Abstract
Cowpea (Vigna unguiculata [L.] Walp) is a vital food security crop in sub-Saharan Africa, including Eswatini. The productivity of the crop is low (<600 kg/ha) in the country due to a lack of improved, locally adapted, and farmer-preferred varieties with biotic and abiotic stress tolerance. The objective of the study was to assess the agronomic performance of newly developed elite cowpea mutants to select best-yielding and adapted pure lines for production and genetic improvement in Eswatini. A total of 30 cowpea genotypes, including 24 newly developed advanced mutant lines, their 3 founder parents and 3 local checks, were profiled for major agronomic traits in two selected sites (Lowveld Experiment and Malkerns Research Stations) using a 6 × 5 alpha lattice design with three replications. A combined analysis of variance revealed that the genotype x location interaction effects were significant (p < 0.05) for germination percentage (DG %), days to flowering (DTF), days to maturity (DMT), number of pods per plant (NPP), pod length (PDL), number of seeds per pod (NSP), hundred seed weight (HSW), and grain yield (GYD). Elite mutant genotypes, including NKL9P7, BRR4P11, SHR9P5, and NKL9P7-2 exhibited higher grain yields at 3158.8 kg/ha, 2651.6 kg/ha, 2627.5 kg/ha, and 2255.8 kg/ha in that order. The highest-yielding mutant, NKL9P7, produced 70%, 61%, and 54% more grain yield than the check varieties Mtilane, Black Eye, and Accession 792, respectively. Furthermore, the selected genotypes displayed promising yield components such as better PDL (varying from 13.1 to 26.3 cm), NPP (15.9 to 26.8), and NSP (9.8 to 16.2). Grain yield had significant positive correlations (p < 0.05) with DG %, NSP, and NPP. The principal component analysis (PCA) revealed that 81.5% of the total genotypic variation was attributable to the assessed quantitative traits. Principal component (PC) 1 accounted for 48.6%, while PC 2 and PC 3 contributed 18.9% and 14% of the overall variation, respectively. Key traits correlated with PC1 were NPP with a loading score of 0.91, NSP (0.83), PDL (0.73), GYD (0.68), HSW (0.58), DMT (−0.60), and DTF (−0.43) in a desirable direction. In conclusion, genotypes NKL9P7, BRR4P11, SHR9P5, NKL9P7-2, Bira, SHR3P4, and SHR2P7 were identified as complementary parents with relatively best yields and local adaptation, making them ideal selections for direct production or breeding. The following traits, NPP, NSP, PDL, GYD, and HSW, offered unique opportunities for genotype selection in the cowpea breeding program in Eswatini.
Keywords:
cowpea; Eswatini; genetic diversity; mutant varieties; phenotypic traits; yield components 1. Introduction
Cowpea (Vigna unguiculata [L.] Walp., 2 n = 2 x = 22) belongs to the family Fabaceae and order Fabales. It is a drought-adapted and protein-rich legume crop adapted to grow in harsh and dry regions, including Eswatini. Cowpea is a predominantly self-pollinating crop with a low genetic diversity level, limiting genetic variation for breeding with traditional selection [,]. Based on environmental adaptation, cowpea is distinguished into two main subspecies: the common cowpea or African cowpea (V. unguiculata ssp. unguiculata) and the asparagus bean or “yardlong” bean (V. unguiculata ssp. sesquipedalis) [,]. The subspecies unguiculata is predominately grown for grain production in sub-Saharan Africa (SSA) [].
Cowpea is an economical source of plant-based protein and minerals []. The dried grains and succulent leaves are excellent sources of protein, ranging from 21 to 33% and 27 to 43%, respectively [,,]. Agronomic significance of cowpea in the dry zones of Africa is attributable to its inherent drought tolerance and adaptation to grow in marginal and infertile soils where other crops fail [,,]. In Africa, cowpea is regarded as a dual-purpose crop serving as human food and livestock fodder []. Promiscuous cowpea types form root nodules and fix atmospheric nitrogen with a symbiotic relationship with rhizobium bacteria, making it a key component crop in traditional intercropping systems with cereals [].
Cowpea is widely produced by smallholder farmers in Southern Africa, including Eswatini []. The sub-region is regarded as a center of the genetic diversity of cowpea []. Omomowo and Babalola [] reported that the average cowpea yield in SSA is about 600 kg/ha, while the potential yield is estimated to be between 1500 to 3000 kg/ha. The low yields of cowpea in SSA are attributable to the use of unimproved landraces, poor agronomic management practices, recurrent drought, poor soil fertility, and biotic constraints (e.g., insect pests, parasitic weeds, and various pathogens). Landrace crop genetic resources are not artificially selected and exhibit low yield potential compared to modern improved varieties. However, farmers prefer landraces because of their adaptability to local conditions, eating quality, better cooking properties, and amenability to making specific traditional dishes. Landraces are low yielders but genetically heterogeneous in flowering, maturity, agronomic and quality traits, as well as in disease and pest resistance, making them gene donors for modern cultivars lacking essential traits.
