Advancements in CRISPR-Based Biosensing for Next-Gen Point of Care Diagnostic Application
Abstract
1. Introduction
2. Benefits of CRISPR-Based Biosensors in Point-of-Care Systems
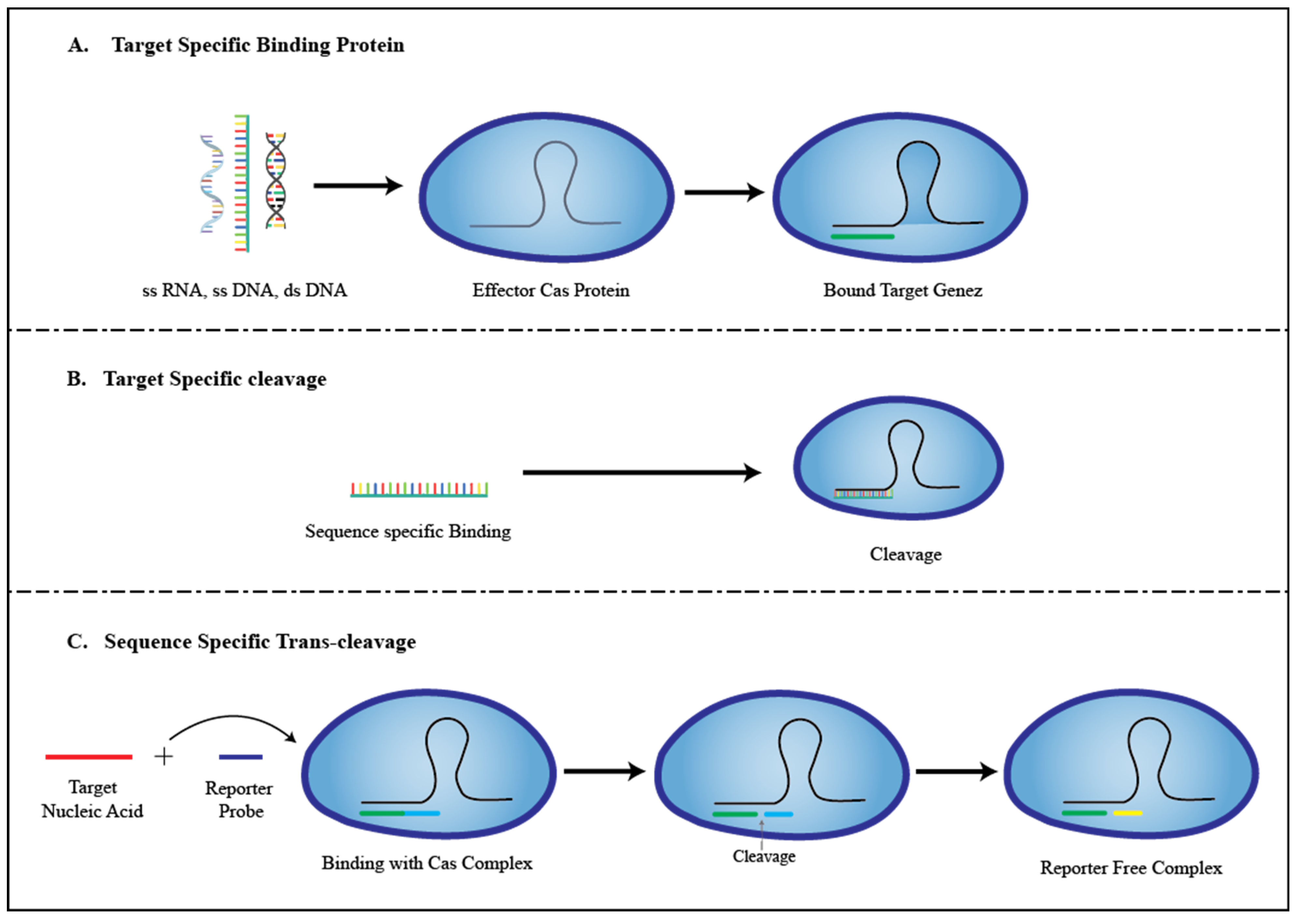
3. Role of CRISPR in the Development of Biosensors
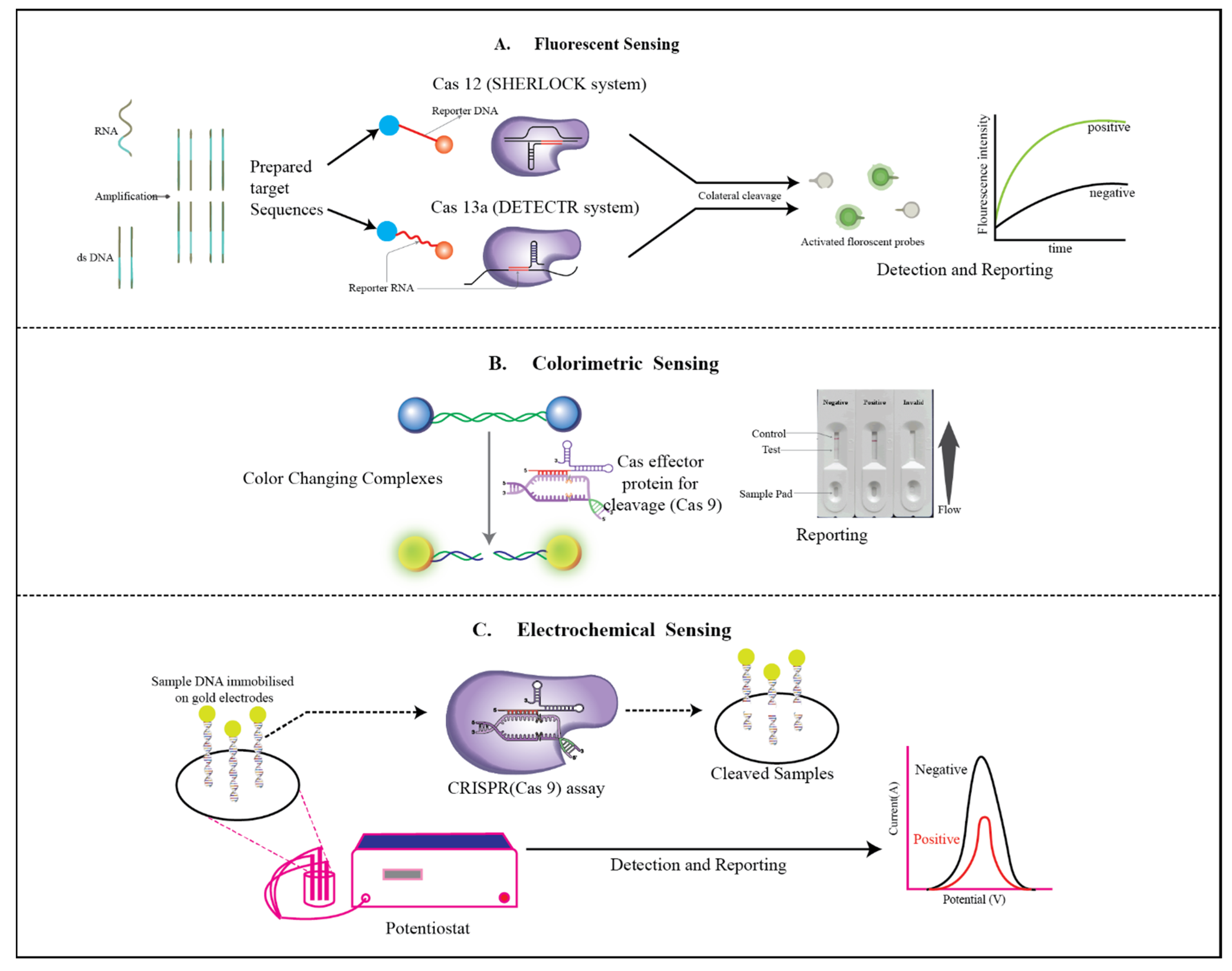
3.1. E-CRISPR
3.2. Colorimetry
3.3. Fluorescent Sensor
4. CRISPR-Based Biosensor in Diagnosis (Diseases)
4.1. Bacterial
4.1.1. Food Poisoning (Escherichia coli, Salmonella, Staphylococcus aureus)
4.1.2. Tuberculosis
4.1.3. Chlamydia
4.1.4. Helicobacter pylori
4.2. Viral
4.2.1. Hepatitis
4.2.2. Human Immunodeficiency Virus (HIV)
4.2.3. Dengue
4.2.4. Human Papillomavirus (HPV)
| Pathogen | CRISPR | Nucleic Acid | Amplification | Readout | LOD | Detection Platform | Ref. |
|---|---|---|---|---|---|---|---|
| Food poisoning (E. coli, Salmonella, S. aureus) | Cas9a | DNA | SDA | Colorimetric | 100 copies | - | [122] |
| Cas12a | dsDNA | HCR | Electrochemical | 20 CFU/mL | [122] | ||
| Cas12a-Ddp | dsDNA | PCR | Microplate | 1 CFU/mL | Dual detection platform | [122] | |
| Cas13a | ssRNA | PCR | Florescence | 1 CFU/mL | CCB-Detection | [122] | |
| Tuberculosis | dCas9 | DNA | PCR | Bioluminescence | ≈3 × 10−21 M | Chimeric dCas9-luciferase | [123] |
| Cas12a | DNA | RPA | Florescence | 1 copy | CRISPR-MTB | [123] | |
| H. pylori | Cas12a | DNA/RNA | RPA | Lateral flow strips, visualization | 5 copies/μL | Lateral flow biosensor | [124] |
| Hepatitis Liver cancer | Cas12a | DNA/RNA | HCR | Gel electrophoresis | 1.5 fM | - | [125] |
| Cas12a | DNA/RNA | LAMP | Colorimetric | 10 aM | AuNP colorimetric | [126] | |
| Cas12a | DNA | - | Colorimetric | 10 pM | MAV-chip | [127] | |
| HIV | dCas9 | DNA | - | Fluorescence | - | Nanoelectrokinetic chip | [128] |
| Cas12a | DNA | - | Resistive pulse | 10 nM | Nanopore sensor (SCAN) | [129] | |
| Zika/Dengue Virus Zika | Cas9 (NASBACC) | RNA | NASBA | Colorimetric | 1 fM of RNA amplicon | NASBACC | [130] |
