Bottom-Up Self-Assembly Based on DNA Nanotechnology
Abstract
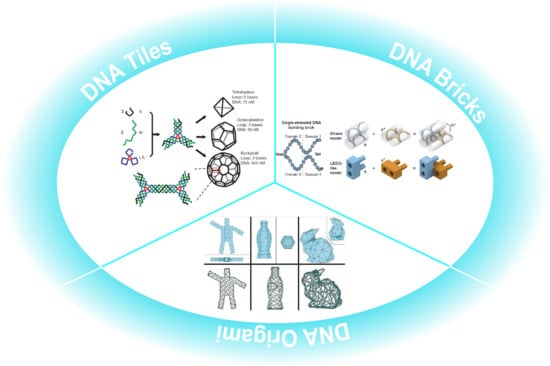
:1. Introduction
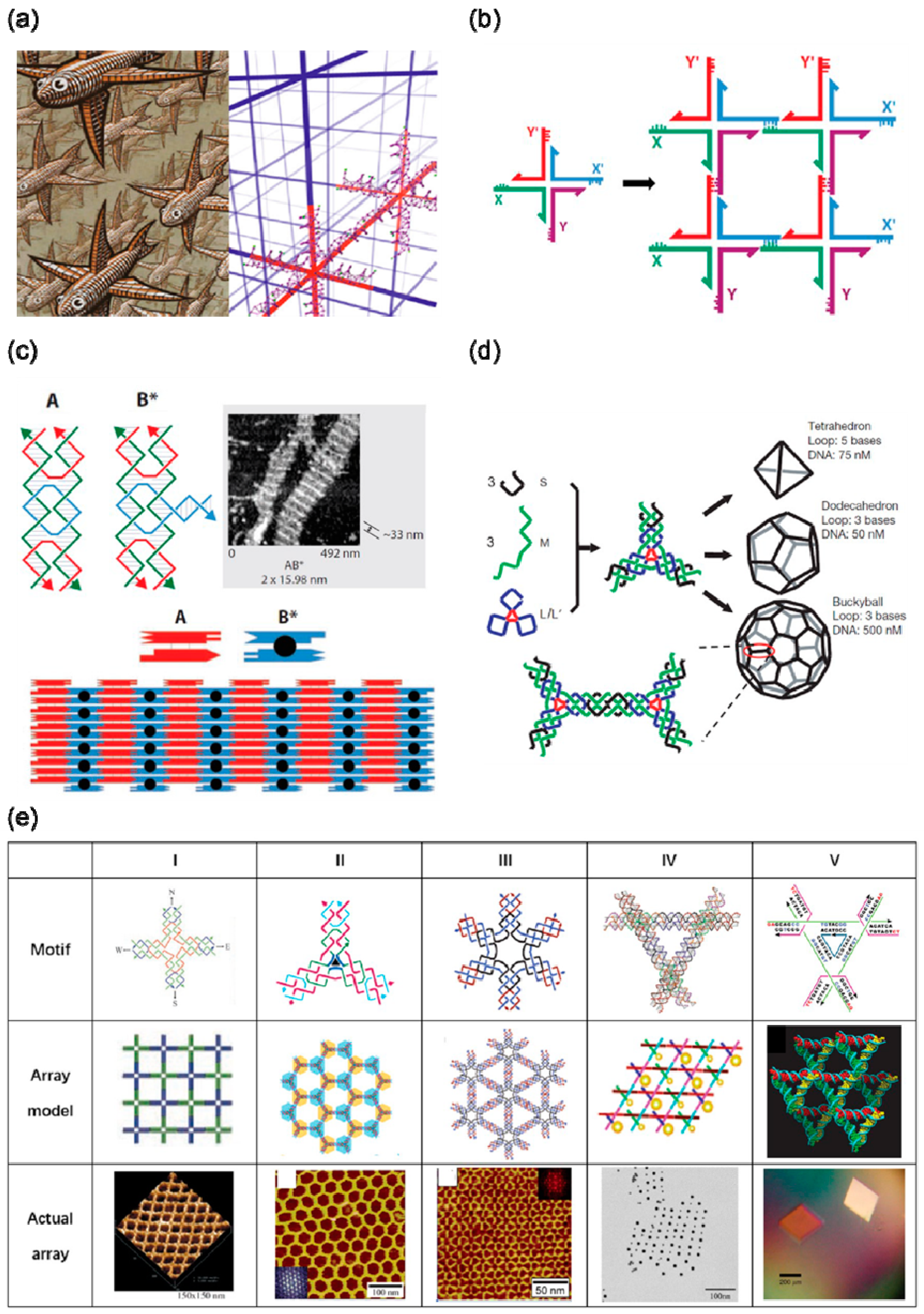
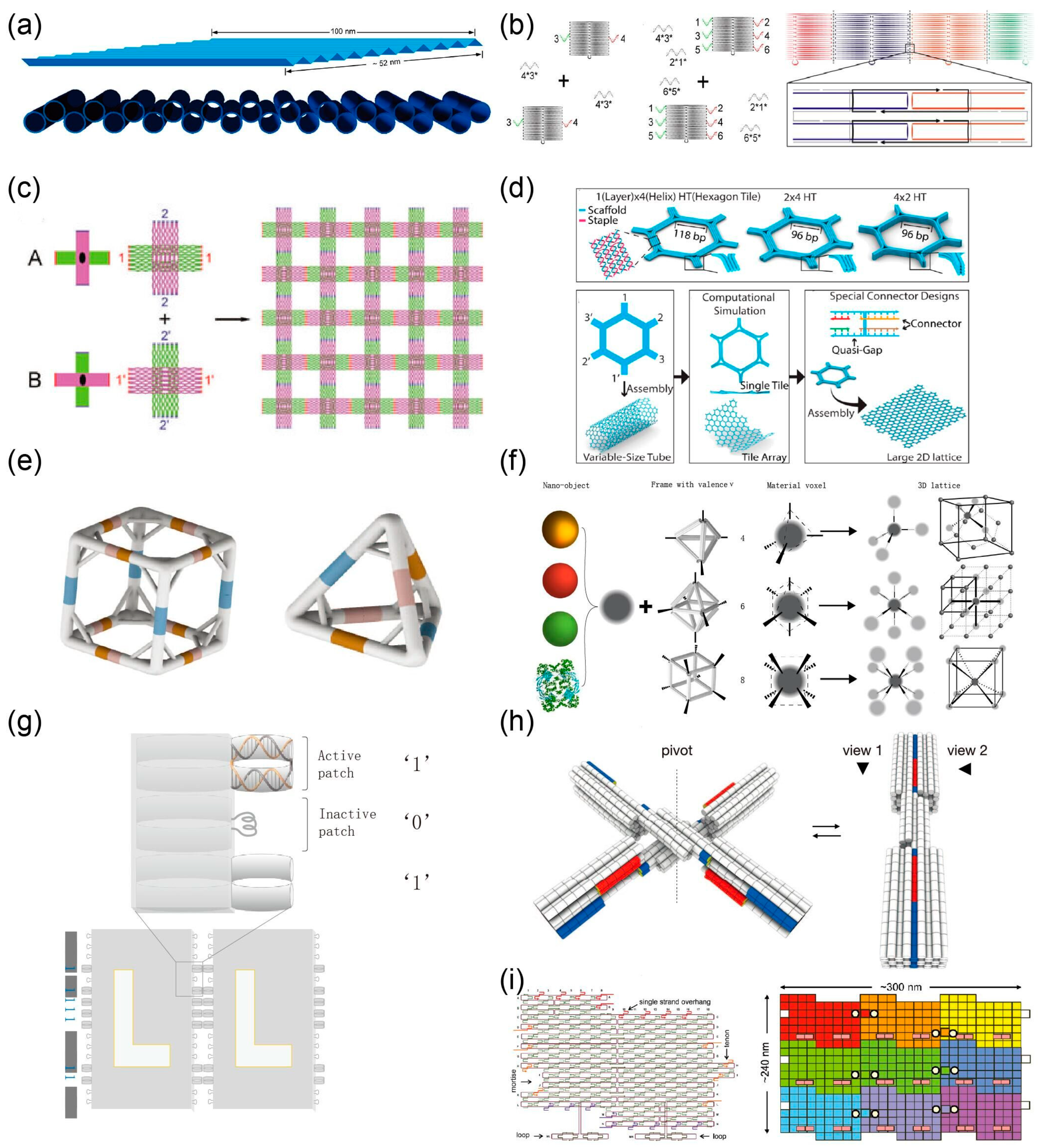
2. Self-Assembly Based on DNA Tile
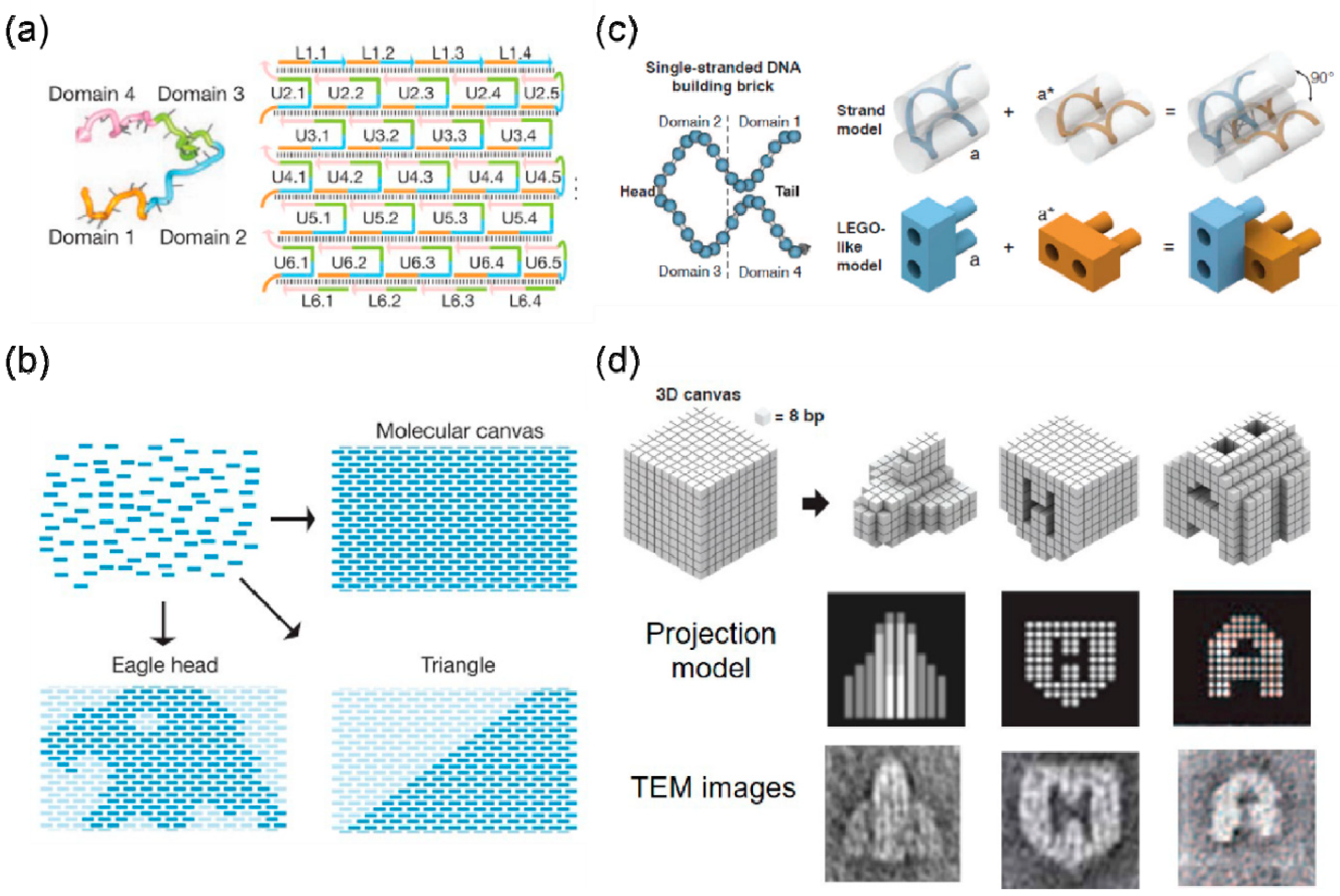
3. Self-Assembly Based on DNA Brick
4. DNA Origami Assembly
4.1. Sticky End Base Pairing
4.2. Blunt Ends Base Stacking
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Seeman, N.C. Nucleic acid junctions and lattices. J. Theory Biol. 1982, 99, 237–247. [Google Scholar] [CrossRef]
- Tian, Y.; Lhermitte, J.R.; Bai, L.; Vo, T.; Xin, H.L.; Li, H.; Li, R.; Fukuto, M.; Yager, K.G.; Kahn, J.S.; et al. Ordered three-dimensional nanomaterials using DNA-prescribed and valence-controlled material voxels. Nat. Mater. 2020, 19, 789–796. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Macfarlane, R.J.; Young, K.L.; Choi, C.H.J.; Hao, L.; Auyeung, E.; Liu, G.; Zhou, X.; Mirkin, C.A. A general approach to DNA-programmable atom equivalents. Nat. Mater. 2013, 12, 741–746. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Lu, F.; Yager, K.G.; van der Lelie, D.; Gang, O. A general strategy for the DNA-mediated self-assembly of functional nanoparticles into heterogeneous systems. Nat. Nanotechnol. 2013, 8, 865–872. [Google Scholar] [CrossRef] [PubMed]
- Hill, H.D.; Macfarlane, R.J.; Senesi, A.J.; Lee, B.; Park, S.Y.; Mirkin, C.A. Controlling the lattice parameters of gold nanoparticle FCC crystals with duplex DNA linkers. Nano Lett. 2008, 8, 2341–2344. [Google Scholar] [CrossRef]
- Winfree, E.; Liu, F.R.; Wenzler, L.A.; Seeman, N.C. Design and self-assembly of two-dimensional DNA crystals. Nature 1998, 394, 539–544. [Google Scholar] [CrossRef]
- Ke, Y.; Ong, L.L.; Shih, W.M.; Yin, P. Three-Dimensional Structures Self-Assembled from DNA Bricks. Science 2012, 338, 1177–1183. [Google Scholar] [CrossRef] [Green Version]
- Rothemund, P.W.K. Folding DNA to create nanoscale shapes and patterns. Nature 2006, 440, 297–302. [Google Scholar] [CrossRef] [Green Version]