Cowpea is a key food security crop in Eswatini after common beans (Phaseolus vulgaries L.) and Bambara groundnut (Vigna subterranea L.). It is a vital source of protein and income for smallholder farmers in the country. However, there has been limited research on cowpea breeding and genetic improvement in the country. In the past, researchers focused solely on evaluating elite lines received from the IITA/Nigeria for yield and adaptability. For instance, varieties designated as IT-16, IT-18, IT-04K-321-2, IT-97K-390-2, and IT-99K-494-4 were released in 2015 because of their tolerance to drought, leaf spot, and bacterial diseases []. However, the released improved varieties are obsolete and were not widely adopted, and many farmers continue growing their landraces, which are low-yielding. Hence, new and improved varieties are yet to be developed and deployed in the country to enhance food security and economic gains from cowpea production.
Eswatini lacks a dedicated cowpea improvement program, and no systematic agronomic and genetic improvement efforts have been undertaken due to limited capacity and research support. New and starter genetic resources must be acquired from other national and international breeding programs that have mainstream cowpea improvement. For instance, cowpea is a major food crop in Namibia. The Ministry of Agriculture, Water, and Land Reform (MAWLR)/Namibia, in partnership with the International Atomic Energy Agency (IAEA), has developed and released improved cowpea mutant varieties []. Eswatini has a drought-prone environment and shares similar agro-ecological conditions with Namibia. Hence, the cowpea varieties developed and adopted in Namibia are useful genetic resources for evaluating and selecting locally adapted, agronomically superior elite mutant lines with market-preferred traits. The cowpea mutant varieties developed in Namibia recorded promising grain yield, early flowering, shorter days to maturity, increased number of seeds per pod, and reduced plant height [], and they can be adapted for large-scale production, particularly in the drought-prone lowveld of Eswatini where maize and other major cereals do not thrive. The most essential yield and yield components of cowpea are early flowering and maturity, longer and straighter pods, increased pods per plant and seeds per pod, heavier seed weight, and high grain yield [,]. In an attempt to select genetically diverse, drought-adapted, and superior cowpea genotypes, diverse entries were acquired from MAWLR/Namibia. There is a need to profile the new genotypes and farmers’ varieties across production environments in Eswatini using farmers’ and market-preferred attributes. This study aimed to assess the agronomic performance of the newly developed cowpea mutants to select the best-yielding and locally adapted pure lines for production and breeding in Eswatini.
2. Materials and Methods
2.1. Plant Material
A total of 30 cowpea genotypes were used in this study (Table 1). Of these, 24 elite mutant lines (M6 generation) were developed through mutation induction using Cobalt 60 gamma rays and 3 founder parents were sourced from the Namibian Ministry of Agriculture, Water, and Land Reform of the Directorate of Agricultural Research and Development. Further, two landraces and one released local variety received from the National Plant Genetic Resources Center (NPGRC)/Eswatini were included in the study as a comparative control. The elite mutant lines exhibit high yield potential and resilience to drought stress and insect pests []. The local landraces are valued for their cooking and eating quality, adaptability, and yield gains under extreme heat and drought stress despite their low yield levels.

Table 1.
Details of the cowpea genotypes used in this study.
2.2. The Study Areas
The experiments were carried out under field conditions between February and May 2023 at two locations, namely the Lowveld Experiment Station (LES) and Malkerns Research Station (MRS). LES and MRS are the main research sites of the Department of Agricultural Research and Specialists Services (DARSS) of the Ministry of Agriculture/Eswatini. The LES is located in the Lubombo Region (−26°, 57.95° S, 31.52° E; 110 m above sea level), with mean daily temperatures ranging between 26.4 and 30.5 °C and an annual total rainfall of 450 mL. The soils are classified as vertisolic [], which are dark brown clay, neutral, and fertile, with prominent swelling and shrinking during the wetting and drying cycles, in that order. The MRS is situated in the Manzini Region (−26.57°, 31.17° S, 31.11° E, altitude is 740 m above sea level), with mean daily temperatures varying between 7.3 and 26.6 °C, and receives an annual total rainfall of 800 to 1460 mL. The soil at the MRS site is characterized by deep red loam, ferralsols, or the Mdutjane soil series and are highly weathered with an oxic horizon [].
2.3. Experimental Design and Procedures
Genotypes were evaluated and compared at both sites using a 6 × 5 alpha lattice design with three replications. Each genotype was sown in a plot with two rows 2 m in length. The area of the experimental unit was 1.4 m2. The inter-row and intra-row spacing was 75 cm and 20 cm, respectively. Two seeds were sown per station at a depth of 2 cm and later thinned to one plant two weeks after emergence. Basal fertilizer with an N:P:K ratio of 2:3:2 was used at a rate of 200 kg ha− 1 before planting as per local recommendations []. Weed control was performed by hand hoes, and insect pests were controlled using the insecticide Decis®, Bayer AG, Lyon, France. The genotypes were field evaluated under rain-fed conditions at both locations. To initiate germination, only supplementary irrigation was provided at th Lowveld Experiment Station because of very high air temperatures during planting.
2.4. Data Collection
Data on economic and agronomic traits were assembled using the descriptors of the International Board for Plant Genetic Resources []. Table 2 presents the list of traits and details of the data collected. Grain yield was expressed in kg ha−1 using the following formula:
where mc represents moisture content at harvest, which was 14% recommended for legumes [], and 10,000 denotes one hectare.