| Cas13a | DNA | RPA | Fluorescent | 9 aM | Droplet microfluidics | [131] | |
| Cas13a | RNA | RT-RPA | Fluorescent/colorimetric | 0.9 aM in saliva | Lateral flow assay | [132] | |
| HPV16/18 | Cas9 (CARP) | DNA | PCR | Electrophoresis/qPCR instruments | 2 pg of DNA amplicon | CARP | [133] |
| Cas12a (DETECTR) | DNA | RPA | Fluorescence | ca. aM of viral DNA | DETECTR | [133] | |
| Cas12a | DNA | RPA | Colorimetric | 0.24 fM | Lateral flow assay | [133] | |
| AaCas12b | DNA | RPA | Fluorescence | 1 × 10−18 M | CDetection | [134] | |
| Cas12a | DNA, Protein | - | Electrochemical | 10−12 M | E-CRISPR | [134] | |
| Cas9 | DNA | qPCR | qPCR | 2 ng | ctPCR3.0 | [135] | |
| Cas12a | DNA | RPA | Fluorescent | 10−100 copies | One-pot reaction | [136] |
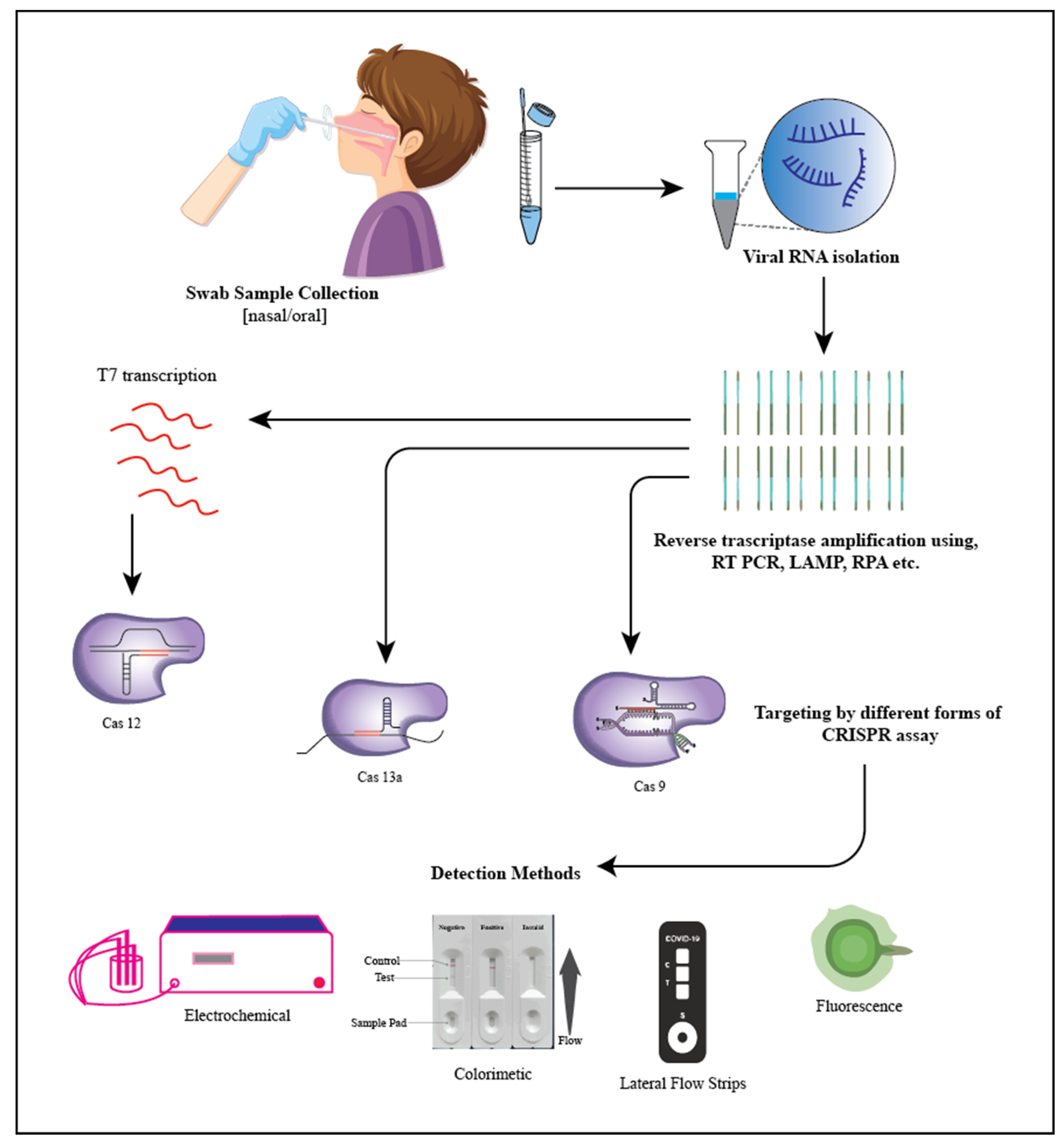
5. SARS-CoV-2 (Severe Acute Respiratory Syndrome Coronavirus 2)
- Spin column-based NAE (nucleic acid extraction) provided the most suitable option for RNA isolation. It plays an important role in ion exchange methods because it offers a stable stationary phase for fast and reliable buffer exchange [142].
- Purification with magnetic beads. Magnetic beads are used to collect viral RNA in this procedure. During the wash and collection, the beads are held in place by an external magnetic field. The magnetic format enables quick sample collection and concentration. Manually capturing and releasing particles, on the other hand, might be time-consuming [143].
- Extracting organically. In this method, a phenol-containing solution is used to homogenize the specimens, which are then centrifuged. The viral RNA is found in the top aqueous phase during centrifugation. The viral RNA is isolated from the upper aqueous phase and recovered during centrifugation, precipitating it using alcohol (elution) and later placing it in a rehydrating medium. This approach is regarded as the finest for RNA extraction; nevertheless, it is tedious, time-consuming, and difficult to automate [143].