- Yan, H.; Park, S.H.; Finkelstein, G.; Reif, J.H.; LaBean, T.H. DNA-templated self-assembly of protein arrays and highly conductive nanowires. Science 2003, 301, 1882–1884. [Google Scholar] [CrossRef]
- Zheng, J.; Birktoft, J.J.; Chen, Y.; Wang, T.; Sha, R.; Constantinou, P.E.; Ginell, S.L.; Mao, C.; Seeman, N.C. From molecular to macroscopic via the rational design of a self-assembled 3D DNA crystal. Nature 2009, 461, 74–77. [Google Scholar] [CrossRef]
- He, Y.; Ye, T.; Su, M.; Zhang, C.; Ribbe, A.E.; Jiang, W.; Mao, C. Hierarchical self-assembly of DNA into symmetric supramolecular polyhedra. Nature 2008, 452, 198–201. [Google Scholar] [CrossRef] [PubMed]
- Ong, L.L.; Hanikel, N.; Yaghi, O.K.; Grun, C.; Strauss, M.T.; Bron, P.; Lai-Kee-Him, J.; Schueder, F.; Wang, B.; Wang, P.; et al. Programmable self-assembly of three-dimensional nanostructures from 10,000 unique components. Nature 2017, 552, 72–77. [Google Scholar] [CrossRef]
- Tian, Y.; Wang, T.; Liu, W.; Xin, H.L.; Li, H.; Ke, Y.; Shih, W.M.; Gang, O. Prescribed nanoparticle cluster architectures and low-dimensional arrays built using octahedral DNA origami frames. Nat. Nanotechnol. 2015, 10, 637–644. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tian, Y.; Zhang, Y.; Wang, T.; Xin, H.L.; Li, H.; Gang, O. Lattice engineering through nanoparticle-DNA frameworks. Nat. Mater. 2016, 15, 654–661. [Google Scholar] [CrossRef]
- Liu, W.; Tagawa, M.; Xin, H.L.; Wang, T.; Emamy, H.; Li, H.; Yager, K.G.; Starr, F.W.; Tkachenko, A.V.; Gang, O. Diamond family of nanoparticle superlattices. Science 2016, 351, 582–586. [Google Scholar] [CrossRef] [Green Version]
- Kallenbach, N.R.; Ma, R.I.; Seeman, N.C. An immobile nucleic acid junction constructed from oligonucleotides. Nature 1983, 305, 829–831. [Google Scholar] [CrossRef]
- Fu, T.-J.; Seeman, N.C. DNA Double-Crossover Molecules. Biochemistry 1993, 32, 3211–3220. [Google Scholar] [CrossRef]
- LaBean, T.H.; Yan, H.; Kopatsch, J.; Liu, F.R.; Winfree, E.; Reif, J.H.; Seeman, N.C. Construction, analysis, ligation, and self-assembly of DNA triple crossover complexes. J. Am. Chem. Soc. 2000, 122, 1848–1860. [Google Scholar] [CrossRef]