Table 2.
Traits evaluated with descriptions and units.
2.5. Data Analysis
The data collected were calculated using the analysis of variance (ANOVA) with the alpha lattice procedure in the Statistical Analysis System (SAS) version 9.4 []. A combined analysis of variance was conducted across locations after homogeneity of variance using the Levene’s test procedure [] and the SAS software version 9.4 program []. Mean values of the test genotypes were compared and separated using the Fischer’s Unprotected Least Significant Difference (LSD) at p ≤ 0.05 significance level.
2.5.1. Estimation of Variance Components
The phenotypic and genotypic variance components of the assessed agronomic trait were calculated from the results of a combined analysis of variance, following the method described by Rahimi and Hernandez []. The sampled genotypes and locations were fixed factors. Variance on genotypic, genotype by location interaction, and phenotypic effects were computed from the expected mean squares of the combined ANOVA as follows:
where σ2p = phenotypic variance, σ2g = genotypic variance, σ2e = environmental variance, and σ2gl = genotype by location interaction variance.
where σ2g = genotypic variance, σ2gl = variance due to genotype by location interaction, msg = the mean square of the genotype, mse = the mean square of error, l = number of locations, and r = number of replications.
σ2p = σ2g + σ2e + σ2gl
2.5.2. Broad-Sense Heritability (H2) and Genetic Advance
The H2 of each trait was estimated according to Alvarado et al. [], using
2.5.3. Estimation of Coefficient of Variability
Coefficients of variation for the genotype (GCV) and phenotype (PCV) were computed following [] as follows:
where σ2g = genotypic variance, σ2p = phenotypic variance, and = grand mean.
Genetic advance (GA) and the genetic advance as a percent of the mean (GAM) were computed following Allard [] as follows:
where K is 2.06 at 5% selection intensity.
GA = K H2 σp
GA (%) denotes a genetic advance as a percent of the mean; GA = genetic advance; = grand mean. GA% was categorized into low (< 10%), moderate (10 to 20%), and high (> 20%) based on Singh and Chaudhary [].
2.5.4. Trait Correlation
Correlation coefficients (r), based on Spearman Rank correlations, were computed using SPSS statistics software 29.0 program [] to determine the magnitude of associations between traits. Furthermore, a correlation matrix between key agronomic traits in cowpea was performed with R Software [].
2.5.5. Principal Component Analysis (PCA) and Genotype Clustering
PCA was performed using R Core Team, version 4.4.3 (2024) [] to identify the most influential principal components (PCs) and their associations with traits. PCs with Eigen values ≥ 1.0 were retained to ascribe the variation in the assessed traits among the test genotypes. Further, PCA biplots were computed using R software version 4.5.1 [] to decipher genotype and trait associations. JMP software Version 17 SAS [] was used for cluster analysis to cluster the test genotypes based on their genetic distance.
3. Results
3.1. Analysis of Variance
The combined ANOVA showed significant (p < 0.05) genotype x location interaction effects for GYD, NSP, DTF, and DG % (Table 3). Significant differences were recorded for DG, DMT, PDL, NPP, NSP, and GYD amongst the test genotypes. Non-significant differences were computed for DTF, PDL, and HSW across locations.

Table 3.
Analysis of variance among 30 cowpea genotypes evaluated for nine agronomic traits across two locations in Eswatini.
3.2. Mean Performance of the Test Genotypes
The mean germination percentage for test genotypes was 68% at LES and 52% at MRS. DTF ranged from 42 days (for the genotype Mtilane) to 59 days (Nakare) (Table 4). The mean DTM of the test genotypes was 68 days. The genotype Mtilane was relatively early, maturing with 59 days. Genotypes NKR4P5 (60 days) and BRR4P11 (61 days) were also identified for their early maturity. The number of pods per plant (NPP) ranged from 16 to 28. Genotypes NKR4P5, NKL9P7, BRR4P11, NKR1P12, and SHR9P5 had the highest NPP (>25 pods plant−1). Pod length significantly varied amongst genotypes. The longest pods were recorded for genotypes SHR3P4-4 (22 cm) and SHR9P5 (21 cm). The genotypes that recorded more seeds per pod were BRR11P2, NKL9P7, NKL9P7-2, and NKR193, with 16.2, 15.6, and 14.9 seeds per pod, respectively. Higher hundred seed weight was recorded for the genotypes NKR8P9 (25.5 g/100 seed), NKR10P5 (24.8 g/100 seed), and NKR9P9 (24.7 g/100 seed). Significant genotype differences existed for GYD, ranging from 171.6 kg ha−1 (for genotype SHL7P1) to 3158.8 kg ha−1 (NKL9P7). The overall mean GYD of test genotypes was 1485.6 kg ha−1. Genotypes BRR4P11 (with mean GYD of 2651.6 kg ha−1), SHR9P5 (2651.6 kg ha−1), NKL9P7-2 (2627.5 kg ha−1), Mtilane (2225.7 kg ha−1), and Bira (2178.8 kg ha−1) were among the top-yielding selections. The founder parent genotype Nakare remained vegetative and did not flower until it dried up. The following test genotypes were selected: NKL9P7, BRR4P11, SHR9P5, NKL9P7-2, Bira, SHR3P4, SHL2P7, NKR1P12, and NKR4P5 based on desirable and contrasting quantitative agronomic traits. The selected genotypes are recommended for production or to develop new populations.

