probe for the two-photon imaging of endogenous superoxide anion (O-2(center dot-)) in live cells and zebrafishes. Talanta 2021, 225, 122020. [Google Scholar] [CrossRef]
- Pak, Y.; Park, S.; Song, G.; Yim, Y.; Kang, H.; Kim, H.; Boufard, J.; Yoon, J. Endoplasmic Reticulum-Targeted Ratiometric N-Heterocyclic Carbene Borane Probe for Two-Photon Microscopic Imaging of Hypochlorous Acid. Anal. Chem. 2018, 90, 12937–12943. [Google Scholar] [CrossRef]
- Ellis, C.; Wang, F.; MacDiarmid, C.; Clark, S.; Lyons, T.; Eide, D. Zinc and the Msc2 zinc transporter protein are required for endoplasmic reticulum function. J. Cell Biol. 2004, 166, 325–335. [Google Scholar] [CrossRef]
- Fang, L.; Trigiante, G.; Crespo-Otero, R.; Hawes, C.; Philpott, M.; Jones, C.; Watkinson, M. Endoplasmic reticulum targeting fluorescent probes to image mobile Zn2+. Chem. Sci. 2019, 10, 10881–10887. [Google Scholar] [CrossRef]
- Lee, M.; Lee, H.; Chang, M.; Kim, H.; Kang, C.; Kim, J. A fluorescent probe for the Fe3+ ion pool in endoplasmic reticulum in liver cells. Dye. Pigment. 2016, 130, 245–250. [Google Scholar] [CrossRef]
- Schneider, L.; Zhang, J. Lysosomal function in macromolecular homeostasis and bioenergetics in Parkinson’s disease. Mol. Neurodegener. 2010, 5, 14. [Google Scholar] [CrossRef] [PubMed]
- Platt, F.; d’Azzo, A.; Davidson, B.; Neufeld, E.; Tifft, C. Lysosomal storage diseases. Nat. Rev. Dis. Prim. 2018, 4, 27. [Google Scholar] [CrossRef]
- Platt, F.; Boland, B.; van der Spoel, A. Lysosomal storage disorders: The cellular impact of lysosomal dysfunction. J. Cell Biol. 2012, 199, 723–734. [Google Scholar] [CrossRef]
- Banik, S.; Pedram, K.; Wisnovsky, S.; Ahn, G.; Riley, N.; Bertozzi, C. Lysosome-targeting chimaeras for degradation of extracellular proteins. Nature 2020, 584, 291–297. [Google Scholar] [CrossRef]
- Zhao, L.; Zhao, J.; Zhong, K.; Tong, A.; Jia, D. Targeted protein degradation: Mechanisms, strategies and application. Signal Transduct. Target. Ther. 2022, 7, 113. [Google Scholar] [CrossRef] [PubMed]
- Mindell, J. Lysosomal Acidification Mechanisms. In Annual Review of Physiology; Julius, D., Clapham, D., Eds.; Annual Reviews: Palo Alto, CA, USA, 2012; Volume 74, pp. 69–86. [Google Scholar]
- Kaufmann, A.; Krise, J. Lysosomal sequestration of amine-containing drugs: Analysis and therapeutic implications. J. Pharm. Sci. 2007, 96, 729–746. [Google Scholar] [CrossRef] [PubMed]
- Stark, M.; Silva, T.; Levin, G.; Machuqueiro, M.; Assaraf, Y. The Lysosomotropic Activity of Hydrophobic Weak Base Drugs is Mediated via Their Intercalation into the Lysosomal Membrane. Cells 2020, 9, 1082. [Google Scholar] [CrossRef] [PubMed]
- Tekpli, X.; Rivedal, E.; Gorria, M.; Landvik, N.; Rissel, M.; Dimanche-Boitrel, M.; Baffet, G.; Holme, J.; Lagadic-Gossmann, D. The B a P-increased intercellular communication via translocation of connexin-93 into gap junctions reduces apoptosis. Toxicol. Appl. Pharmacol. 2010, 242, 231–240. [Google Scholar] [CrossRef]
- DeVorkin, L.; Gorski, S. LysoTracker staining to aid in monitoring autophagy in Drosophila. Cold Spring Harb. Protoc. 2014, 2014, 951–958. [Google Scholar] [CrossRef]
- Fogel, J.; Thein, T.; Mariani, F. Use of LysoTracker to Detect Programmed Cell Death in Embryos and Differentiating Embryonic Stem Cells. JoVE J. Vis. Exp. 2012, 68, e4254. [Google Scholar]
- Kurz, T.; Terman, A.; Gustafsson, B.; Brunk, U. Lysosomes in iron metabolism, ageing and apoptosis. Histochem. Cell Biol. 2008, 129, 389–406. [Google Scholar] [CrossRef]