- Li, W.; Yang, Y.; Jiang, S.; Yan, H.; Liu, Y. Controlled Nucleation and Growth of DNA Tile Arrays within Prescribed DNA Origami Frames and Their Dynamics. J. Am. Chem. Soc. 2014, 136, 3724–3727. [Google Scholar] [CrossRef]
- Yan, H.; Zhang, X.P.; Shen, Z.Y.; Seeman, N.C. A robust DNA mechanical device controlled by hybridization topology. Nature 2002, 415, 62–65. [Google Scholar] [CrossRef]
- Wang, W.; Chen, S.; An, B.; Huang, K.; Bai, T.; Xu, M.; Bellot, G.; Ke, Y.; Xiang, Y.; Wei, B. Complex wireframe DNA nanostructures from simple building blocks. Nat. Commun. 2019, 10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Seeman, N.C.; Gang, O. Three-dimensional molecular and nanoparticle crystallization by DNA nanotechnology. MRS Bull. 2017, 42, 904–912. [Google Scholar] [CrossRef]
- He, Y.; Chen, Y.; Liu, H.P.; Ribbe, A.E.; Mao, C.D. Self-assembly of hexagonal DNA two-dimensional (2D) arrays. J. Am. Chem. Soc. 2005, 127, 12202–12203. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Tian, Y.; Ribbe, A.E.; Mao, C. Highly connected two-dimensional crystals of DNA six-point-stars. J. Am. Chem. Soc. 2006, 128, 15978–15979. [Google Scholar] [CrossRef] [PubMed]
- Zheng, J.; Constantinou, P.E.; Micheel, C.; Alivisatos, A.P.; Kiehl, R.A.; Seeman, N.C. Two-dimensional nanoparticle arrays show the organizational power of robust DNA motifs. Nano Lett. 2006, 6, 1502–1504. [Google Scholar] [CrossRef] [Green Version]
- Park, S.H.; Yin, P.; Liu, Y.; Reif, J.H.; LaBean, T.H.; Yan, H. Programmable DNA self-assemblies for nanoscale organization of ligands and proteins. Nano Lett. 2005, 5, 729–733. [Google Scholar] [CrossRef]
- Li, H.Y.; Park, S.H.; Reif, J.H.; LaBean, T.H.; Yan, H. DNA-templated self-assembly of protein and nanoparticle linear arrays. J. Am. Chem. Soc. 2004, 126, 418–419. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, J.P.; Liu, Y.; Ke, Y.G.; Yan, H. Periodic square-like gold nanoparticle arrays templated by self-assembled 2D DNA nanogrids on a surface. Nano Lett. 2006, 6, 248–251. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.; Liu, Y.; Yan, H. Complex Archimedean Tiling Self-Assembled from DNA Nanostructures. J. Am. Chem. Soc. 2013, 135, 7458–7461. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.; Wang, M.S.; Deng, Z.X.; Walulu, R.; Mao, C.D. Tensegrity: Construction of rigid DNA triangles with flexible four-arm DNA junctions. J. Am. Chem. Soc. 2004, 126, 2324–2325. [Google Scholar] [CrossRef]
- Zhang, C.; Su, M.; He, Y.; Zhao, X.; Fang, P.-a.; Ribbe, A.E.; Jiang, W.; Mao, C. Conformational flexibility facilitates self-assembly of complex DNA nanostructures. Proc. Natl. Acad. Sci. USA 2008, 105, 10665–10669. [Google Scholar] [CrossRef] [Green Version]
- He, Y.; Tian, Y.; Chen, Y.; Deng, Z.X.; Ribbe, A.E.; Mao, C.D. Sequence symmetry as a tool for designing DNA nanostructures. Angew. Chem. Int. Ed. 2005, 44, 6694–6696. [Google Scholar] [CrossRef]
- Liu, L.; Li, Z.; Li, Y.; Mao, C. Rational Design and Self-Assembly of Two-Dimensional, Dodecagonal DNA Quasicrystals. J. Am. Chem. Soc. 2019, 141, 4248–4251. [Google Scholar] [CrossRef]
- Tian, C.; Li, X.; Liu, Z.; Jiang, W.; Wang, G.; Mao, C. Directed Self-Assembly of DNA Tiles into Complex Nanocages. Angew. Chem. Int. Ed. 2014, 53, 8041–8044. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Liu, Z.; Yu, G.; Jiang, W.; Mao, C. Self-Assembly of Molecule-like Nanoparticle Clusters Directed by DNA Nanocages. J. Am. Chem. Soc. 2015, 137, 4320–4323. [Google Scholar] [CrossRef]
- Liu, Z.; Tian, C.; Yu, J.; Li, Y.; Jiang, W.; Mao, C. Self-Assembly of Responsive Multilayered DNA Nanocages. J. Am. Chem. Soc. 2015, 137, 1730–1733. [Google Scholar] [CrossRef]
- Zhang, C.; Tian, C.; Guo, F.; Liu, Z.; Jiang, W.; Mao, C. DNA-Directed Three-Dimensional Protein Organization. Angew. Chem. Int. Ed. 2012, 51, 3382–3385. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Zhao, Y.; Li, Z.; Wang, Y.; Sha, R.; Seeman, N.C.; Mao, C. Modulating Self-Assembly of DNA Crystals with Rationally Designed Agents. Angew. Chem. Int. Ed. 2018, 57, 16529–16532. [Google Scholar] [CrossRef]
