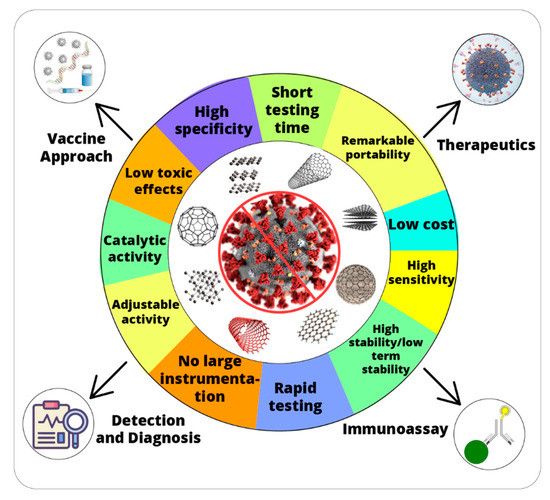
Abstract
The current coronavirus disease 2019 (COVID-19) outbreak is considered as one of the biggest public health challenges and medical emergencies of the century. A global health emergency demands an urgent development of rapid diagnostic tools and advanced therapeutics for the mitigation of COVID-19. To cope with the current crisis, nanotechnology offers a number of approaches based on abundance and versatile functioning. Despite major developments in early diagnostics and control of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), there is still a need to find effective nanomaterials with low cost, high stability and easy use. Nanozymes are nanomaterials with innate enzyme-like characteristics and exhibit great potential for various biomedical applications such as disease diagnosis and anti-viral agents. Overall the potential and contribution of nanozymes in the fight against SARS-CoV-2 infection i.e., rapid detection, inhibition of the virus at various stages, and effective vaccine development strategies, is not fully explored. This paper discusses the utility and potential of nanozymes from the perspective of COVID-19. Moreover, future research directions and potential applications of nanozymes are highlighted to overcome the challenges related to early diagnosis and therapeutics development for the SARS-CoV-2. We anticipate the current perspective will play an effective role in the existing response to the COVID-19 crisis.
1. Introduction
In the 20th century, millions of deaths occurred due to three influenza pandemics [1]. The first deadly pandemic named ‘Spanish flu’ occurred in 1918, which was caused by H1N1 influenza A strain. The estimated number of deaths from this virus were more than 40 million [2]. In 1957 another global pandemic called ‘Asian Flu’ [3,4] first originated in the Yunan province of China [4]. This virus was a mutated form of avian (H2N2) and already present human influenza viruses strains [3]. In mid-1968, the Hong Kong flu pandemic occurred, which was caused by the H3N2 influenza strain and afterward spread all over the globe [1,4]. The death toll of this pandemic was approximately 700,000 [4]. Recently, in late December 2019, a novel coronavirus (CoV) outbreak was first reported in Wuhan, China. The virus first disseminated in different parts of the world and eventually gained the status of a pandemic. The World Health Organization (WHO) announced this outbreak as a worldwide emergency on 30 January 2020 [5]. Etiologically, this illness has been caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) [6,7]. Up to April 2021 more than 2.91 million deaths have been reported.
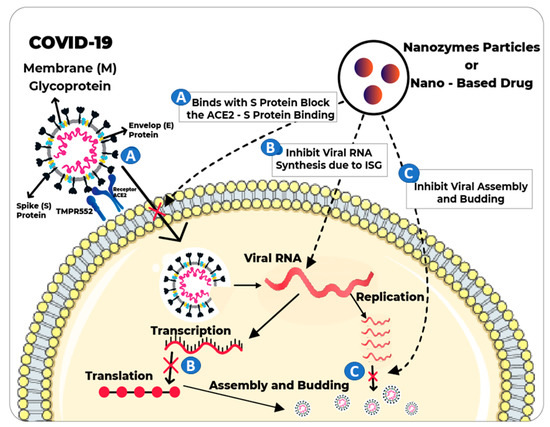
SARS-CoV-2 is an enveloped, positive sense, single-stranded RNA beta-Covid encoding 3 non-structural (3-chymotrypsin-like protease, papain-like protease, helicase, and RNA-dependent RNA polymerase), structural (spike glycoprotein) and accessory proteins [8]. SARS-CoV-2 is highly infectious and human-to-human transmission has been frequently reported. The world economy is facing a long lasting downturn due to mandatory quarantine and lockdowns. The implications of the pandemic are worsening day by day which poses serious challenges for health services. One serious concern of healthcare professionals is to develop, disseminate and deploy safe and effective vaccines against COVID-19 [9]. Until now, few SARS-CoV-2 vaccines have been approved, with more expected to be licensed in Yet having licensed vaccines, efficacy, large scale production, affordable price and rapid availability to local communities remain big challenges in controlling the SARS-CoV-2 pandemic. Reinfections and uncertainties in preliminary data for some vaccines indicate that the immunization process needs further investigation.