Table 4.
Mean values of grain yield and related traits of 30 cowpea genotypes based on grain yield (kg/ha) evaluated at two locations in Eswatini.
3.3. Variance Components, Broad-Sense Heritability and Genetic Advance
The variance components, phenotypic coefficient of variation (PCV) and genotypic coefficient of variation (GCV), heritability, and genetic advance estimates of the agronomic traits from the two study sites are presented in Table 5. The GCV values varied between 1.4 and 43.65%, while the PCV varied from 11.85 to 88.93%. Larger variances between GCV and PCV values were recorded for all studied traits. Low heritability (≤30) estimates were recorded for the number of days to flowering, number of days to maturity, number of seeds per pod, and hundred seed weight. Moderate heritability values (30–60%) were recorded for pods per plant and grain yield, while higher heritability (>60%) values were observed for pod length. These can be used for further selection and breeding gains. The genotypic variance accounted for ≥30% of the total variation for grain yield. Genetic advance ranged from 0 to 56.1% with the lowest estimates computed for days to flowering (0.34%), days to maturity (2.51%), hundred seed weight (5.1%), and seeds per pod (17.1%). High GA% were calculated for grain yield (56.1%), pod length (48.5%), and number of pods per plant (35.7%), respectively.

Table 5.
Genetic parameters for yield and yield components in 30 cowpea genotypes assessed across two locations in Eswatini.
3.4. Trait Correlations
Figure 1 summarizes the correlation coefficients among the assessed traits. Grain yield exhibited positive and significant (p < 0.05) correlations with NSP (r = 0.60), NPP (r = 0.66) and DG % (r = 0.50). Significant (p < 0.05) and positive correlations were noted for NPP and NSP (r = 0.78), NPP and PDL (0.50), PDL and HSW (0.63), DMT and DTF (0.47), as well as NPP and HSW (0.39). DMT and PDL exhibited a strong negative correlation (−0.59).
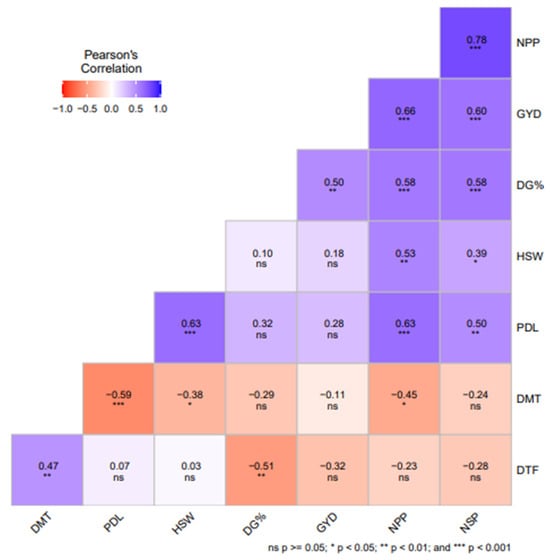
Figure 1.
Correlation coefficients of grain yield and associated traits among 30 cowpea genotypes evaluated in two sites in Eswatini. Note: The color variation shows the extent of correlations; deep blue shows strong and positive correlations, while deep red shows strong negative correlation. *, **, and *** denote significance associations at 0.05, 0.01, and <0.001 probability values, respectively. ns = Not significant; DG % = Percent seed germination; DTF = Days to 50% flowering; DMT = Days to maturity; PDL = Pod length (cm); NPP = Number of Pods per plant; NSP = Number of Seeds per pod; HSW = Hundred seed weight (g/100 seed); GYD = Grain yield (kg ha−1).
3.5. Principal Component (PC) and Biplot Analyses
The first three PCs accounted for 81.5% of the total variation in the studied traits (Table 6). The first three Eigen values ranged between 3.89 and 1.12, signifying greater geneotype variation among the traits (Table 6). The first principal component (PC1) accounted for 48.6%, while PC2 and PC3 contributed to 18.9 and 14% of the total variation, respectively. The highest contributing traits correlated with PC1 were NPP (with a loading score of 0.91), NSP (0.83), PDL (0.73), GYD (0.68), and HSW (0.58). The loadings on PC2 were mostly contributed by DTF (0.66), PDL (0.56), and HSW (0.63). The largest contribution correlated with PC3 were observed from DMT (0.73) and DTF (0.52).

Table 6.
Eigen values, explained variances, and loading scores of eight traits in 30 cowpea genotypes assessed across two locations in Eswatini.
The principal component biplot illustrated the correlation between the assessed traits and genotypes (Figure 2). The narrow angles between vectors (for HSW and PDL, NPP and NSP, as well as GYD, NSP, and DG %) resolved strong positive correlations. Most assesed genotypes were scattered with positive trends in the first principal component, with genotypes NKR1P12, SHR3P4, NKR4P5, BRR11P11, BRR4P11, Black eye, Accession 792, SHR9P5, BEE11P2, SHL2P4, Bira, NKL9P7, Mtilane, and SHR10P12 outperforming in grain yield and yield components.