6. Future Prospects
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Price, C.P. Regular review: Point of care testing. BMJ 2001, 322, 1285–1288. [Google Scholar] [CrossRef] [PubMed]
- Louie, R.F.; Tang, Z.; Shelby, M.D.G.; Kost, G.J. Point-of-Care Testing: Millennium Technology for Critical Care. Lab. Med. 2000, 31, 402–408. [Google Scholar] [CrossRef]
- Bonini, A.; Poma, N.; Vivaldi, F.; Kirchhain, A.; Salvo, P.; Bottai, D.; Tavanti, A.; Di Francesco, F. Advances in biosensing: The CRISPR/Cas system as a new powerful tool for the detection of nucleic acids. J. Pharm. Biomed. Anal. 2021, 192, 113645. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Shi, Y.; Chen, Y.; Yang, Z.; Wu, H.; Zhou, Z.; Li, J.; Ping, J.; He, L.; Shen, H.; et al. Contamination-free visual detection of SARS-CoV-2 with CRISPR/Cas12a: A promising method in the point-of-care detection. Biosens. Bioelectron. 2020, 169, 112642. [Google Scholar] [CrossRef]
- Li, Z.; Zhao, W.; Ma, S.; Li, Z.; Yao, Y.; Fei, T. A chemical-enhanced system for CRISPR-Based nucleic acid detection. Biosens. Bioelectron. 2021, 192, 113493. [Google Scholar] [CrossRef]
- Xu, W.; Jin, T.; Dai, Y.; Liu, C.C. Surpassing the detection limit and accuracy of the electrochemical DNA sensor through the application of CRISPR Cas systems. Biosens. Bioelectron. 2020, 155, 112100. [Google Scholar] [CrossRef]
- Qing, M.; Chen, S.L.; Sun, Z.; Fan, Y.; Luo, H.Q.; Li, N.B. Universal and Programmable Rolling Circle Amplification-CRISPR/Cas12a-Mediated Immobilization-Free Electrochemical Biosensor. Anal. Chem. 2021, 93, 7499–7507. [Google Scholar] [CrossRef]
- Yao, R.; Liu, D.; Jia, X.; Zheng, Y.; Liu, W.; Xiao, Y. CRISPR-Cas9/Cas12a biotechnology and application in bacteria. Synth. Syst. Biotechnol. 2018, 3, 135–149. [Google Scholar] [CrossRef]
- Paul, B.; Montoya, G. CRISPR-Cas12a: Functional overview and applications. Biomed. J. 2020, 43, 8–17. [Google Scholar] [CrossRef]
- Hinge, V.R.; Chavhan, R.L.; Kale, S.P.; Suprasanna, P.; Kadam, U.S. Engineering Resistance Against Viruses in Field Crops Using CRISPRCas9. Curr. Genom. 2021, 22, 214–231. [Google Scholar] [CrossRef]
- Kostyusheva, A.; Brezgin, S.; Babin, Y.; Vasilyeva, I.; Glebe, D.; Kostyushev, D.; Chulanov, V. CRISPR-Cas systems for diagnosing infectious diseases. Methods 2022, 203, 431–446. [Google Scholar] [CrossRef] [PubMed]
- Katzmeier, F.; Aufinger, L.; Dupin, A.; Quintero, J.; Lenz, M.; Bauer, L.G.; Klumpe, S.; Sherpa, D.; Dürr, B.; Honemann, M.; et al. A low-cost fluorescence reader for in vitro transcription and nucleic acid detection with Cas13a. PLoS ONE 2019, 14, e0220091. [Google Scholar] [CrossRef] [PubMed]
- Batista, A.C.; Pacheco, L.G. Detecting pathogens with Zinc-Finger, TALE and CRISPR- based programmable nucleic acid binding proteins. J. Microbiol. Methods 2018, 152, 98–104. [Google Scholar] [CrossRef] [PubMed]
- Yuan, M.; Ding, R.; Chen, S.; Duan, G. Advances in Field Detection Based on CRISPR/Cas System. ACS Synth. Biol. 2021, 10, 2824–2832. [Google Scholar] [CrossRef] [PubMed]
- Bhalla, N.; Jolly, P.; Formisano, N.; Estrela, P. Introduction to biosensors. Essays Biochem. 2016, 60, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Campbell, A.S.; de Ávila, B.E.-F.; Wang, J. Wearable biosensors for healthcare monitoring. Nat. Biotechnol. 2019, 37, 389–406. [Google Scholar] [CrossRef]
- Chen, M.; Luo, D. A CRISPR Path to Cutting-Edge Materials. N. Engl. J. Med. 2020, 382, 85–88. [Google Scholar] [CrossRef]
- Kumar, S.; Tripathy, S.; Jyoti, A.; Singh, S.G. Recent advances in biosensors for diagnosis and detection of sepsis: A comprehensive review. Biosens. Bioelectron. 2018, 124–125, 205–215. [Google Scholar] [CrossRef]
- Bao, M.; Chen, Q.; Xu, Z.; Jensen, E.C.; Liu, C.; Waitkus, J.T.; Yuan, X.; He, Q.; Qin, P.; Du, K. Challenges and Opportunities for Clustered Regularly Interspaced Short Palindromic Repeats Based Molecular Biosensing. ACS Sens. 2021, 6, 2497–2522. [Google Scholar] [CrossRef]
- Bonini, A.; Poma, N.; Vivaldi, F.; Biagini, D.; Bottai, D.; Tavanti, A.; Di Francesco, F. A label-free impedance biosensing assay based on CRISPR/Cas12a collateral activity for bacterial DNA detection. J. Pharm. Biomed. Anal. 2021, 204, 114268. [Google Scholar] [CrossRef]
- Dai, Y.; Wu, Y.; Liu, G.; Gooding, J.J. CRISPR Mediated Biosensing Toward Understanding Cellular Biology and Point-of-Care Diagnosis. Angew. Chem. Int. Ed. 2020, 59, 20754–20766. [Google Scholar] [CrossRef] [PubMed]
- Cho, S.; Shin, J.; Cho, B.-K. Applications of CRISPR/Cas System to Bacterial Metabolic Engineering. Int. J. Mol. Sci. 2018, 19, 1089. [Google Scholar] [CrossRef] [PubMed]
- Van Der Oost, J.; Westra, E.R.; Jackson, R.N.; Wiedenheft, B. Unravelling the structural and mechanistic basis of CRISPR–Cas systems. Nat. Rev. Microbiol. 2014, 12, 479–492. [Google Scholar] [CrossRef] [PubMed]
- Safari, F.; Hatam, G.; Behbahani, A.B.; Rezaei, V.; Barekati-Mowahed, M.; Petramfar, P.; Khademi, F. CRISPR System: A High-throughput Toolbox for Research and Treatment of Parkinson’s Disease. Cell. Mol. Neurobiol. 2020, 40, 477–493. [Google Scholar] [CrossRef] [PubMed]
- Bruch, R.; Urban, G.A.; Dincer, C. CRISPR/Cas Powered Multiplexed Biosensing. Trends Biotechnol. 2019, 37, 791–792. [Google Scholar] [CrossRef]
- Gootenberg, J.S.; Abudayyeh, O.O.; Lee, J.W.; Essletzbichler, P.; Dy, A.J.; Joung, J.; Verdine, V.; Donghia, N.; Daringer, N.M.; Freije, C.A.; et al. Nucleic acid detection with CRISPR-Cas13a/C2c2. Science 2017, 356, 438–442. [Google Scholar] [CrossRef]
- Joung, J.; Ladha, A.; Saito, M.; Segel, M.; Bruneau, R.; Huang, M.W.; Kim, N.-G.; Yu, X.; Li, J.; Walker, B.D.; et al. Point-of-care testing for COVID-19 using SHERLOCK diagnostics. medRxiv 2020. [Google Scholar] [CrossRef]
- Xie, S.; Ji, Z.; Suo, T.; Li, B.; Zhang, X. Advancing sensing technology with CRISPR: From the detection of nucleic acids to a broad range of analytes—A review. Anal. Chim. Acta 2021, 1185, 338848. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Ji, S.; Koh, H.R. CRISPR as a Diagnostic Tool. Biomolecules 2021, 11, 1162. [Google Scholar] [CrossRef]
- Zhang, D.; Yan, Y.; Que, H.; Yang, T.; Cheng, X.; Ding, S.; Zhang, X.; Cheng, W. CRISPR/Cas12a-Mediated Interfacial Cleaving of Hairpin DNA Reporter for Electrochemical Nucleic Acid Sensing. ACS Sens. 2020, 5, 557–562. [Google Scholar] [CrossRef]