Meanwhile, reports about the emergence of new variants of coronavirus are continuously rising [10]. Early diagnosis and quarantine to cut off the source of infection are the most effective control strategies for disease outbreaks, including the COVID-19 pandemic [11,12]. Diagnosis of SARS-CoV-2 might be influenced by epidemiological history, clinical features, imageology and pathogenic index. Development of effective antiviral agents is hindered by the ability of SARS-CoV-2 to grow in the host cells without keeping its genome. Thus, there is an urgent need to expand testing capacities, deploy effective therapeutics, and develop safe vaccines that provide long lasting immunity [13]. Reverse transcription polymerase chain reaction (RT-PCR) is the most common molecular method used for early detection of SARS-CoV-Some other nucleic acid-based techniques, such as CRISPR (clustered regularly interspaced short palindromic repeats), microarrays, high-throughput qPCR (HT-qPCR) and loop-mediated isothermal amplification (LAMP) are also favorable options for detecting SARS-CoV-2 in clinical as well as in environmental samples [14,15,16]. Despite reliable results, genome extraction, amplification and data analysis require sophisticated biosafety labs, skilled personnel that make nucleic acid testing costly and unsuitable for under-developed countries [17]. Alternatively, antibody testing is chosen for detecting IgM or/and IgG antibodies produced after exposure to SARS-CoV-2 [18]. Typically, production of antibodies occurs 10–15 days post infection, so early screening and diagnosis of SARS-CoV-2 could not be possible with this technique [19]. The WHO has recommended the use of rapid antigen diagnostic tests that meet at least 80% sensitivity and 97% specificity for the active SARS-CoV-2 infections [20]. A detailed overview of various currently used diagnostic techniques, with detection limits, specificity, processing time is presented in Table 1. It could be inferred that extensive efforts are required to improve the early diagnosis of SARS-CoV-2, which will improve the therapeutic decision-making and will further decrease the intensity of illness and duration of hospital stay.

Table 1.
Different diagnostic techniques currently being used for severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2).
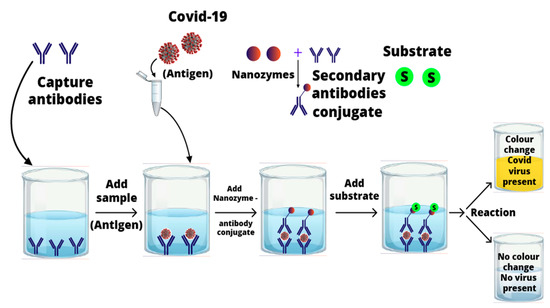
Various biosensing techniques have been developed for rapid, reliable, and sensitive detection of biomolecules (biomarker) for gauging virulence, pathogenicity, and microbial load [21,22]. Biosensing methods use natural enzymes such as horseradish peroxidases to catalyze different colorimetric reactions in the presence of substrates. Regardless of their novel catalytic effectiveness, natural enzymes have some limits for industrial application, such as low stability in harsh environmental conditions, and relatively high costs for preparation, purification, and storage [23]. Therefore, over the past few decades, researchers have made an intense effort to develop artificial enzymes for a wide range of applications [24]. Recently, nanomaterial-based enzyme mimetics (nanozymes) have revolutionized the fields of diagnosis and therapeutics [24].
Nanozymes have been frequently employed in various biomedical applications such as disease diagnosis, cancer therapy and anti-viral agents. Therefore, inspired by the unique characteristics of nanozymes, it is assumed they have the potential to overcome the challenges related to the early diagnosis and therapeutic developments for SARS-CoV-2 infections. According to the author’s knowledge, almost no efforts have been devoted to reviewing the vast potential of nanozymes to combat the COVID-19 infection. Thus, in this perspective, we present a comprehensive study of recent updates on nanozymes and their possible applications for detection and treatment for the SARS-CoV-2. We anticipate the current article will pave the way toward the development of rapid and sensitive diagnostics. Moreover, our findings will play a very effective role in the welfare of humans and the medical community in the COVID-19 crisis.
4. Conclusions and Recommendations
Nanotechnology is expected to play a critical role in the fight against COVID-19. Various nanomaterials have been utilized for therapeutics, building rapid diagnostic test kits, inhibition of virus replication and vaccines. Morphological and physicochemical similarities of nanozymes with SARS-CoV-2 mean they can provide powerful tools to interfere with the viral life cycle. The multi-functionality of nanozymes will significantly promote the proficient treatment against SARS-CoV-2. Nanozymes can directly inhibit the entry of the virus into the host cells by blocking the attachment or inhibiting the viral replication. IONzymes have to an extraordinary extent improved the defensive capacity of PPEs like facemasks, specifically by halting the viral actions. Hence, efficient antiviral strategies are vital to minimize viral proliferation, cellular damages induced by viral invasions and mutation frequency, which otherwise may result in therapeutic resistance. Future research projects must explore the different combinations of biocompatible nanozymes to broaden the antiviral spectrum against SARS-CoV-2 and other human-infecting viruses. Moreover, efficacy of SARS-CoV-2 vaccines could be improved through the addition of immunomodulating nanozymes or using them as adjuvants. Nanozyme-based virus-like particles (VLPs) that mimic the SARS-CoV-2 can induce long-lasting immunity by avoiding exposure to virulent components. High thermostability and large-scale production of SARS-CoV-2 vaccines along with availability to all countries is the target area for future applications. In future, nanozymes can also be employed for lessening the impact of other global challenges like antimicrobial resistance [98,99]. Despite the incredible features of nanozymes, nanotoxicity and low selectivity are the limitations for biomedical applications [100]. Therefore, more deep research could improve the efficacy of antiviral medications and reduce their side effects. In summary, nanozyme/nanomaterial-based therapeutics are expected to play a frontline role in tackling this outbreak.
Author Contributions
J.A. conceptualization, data analysis and wrote first draft of the manuscript, S.N.E. and A.A. revised the tabulated data and helped in graphical work. R.A. helped in data collection and drawing figures. H.W. critically revised article and final approval of the manuscript to be published. M.M.M. provided funding and supervision. He also critically revised the article and provided final approval of the manuscript to be published. All authors have read and agreed to the published version of the manuscript.
Funding
This work was funded by Abu Dhabi Department of Education and Knowledge (ADEK) through ADEK Award for Research Excellence (AARE) funding program (award # AARE19-047) and the National Water and Energy Center at United Arab Emirates University, UAE (Grant No. 21N226).