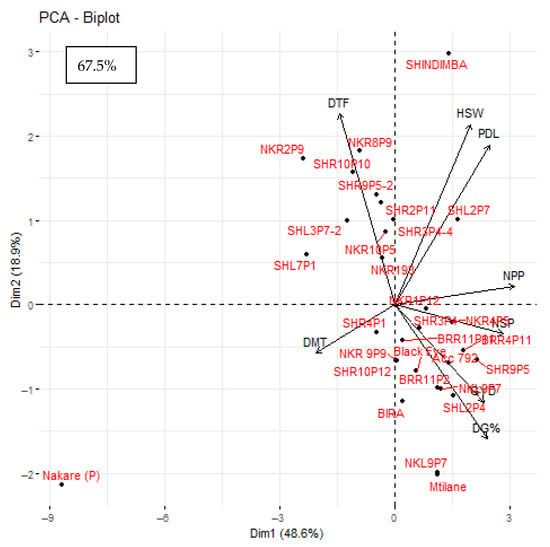
Figure 2.
Biplot depicting the association of eight traits in 30 cowpea genotypes assessed in two locations in Eswatini. Note: DG % = Percent seed germination; DTF = Days to 50% flowering; DMT = Days to maturity; PDL = Pod length (cm); NPP = Number of Pods per plant; NSP = Number of Seeds per pod; HSW = Hundred seed weight (grams/100 seed); GYD = Grain yield (kg ha−1). PC-1 and PC-2: principal component 1 and principal component 2, respectively.
3.6. Cluster Analysis for Agronomic Traits
Phenotypic diversity and genotype grouping were further assessed using cluster analysis. The analysis revealed the test genotypes into two distinct major clusters, I and II. Cluster II had two sub-clusters designated as II-a and II-b (Figure 3). Cluster II consisted of 29 genotypes, allocating 18 genotypes in II-a and 11 genotypes in II-b. Sub-cluster II-a constituted all three check genotypes (Accession 792, Mtilane and Black Eye) acquired from the genebank of Eswatini and two founder mutant parent lines (Bira and Shindimba) from MAWLR/Namibia. Also, elite mutants such as BRR11P2, BRR11P11, NKR9P9, SHR10P12, NKR1P12, NKR4P5, BRR4P11, SHR9P5, SHR3P4, and NKL9P7 were grouped in II-a. Sub-cluster II-b constituted mutant genotypes only (NKR10P5, NKR8P9, SHR10P10, SHR2P11, NKR2P9, SHL3P7-2, NKR193, SHR3P4-4, SHR9P5-2, SHL7P1, and SHR4P1). Interestingly, Cluster I only constituted one genotype, Nakare.
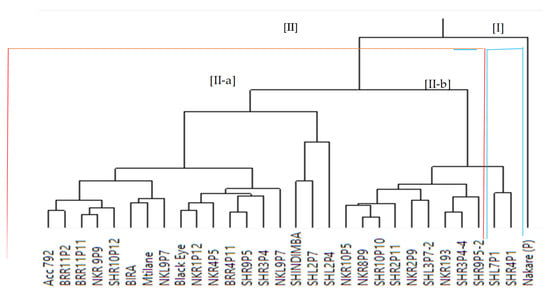
Figure 3.
Hierarchical clustering using Ward’s method showing phenotypic groupings of 30 cowpea genotypes assessed based on yield and yield components. Note: I and II indicate the two main clusters, while II-a and II-b are sub-clusters of cluster II.
4. Discussion
4.1. Agronomic Performance of Elite Cowpea Mutants
Cowpea is a key legume crop for both human consumption and livestock feed, and it is widely grown in semi-arid regions globally, including Eswatini. However, the average yield potential of the crop in Eswatini remains low (600 kg/ha) compared to reports of other cowpea-producing countries such as Nigeria (1500 to 3000 kg/ha), Ethiopia (2200–3200 kg/ha), and Egypt (2666 kg/ha) []. This highlights the yield gap and potential for improvement in Eswatini and elsewhere. The low yield is attributed to the unavailability of improved seeds and several biotic stresses (e.g., bacterial and viral diseases, root-knot nematodes, parasitic weeds, insect pests, notably aphids, and abiotic stresses, such as severe drought, salinity, heat stress, and low soil fertility) [].
The current study profiled 30 cowpea genotypes, including 24 newly developed advanced mutant lines, their 3 founder parents and 3 local checks for major agronomic traits in two locations in Eswatini. There were significant differences among the genotypes for the assessed agronomic traits, which indicated the existence of a considerable variation for the selection of adapted and stable yielding lines. Viswanatha and Yogeesh [] reported related findings for cowpea genotypes evaluated in India.
In this study, the LES site (with a mean yield of 1537 kg/ha) is a high-yielding environment relative to MRS site (1433 kg/ha) probably due to favorable conditions such as better soil fertility. The MRS site is a relatively wet midveld and susceptible to high nutrient leaching and soil acidity. Cowpea grows vegetatively under heavy rain, as recorded at the MRS experiment in 2023. Developmental stages and biomass production exhibit phenotypic plasticity in response to soil variation and climatic factors [,]. In this study, test genotypes NKL9P7 (with grain yield of 3158.8 kg/ha), BRR4P11 (2651.6 kg/ha), SHR9P5 (2627.5 kg/ha), and NKL9P7-2 (2255.8 kg/ha) were top yielders, making them ideal selections for production in Eswatini. The lowest yield was obtained from SHL7P1 (171.6 kg/ha). Quantitative traits are conditioned by polygenes, each with a minor effect []. Consequently, genotype selections across multiple growing environments are essential to ensure selection gains and for sustainable production [,,].