- Rizzollo, F.; More, S.; Vangheluwe, P.; Agostinis, P. The lysosome as a master regulator of iron metabolism. Trends Biochem. Sci. 2021, 46, 960–975. [Google Scholar] [CrossRef] [PubMed]
- Kazan, H.; Urfali-Mamatoglu, C.; Gunduz, U. Iron metabolism and drug resistance in cancer. Biometals 2017, 30, 629–641. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Liu, F.; Pu, C.; Tong, Z.; Wang, M.; Wang, J. Galactose-imidazole mediated dual-targeting fluorescent probe for detecting Fe3+ in the lysosomes of hepatocytes: Design, synthesis and evaluation. Biosens. Bioelectron. 2022, 204, 114083. [Google Scholar] [CrossRef] [PubMed]
- Giuffrida, M.; Sfrazzetto, G.; Satriano, C.; Zimbone, S.; Tomaselli, G.; Copani, A.; Rizzarelli, E. A New Ratiometric Lysosomal Copper(II) Fluorescent Probe To Map a Dynamic Metallome in Live Cells. Inorg. Chem. 2018, 57, 2365–2368. [Google Scholar] [CrossRef]
- Chen, Y.; Long, Z.; Wang, C.; Zhu, J.; Wang, S.; Liu, Y.; Wei, P.; Yi, T. A lysosome-targeted near-infrared fluorescent probe for cell imaging of Cu2+. Dye. Pigment. 2022, 204, 110472. [Google Scholar] [CrossRef]
- Zhang, S.; Ong, C.; Shen, H. Critical roles of intracellular thiols and calcium in parthenolide-induced apoptosis in human colorectal cancer cells. Cancer Lett. 2004, 208, 143–153. [Google Scholar] [CrossRef]
- Jung, H.; Chen, X.; Kim, J.; Yoon, J. Recent progress in luminescent and colorimetric chemosensors for detection of thiols. Chem. Soc. Rev. 2013, 42, 6019–6031. [Google Scholar] [CrossRef]
- Mao, G.-J.; Liang, Z.-Z.; Bi, J.; Zhang, H.; Meng, H.-M.; Su, L.; Gong, Y.-J.; Feng, S.; Zhang, G. A near-infrared fluorescent probe based on photostable Si-rhodamine for imaging hypochlorous acid during lysosome-involved inflammatory response. Anal. Chim. Acta 2019, 1048, 143–153. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Ren, J.; Bao, X.; Gao, W.; Wu, C.; Zhao, Y. pH-Switchable Fluorescent Probe for Spatially-Confined Visualization of Intracellular Hydrogen Peroxide. Anal. Chem. 2016, 88, 5865–5870. [Google Scholar] [CrossRef] [PubMed]
- Zhou, T.; Wang, J.; Xu, J.; Zheng, C.; Niu, Y.; Wang, C.; Xu, F.; Yuan, L.; Zhao, X.; Liang, L.; et al. A Smart Fluorescent Probe for NO Detection and Application in Myocardial Fibrosis Imaging. Anal. Chem. 2020, 92, 5064–5072. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.; Xiao, Y.; Jin, L. A Lysosome-Targetable and Two-Photon Fluorescent Probe for Monitoring Endogenous and Exogenous Nitric Oxide in Living Cells. J. Am. Chem. Soc. 2012, 134, 17486–17489. [Google Scholar] [CrossRef]
- Wang, K.; Guo, R.; Chen, X.-Y.; Yang, Y.-S.; Qiao, L.-Q.; Wang, M.-L. Multifunctional lysosome-targetable fluorescent probe for imaging peroxynitrite in acute liver injury model. Chem. Eng. J. 2022, 455, 140491. [Google Scholar] [CrossRef]
- Zhang, H.; Xu, L.; Chen, W.; Huang, J.; Huang, C.; Sheng, J.; Song, X. A Lysosome-Targetable Fluorescent Probe for Simultaneously Sensing Cys/Hcy, GSH, and H2S from Different Signal Patterns. ACS Sens. 2018, 3, 2513–2517. [Google Scholar] [CrossRef]
- Wang, L.; Xiao, Y.; Tian, W. Deng, Activatable Rotor for Quantifying Lysosomal Viscosity in Living Cells. J. Am. Chem. Soc. 2013, 135, 2903–2906. [Google Scholar] [CrossRef]
- Shi, D.; Hu, L.; Li, X.; Liu, W.; Gao, Y.; Li, X.; Jiang, B.; Xia, C.; Guo, Y.; Li, J. Lysosomal polarity increases with aging as revealed by a lysosome- targetable near -infrared fluorescent probe. Sens. Actuator B Chem. 2020, 319, 128302. [Google Scholar] [CrossRef]
- Futerman, A.; van Meer, G. The cell biology of lysosomal storage disorders. Nat. Rev. Mol. Cell Biol. 2004, 5, 554–565. [Google Scholar] [CrossRef]