Data Availability Statement
Not applicable we did not have conducted any research experimental work in this study and don’t have any data avaialable or archived datasets or supplementary data.
Acknowledgments
We are thankful to the University of Sialkot, Pakistan for the support during this work.
Conflicts of Interest
Authors declares the no conflict of interest.
References
- Kilbourne, E.D. Influenza pandemics of the 20th century. Emerg. Infect. Dis. 2006, 12, 9–14. [Google Scholar] [CrossRef] [PubMed]
- Taubenberger, J.K. The origin and virulence of the 1918 “Spanish” influenza virus. Proc. Am. Philos. Soc. 2006, 150, 86–112. [Google Scholar] [PubMed]
- Greenberg, M.; Jacobziner, H.; Pakter, J.; Weisl, B.A. Maternal mortality in the epidemic of Asian influenza, New York City, 1957. Am. J. Obstet. Gynecol. 1958, 76, 897–902. [Google Scholar] [CrossRef]
- Rajagopal, S.; Treanor, J. Pandemic (avian) influenza. Semin. Respir. Crit. Care Med. 2007, 28, 159–170. [Google Scholar] [CrossRef] [PubMed]
- Meng, L.; Hua, F.; Bian, Z. Coronavirus Disease 2019 (COVID-19): Emerging and Future Challenges for Dental and Oral Medicine. J. Dent. Res. 2020, 99, 481–487. [Google Scholar] [CrossRef]
- Su Eun Park Epidemiology, virology, and clinical features of severe acute respiratory syndrome-coronavirus-2. Clin. Exp. Pediatr. 2020, 63, 119–124. [CrossRef] [PubMed]
- Harapan, H.; Itoh, N.; Yufika, A.; Winardi, W.; Keam, S.; Te, H.; Megawati, D.; Hayati, Z.; Wagner, A.L.; Mudatsir, M. Coronavirus disease 2019 (COVID-19): A literature review. J. Infect. Public Health 2020, 13, 667–673. [Google Scholar] [CrossRef]
- Li, G.; De Clercq, E. Therapeutic options for the 2019 novel coronavirus (2019-nCoV). Nat. Rev. Drug Discov. 2020, 19, 149–150. [Google Scholar] [CrossRef]
- Forman, R.; Shah, S.; Jeurissen, P.; Jit, M.; Mossialos, E. COVID-19 vaccine challenges: What have we learned so far and what remains to be done? Health Policy 2021, 125, 553–567. [Google Scholar] [CrossRef]
- Ledford, H. How ‘killer’ T cells could boost COVID immunity in face of new variants. Nature 2021, 2–7. [Google Scholar] [CrossRef]
- Samhan, F.A.; Stedtfeld, T.M.; Waseem, H.; Williams, M.R.; Stedtfeld, R.D.; Hashsham, S.A. On-filter direct amplification of Legionella pneumophila for rapid assessment of its abundance and viability. Water Res. 2017, 121. [Google Scholar] [CrossRef] [PubMed]
- Girum, T.; Lentiro, K.; Geremew, M.; Migora, B.; Shewamare, S. Global strategies and effectiveness for COVID-19 prevention through contact tracing, screening, quarantine, and isolation: A systematic review. Trop. Med. Health 2020, 48, 1–15. [Google Scholar] [CrossRef]
- Chakravarty, M.; Vora, A. Nanotechnology-based antiviral therapeutics. Drug Deliv. Transl. Res. 2020, 1–40. [Google Scholar] [CrossRef] [PubMed]
- Hou, T.; Zeng, W.; Yang, M.; Chen, W.; Ren, L.; Ai, J.; Wu, J.; Liao, Y.; Gou, X.; Li, Y.; et al. Development and evaluation of a rapid CRISPR-based diagnostic for COVID-19. PLoS Pathog. 2020, 16, e1008705. [Google Scholar] [CrossRef] [PubMed]
- Waseem, H.; Saleem ur Rehman, H.; Ali, J.; Iqbal, M.J.; Ali, M.I. Antibiotics and Antimicrobial Resistance Genes in the Environment; Elsevier: Amsterdam, The Netherlands, 2020; pp. 206–222. [Google Scholar]
- Munir, A.; Waseem, H.; Williams, M.R.; Stedtfeld, R.D.; Gulari, E.; Tiedje, J.M.; Hashsham, S.A. Modeling hybridization kinetics of gene probes in a DNA biochip using FEMLAB. Microarrays 2017, 6, 9. [Google Scholar] [CrossRef]
- Waseem, H.; Jameel, S.; Ali, J.; Ur Rehman, H.S.; Tauseef, I.; Farooq, U.; Jamal, A.; Ali, M.I. Contributions and challenges of high throughput qPCR for determining antimicrobial resistance in the environment: A critical review. Molecules 2019, 24, 163. [Google Scholar] [CrossRef]
- Yu, C.Y.; Chan, K.G.; Yean, C.Y.; Ang, G.Y. Nucleic Acid-Based Diagnostic Tests for the Detection SARS-CoV-2: An Update. Diagnostics 2021, 11, 53. [Google Scholar] [CrossRef]
- De Gasparo, R.; Pedotti, M.; Simonelli, L.; Nickl, P.; Muecksch, F.; Cassaniti, I.; Percivalle, E.; Lorenzi, J.C.C.; Mazzola, F.; Magrì, D.; et al. Bispecific IgG neutralizes SARS-CoV-2 variants and prevents escape in mice. Nature 2021. [Google Scholar] [CrossRef]
- Ayove, T.; Houniei, W.; Wangnapi, R.; Bieb, S.V.; Kazadi, W.; Luke, L.-N.; Manineng, C.; Moses, P.; Paru, R.; Esfandiari, J.; et al. Sensitivity and specificity of a rapid point-of-care test for active yaws: A comparative study. Lancet Glob. Heal. 2014, 2, e415–e421. [Google Scholar] [CrossRef]