The cowpea genotypes evaluated in this study showed significant variation in days to flowering and maturity. Early maturity is associated with drought escape []; consequently, early-maturing genotypes are attractive for breeders and farmers because they fit the short rainy periods, and escape certain pests and diseases to guarantee the expected economic yield or breeding opportunities. According to Angelova and Stoilova [] as well as Hall [], early maturity in cowpea could help escape terminal drought and reduce the incidence of pests and diseases, which arise in the later stages of the cropping season. Horn et al. [] reported that short flowering and maturity periods are farmer-preferred traits of cowpea in southern Africa, enabling them to avoid drought-induced hunger. Dugje et al. [] classified cowpea maturity into extra-early (60 days), early (61–80 days), and late (>80 days) groups. Thus, genotypes NKR4P5 and Mtilane were extra-early maturing, while other genotypes were early maturing, except for Nakare, which was late maturing (84 days). Ten of the earliest maturing genotypes evaluated in this study were the highest-yielding (Table 4), making them ideal candidate genotypes as a food source for drought-prone areas. NKL9P7, NKL9P7-2, BRR11P2, and Black eye expressed the highest number of seeds per pod. This could be attributed to increased pod length by these genotypes, agreeing with Nkoana et al. []. Seed weight, closely linked to seed size, is recommended as an indirect selection for enhanced grain yield response in cowpea. Horn et al. [] reported a high yield potential for cowpea genotypes reaching 3 t/ha. In the present study, genotype NKL9P7 yielded 3158.8 kg/ha. Also, other mutant genotypes (e.g., BRR4P11, SHR9P5 and NKL9P7-2) recorded more than 2 t/ha, which can be attributed to genetic gains from effective mutation induction.
4.2. Variance Components and Heritability of Quantitative Agronomic Traits
The results from this study suggest that the evaluated genotypes displayed adequate variation for genotype selection with better DMT, PDL, NPP, NSP, HSW, and GYD. Other studies [,,,,] reported high PCV and GCV for cowpea grain yield, days to maturity, pod length, number of pods per plant, number of seeds per pod, and hundred seed weight. Owusu et al. [] reported high values for the number of pods per plant, number of seeds per pod, and hundred seed weight. In the present study, traits such as days to maturity, number of pods per plant, number of seeds per pod, and hundred seed weight were assessed, and PCV was higher than GCV values. The high PCV values suggest that environmental factors rather than genetic factors influence the observed variation. Low-to-moderate variability (GVC) was observed for DTF and DMT. Heritability refers to the proportion of the phenotypic variance attributed to genetic variance. The moderate broad-sense heritability values for grain yield (39%) and the number of pods per plant (49%), indicating that genetic factors accounted for a moderate proportion of the observed phenotypic variation. This suggests that environmental effects and genotype x environment interactions considerably influenced yield expression in the test populations. Direct selection of theses traits can be less effective in early generations. In this regard, indirect selection based on traits with higher heritability or marker-assisted selection could be more effective. The moderate heritability value for grain yield corroborates with the findings of Mbuma et al. [] and Owusu et al. []. The high heritability value for pod length (almost at 100%) suggests that pod length is predominantly controlled by genetic factors, making it a reliable trait for selection in a breeding program. Ajayi et al. [] reported high broad-sense heritability for pod length of 98.1%. Genetic advance (GA) is the degree of gain obtained in a trait under a given selection pressure. High GA was recorded for pod length (48.5%), number of pods per plant (35.7%), and grain yield (56.1%) (Table 5). Related results were reported by Manju and Jayamani [], Zaki and Radwan [], as well as Meenatchi et al. []. Pod length, number of pods per plant, and grain yield in cowpea are ideal traits for selection in breeding programs aimed at improving grain yield. Selection response is related to the magnitude of GCV, broad-sense heritability, and genetic advance []. However, it is imperative to note that high heritability alone is insufficient for efficient selection in advanced generations unless it is accompanied by substantial genetic advance []. Therefore, heritability estimates coupled with high genetic advance observed for most yield components (PDL–48.5%, NPP–35.7%, and GYD–56.1%) indicate that promising and elite mutant lines could be selected for further evaluation, selection, and release.
4.3. Correlations, Principal Component (PC), and Biplot Analyses Among Quantitative Traits
The trend and magnitude of correlations among yield and yield components are crucial when designing selections. Selection of one trait may positively or negatively affect the expression of other traits []. The magnitude of trait association with economic traits such as grain yield affects selection strategies []. Higher contribution and strong association of PDL, NPP, NSP, HSW, and GYD with PC1 as well as DTF, PDL, and HSW with PC2 suggest that these traits account for genetic variation and selection gains. Arora et al. [] and Walle et al. (2019) [] reported a strong association of NPP and GYD with PC1 in cowpea, which corroborates the results in the current study. Walle et al. [] and Nkoana et al. [] identified and reported these traits as critical yield-influencing attributes. Mwadzingeni et al. [] found that a strong correlation of traits in discriminating genotypes can be observed from traits with small angles between dimension vectors in the same direction. The same inference was drawn for PC1 traits. Mbuma et al. [] reported a significant positive correlation between grain yield and the number of seeds per pod, indicating that selecting cowpea genotypes based on such traits could effectively maximize grain yield.