- Liu, C.; Zhao, T.; He, S.; Zhao, L.; Zeng, X. A lysosome-targeting viscosity-sensitive fluorescent probe based on a novel functionalised near-infrared xanthene-indolium dye and its application in living cells. J. Mater. Chem. B 2020, 8, 8838–8844. [Google Scholar] [CrossRef]
- Chen, Y.; Zhu, Z.; Liu, X.; Jiang, Y.; Shen, J. Lysosome-targeting benzothiazole-based fluorescent probe for imaging viscosity and hypochlorite levels in living cells and zebrafish. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2022, 275, 121141. [Google Scholar] [CrossRef]
- Shi, W.-J.; Chen, R.; Yang, J.; Wei, Y.-F.; Guo, Y.; Wang, Z.-Z.; Yan, J.-w.; Niu, L. Novel Meso-Benzothiazole-Substituted BODIPY-Based AIE Fluorescent Rotor for Imaging Lysosomal Viscosity and Monitoring Autophagy. Anal. Chem. 2022, 94, 14707–14715. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Yan, H.; Wu, J.; Wang, Z.; He, S.; Zhao, L.; Zeng, X. Lysosomes-targeting near-infrared fluorescent probe for the detection of pH in living cells. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2022, 278, 121368. [Google Scholar] [CrossRef]
- Rutkowski, D.; Kaufman, R. A trip to the ER: Coping with stress. Trends Cell Biol. 2004, 14, 20–28. [Google Scholar] [CrossRef] [PubMed]
- Nargund, A.; Pellegrino, M.; Fiorese, C.; Baker, B.; Haynes, C. Mitochondrial Import Efficiency of ATFS-1 Regulates Mitochondrial UPR Activation. Science 2012, 337, 587–590. [Google Scholar] [CrossRef]
- Rowland, A.; Voeltz, G. Endoplasmic reticulum-mitochondria contacts: Function of the junction. Nat. Rev. Mol. Cell Biol. 2012, 13, 607–615. [Google Scholar] [CrossRef]
- Phillips, M.; Voeltz, G. Structure and function of ER membrane contact sites with other organelles. Nat. Rev. Mol. Cell Biol. 2016, 17, 69–82. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Giordano, S.; Zhang, J. Autophagy, mitochondria and oxidative stress: Cross-talk and redox signalling. Biochem. J. 2012, 441, 523–540. [Google Scholar] [CrossRef]
- Tian, M.; Sun, J.; Dong, B.; Lin, W. Dynamically Monitoring Cell Viability in a Dual-Color Mode: Construction of an Aggregation/Monomer-Based Probe Capable of Reversible Mitochondria-Nucleus Migration. Angew. Chem. Int. Ed. 2018, 57, 16506–16510. [Google Scholar] [CrossRef] [PubMed]
- Kong, X.; Yin, J.; Li, M.; Zhu, L.; Dong, B.; Ma, Y.; Lin, W. Simultaneously imaging of SO2 in lysosomes and mitochondria based on a dual organelle-targeted fluorescent probe. Sens. Actuators B Chem. 2019, 292, 80–87. [Google Scholar] [CrossRef]
- Ho, P.-Y.; Chou, T.; Kam, C.; Huang, W.; He, Z.; Ngan, A.; Chen, S. A dual organelle-targeting mechanosensitive probe. Sci. Adv. 2023, 9, eabn5390. [Google Scholar] [CrossRef]
- Shan, Y.-M.; Yu, K.-K.; Wang, N.; Yu, F.-Y.; Li, K.; Liu, Y.-H.; Yu, X.-Q. Assessing ClO- level during ER stress and cellular senescence through a ratio fluorescent probe with dual organelle targeting ability. Sens. Actuator B Chem. 2022, 358, 131383. [Google Scholar] [CrossRef]
- Dai, F.; Zhang, S.; Zhou, B.; Duan, D.-C.; Liu, J.; Zheng, Y.-L.; Chen, H.; Zhang, X.; Zhang, Y. Cellular and Intravital Imaging of NAD(P)H by a Red-Emitting Quinolinium-Based Fluorescent Probe that Features a Shift of Its Product from Mitochondria to the Nucleus. Anal. Chem. 2022, 95, 1335–1342. [Google Scholar] [CrossRef]
- Wang, H.; Sun, Y.; Lin, X.; Feng, W.; Li, Z.; Yu, M. Multi-organelle-targeting pH-dependent NIR fluorescent probe for lysosomal viscosity. Chin. Chem. Lett. 2023, 34, 107626. [Google Scholar] [CrossRef]