- Waseem, H.; Williams, M.R.; Stedtfeld, T.; Chai, B.; Stedtfeld, R.D.; Cole, J.R.; Tiedje, J.M.; Hashsham, S.A. Virulence factor activity relationships (VFARs): A bioinformatics perspective. Environ. Sci. Process. Impacts 2017, 19, 247–260. [Google Scholar] [CrossRef]
- Waseem, H.; Ali, J.; Syed, J.H.; Jones, K.C. Establishing the relationship between molecular biomarkers and biotransformation rates: Extension of knowledge for dechlorination of polychlorinated dibenzo-p-dioxins and furans (PCDD/Fs). Environ. Pollut. 2020, 263, 114676. [Google Scholar] [CrossRef]
- Bilal, M.; Barceló, D.; Iqbal, H.M.N. Nanostructured materials for harnessing the power of horseradish peroxidase for tailored environmental applications. Sci. Total Env. 2020, 749, 142360. [Google Scholar] [CrossRef] [PubMed]
- Shin, H.Y.; Park, T.J.; Kim, M. Il Recent Research Trends and Future Prospects in Nanozymes. J. Nanomater. 2015, 2015, 756278. [Google Scholar] [CrossRef]
- Poon, L.L.M.; Chan, K.H.; Wong, O.K.; Yam, W.C.; Yuen, K.Y.; Guan, Y.; Lo, Y.M.D.; Peiris, J.S.M. Early diagnosis of SARS Coronavirus infection by real time RT-PCR. J. Clin. Virol. 2003, 28, 233–238. [Google Scholar] [CrossRef] [PubMed]
- Vogels, C.B.F.; Brackney, D.E.; Wang, J.; Kalinich, C.C.; Ott, I.M.; Kudo, E.; Lu, P.; Venkataraman, A.; Tokuyama, M.; Moore, A.J.; et al. SalivaDirect: Simple and sensitive molecular diagnostic test for SARS-CoV-2 surveillance. medRxiv 2020. [Google Scholar] [CrossRef]
- Weissleder, R.; Lee, H.; Ko, J.; Pittet, M.J. COVID-19 diagnostics in context. Sci. Transl. Med. 2020, 12. [Google Scholar] [CrossRef]
- Suo, T.; Liu, X.; Feng, J.; Guo, M.; Hu, W.; Guo, D.; Ullah, H.; Yang, Y.; Zhang, Q.; Wang, X.; et al. ddPCR: A more accurate tool for SARS-CoV-2 detection in low viral load specimens. Emerg. Microbes Infect. 2020, 9, 1259–1268. [Google Scholar] [CrossRef]
- Dao Thi, V.L.; Herbst, K.; Boerner, K.; Meurer, M.; Kremer, L.P.M.; Kirrmaier, D.; Freistaedter, A.; Papagiannidis, D.; Galmozzi, C.; Stanifer, M.L.; et al. A colorimetric RT-LAMP assay and LAMP-sequencing for detecting SARS-CoV-2 RNA in clinical samples. Sci. Transl. Med. 2020, 12, eabc7075. [Google Scholar] [CrossRef]
- Flynn, M.J.; Snitser, O.; Flynn, J.; Green, S.; Yelin, I.; Szwarcwort-Cohen, M.; Kishony, R.; Elowitz, M.B. A simple direct RT-LAMP SARS-CoV-2 saliva diagnostic. medRxiv 2020. [Google Scholar] [CrossRef]
- Xia, S.; Chen, X. Single-copy sensitive, field-deployable, and simultaneous dual-gene detection of SARS-CoV-2 RNA via modified RT-RPA. Cell Discov. 2020, 6, 37. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Yu, L.; Liu, C.; Ye, S.; Chen, W.; Li, D.; Huang, W. One-tube SARS-CoV-2 detection platform based on RT-RPA and CRISPR/Cas12a. J. Transl. Med. 2021, 19, 1–10. [Google Scholar] [CrossRef]
- El Wahed, A.A.; Patel, P.; Maier, M.; Pietsch, C.; Rüster, D.; Böhlken-Fascher, S.; Kissenkötter, J.; Behrmann, O.; Frimpong, M.; Diagne, M.M.; et al. Suitcase Lab for Rapid Detection of SARS-CoV-2 Based on Recombinase Polymerase Amplification Assay. Anal. Chem. 2021, 93, 2627–2634. [Google Scholar] [CrossRef] [PubMed]
- Daher, R.K.; Stewart, G.; Boissinot, M.; Bergeron, M.G. Recombinase polymerase amplification for diagnostic applications. Clin. Chem. 2016, 62, 947–958. [Google Scholar] [CrossRef]
- Chaibun, T.; Puenpa, J.; Ngamdee, T.; Boonapatcharoen, N.; Athamanolap, P.; O’Mullane, A.P.; Vongpunsawad, S.; Poovorawan, Y.; Lee, S.Y.; Lertanantawong, B. Rapid electrochemical detection of coronavirus SARS-CoV-2. Nat. Commun. 2021, 12, 802. [Google Scholar] [CrossRef]
- De Paz, H.D.; Brotons, P.; Muñoz-Almagro, C. Molecular isothermal techniques for combating infectious diseases: Towards low-cost point-of-care diagnostics. Expert Rev. Mol. Diagn. 2014, 14, 827–843. [Google Scholar] [CrossRef] [PubMed]
- Broughton, J.P.; Deng, X.; Yu, G.; Fasching, C.L.; Servellita, V.; Singh, J.; Miao, X.; Streithorst, J.A.; Granados, A.; Sotomayor-Gonzalez, A.; et al. CRISPR–Cas12-based detection of SARS-CoV-2. Nat. Biotechnol. 2020, 38, 870–874. [Google Scholar] [CrossRef] [PubMed]
- Taylor, M. CRISPR-Based COVID Test Is Rapid, Accurate and Costs Less Than $1. Available online: https://www.laboratoryequipment.com/567702-CRISPR-based-COVID-Test-is-Rapid-Accurate-and-Costs-Less-Than-1/ (accessed on 21 March 2021).