Kutu [] reported that influential traits in cowpea genotype selection include the number of pods per plant, number of seeds per pod, hundred seed weight, and grain yield. The positive association between pod length and the number of seeds per pod is one of the factors worth considering when selecting test genotypes to advance in a breeding program. In this study, genotypes such as NKL9P7 and BRR4P11, which had the highest number of seeds per pod (15.6; 14.6) and pod length (18 cm; 18.9 cm), yielded 3158.8 and 2651.6 kg/ha, respectively. The same results corroborated the findings of Nkoana et al. [] and Gerrano et al. [], where the increased length of the pods by test genotypes resulted in increased yields. Previous reports validated that variation in the number of pods and ultimate yield are influenced by both the timing and the initial rate of flower production and genotype []. A genotype-trait biplot was generated to enable the visualization and simultaneous selection of genotypes for many traits. Strong correlations were recorded among the number of pods per plant, pod length, number of seeds per pod, and grain yield. These traits could be used for enhanced grian yield in cowpea. Nkoana et al. [] and Walle et al. [] also reported a significant correlation among these traits. The study identified the following genotypes for their high grain yield: NKL9P7, BRR4P11, SHR9P5, NKL9P7-2, Mtilane, and Bira. These can be recommended for breeding or production in the study areas and similar agro-ecologies.
4.4. Classfication of Genotypes Based on Agronomic Traits
Cluster analysis is a key non-parametric statistical technique used to classify genotypes into distinct groups based on their similarity using phenotypic or molecular markers. It facilitates the identification of diverse genotypes to select contrasting traits and lines for breeding and production []. In the present study, the cluster analysis delineated the genotypes into two main groups I and II. Cluster II was further divided into sub-clusters II-a and II-b (Figure 3). Genotypes grouped under sub-cluster II-a were categorized under the top ten (BRR11P2, BRR11P11, NKR9P9, SHR10P12, NKR1P12, NKR4P5, BRR4P11, SHR9P5, SHR3P4, and NKL9P7) high-yielding genotypes (>1.8 t/ha), making them ideal lines for breeding and direct production in Eswatini or related agro-ecologies. Founder parent genotypes (Bira and Shindimba) and one local check (Black eye), which were grouped under II-a, are all IITA varieties that were evaluated for adaptability and released in Eswatini and Namibia, respectively. Nkhoma et al. [] documented that these groupings could be attributed to tracing the origin of genotypes and the availability of germplasm exchange between and among cowpea breeding programs in sub-Saharan Africa, particularly in southern Africa. The agro-ecological conditions of Eswatini and Namibia are common. Also, the exchange of germplasm between the two countries cannot be ruled out. This has partly limited the population structure analysis without distinguishing the genotypes based on geographical sources, which agrees with the report of Chen et al. []. The grouping of founder parent genotypes with their derivative mutants (Figure 3) suggests that the irradiation and subsequent mutation events were minor, and the genetic background largely remained the same for most of the assessed mutant lines when compared with the founder parents. Exchange of genetic resources is crucial for plant-breeding research and cultivar development, which relies on genetic varaition [].
5. Conclusions
The current study assessed the agronomic performance of the newly developed cowpea mutants to select the best-yielding and locally adapted pure lines for production and breeding in Eswatini. The principal component analysis identified the following principal traits for enhancing grain yield in cowpea: NPP, PDL, NSP, DTF, and HSW. The study identified the following mutant genotypes NKL9P7, BRR4P11, SHR9P5, NKL9P7-2 SHR3P4, SHR2P7, and Parent (Bira) as genetically unique, high-yielding, and promising pure lines for breeding and production in Eswatini.
Author Contributions
Conceptualization, K.A.K.M. and H.S.; methodology, K.A.K.M. and H.S.; investigation, K.A.K.M.; data collection, K.A.K.M.; formal analysis, K.A.K.M., S.A. and A.N.; writing—original draft, K.A.K.M. visualization, K.A.K.M.; software, K.A.K.M., S.A. and A.N.; funding acquisition, H.S.; resources, H.S.; supervision, H.S.; validation, K.A.K.M., H.S., S.A. and A.N.; writing—review and editing, H.S.; data curation, S.A. and A.N. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the Ministry of Agriculture, Department of Agricultural Research and Specialists Services of Eswatini, and the African Center for Crop Improvement at the University of KwaZulu-Natal.
Institutional Review Board Statement
Not applicable.
Data Availability Statement
The datasets generated and analyzed during this present study are included in this published article and any other data can be availed by the corresponding author upon request.