- Akkilic, N.; Geschwindner, S.; Höök, F. Single-molecule biosensors: Recent advances and applications. Biosens. Bioelectron. 2020, 151, 111944. [Google Scholar] [CrossRef] [PubMed]
- Thévenot, D.R.; Toth, K.; Durst, R.A.; Wilson, G.S. Electrochemical biosensors: Recommended definitions and classification. Biosens. Bioelectron. 2001, 16, 121–131. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Li, S.; Wang, J.; Liu, G. CRISPR/Cas Systems towards Next-Generation Biosensing. Trends Biotechnol. 2019, 37, 730–743. [Google Scholar] [CrossRef] [PubMed]
- Dai, Y.; Somoza, R.; Wang, L.; Welter, J.F.; Li, Y.; Caplan, A.; Liu, C.C. Exploring the Trans-Cleavage Activity of CRISPR-Cas12a (cpf1) for the Development of a Universal Electrochemical Biosensor. Angew. Chem. Int. Ed. 2019, 58, 17399–17405. [Google Scholar] [CrossRef]
- Kadam, U.S.; Hong, J.C. Advances in aptameric biosensors designed to detect toxic contaminants from food, water, human fluids, and the environment. Trends Environ. Anal. Chem. 2022, 36, e00184. [Google Scholar] [CrossRef]
- Gootenberg, J.S.; Abudayyeh, O.O.; Kellner, M.J.; Joung, J.; Collins, J.J.; Zhang, F. Multiplexed and portable nucleic acid detection platform with Cas13, Cas12a, and Csm6. Science 2018, 360, 439–444. [Google Scholar] [CrossRef]
- Murugan, K.; Babu, K.; Sundaresan, R.; Rajan, R.; Sashital, D.G. The Revolution Continues: Newly Discovered Systems Expand the CRISPR-Cas Toolkit. Mol. Cell 2017, 68, 15–25. [Google Scholar] [CrossRef]
- Wu, H.; Chen, X.; Zhang, M.; Wang, X.; Chen, Y.; Qian, C.; Wu, J.; Xu, J. Versatile detection with CRISPR/Cas system from applications to challenges. TrAC Trends Anal. Chem. 2021, 135, 116150. [Google Scholar] [CrossRef]
- Swetha, P.D.P.; Sonia, J.; Sapna, K.; Prasad, K.S. Towards CRISPR powered electrochemical sensing for smart diagnostics. Curr. Opin. Electrochem. 2021, 30, 100829. [Google Scholar] [CrossRef]
- Zhang, Y.; Wu, Y.; Wu, Y.; Chang, Y.; Liu, M. CRISPR-Cas systems: From gene scissors to programmable biosensors. TrAC Trends Anal. Chem. 2021, 137, 116210. [Google Scholar] [CrossRef]
- Wang, J. Electrochemical biosensors: Towards point-of-care cancer diagnostics. Biosens. Bioelectron. 2006, 21, 1887–1892. [Google Scholar] [CrossRef] [PubMed]
- Lazaro, A.; Boada, M.; Villarino, R.; Girbau, D. Color Measurement and Analysis of Fruit with a Battery-Less NFC Sensor. Sensors 2019, 19, 1741. [Google Scholar] [CrossRef] [PubMed]
- Moon, J.; Kwon, H.-J.; Yong, D.; Lee, I.-C.; Kim, H.; Kang, H.; Lim, E.-K.; Lee, K.-S.; Jung, J.; Park, H.G.; et al. Colorimetric Detection of SARS-CoV-2 and Drug-Resistant pH1N1 Using CRISPR/dCas9. ACS Sens. 2020, 5, 4017–4026. [Google Scholar] [CrossRef] [PubMed]
- Cao, Y.; Wu, J.; Pang, B.; Zhang, H.; Le, X.C. CRISPR/Cas12a-mediated gold nanoparticle aggregation for colorimetric detection of SARS-CoV-2. Chem. Commun. 2021, 57, 6871–6874. [Google Scholar] [CrossRef]
- Shi, Y.; Fu, X.; Yin, Y.; Peng, F.; Yin, X.; Ke, G.; Zhang, X. CRISPR-Cas12a System for Biosensing and Gene Regulation. Chem. Asian J. 2021, 16, 857–867. [Google Scholar] [CrossRef] [PubMed]
- Cheng, M.; Xiong, E.; Tian, T.; Zhu, D.; Ju, H.-Q.; Zhou, X. A CRISPR-driven colorimetric code platform for highly accurate telomerase activity assay. Biosens. Bioelectron. 2021, 172, 112749. [Google Scholar] [CrossRef]
- Jiang, Y.; Hu, M.; Liu, A.-A.; Lin, Y.; Liu, L.; Yu, B.; Zhou, X.; Pang, D.-W. Detection of SARS-CoV-2 by CRISPR/Cas12a-Enhanced Colorimetry. ACS Sens. 2021, 6, 1086–1093. [Google Scholar] [CrossRef]
- Nalefski, E.A.; Patel, N.; Leung, P.J.; Islam, Z.; Kooistra, R.M.; Parikh, I.; Marion, E.; Knott, G.J.; Doudna, J.A.; Le Ny, A.-L.M.; et al. Kinetic analysis of Cas12a and Cas13a RNA-Guided nucleases for development of improved CRISPR-Based diagnostics. iScience 2021, 24, 102996. [Google Scholar] [CrossRef]
- Chen, J.S.; Ma, E.; Harrington, L.B.; Da Costa, M.; Tian, X.; Palefsky, J.M.; Doudna, J.A. CRISPR-Cas12a target binding unleashes indiscriminate single-stranded DNase activity. Science 2018, 360, 436–439. [Google Scholar] [CrossRef]
- Zetsche, B.; Gootenberg, J.S.; Abudayyeh, O.O.; Slaymaker, I.M.; Makarova, K.S.; Essletzbichler, P.; Volz, S.E.; Joung, J.; van der Oost, J.; Regev, A.; et al. Cpf1 Is a Single RNA-Guided Endonuclease of a Class 2 CRISPR-Cas System. Cell 2015, 163, 759–771. [Google Scholar] [CrossRef]
- Mustafa, M.I.; Makhawi, A.M. SHERLOCK and DETECTR: CRISPR-Cas Systems as Potential Rapid Diagnostic Tools for Emerging Infectious Diseases. J. Clin. Microbiol. 2021, 59, e00745. [Google Scholar] [CrossRef] [PubMed]
- Kellner, M.J.; Koob, J.G.; Gootenberg, J.S.; Abudayyeh, O.O.; Zhang, F. SHERLOCK: Nucleic acid detection with CRISPR nucleases. Nat. Protoc. 2019, 14, 2986–3012, Erratum in Nat. Protoc. 2020, 15, 1311. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Horby, P.W.; Hayden, F.G.; Gao, G.F. A novel coronavirus outbreak of global health concern. Lancet 2020, 395, 470–473, Erratum in Lancet 2020, 31986257. [Google Scholar] [CrossRef] [PubMed]
- Yin, L.; Man, S.; Ye, S.; Liu, G.; Ma, L. CRISPR-Cas based virus detection: Recent advances and perspectives. Biosens. Bioelectron. 2021, 193, 113541. [Google Scholar] [CrossRef]
- Zhu, N.; Zhang, D.; Wang, W.; Li, X.; Yang, B.; Song, J.; Zhao, X.; Huang, B.; Shi, W.; Lu, R.; et al. A Novel Coronavirus from Patients with Pneumonia in China, 2019. N. Engl. J. Med. 2020, 382, 727–733. [Google Scholar] [CrossRef]
- Bhattacharyya, R.P.; Thakku, S.G.; Hung, D.T. Harnessing CRISPR Effectors for Infectious Disease Diagnostics. ACS Infect. Dis. 2018, 4, 1278–1282. [Google Scholar] [CrossRef]
- Quan, J.; Langelier, C.; Kuchta, A.; Batson, J.; Teyssier, N.; Lyden, A.; Caldera, S.; McGeever, A.; Dimitrov, B.; King, R.; et al. FLASH: A next-generation CRISPR diagnostic for multiplexed detection of antimicrobial resistance sequences. Nucleic Acids Res. 2019, 47, e83. [Google Scholar] [CrossRef]
- Ma, L.; Peng, L.; Yin, L.; Liu, G.; Man, S. CRISPR-Cas12a-Powered Dual-Mode Biosensor for Ultrasensitive and Cross-validating Detection of Pathogenic Bacteria. ACS Sens. 2021, 6, 2920–2927. [Google Scholar] [CrossRef]
- Li, Z.; Wei, J.; Di, D.; Wang, X.; Li, C.; Li, B.; Qiu, Y.; Liu, K.; Gu, F.; Tong, M.; et al. Rapid and accurate detection of African swine fever virus by DNA endonuclease-targeted CRISPR trans reporter assay. Acta Biochim. Biophys. Sin. 2020, 52, 1413–1419. [Google Scholar] [CrossRef]