- Ohayon, Y.P.; Hernandez, C.; Chandrasekaran, A.R.; Wang, X.; Abdallah, H.O.; Jong, M.A.; Mohsen, M.G.; Sha, R.; Birktoft, J.J.; Lukeman, P.S.; et al. Designing Higher Resolution Self-Assembled 3D DNA Crystals via Strand Terminus Modifications. ACS Nano 2019, 13, 7957–7965. [Google Scholar] [CrossRef]
- Li, Z.; Liu, L.; Zheng, M.; Zhao, J.; Seeman, N.C.; Mao, C. Making Engineered 3D DNA Crystals Robust. J. Am. Chem. Soc. 2019, 141, 15850–15855. [Google Scholar] [CrossRef]
- Hao, Y.; Kristiansen, M.; Sha, R.; Birktoft, J.J.; Hernandez, C.; Mao, C.; Seeman, N.C. A device that operates within a self-assembled 3D DNA crystal. Nat. Chem. 2017, 9, 824–827. [Google Scholar] [CrossRef] [PubMed]
- Wei, B.; Dai, M.; Yin, P. Complex shapes self-assembled from single-stranded DNA tiles. Nature 2012, 485, 623–626. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ke, Y.; Ong, L.L.; Sun, W.; Song, J.; Dong, M.; Shih, W.M.; Yin, P. DNA brick crystals with prescribed depths. Nat. Chem. 2014, 6, 994–1002. [Google Scholar] [PubMed] [Green Version]
- Qian, L.; Wang, Y.; Zhang, Z.; Zhao, J.; Pan, D.; Zhang, Y.; Liu, Q.; Fan, C.; Hu, J.; He, L. Analogic China map constructed by DNA. Chin. Sci. Bull. 2006, 51, 2973–2976. [Google Scholar] [CrossRef]
- Andersen, E.S.; Dong, M.; Nielsen, M.M.; Jahn, K.; Lind-Thomsen, A.; Mamdouh, W.; Gothelf, K.V.; Besenbacher, F.; Kjems, J. DNA origami design of dolphin-shaped structures with flexible tails. ACS Nano 2008, 2, 1213–1218. [Google Scholar] [CrossRef] [PubMed]
- Douglas, S.M.; Dietz, H.; Liedl, T.; Hogberg, B.; Graf, F.; Shih, W.M. Self-assembly of DNA into nanoscale three-dimensional shapes. Nature 2009, 459, 1154. [Google Scholar] [CrossRef]
- Ke, Y.; Douglas, S.M.; Liu, M.; Sharma, J.; Cheng, A.; Leung, A.; Liu, Y.; Shih, W.M.; Yan, H. Multilayer DNA Origami Packed on a Square Lattice. J. Am. Chem. Soc. 2009, 131, 15903–15908. [Google Scholar] [CrossRef] [Green Version]