- Yip, C.C.; Sridhar, S.; Leung, K.-H.; Ng, A.C.; Chan, K.-H.; Chan, J.F.; Tsang, O.T.; Hung, I.F.; Cheng, V.C.; Yuen, K.-Y.; et al. Development and Evaluation of Novel and Highly Sensitive Single-Tube Nested Real-Time RT-PCR Assays for SARS-CoV-2 Detection. Int. J. Mol. Sci. 2020, 21, 5674. [Google Scholar] [CrossRef]
- Chan, J.F.-W.; Yip, C.C.-Y.; To, K.K.-W.; Tang, T.H.-C.; Wong, S.C.-Y.; Leung, K.-H.; Fung, A.Y.-F.; Ng, A.C.-K.; Zou, Z.; Tsoi, H.-W.; et al. Improved molecular diagnosis of COVID-19 by the novel, highly sensitive and specific COVID-19-RdRp/Hel real-time reverse transcription-PCR assay validated in vitro and with clinical specimens. J. Clin. Microbiol. 2020, 58, 5. [Google Scholar] [CrossRef]
- Brandsma, E.; Verhagen, H.J.M.P.; van de Laar, T.J.W.; Claas, E.C.J.; Cornelissen, M.; van den Akker, E. Rapid, Sensitive, and Specific Severe Acute Respiratory Syndrome Coronavirus 2 Detection: A Multicenter Comparison Between Standard Quantitative Reverse-Transcriptase Polymerase Chain Reaction and CRISPR-Based DETECTR. J. Infect. Dis. 2021, 223, 206–213. [Google Scholar] [CrossRef]
- Mercier-Delarue, S.; Vray, M.; Plantier, J.C.; Maillard, T.; Adjout, Z.; de Olivera, F.; Schnepf, N.; Maylin, S.; Simon, F.; Delaugerre, C. Higher Specificity of Nucleic Acid Sequence-Based Amplification Isothermal Technology than of Real-Time PCR for Quantification of HIV-1 RNA on Dried Blood Spots. J. Clin. Microbiol. 2014, 52, 52–56. [Google Scholar] [CrossRef][Green Version]
- Wu, Q.; Suo, C.; Brown, T.; Wang, T.; Teichmann, S.A.; Bassett, A.R. INSIGHT: A population-scale COVID-19 testing strategy combining point-of-care diagnosis with centralized high-throughput sequencing. Sci. Adv. 2021, 7. [Google Scholar] [CrossRef]
- Kilic, T.; Weissleder, R.; Lee, H. Molecular and Immunological Diagnostic Tests of COVID-19: Current Status and Challenges. iScience 2020, 23, 101406. [Google Scholar] [CrossRef]
- Elisa Test for Covid-19, We Clear 10 Of The Most Common Doubts. Available online: https://lifelength.com/elisa-test-for-covid-19-we-clear-10-of-the-most-common-doubts/ (accessed on 21 March 2021).
- Michel, M.; Bouam, A.; Edouard, S.; Fenollar, F.; Di Pinto, F.; Mège, J.L.; Drancourt, M.; Vitte, J. Evaluating ELISA, Immunofluorescence, and Lateral Flow Assay for SARS-CoV-2 Serologic Assays. Front. Microbiol. 2020, 11, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Edouard, S.; Colson, P.; Melenotte, C.; Di Pinto, F.; Thomas, L.; La Scola, B.; Million, M.; Tissot-Dupont, H.; Gautret, P.; Stein, A.; et al. Evaluating the serological status of COVID-19 patients using an indirect immunofluorescent assay, France. Eur. J. Clin. Microbiol. Infect. Dis. 2021, 40, 361–371. [Google Scholar] [CrossRef] [PubMed]
- Pinto, D.; Park, Y.-J.; Beltramello, M.; Walls, A.C.; Tortorici, M.A.; Bianchi, S.; Jaconi, S.; Culap, K.; Zatta, F.; De Marco, A.; et al. Cross-neutralization of SARS-CoV-2 by a human monoclonal SARS-CoV antibody. Nature 2020, 583, 290–295. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Tang, B.; Wang, Y.; Gu, B. Since January 2020 Elsevier has created a COVID-19 resource centre with free information in English and Mandarin on the novel coronavirus COVID- 19. The COVID-19 resource centre is hosted on Elsevier Connect, the company’s public news and information. Clin. Chim. Acta 2020, 510, 35–46. [Google Scholar] [CrossRef] [PubMed]
- Fenollar, F.; Bouam, A.; Ballouche, M.; Fuster, L.; Prudent, E.; Colson, P.; Tissot-Dupont, H.; Million, M.; Drancourt, M.; Raoult, D.; et al. Evaluation of the panbio COVID-19 rapid antigen detection test device for the screening of patients with COVID-19. J. Clin. Microbiol. 2021, 59, 19–21. [Google Scholar] [CrossRef]
- Eshghifar, N.; Busheri, A.; Shrestha, R.; Beqaj, S. Evaluation of analytical performance of seven rapid antigen detection kits for detection of SARS-CoV-2 virus. Int. J. Gen. Med. 2021, 14, 435–440. [Google Scholar] [CrossRef]
- Abbott PanbioTM: COVID-19 Ag Rapid Test Device 41FK10. Available online: https://www.meteka.com/wp-content/uploads/2020/10/Testanleitung_Abbott.pdf (accessed on 15 March 2021).