- Escalona-Noguero, C.; López-Valls, M.; Sot, B. CRISPR/Cas technology as a promising weapon to combat viral infections. Bioessays 2021, 43, e2000315. [Google Scholar] [CrossRef]
- Nishibuchi, M. Food poisoning—Importance of international perspective. Nihon Rinsho. Jpn. J. Clin. Med. 2012, 70, 1280. (In Japsnese) [Google Scholar]
- Rosegrant, M.W.; Cline, S.A. Global Food Security: Challenges and Policies. Science 2003, 302, 1917–1919. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Bu, S.; Feng, J.; Wei, H.; Wang, Z.; Li, X.; Zhou, H.; He, X.; Wan, J. Electrochemical biosensor for detecting pathogenic bacteria based on a hybridization chain reaction and CRISPR-Cas12a. Anal. Bioanal. Chem. 2021, 414, 1073–1080. [Google Scholar] [CrossRef] [PubMed]
- De Freitas, C.G.; Santana, P.; da Silva, P.H.C.; Gonçalves, V.S.P.; Barros, M.D.A.F.; Torres, F.A.G.; Murata, L.S.; Perecmanis, S. PCR multiplex for detection of Salmonella Enteritidis, Typhi and Typhimurium and occurrence in poultry meat. Int. J. Food Microbiol. 2010, 139, 15–22. [Google Scholar] [CrossRef] [PubMed]
- Fu, X.; Sun, J.; Ye, Y.; Zhang, Y.; Sun, X. A rapid and ultrasensitive dual detection platform based on Cas12a for simultaneous detection of virulence and resistance genes of drug-resistant Salmonella. Biosens. Bioelectron. 2022, 195, 113682. [Google Scholar] [CrossRef]
- Sun, X.; Wang, Y.; Zhang, L.; Liu, S.; Zhang, M.; Wang, J.; Ning, B.; Peng, Y.; He, J.; Hu, Y.; et al. CRISPR-Cas9 Triggered Two-Step Isothermal Amplification Method for E. coli O157:H7 Detection Based on a Metal–Organic Framework Platform. Anal. Chem. 2020, 92, 3032–3041. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Yin, L.; Dong, Y.; Peng, L.; Liu, G.; Man, S.; Ma, L. CRISPR-Cas13a based bacterial detection platform: Sensing pathogen Staphylococcus aureus in food samples. Anal. Chim. Acta 2020, 1127, 225–233. [Google Scholar] [CrossRef] [PubMed]
- Yan, M.-Y.; Li, S.-S.; Ding, X.-Y.; Guo, X.-P.; Jin, Q.; Sun, Y.-C. A CRISPR-Assisted Nonhomologous End-Joining Strategy for Efficient Genome Editing in Mycobacterium tuberculosis. mBio 2020, 11, e02364-19. [Google Scholar] [CrossRef]
- Wei, J.; Lu, N.; Li, Z.; Wu, X.; Jiang, T.; Xu, L.; Yang, C.; Guo, S. The Mycobacterium tuberculosis CRISPR-Associated Cas1 Involves Persistence and Tolerance to Anti-Tubercular Drugs. BioMed Res. Int. 2019, 2019, 7861695. [Google Scholar] [CrossRef]
- Grüschow, S.; Athukoralage, J.; Graham, S.; Hoogeboom, T.; White, M.F. Cyclic oligoadenylate signalling mediates Mycobacterium tuberculosis CRISPR defence. Nucleic Acids Res. 2019, 47, 9259–9270. [Google Scholar] [CrossRef]
- Ai, J.-W.; Zhou, X.; Xu, T.; Yang, M.; Chen, Y.; He, G.-Q.; Pan, N.; Cai, Y.; Li, Y.; Wang, X.; et al. CRISPR-based rapid and ultra-sensitive diagnostic test for Mycobacterium tuberculosis. Emerg. Microbes Infect. 2019, 8, 1361–1369. [Google Scholar] [CrossRef] [PubMed]
- Bertelli, C.; Cissé, O.H.; Rusconi, B.; Kebbi-Beghdadi, C.; Croxatto, A.; Goesmann, A.; Collyn, F.; Greub, G. CRISPR System Acquisition and Evolution of an Obligate Intracellular Chlamydia-Related Bacterium. Genome Biol. Evol. 2016, 8, 2376–2386. [Google Scholar] [CrossRef] [PubMed]
- Ouellette, S.P. Feasibility of a Conditional Knockout System for Chlamydia Based on CRISPR Interference. Front. Cell. Infect. Microbiol. 2018, 8, 59. [Google Scholar] [CrossRef] [PubMed]
- Ouellette, S.P.; Blay, E.A.; Hatch, N.D.; Fisher-Marvin, L.A. CRISPR Interference To Inducibly Repress Gene Expression in Chlamydia trachomatis. Infect. Immun. 2021, 89, e0010821. [Google Scholar] [CrossRef]
- Bangpanwimon, K.; Sottisuporn, J.; Mittraparp-Arthorn, P.; Ueaphatthanaphanich, W.; Rattanasupar, A.; Pourcel, C.; Vuddhakul, V. CRISPR-like sequences in Helicobacter pylori and application in genotyping. Gut Pathog. 2017, 9, 65. [Google Scholar] [CrossRef]
- Waskito, L.A.; Salama, N.R.; Yamaoka, Y. Pathogenesis of Helicobacter pylori infection. Helicobacter 2018, 23 (Suppl. 1), e12516. [Google Scholar] [CrossRef]
- Kao, C.-Y.; Sheu, B.-S.; Wu, J.-J. Helicobacter pylori infection: An overview of bacterial virulence factors and pathogenesis. Biomed. J. 2016, 39, 14–23. [Google Scholar] [CrossRef]
- García-Zea, J.A.; De La Herrán, R.; Rodríguez, F.R.; Navajas-Pérez, R.; Rejón, C.R. Detection and variability analyses of CRISPR-like loci in the H. pylori genome. PeerJ 2019, 7, e6221. [Google Scholar] [CrossRef]
- Zawilak-Pawlik, A.; Zakrzewska-Czerwińska, J. Recent Advances in Helicobacter pylori Replication: Possible Implications in Adaptation to a Pathogenic Lifestyle and Perspectives for Drug Design. Curr. Top Microbiol. Immunol. 2017, 400, 73–103. [Google Scholar] [CrossRef]
- Wang, Y.-K.; Kuo, F.-C.; Liu, C.-J.; Wu, M.-C.; Shih, H.-Y.; Wang, S.S.; Wu, J.-Y.; Kuo, C.-H.; Huang, Y.-K.; Wu, D.-C. Diagnosis of Helicobacter pylori infection: Current options and developments. World J. Gastroenterol. 2015, 21, 11221–11235. [Google Scholar] [CrossRef]
- Khalilpour, A.; Kazemzadeh-Narbat, M.; Tamayol, A.; Oklu, R.; Khademhosseini, A. Biomarkers and diagnostic tools for detection of Helicobacter pylori. Appl. Microbiol. Biotechnol. 2016, 100, 4723–4734. [Google Scholar] [CrossRef] [PubMed]
- Yee, J.K. Helicobacter pylori colonization of the oral cavity: A milestone discovery. World J. Gastroenterol. 2016, 22, 641–648. [Google Scholar] [CrossRef] [PubMed]
- Kawai, S.; Arai, K.; Lin, Y.; Nishiyama, T.; Sasakabe, T.; Wang, C.; Miwa, H.; Kikuchi, S. Comparison of the detection of Helicobacter pylori infection by commercially available serological testing kits and the 13C-urea breath test. J. Infect. Chemother. 2019, 25, 769–773. [Google Scholar] [CrossRef] [PubMed]
- Kosunen, T.; Seppäla, K.; Sarna, S.; Sipponen, P. Diagnostic value of decreasing IgG, IgA, and IgM antibody titres after eradication of Helicobacter pylori. Lancet 1992, 339, 893–895. [Google Scholar] [CrossRef] [PubMed]
- Beer-Davidson, G.; Hindiyeh, M.; Muhsen, K. Detection of Helicobacter pylori in stool samples of young children using real-time polymerase chain reaction. Helicobacter 2018, 23, e12450. [Google Scholar] [CrossRef] [PubMed]