- Zhuang, J.; Li, N.; Zhang, Y.; Li, B.; Tang, B. Esterase-Activated Theranostic Prodrug for Dual Organelles Targeted Imaging and Synergetic Chemo-Photodynamic Cancer Therapy. CCS Chem. 2022, 3, 1028–1043. [Google Scholar] [CrossRef]
- Ge, E.; Tian, M.; Lin, W. A unique fluorescent probe for visualization of cell death via its subcellular immigration from lysosomes to nucleus. Sens. Actuators B Chem. 2021, 347, 130656. [Google Scholar] [CrossRef]
- Yapici, N.; Gao, X.; Yan, X.; Hou, S.; Jockusch, S.; Lesniak, L.; Gibson, K.; Bi, L. Novel Dual-Organelle-Targeting Probe (RCPP) for Simultaneous Measurement of Organellar Acidity and Alkalinity in Living Cells. ACS Omega 2021, 6, 31447–31456. [Google Scholar] [CrossRef]
- Huang, Y.; Zhang, Y.; Huo, F. A dual-targeted organelles SO2 specific probe for bioimaging in related diseases and food analysis. Chem. Eng. J. 2022, 433, 133750. [Google Scholar] [CrossRef]
- Yang, Z.; Li, L.; Ling, J.; Liu, T.; Huang, X.; Ying, Y.; Zhao, Y.; Lei, K.; Chen, L.; Chen, Z. Cyclooctatetraene-conjugated cyanine mitochondrial probes minimize phototoxicity in fluorescence and nanoscopic imaging. Chem. Sci. 2020, 11, 8506–8516. [Google Scholar] [CrossRef]
- Horton, K.; Stewart, K.; Fonseca, S.; Guo, Q.; Kelley, S. Mitochondria-penetrating peptides. Chem. Biol. 2008, 15, 375–382. [Google Scholar] [CrossRef] [PubMed]
- Jean, S.; Ahmed, M.; Lei, E.; Wisnovsky, S.; Kelley, S. Peptide-Mediated Delivery of Chemical Probes and Therapeutics to Mitochondria. Acc. Chem. Res. 2016, 49, 1893–1902. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Gao, H.; Liu, R.; Chen, C.; Zeng, S.; Liu, Q.; Ding, D. Endoplasmic reticulum targeted AIE bioprobe as a highly efficient inducer of immunogenic cell death. Sci. China Chem. 2020, 63, 1428–1434. [Google Scholar] [CrossRef]
- Lange, A.; Mills, R.; Lange, C.; Stewart, M.; Devine, S.; Corbett, A. Classical nuclear localization signals: Definition, function, and interaction with importin alpha. J. Biol. Chem. 2007, 282, 5101–5105. [Google Scholar] [CrossRef]
- Wang, Y.; Li, C.; Zhuo, J.; Hui, H.; Zhou, B.; Tian, J. The Detection of Divalent Iron and Reactive Oxygen Species During Ferroptosis with the Use of a Dual-Reaction Turn-On Fluorescent Probe. Mol. Imaging Biol. 2022, 1–12. [Google Scholar] [CrossRef]
- Kleele, T.; Rey, T.; Winter, J.; Zaganelli, S.; Mahecic, D.; Lambert, H.; Ruberto, F.; Nemir, M.; Wai, T.; Pedrazzini, T.; et al. Distinct fission signatures predict mitochondrial degradation or biogenesis. Nature 2021, 593, 435–439. [Google Scholar] [CrossRef]
- Colom, A.; Derivery, E.; Soleimanpour, S.; Tomba, C.; Molin, M.D.; Sakai, N.; Gonzalez-Gaitan, M.; Matile, S.; Roux, A. A fluorescent membrane tension probe. Nat. Chem. 2018, 10, 1118–1125. [Google Scholar] [CrossRef]
- Piazzolla, F.; Mercier, V.; Assies, L.; Sakai, N.; Roux, A.; Matile, S. Fluorescent Membrane Tension Probes for Early Endosomes. Angew. Chem. Int. Ed. 2021, 60, 12258–12263. [Google Scholar] [CrossRef]
- Garcia-Calvo, J.; Lopez-Andarias, J.; Maillard, J.; Mercier, V.; Roffay, C.; Roux, A.; Fuerstenberg, A.; Sakai, N.; Matile, S. HydroFlipper membrane tension probes: Imaging membrane hydration and mechanical compression simultaneously in living cells. Chem. Sci. 2022, 13, 2086–2093. [Google Scholar] [CrossRef]
- Chen, X.-X.; Bayard, F.; Gonzalez-Sanchis, N.; Pamungkas, K.; Sakai, N.; Matile, S. Fluorescent Flippers: Small-Molecule Probes to Image Membrane Tension in Living Systems. Angew. Chem. 2023, e202217868. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).