- Grossberg, A.N.; Koza, L.A.; Ledreux, A.; Prusmack, C.; Krishnamurthy, H.K.; Jayaraman, V.; Granholm, A.C.; Linseman, D.A. A multiplex chemiluminescent immunoassay for serological profiling of COVID-19-positive symptomatic and asymptomatic patients. Nat. Commun. 2021, 12. [Google Scholar] [CrossRef]
- Mendoza, R.; Silver, M.; Zuretti, A.R.; Christian, M.; Das, B.; Norin, A.J.; Borgen, P.; Libien, J.; Bluth, M.H. Correlation of Automated Chemiluminescent Method with Enzyme-Linked Immunosorbent Assay (ELISA) Antibody Titers in Convalescent COVID-19 Plasma Samples: Development of Rapid, Cost-Effective Semi-Quantitative Diagnostic Methods. J. Blood Med. 2021, 12, 157–164. [Google Scholar] [CrossRef]
- He, Q.; Chong, K.H.; Chng, H.H.; Leung, B.; Ling, A.E.; Wei, T.; Chan, S.W.; Ooi, E.E.; Kwang, J. Development of a Western Blot Assay for Detection of Antibodies against Coronavirus Causing Severe Acute Respiratory Syndrome. Clin. Diagn. Lab. Immunol. 2004, 11, 417–422. [Google Scholar] [CrossRef]
- Wu, J.-L.; Tseng, W.-P.; Lin, C.-H.; Lee, T.-F.; Chung, M.-Y.; Huang, C.-H.; Chen, S.-Y.; Hsueh, P.-R.; Chen, S.-C. Four point-of-care lateral flow immunoassays for diagnosis of COVID-19 and for assessing dynamics of antibody responses to SARS-CoV-2. J. Infect. 2020, 81, 435–442. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez-moncayo, R.; Cedillo-alcantar, D.F.; Guevara-pantoja, P.E.; Chavez-pineda, O.G.; Hernandez-ortiz, J.A.; Amador-hernandez, J.U.; Rojas-velasco, G.; Sanchez-muñoz, F.; Manzur-sandoval, D.; Patino-lopez, L.D.; et al. A high-throughput multiplexed microfluidic device for COVID-19 serology assays. Lab Chip 2021, 21, 93–104. [Google Scholar] [CrossRef] [PubMed]
- Benzigar, M.R.; Bhattacharjee, R.; Baharfar, M.; Liu, G. Current methods for diagnosis of human coronaviruses: Pros and cons. Anal. Bioanal. Chem. 2021, 413, 2311–2330. [Google Scholar] [CrossRef] [PubMed]
- Pohanka, M. Point-of-Care Diagnoses and Assays Based on Lateral Flow Test. Int. J. Anal. Chem. 2021. [Google Scholar] [CrossRef]
- EUA Authorized Serology Test Performance. Available online: https://www.fda.gov/medical-devices/coronavirus-disease-2019-covid-19-emergency-use-authorizations-medical-devices/eua-authorized-serology-test-performance (accessed on 15 March 2021).
- Liang, Y.; Yan, H.; Huang, L.; Zhao, J.; Wang, H.; Kang, M.; Wan, Z.; Shui, J.; Tang, S. A luciferase immunosorbent assay for quantitative detection of IgG antibodies against SARS-CoV-2 nucleoprotein. J. Virol. Methods 2021, 292, 114141. [Google Scholar] [CrossRef]
- Zhang, C.; Zhou, L.; Liu, H.; Zhang, S.; Tian, Y.; Huo, J.; Zhang, Y.; Wei, B.; Xu, D.; Hu, J.; et al. Establishing a high sensitivity detection method for SARS-CoV-2 IgM / IgG and developing a clinical application of this method. Emerg. Microbes Infect. 2020, 9, 2020–2029. [Google Scholar] [CrossRef]
- Celltrion to Launch Both Antigen and Antibody Testing Kits in the U.S. Available online: https://www.pharmasalmanac.com/articles/celltrion-to-launch-both-antigen-and-antibody-testing-kits-in-the-u.s. (accessed on 15 March 2021).