- Mentis, A.; Lehours, P.; Mégraud, F. Epidemiology and Diagnosis of Helicobacter pylori infection. Helicobacter 2015, 20, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Vidic, J.; Manzano, M. Electrochemical biosensors for rapid pathogen detection. Curr. Opin. Electrochem. 2021, 29, 100750. [Google Scholar] [CrossRef]
- Saxena, K.; Chauhan, N.; Jain, U. Advances in diagnosis of Helicobacter pylori through biosensors: Point of care devices. Anal. Biochem. 2021, 630, 114325. [Google Scholar] [CrossRef]
- Liu, Z.; Su, X. A novel fluorescent DNA sensor for ultrasensitive detection of Helicobacter pylori. Biosens. Bioelectron. 2017, 87, 66–72. [Google Scholar] [CrossRef]
- Ly, S.Y.; Yoo, H.-S.; Choa, S.H. Diagnosis of Helicobacter pylori bacterial infections using a voltammetric biosensor. J. Microbiol. Methods 2011, 87, 44–48. [Google Scholar] [CrossRef]
- Ali, M.M.; Wolfe, M.; Tram, K.; Gu, J.; Filipe, C.D.M.; Li, Y.; Brennan, J.D. A DNAzyme-Based Colorimetric Paper Sensor for Helicobacter pylori. Angew. Chem. Int. Ed. 2019, 58, 9907–9911. [Google Scholar] [CrossRef]
- Del Pozo, M.V.; Alonso, C.; Pariente, F.; Lorenzo, E. DNA Biosensor for Detection of Helicobacter pylori Using Phen-dione as the Electrochemically Active Ligand in Osmium Complexes. Anal. Chem. 2005, 77, 2550–2557. [Google Scholar] [CrossRef] [PubMed]
- Hajihosseini, S.; Nasirizadeh, N.; Hejazi, M.S.; Yaghmaei, P. An electrochemical DNA biosensor based on Oracet Blue as a label for detection of Helicobacter pylori. Int. J. Biol. Macromol. 2016, 91, 911–917. [Google Scholar] [CrossRef] [PubMed]
- Dindar, B.; Karakuş, E.; Abasıyanık, F. New Urea Biosensor Based on Urease Enzyme Obtained from Helycobacter pylori. Appl. Biochem. Biotechnol. 2011, 165, 1308–1321. [Google Scholar] [CrossRef] [PubMed]
- Vangah, S.J.; Katalani, C.; Boone, H.A.; Hajizade, A.; Sijercic, A.; Ahmadian, G. CRISPR-Based Diagnosis of Infectious and Noninfectious Diseases. Biol. Proced. Online 2020, 22, 22. [Google Scholar] [CrossRef] [PubMed]
- Qiu, E.; Jin, S.; Xiao, Z.; Chen, Q.; Wang, Q.; Liu, H.; Xie, C.; Chen, C.; Li, Z.; Han, S. CRISPR-based detection of Helicobacter pylori in stool samples. Helicobacter 2021, 26, e12828. [Google Scholar] [CrossRef] [PubMed]
- Gregorio, G.V.; Mieli-Vergani, G.; Mowat, A.P. Viral hepatitis. Arch. Dis. Child. 1994, 70, 343–348. [Google Scholar] [CrossRef]
- Kumar, H.; Kamar, N.; Kumar, D. Hepatitis E: Current Status in India and Other Asian Countries. J. Pure Appl. Microbiol. 2019, 13, 141–159. [Google Scholar] [CrossRef]
- Pardee, M. Diagnosis and Management of Hepatitis B and C. Nurs. Clin. N. Am. 2019, 54, 277–284. [Google Scholar] [CrossRef]
- Shariati, M.; Sadeghi, M. Ultrasensitive DNA biosensor for hepatitis B virus detection based on tin-doped WO3/In2O3 heterojunction nanowire photoelectrode under laser amplification. Anal. Bioanal. Chem. 2020, 412, 5367–5377. [Google Scholar] [CrossRef]
- Yao, C.-Y. Biosensors for hepatitis B virus detection. World J. Gastroenterol. 2014, 20, 12485–12492. [Google Scholar] [CrossRef] [PubMed]
- Manzano, M.; Viezzi, S.; Mazerat, S.; Marks, R.S.; Vidic, J. Rapid and label-free electrochemical DNA biosensor for detecting hepatitis A virus. Biosens. Bioelectron. 2018, 100, 89–95. [Google Scholar] [CrossRef] [PubMed]
- Riedel, T.; Surman, F.; Hageneder, S.; Pop-Georgievski, O.; Noehammer, C.; Hofner, M.; Brynda, E.; Rodriguez-Emmenegger, C.; Dostálek, J. Hepatitis B plasmonic biosensor for the analysis of clinical serum samples. Biosens. Bioelectron. 2016, 85, 272–279. [Google Scholar] [CrossRef] [PubMed]
- Timurdogan, E.; Alaca, B.E.; Kavakli, I.H.; Urey, H. MEMS biosensor for detection of Hepatitis A and C viruses in serum. Biosens. Bioelectron. 2011, 28, 189–194. [Google Scholar] [CrossRef]
- Chowdhury, A.D.; Takemura, K.; Li, T.-C.; Suzuki, T.; Park, E.Y. Electrical pulse-induced electrochemical biosensor for hepatitis E virus detection. Nat. Commun. 2019, 10, 3737. [Google Scholar] [CrossRef]
- Tang, D.; Tang, J.; Su, B.; Ren, J.; Chen, G. Simultaneous determination of five-type hepatitis virus antigens in 5min using an integrated automatic electrochemical immunosensor array. Biosens. Bioelectron. 2010, 25, 1658–1662. [Google Scholar] [CrossRef]
- Chen, X.; Tan, Y.; Wang, S.; Wu, X.; Liu, R.; Yang, X.; Wang, Y.; Tai, J.; Li, S. A CRISPR-Cas12b–Based Platform for Ultrasensitive, Rapid, and Highly Specific Detection of Hepatitis B Virus Genotypes B and C in Clinical Application. Front. Bioeng. Biotechnol. 2021, 9, 743322. [Google Scholar] [CrossRef]
- Gong, S.; Zhang, S.; Wang, X.; Li, J.; Pan, W.; Li, N.; Tang, B. Strand Displacement Amplification Assisted CRISPR-Cas12a Strategy for Colorimetric Analysis of Viral Nucleic Acid. Anal. Chem. 2021, 93, 15216–15223. [Google Scholar] [CrossRef]
- Wang, S.; Li, H.; Kou, Z.; Ren, F.; Jin, Y.; Yang, L.; Dong, X.; Yang, M.; Zhao, J.; Dong, N.; et al. Highly sensitive and specific detection of hepatitis B virus DNA and drug resistance mutations utilizing the PCR-based CRISPR-Cas13a system. Clin. Microbiol. Infect. 2021, 27, 443–450. [Google Scholar] [CrossRef]
- Choi, J.-H.; Shin, M.; Yang, L.; Conley, B.; Yoon, J.; Lee, S.-N.; Lee, K.-B.; Choi, J.-W. Clustered Regularly Interspaced Short Palindromic Repeats-Mediated Amplification-Free Detection of Viral DNAs Using Surface-Enhanced Raman Spectroscopy-Active Nanoarray. ACS Nano 2021, 15, 13475–13485. [Google Scholar] [CrossRef]
- Ding, R.; Long, J.; Yuan, M.; Zheng, X.; Shen, Y.; Jin, Y.; Yang, H.; Li, H.; Chen, S.; Duan, G. CRISPR/Cas12-Based Ultra-Sensitive and Specific Point-of-Care Detection of HBV. Int. J. Mol. Sci. 2021, 22, 4842. [Google Scholar] [CrossRef] [PubMed]
- Herrera-Carrillo, E.; Gao, Z.; Berkhout, B. CRISPR therapy towards an HIV cure. Brief. Funct. Genom. 2020, 19, 201–208. [Google Scholar] [CrossRef] [PubMed]
- Saayman, S.; Ali, S.; Morris, K.; Weinberg, M. The therapeutic application of CRISPR/Cas9 technologies for HIV. Expert Opin. Biol. Ther. 2015, 15, 819–830. [Google Scholar] [CrossRef] [PubMed]
- Yin, L.; Zhao, F.; Sun, H.; Wang, Z.; Huang, Y.; Zhu, W.; Xu, F.; Mei, S.; Liu, X.; Zhang, D.; et al. CRISPR-Cas13a Inhibits HIV-1 Infection. Mol. Ther. Nucleic Acids 2020, 21, 147–155. [Google Scholar] [CrossRef]
- Li, H.; Wang, S.; Dong, X.; Li, Q.; Li, M.; Li, J.; Guo, Y.; Jin, X.; Zhou, Y.; Song, H.; et al. CRISPR-Cas13a Cleavage of Dengue Virus NS3 Gene Efficiently Inhibits Viral Replication. Mol. Ther. Nucleic Acids 2020, 19, 1460–1469. [Google Scholar] [CrossRef]