- Ke, Y.; Voigt, N.V.; Gothelf, K.V.; Shih, W.M. Multilayer DNA Origami Packed on Hexagonal and Hybrid Lattices. J. Am. Chem. Soc. 2012, 134, 1770–1774. [Google Scholar] [CrossRef] [Green Version]
- Hong, F.; Jiang, S.; Wang, T.; Liu, Y.; Yan, H. 3D Framework DNA Origami with Layered Crossovers. Angew. Chem. Int. Ed. 2016, 55, 12832–12835. [Google Scholar] [CrossRef]
- Dietz, H.; Douglas, S.M.; Shih, W.M. Folding DNA into Twisted and Curved Nanoscale Shapes. Science 2009, 325, 725–730. [Google Scholar] [CrossRef] [Green Version]
- Han, D.; Pal, S.; Nangreave, J.; Deng, Z.; Liu, Y.; Yan, H. DNA Origami with Complex Curvatures in Three-Dimensional Space. Science 2011, 332, 342–346. [Google Scholar] [CrossRef] [Green Version]
- Veneziano, R.; Ratanalert, S.; Zhang, K.; Zhang, F.; Yan, H.; Chiu, W.; Bathe, M. Designer nanoscale DNA assemblies programmed from the top down. Science 2016, 352, 1534. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Benson, E.; Mohammed, A.; Gardell, J.; Masich, S.; Czeizler, E.; Orponen, P.; Hogberg, B. DNA rendering of polyhedral meshes at the nanoscale. Nature 2015, 523, 441–444. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Andersen, E.S.; Dong, M.; Nielsen, M.M.; Jahn, K.; Subramani, R.; Mamdouh, W.; Golas, M.M.; Sander, B.; Stark, H.; Oliveira, C.L.P.; et al. Self-assembly of a nanoscale DNA box with a controllable lid. Nature 2009, 459, 73–76. [Google Scholar] [CrossRef] [Green Version]
- Li, Z.; Liu, M.; Wang, L.; Nangreave, J.; Yan, H.; Liu, Y. Molecular Behavior of DNA Origami in Higher-Order Self-Assembly. J. Am. Chem. Soc. 2010, 132, 13545–13552. [Google Scholar] [CrossRef] [Green Version]
- Jungmann, R.; Scheible, M.; Kuzyk, A.; Pardatscher, G.; Castro, C.E.; Simmel, F.C. DNA origami-based nanoribbons: Assembly, length distribution, and twist. Nanotechnology 2011, 22. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Zhong, H.; Wang, R.; Seeman, N.C. Crystalline Two-Dimensional DNA-Origami Arrays. Angew. Chem. Int. Ed. 2011, 50, 264–267. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, P.; Gaitanaros, S.; Lee, S.; Bathe, M.; Shih, W.M.; Ke, Y. Programming Self-Assembly of DNA Origami Honeycomb Two-Dimensional Lattices and Plasmonic Metamaterials. J. Am. Chem. Soc. 2016, 138, 7733–7740. [Google Scholar] [CrossRef]