- Kevadiya, B.D.; Machhi, J.; Herskovitz, J.; Oleynikov, M.D.; Blomberg, W.R.; Bajwa, N.; Soni, D.; Das, S.; Hasan, M.; Patel, M.; et al. Diagnostics for SARS-CoV-2 infections. Nat. Mater. 2021, 20, 593–605. [Google Scholar] [CrossRef]
- Imai, K.; Tabata, S.; Ikeda, M.; Noguchi, S.; Kitagawa, Y.; Matuoka, M.; Miyoshi, K.; Tarumoto, N.; Sakai, J.; Ito, T.; et al. Clinical evaluation of an immunochromatographic IgM/IgG antibody assay and chest computed tomography for the diagnosis of COVID-19. J. Clin. Virol. 2020, 128, 104393. [Google Scholar] [CrossRef]
- Ali, J.; Wang, L.; Waseem, H.; Djellabi, R.; Oladoja, N.A.; Pan, G. FeS@rGO nanocomposites as electrocatalysts for enhanced chromium removal and clean energy generation by microbial fuel cell. Chem. Eng. J. 2020, 384, 123335. [Google Scholar] [CrossRef]
- Ali, J.; Ali, N.; Wang, L.; Waseem, H.; Pan, G. Revisiting the mechanistic pathways for bacterial mediated synthesis of noble metal nanoparticles. J. Microbiol. Methods 2019, 159, 18–25. [Google Scholar] [CrossRef] [PubMed]
- Ali, J.; Wang, L.; Waseem, H.; Sharif, H.M.A.; Djellabi, R.; Zhang, C.; Pan, G. Bioelectrochemical recovery of silver from wastewater with sustainable power generation and its reuse for biofouling mitigation. J. Clean. Prod. 2019, 235, 1425–1437. [Google Scholar] [CrossRef]
- Djellabi, R.; Ali, J.; Zhao, X.; Saber, A.N.; Yang, B. CuO NPs incorporated into electron-rich TCTA@PVP photoactive polymer for the photocatalytic oxidation of dyes and bacteria inactivation. J. Water Process Eng. 2020, 36, 101238. [Google Scholar] [CrossRef]
- Nguyen, Q.H.; Kim, M. Il Trends in Analytical Chemistry Nanomaterial-mediated paper-based biosensors for colorimetric pathogen detection. Trends Anal. Chem. 2020, 132, 116038. [Google Scholar] [CrossRef]
- Liu, D.; Ju, C.; Han, C.; Shi, R.; Chen, X.; Duan, D. Ultra-sensitive nanozyme-based chemiluminescence paper test for rapid diagnosis of SARS-CoV-2 infection. bioRxiv 2020. [Google Scholar] [CrossRef]
- Duan, D.; Fan, K.; Zhang, D.; Tan, S.; Liang, M.; Liu, Y.; Zhang, J.; Zhang, P.; Liu, W.; Qiu, X.; et al. Biosensors and Bioelectronics Nanozyme-strip for rapid local diagnosis of Ebola. Biosens. Bioelectron. 2015, 74, 134–141. [Google Scholar] [CrossRef] [PubMed]
- Oh, S.; Kim, J.; Tran, V.T.; Lee, D.K.; Ahmed, S.R.; Hong, J.C.; Lee, J.; Park, E.Y.; Lee, J. Magnetic Nanozyme-Linked Immunosorbent Assay for Ultrasensitive Influenza A Virus Detection. ACS Appl. Mater. Interfaces 2018, 10, 12534–12543. [Google Scholar] [CrossRef] [PubMed]
- Long, L.; Cai, R.; Liu, J.; Wu, X. A Novel Nanoprobe Based on Core–Shell Au@ Pt@ Mesoporous SiO2 Nanozyme With Enhanced Activity and Stability for Mumps Virus Diagnosis. Front. Chem. 2020, 8, 1–9. [Google Scholar] [CrossRef]
- Goodacre, R.; Behera, B.K.; Bansal, V. Ultrasensitive Colorimetric Detection of Murine Norovirus Using NanoZyme Aptasensor. Anal. Chem. 2019, 91, 3270–3276. [Google Scholar] [CrossRef]
- Zhao, Y.; Ding, B.; Xiao, X.; Jiang, F.; Wang, M.; Hou, Z.; Xing, B.; Teng, B.; Cheng, Z.; Ma, P.; et al. Virus-Like Fe3O4@ Bi2S3 Nanozymes with Resistance-Free Apoptotic Hyperthermia-Augmented Nanozymitic Activity for Enhanced Synergetic Cancer Therapy. ACS Appl. Mater. Interfaces 2020, 12, 11320–11328. [Google Scholar] [CrossRef]
- Qin, T.; Ma, R.; Yin, Y.; Miao, X.; Chen, S.; Fan, K.; Xi, J.; Liu, Q. Catalytic inactivation of influenza virus by iron oxide nanozyme. Theranostics 2019, 23, 6920–6935. [Google Scholar] [CrossRef] [PubMed]
- Rana, S.; Nazar, U.; Ali, J.; ul Ain Ali, Q.; Ahmad, N.M.; Sarwar, F.; Waseem, H.; Jamil, S.U.U. Improved antifouling potential of polyether sulfone polymeric membrane containing silver nanoparticles: Self-cleaning membranes. Environ. Technol. 2018, 39, 1413–1421. [Google Scholar] [CrossRef] [PubMed]
- Ruiz-hitzky, E.; Darder, M.; Wicklein, B.; Ruiz-garcia, C.; Martín-sampedro, R.; Real, G.; Aranda, P. Nanotechnology Responses to COVID-19. Adv. Healthc. Mater. 2020, 2, 1–26. [Google Scholar] [CrossRef] [PubMed]
- Chauhan, G.; Madou, M.J.; Kalra, S.; Chopra, V.; Ghosh, D.; Martinez-Chapa, S.O. Nanotechnology for COVID-19: Therapeutics and Vaccine Research. ACS Nano 2020, 14, 7760–7782. [Google Scholar] [CrossRef] [PubMed]
- Niu, X.; Cheng, N.; Ruan, X.; Du, D.; Lin, Y. Review—Nanozyme-Based Immunosensors and Immunoassays: Recent Developments and Future Trends Review. J. Electrochem. Soc. 2020, 167, 037508. [Google Scholar] [CrossRef]