- Kistler, K.E.; Vosshall, L.B.; Matthews, B.J. Genome Engineering with CRISPR-Cas9 in the Mosquito Aedes aegypti. Cell Rep. 2015, 11, 51–60. [Google Scholar] [CrossRef]
- Carlin, A.F.; Shresta, S. Genome-wide approaches to unravelling host–virus interactions in Dengue and Zika infections. Curr. Opin. Virol. 2019, 34, 29–38. [Google Scholar] [CrossRef]
- Gong, J.; Zhang, G.; Wang, W.; Liang, L.; Li, Q.; Liu, M.; Xue, L.; Tang, G. A simple and rapid diagnostic method for 13 types of high-risk human papillomavirus (HR-HPV) detection using CRISPR-Cas12a technology. Sci. Rep. 2021, 11, 12800. [Google Scholar] [CrossRef]
- Maver, P.; Poljak, M. Primary HPV-based cervical cancer screening in Europe: Implementation status, challenges, and future plans. Clin. Microbiol. Infect. 2020, 26, 579–583. [Google Scholar] [CrossRef]
- Li, Z.; Ding, X.; Yin, K.; Xu, Z.; Cooper, K.; Liu, C. Electric field-enhanced electrochemical CRISPR biosensor for DNA detection. Biosens. Bioelectron. 2021, 192, 113498. [Google Scholar] [CrossRef]
- Tsou, J.-H.; Leng, Q.; Jiang, F. A CRISPR Test for Detection of Circulating Nuclei Acids. Transl. Oncol. 2019, 12, 1566–1573. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Shen, X.; Wang, T.; Chen, P.; Qi, N.; Yin, B.-C.; Ye, B.-C. A lateral flow strip combined with Cas9 nickase-triggered amplification reaction for dual food-borne pathogen detection. Biosens. Bioelectron. 2020, 165, 112364. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Qian, L.; Wei, W.; Wang, Y.; Wang, B.; Lin, P.; Liu, W.; Yixuan, Y.; Li, X.; Liu, D.; et al. Paired Design of dCas9 as a Systematic Platform for the Detection of Featured Nucleic Acid Sequences in Pathogenic Strains. ACS Synth. Biol. 2017, 6, 211–216. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Wang, J.; Hu, X.; Tang, X.; Zhang, C. A rapid and high-throughput Helicobacter pylori RPA-CRISPR/Cas12a-based nucleic acid detection system. Clin. Chim. Acta 2022. [Google Scholar] [CrossRef]
- Kachwala, M.J.; Smith, C.W.; Nandu, N.; Yigit, M.V. Reprogrammable Gel Electrophoresis Detection Assay Using CRISPR-Cas12a and Hybridization Chain Reaction. Anal. Chem. 2021, 93, 1934–1938. [Google Scholar] [CrossRef]
- Zhou, R.; Li, Y.; Dong, T.; Tang, Y.; Li, F. A sequence-specific plasmonic loop-mediated isothermal amplification assay with orthogonal color readouts enabled by CRISPR Cas12a. Chem. Commun. 2020, 56, 3536–3538. [Google Scholar] [CrossRef]
- Shao, N.; Han, X.; Song, Y.; Zhang, P.; Qin, L. CRISPR-Cas12a Coupled with Platinum Nanoreporter for Visual Quantification of SNVs on a Volumetric Bar-Chart Chip. Anal. Chem. 2019, 91, 12384–12391. [Google Scholar] [CrossRef]
- Lee, H.; Choi, J.; Jeong, E.; Baek, S.; Kim, H.C.; Chae, J.-H.; Koh, Y.; Seo, S.W.; Kim, J.-S.; Kim, S.J. dCas9-mediated Nanoelectrokinetic Direct Detection of Target Gene for Liquid Biopsy. Nano Lett. 2018, 18, 7642–7650. [Google Scholar] [CrossRef]
- Nouri, R.; Jiang, Y.; Lian, X.L.; Guan, W. Sequence-Specific Recognition of HIV-1 DNA with Solid-State CRISPR-Cas12a-Assisted Nanopores (SCAN). ACS Sens. 2020, 5, 1273–1280. [Google Scholar] [CrossRef]
- Pardee, K.; Green, A.A.; Takahashi, M.K.; Braff, D.; Lambert, G.; Lee, J.W.; Ferrante, T.; Ma, D.; Donghia, N.; Fan, M.; et al. Rapid, Low-Cost Detection of Zika Virus Using Programmable Biomolecular Components. Cell 2016, 165, 1255–1266. [Google Scholar] [CrossRef]
- Ackerman, C.M.; Myhrvold, C.; Thakku, S.G.; Freije, C.A.; Metsky, H.C.; Yang, D.K.; Ye, S.H.; Boehm, C.K.; Kosoko-Thoroddsen, T.-S.F.; Kehe, J.; et al. Massively multiplexed nucleic acid detection with Cas13. Nature 2020, 582, 277–282. [Google Scholar] [CrossRef] [PubMed]
- Myhrvold, C.; Freije, C.A.; Gootenberg, J.S.; Abudayyeh, O.O.; Metsky, H.C.; Durbin, A.F.; Kellner, M.J.; Tan, A.L.; Paul, L.M.; Parham, L.A.; et al. Field-deployable viral diagnostics using CRISPR-Cas13. Science 2018, 360, 444–448. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Wang, Q.; Xu, X.; Xia, Q.; Long, F.; Li, W.; Shui, Y.; Xia, X.; Wang, J. Detection of target DNA with a novel Cas9/sgRNAs-associated reverse PCR (CARP) technique. Anal. Bioanal. Chem. 2018, 410, 2889–2900. [Google Scholar] [CrossRef] [PubMed]
- Teng, F.; Guo, L.; Cui, T.; Wang, X.-G.; Xu, K.; Gao, Q.; Zhou, Q.; Li, W. CDetection: CRISPR-Cas12b-based DNA detection with sub-attomolar sensitivity and single-base specificity. Genome Biol. 2019, 20, 132. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Xia, Q.; Wang, Q.; Xia, X.; Wang, J. Detecting and typing target DNA with a novel CRISPR-typing PCR (ctPCR) technique. Anal. Biochem. 2018, 561–562, 37–46. [Google Scholar] [CrossRef]
- Yin, K.; Ding, X.; Li, Z.; Zhao, H.; Cooper, K.; Liu, C. Dynamic Aqueous Multiphase Reaction System for One-Pot CRISPR-Cas12a-Based Ultrasensitive and Quantitative Molecular Diagnosis. Anal. Chem. 2020, 92, 8561–8568. [Google Scholar] [CrossRef]
- Rai, P.; Kumar, B.K.; Deekshit, V.K.; Karunasagar, I.; Karunasagar, I. Detection technologies and recent developments in the diagnosis of COVID-19 infection. Appl. Microbiol. Biotechnol. 2021, 105, 441–455. [Google Scholar] [CrossRef]
- Yesudhas, D.; Srivastava, A.; Gromiha, M.M. COVID-19 outbreak: History, mechanism, transmission, structural studies and therapeutics. Infection 2021, 49, 199–213. [Google Scholar] [CrossRef]
- Corman, V.M.; Landt, O.; Kaiser, M.; Molenkamp, R.; Meijer, A.; Chu, D.K.W.; Bleicker, T.; Brünink, S.; Schneider, J.; Schmidt, M.L.; et al. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Eurosurveillance 2020, 25, 2000045, Erratum in Euro Surveill. 2021, 26. [Google Scholar] [CrossRef]
- Behera, B.C.; Mishra, R.R.; Thatoi, H. Recent biotechnological tools for diagnosis of corona virus disease: A review. Biotechnol. Prog. 2021, 37, e3078. [Google Scholar] [CrossRef]
- McCormick-Baw, C.; Morgan, K.; Gaffney, D.; Cazares, Y.; Jaworski, K.; Byrd, A.; Molberg, K.; Cavuoti, D. Saliva as an Alternate Specimen Source for Detection of SARS-CoV-2 in Symptomatic Patients Using Cepheid Xpert Xpress SARS-CoV-2. J. Clin. Microbiol. 2020, 58, e01109. [Google Scholar] [CrossRef] [PubMed]
- Ali, N.; Rampazzo, R.; Costa, A.D.T.; Krieger, M.A. Current Nucleic Acid Extraction Methods and Their Implications to Point-of-Care Diagnostics. BioMed Res. Int. 2017, 2017, 9306564. [Google Scholar] [CrossRef]