- Iinuma, R.; Ke, Y.; Jungmann, R.; Schlichthaerle, T.; Woehrstein, J.B.; Yin, P. Polyhedra Self-Assembled from DNA Tripods and Characterized with 3D DNA-PAINT. Science 2014, 344, 65–69. [Google Scholar] [CrossRef] [Green Version]
- Woo, S.; Rothemund, P.W.K. Programmable molecular recognition based on the geometry of DNA nanostructures. Nat. Chem. 2011, 3, 620–627. [Google Scholar] [CrossRef]
- Gerling, T.; Wagenbauer, K.F.; Neuner, A.M.; Dietz, H. Dynamic DNA devices and assemblies formed by shape-complementary, non-base pairing 3D components. Science 2015, 347, 1446–1452. [Google Scholar] [CrossRef]
- Rajendran, A.; Endo, M.; Katsuda, Y.; Hidaka, K.; Sugiyama, H. Programmed Two-Dimensional Self-Assembly of Multiple DNA Origami Jigsaw Pieces. Acs Nano 2011, 5, 665–671. [Google Scholar] [CrossRef] [PubMed]
- Endo, M.; Sugita, T.; Katsuda, Y.; Hidaka, K.; Sugiyama, H. Programmed-Assembly System Using DNA Jigsaw Pieces. Chemistry 2010, 16, 5362–5368. [Google Scholar] [CrossRef] [PubMed]
- Endo, M.; Sugita, T.; Rajendran, A.; Katsuda, Y.; Emura, T.; Hidaka, K.; Sugiyama, H. Two-dimensional DNA origami assemblies using a four-way connector. Chem. Commun. 2011, 47, 3213–3215. [Google Scholar] [CrossRef]
- Sherman, W.B.; Seeman, N.C. A precisely controlled DNA biped walking device. Nano Lett. 2004, 4, 1801. [Google Scholar] [CrossRef] [Green Version]
- Mao, C.D.; Sun, W.Q.; Shen, Z.Y.; Seeman, N.C. A nanomechanical device based on the B-Z transition of DNA. Nature 1999, 397, 144–146. [Google Scholar] [CrossRef]
- Yurke, B.; Turberfield, A.J.; Mills, A.P.; Simmel, F.C.; Neumann, J.L. A DNA-fuelled molecular machine made of DNA. Nature 2000, 406, 605–608. [Google Scholar] [CrossRef]
- Gu, H.Z.; Chao, J.; Xiao, S.J.; Seeman, N.C. A proximity-based programmable DNA nanoscale assembly line. Nature 2010, 465, 202–205. [Google Scholar] [CrossRef]