- Bu, T.; Huang, Q.; Yan, L.; Huang, L.; Zhang, M.; Yang, Q. SC. Ultra technically-simple and sensitive detection for Salmonella Enteritidis by immunochromatographic assay based on gold growth. Food Control 2017, 84, 536–543. [Google Scholar] [CrossRef]
- Liu, R.; He, L.; Hu, Y.; Luo, Z.; Zhang, J. A serological aptamer-assisted proximity ligation assay for COVID-19 diagnosis and seeking neutralizing aptamers. Chem. Sci. 2020, 11, 12157–12164. [Google Scholar] [CrossRef]
- Yoo, S.M.; Kim, D.; Lee, S.Y. Aptamer-functionalized localized surface plasmon resonance sensor for the multiplexed detection of different bacterial species. Talanta 2015, 132, 112–117. [Google Scholar] [CrossRef]
- Iqbal, H.; Parra, R. The Emergence of Novel-Coronavirus and its Replication Cycle-An Overview. J. Pure Appl. Microbiol. 2020, 14, 13–16. [Google Scholar] [CrossRef]
- Wang, Q.; Zhang, Y.; Wu, L.; Niu, S.; Song, C.; Zhang, Z.; Lu, G.; Qiao, C.; Hu, Y.; Yuen, K.Y.; et al. Structural and Functional Basis of SARS-CoV-2 Entry by Using Human ACE2 ll ll Article Structural and Functional Basis of SARS-CoV-2 Entry by Using Human ACE2. Cell 2020, 181, 894–904. [Google Scholar] [CrossRef]
- Palestino, G.; García-Silva, I.; González-Ortega, O.; Rosales-Mendoza, S. Can nanotechnology help in the fight against COVID-19? Expert Rev. Anti. Infect. Ther. 2020, 18, 849–864. [Google Scholar] [CrossRef] [PubMed]
- Iannazzo, D.; Pistone, A.; Ferro, S.; De Luca, L.; Monforte, A.M.; Romeo, R.; Buemi, M.R.; Pannecouque, C. Graphene Quantum Dots Based Systems as HIV Inhibitors. Bioconjug. Chem. 2018, 29, 3084–3093. [Google Scholar] [CrossRef] [PubMed]
- Medhi, R.; Srinoi, P.; Ngo, N.; Tran, H.; Lee, T.R. Nanoparticle-Based Strategies to Combat COVID-19. ACS Appl. Nano Mater. 2020, 9, 8557–8580. [Google Scholar] [CrossRef]
- Du, T.; Liang, J.; Dong, N.; Lu, J.; Fu, Y.; Fang, L.; Xiao, S.; Han, H. Glutathione-Capped Ag2S Nanoclusters Inhibit Coronavirus Proliferation through Blockage of Viral RNA Synthesis and Budding. ACS Appl. Mater. Interfaces 2018, 10, 4369–4378. [Google Scholar] [CrossRef]
- Sun, H.; Zhou, Y.; Ren, J.; Qu, X. Carbon Nanozymes: Enzymatic Properties, Catalytic Mechanism, and Applications. Angew. Chemie Int. Ed. 2018, 57, 9224–9237. [Google Scholar] [CrossRef] [PubMed]
- Du, T.; Liang, J.; Dong, N.; Liu, L.; Fang, L.; Xiao, S.; Han, H. Carbon dots as inhibitors of virus by activation of type I interferon response. Carbon 2016, 110, 278–285. [Google Scholar] [CrossRef]
- Cagno, V.; Andreozzi, P.; D’Alicarnasso, M.; Jacob Silva, P.; Mueller, M.; Galloux, M.; Le Goffic, R.; Jones, S.T.; Vallino, M.; Hodek, J.; et al. Broad-spectrum non-toxic antiviral nanoparticles with a virucidal inhibition mechanism. Nat. Mater. 2018, 17, 195–203. [Google Scholar] [CrossRef]
- Mackay, F.; Figgett, W.A.; Saulep, D.; Lepage, M.; Hibbs, M.L. B-cell stage and context-dependent requirements for survival signals from BAFF and the B-cell receptor. Immunol. Rev. 2010, 237, 205–225. [Google Scholar] [CrossRef]
- Bettini, E.; Locci, M. SARS-CoV-2 mRNA Vaccines: Immunological mechanism and beyond. Vaccines 2021, 9, 147. [Google Scholar] [CrossRef]
- Jindal, A.; Sarkar, S.; Alam, A. Nanomaterials-Mediated Immunomodulation for Cancer Therapeutics. Front. Chem. 2021, 9, 46. [Google Scholar] [CrossRef]
- Patterson, D.P.; Rynda-Apple, A.; Harmsen, A.L.; Harmsen, A.G.; Douglas, T. Biomimetic antigenic nanoparticles elicit controlled protective immune response to influenza. ACS Nano 2013, 7, 3036–3044. [Google Scholar] [CrossRef] [PubMed]
- Waseem, H.; Williams, M.R.; Jameel, S.; Hashsham, S.A. Antimicrobial Resistance in the Environment. Water Environ. Res. 2018, 90, 865–884. [Google Scholar] [CrossRef] [PubMed]
- Waseem, H.; Ali, J.; Jamal, A.; Ali, M.I. Potential Dissemination of Antimicrobial Resistance from Small Scale Poultry Slaughterhouses in Pakistan. Appl. Ecol. Environ. Res. 2019, 17, 3049–3063. [Google Scholar] [CrossRef]
- Waseem, H.; Williams, M.R.; Stedtfeld, R.D.; Stedtfeld, T.M.; Shanker, R.; Hashsham, S.A. Chapter 8: Organ-on-chip Systems: An Emerging Platform for Toxicity Screening of Chemicals, Pharmaceuticals, and Nanomaterials. Nanotoxicology: Experimental and Computational Perspectives; Dhawan, A., Anderson, D., Shanker, R., Eds.; Royal Society of Chemistry: Cambridge, UK, 2018; pp. 203–231. [Google Scholar]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).