- Ravi, N.; Cortade, D.L.; Ng, E.; Wang, S.X. Diagnostics for SARS-CoV-2 detection: A comprehensive review of the FDA-EUA COVID-19 testing landscape. Biosens. Bioelectron. 2020, 165, 112454. [Google Scholar] [CrossRef] [PubMed]
- Nouri, R.; Tang, Z.; Dong, M.; Liu, T.; Kshirsagar, A.; Guan, W. CRISPR-based detection of SARS-CoV-2: A review from sample to result. Biosens. Bioelectron. 2021, 178, 113012. [Google Scholar] [CrossRef] [PubMed]
- Xiong, Y.; Zhang, J.; Yang, Z.; Mou, Q.; Ma, Y.; Xiong, Y.; Lu, Y. Functional DNA Regulated CRISPR-Cas12a Sensors for Point-of-Care Diagnostics of Non-Nucleic-Acid Targets. J. Am. Chem. Soc. 2020, 142, 207–213. [Google Scholar] [CrossRef]
- Lucia, C.; Federico, P.B.; Alejandra, G.C. An ultrasensitive, rapid, and portable coronavirus SARS-CoV-2 sequence detection method based on CRISPR-Cas12. bioRxiv 2020. [Google Scholar] [CrossRef]
- Ding, X.; Bin, P.; Wu, W.; Chang, Y.; Zhu, G. Tryptophan Metabolism, Regulatory T Cells, and Inflammatory Bowel Disease: A Mini Review. Mediat. Inflamm. 2020, 2020, 9706140. [Google Scholar] [CrossRef]
- Broughton, J.P.; Deng, X.; Yu, G.; Fasching, C.L.; Servellita, V.; Singh, J.; Miao, X.; Streithorst, J.A.; Granados, A.; Sotomayor-Gonzalez, A.; et al. CRISPR–Cas12-based detection of SARS-CoV-2. Nat. Biotechnol. 2020, 38, 870–874. [Google Scholar] [CrossRef]
- Ali, Z.; Aman, R.; Mahas, A.; Rao, G.S.; Tehseen, M.; Marsic, T.; Salunke, R.; Subudhi, A.K.; Hala, S.M.; Hamdan, S.M.; et al. iSCAN: An RT-LAMP-coupled CRISPR-Cas12 module for rapid, sensitive detection of SARS-CoV-2. Virus Res. 2020, 288, 198129. [Google Scholar] [CrossRef]
- Guo, L.; Liu, W.; Li, Z.; Li, L. An adaptive sliding mode control strategy for the heading control of autonomous underwater vehicles. In Proceedings of the Global Oceans 2020: Singapore—U.S. Gulf Coast, Biloxi, MS, USA, 5–30 October 2020; pp. 1–4. [Google Scholar]
- Rauch, J.N.; Valois, E.; Solley, S.C.; Braig, F.; Lach, R.S.; Audouard, M.; Ponce-Rojas, J.C.; Costello, M.S.; Baxter, N.J.; Kosik, K.S.; et al. A Scalable, Easy-to-Deploy Protocol for Cas13-Based Detection of SARS-CoV-2 Genetic Material. J. Clin. Microbiol. 2021, 59, e02402. [Google Scholar] [CrossRef]
- Hou, T.; Zeng, W.; Yang, M.; Chen, W.; Ren, L.; Ai, J.; Wu, J.; Liao, Y.; Gou, X.; Li, Y.; et al. Development and evaluation of a rapid CRISPR-based diagnostic for COVID-19. PLoS Pathog. 2020, 16, e1008705. [Google Scholar] [CrossRef] [PubMed]
- Patchsung, M.; Jantarug, K.; Pattama, A.; Aphicho, K.; Suraritdechachai, S.; Meesawat, P.; Sappakhaw, K.; Leelahakorn, N.; Ruenkam, T.; Wongsatit, T.; et al. Clinical validation of a Cas13-based assay for the detection of SARS-CoV-2 RNA. Nat. Biomed. Eng. 2020, 4, 1140–1149. [Google Scholar] [CrossRef] [PubMed]
- Hayden, C.; Metsky, C.A.; Freije, T.; Solveig, F.; Kosoko-Thoroddsen, P.C.; Sabeti, C. Myhrvold CRISPR-based surveillance for COVID-19 using genomically-comprehensive machine learning design. bioRxiv 2020. [Google Scholar]
- Arizti-Sanz, J.; Freije, C.A.; Stanton, A.C.; Boehm, C.K.; Petros, B.A.; Siddiqui, S.; Shaw, B.M.; Adams, G.; Kosoko-Thoroddsen, T.F.; Kemball, M.E.; et al. Integrated sample inactivation, amplification, and Cas13-based detection of SARS-CoV-2. bioRxiv 2020, 28, 119131, Preprint. Update in Nat. Commun. 2020, 11, 5921. [Google Scholar]
- Aman, R.; Mahas, A.; Mahfouz, M. Nucleic Acid Detection Using CRISPR/Cas Biosensing Technologies. ACS Synth. Biol. 2020, 9, 1226–1233. [Google Scholar] [CrossRef] [PubMed]
- Diego, J.G.-B.; Fernández-Soto, P.; Muro, A. The Future of Point-of-Care Nucleic Acid Amplification Diagnostics after COVID-19: Time to Walk the Walk. Int. J. Mol. Sci. 2022, 23, 14110. [Google Scholar] [CrossRef]
- Kaminski, M.M.; Abudayyeh, O.O.; Gootenberg, J.S.; Zhang, F.; Collins, J.J. CRISPR-based diagnostics. Nat. Biomed. Eng. 2021, 5, 643–656. [Google Scholar] [CrossRef]
- Qi, Y.; Li, K.; Li, Y.; Guo, D.; Xu, J.; Li, Y.; Gong, W. CRISPR-Based Diagnostics: A Potential Tool to Address the Diagnostic Challenges of Tuberculosis. Pathogens 2022, 11, 1211. [Google Scholar] [CrossRef]
| CRISPR Effector Protein | Targeted Gene | Preamplification Method | Signal Readout | Assay Reaction Time | LOD | Reference |
|---|---|---|---|---|---|---|
| Cas9a | N1, N2, and N3 genes | RT-PCR | Colorimetric | Not reported | 140 pM | [145] |
| Cas12a | ORF1ab | RT-RPA | Colorimetric and fluorescence | 1 h | 10 copies/μL | [146] |
| Cas12a | N gene | RT-RPA | Fluorescence | 40 min | 10 copies/μL | [147] |
| Cas12a | N gene and E gene | RT-LAMP | Colorimetric | 30–40 min | 10 copies/μL | [148] |
| Cas12a and Cas12b | N gene and E gene | RT-LAMP | Fluorescence | 40 min | 10 copies/μL | [149] |
| Cas12b | N gene | RT-RAA | Colorimetric and fluorescence | 1 h | 10 copies/μL | [150] |
| Cas13a | N gene | RT-PCR | Colorimetric | Not reported | 10 copies/μL | [151] |
| Cas13a | ORF1ab and N | RT-RPA | Fluorescence | 40 min | 1 copy/μL | [152] |
| Cas13a | Orf1ab, S, and N | RT-RPA | Colorimetric and fluorescence | 35–70 min | 10 copies/μL | [153] |
| Cas13a | Not reported | RT-RPA | Colorimetric | 50 min | 10 copies/μL | [154] |
| Cas13a | ORF1a | RT-RPA | Colorimetric and fluorescence | 1 h | 10 copies/μL | [155] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kumaran, A.; Jude Serpes, N.; Gupta, T.; James, A.; Sharma, A.; Kumar, D.; Nagraik, R.; Kumar, V.; Pandey, S. Advancements in CRISPR-Based Biosensing for Next-Gen Point of Care Diagnostic Application. Biosensors 2023, 13, 202. https://doi.org/10.3390/bios13020202
Kumaran A, Jude Serpes N, Gupta T, James A, Sharma A, Kumar D, Nagraik R, Kumar V, Pandey S. Advancements in CRISPR-Based Biosensing for Next-Gen Point of Care Diagnostic Application. Biosensors. 2023; 13(2):202. https://doi.org/10.3390/bios13020202
Chicago/Turabian StyleKumaran, Akash, Nathan Jude Serpes, Tisha Gupta, Abija James, Avinash Sharma, Deepak Kumar, Rupak Nagraik, Vaneet Kumar, and Sadanand Pandey. 2023. "Advancements in CRISPR-Based Biosensing for Next-Gen Point of Care Diagnostic Application" Biosensors 13, no. 2: 202. https://doi.org/10.3390/bios13020202
APA StyleKumaran, A., Jude Serpes, N., Gupta, T., James, A., Sharma, A., Kumar, D., Nagraik, R., Kumar, V., & Pandey, S. (2023). Advancements in CRISPR-Based Biosensing for Next-Gen Point of Care Diagnostic Application. Biosensors, 13(2), 202. https://doi.org/10.3390/bios13020202