- Douglas, S.M.; Bachelet, I.; Church, G.M. A Logic-Gated Nanorobot for Targeted Transport of Molecular Payloads. Science 2012, 335, 831–834. [Google Scholar] [CrossRef] [PubMed]
- Lin, C.; Jungmann, R.; Leifer, A.M.; Li, C.; Levner, D.; Church, G.M.; Shih, W.M.; Yin, P. Submicrometre geometrically encoded fluorescent barcodes self-assembled from DNA. Nat. Chem. 2012, 4, 832–839. [Google Scholar] [CrossRef]
- Praetorius, F.; Kick, B.; Behler, K.L.; Honemann, M.N.; Weuster-Botz, D.; Dietz, H. Biotechnological mass production of DNA origami. Nature 2017, 552, 84–87. [Google Scholar] [CrossRef]
- Kielar, C.; Xin, Y.; Shen, B.; Kostiainen, M.A.; Grundmeier, G.; Linko, V.; Keller, A. On the Stability of DNA Origami Nanostructures in Low-Magnesium Buffers. Angew. Chem. Int. Ed. 2018, 57, 9470–9474. [Google Scholar] [CrossRef]

| Self-Assembly Method | DNA Tiles | DNA Bricks | DNA Origami |
|---|---|---|---|
| Starting Materials | Several Short Single-Stranded Oligonucleotides | A Single Short Oligonucleotide | Hundreds of Short Single-Stranded Oligonucleotides and A Long Oligonucleotide |
| Strategies of Scale-Up | Sticky Ends Base Pairing | Sticky Ends Base Pairing | Sticky Ends Base Pairing and Blunt Ends Base Stacking |
| Resulting Assembly | 2D Array or 3D Polyhedral Frame | Arbitrary and Discrete 3D DNA Structures | 1D, 2D or 3D Arbitrary Structures |
| Assembly Dimension | Up to 1 mm | Approximately 100 nm | Approximately 10 μm |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yan, X.; Huang, S.; Wang, Y.; Tang, Y.; Tian, Y. Bottom-Up Self-Assembly Based on DNA Nanotechnology. Nanomaterials 2020, 10, 2047. https://doi.org/10.3390/nano10102047
Yan X, Huang S, Wang Y, Tang Y, Tian Y. Bottom-Up Self-Assembly Based on DNA Nanotechnology. Nanomaterials. 2020; 10(10):2047. https://doi.org/10.3390/nano10102047
Chicago/Turabian StyleYan, Xuehui, Shujing Huang, Yong Wang, Yuanyuan Tang, and Ye Tian. 2020. "Bottom-Up Self-Assembly Based on DNA Nanotechnology" Nanomaterials 10, no. 10: 2047. https://doi.org/10.3390/nano10102047