Acknowledgments
The authors are very thankful to the Ministry of Agriculture, Department of Agricultural Research and Specialists Services (DARSS) of Eswatini for the financial and technical support. The Ministry of Agriculture, Water, and Land Reform (MAWLR) of Namibia and the National Plant Genetic Resource Center (NPGRC) of Eswatini, are sincerely thanked for providing the germplasm used in the current study.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Coulibaly, S.; Pasquet, R.S.; Papa, R.; Gepts, P. AFLP analysis of the phenetic organization and genetic diversity of Vigna unguiculata L. Walp. reveals extensive gene flow between wild and domesticated types. Theor. Appl. Genet. 2002, 104, 358–366. [Google Scholar] [CrossRef]
- Kouam, E.B.; Pasquet, R.S.; Campagne, P.; Tignegre, J.B.; Thoen, K.; Gaudin, R.; Ouedraogo, J.T.; Salifu, A.B.; Muluvi, G.M.; Gepts, P. Genetic structure and mating system of wild cowpea populations in West Africa. BMC Plant Biol. 2012, 12, 113. [Google Scholar] [CrossRef]
- Singh, B.B.; Ajeigbe, H.A.; Tarawali, S.A.; Fernandez-Rivera, S.; Abubakar, M. Improving the production and utilization of cowpea as food and fodder. Field Crops Res. 2003, 84, 169–177. [Google Scholar] [CrossRef]
- Hu, T.; Xu, P. The Genetic Driving Force for the Grain-Vegetable Cowpea Diversification: A Focus on the Pod Length. J. Cell Signal. 2017, 2, 1–2. [Google Scholar] [CrossRef]
- Boukar, O.; Abberton, M.; Oyatomi, O.; Togola, A.; Tripathi, L.; Fatokun, C. Introgression breeding in cowpea (Vigna unguiculata [L.] Walp.). Front. Plant Sci. 2020, 11, 567425. [Google Scholar] [CrossRef]
- Ajeigbe, H.A.; Muhammad, M.I.; Singh, B.B. Potential of Triple Cropping of Wheat-Cowpea-Rice with Supplemental Irrigation in Sudan Savanna Zone of Nigeria. In Proceedings of the 46th Annual Conference of the Agricultural Society of Nigeria, Kano, Nigeria, 5–9 November 2012. [Google Scholar]
- Ahenkora, K.; Adu Dapaah, H.K.; Agyemang, A. Selected nutritional components and sensory attributes of cowpea (Vigna unguiculata [L.] Walp) leaves. Plant Foods Hum. Nutr. 1998, 52, 221–229. [Google Scholar] [CrossRef]
- Boukar, O.; Massawe, F.; Muranaka, S.; Franco, J.; Maziya-Dixon, B.; Singh, B.; Fatokun, C. Evaluation of cowpea germplasm lines for protein and mineral concentrations in grains. Plant Genet. Resour. 2011, 9, 515–522. [Google Scholar] [CrossRef]
- Ddamulira, G.; Santos, C.A.F.; Obuo, P.; Alanyo, M.; Lwanga, C.K. Grain yield and protein content of Brazilian cowpea genotypes under diverse Ugandan environments. Am. J. Plant Sci. 2015, 6, 2074–2084. [Google Scholar] [CrossRef]
- Ehlers, J.D.; Hall, A.E. Cowpea (Vigna unguiculata L. Walp.). Field Crops Res. 1997, 53, 187–204. [Google Scholar] [CrossRef]
- Ewansiha, S.U.; Singh, B.B. Relative drought tolerance of important herbaceous legumes and cereals in the moist and semi-arid regions of West Africa. J. Food Agric. Environ. 2006, 4, 188. [Google Scholar]
- Fatokun, C.A.; Boukar, O.; Muranaka, S. Evaluation of cowpea (Vigna unguiculata (L.) Walp.) germplasm lines for tolerance to drought. Plant. Genet. Resour. 2012, 10, 171–176. [Google Scholar] [CrossRef]
- Kyei-Boahen, S.; Savala, C.E.; Chikoye, D.; Abaidoo, R. Growth and yield responses of cowpea to inoculation and phosphorus fertilization in different environments. Front. Plant Sci. 2017, 8, 646. [Google Scholar] [CrossRef]
- Horn, L.; Shimelis, H.; Laing, M. Participatory Appraisal of Production Constraints, Preferred Traits and Farming Systems of Cowpea in Northern Namibia: Implications for Breeding. Legume Res. 2015, 38, 691–700. [Google Scholar] [CrossRef]
- Fatokun, C.; Girma, G.; Abberton, M.; Gedil, M.; Unachukwu, N.; Oyatomi, O.; Yusuf, M.; Rabbi, I.; Boukar, O. Genetic diversity and population structure of a mini-core subset from the world cowpea (Vigna unguiculata (L.) Walp.) germplasm collection. Sci. Rep. 2018, 8, 16035. [Google Scholar] [CrossRef]
- Omomowo, O.I.; Babalola, O.O. Constraints and Prospects of Improving Cowpea Productivity to Ensure Food, Nutritional Security and Environmental Sustainability. Front. Plant Sci. 2021, 12, 751731. [Google Scholar] [CrossRef]
- Gondwe, T.M.; Alamu, E.O.; Mdziniso, P.; Maziya-Dixon, B. Cowpea (Vigna unguiculata (L.) Walp) for food security: An evaluation of end-user traits of improved varieties in Swaziland. Sci. Rep. 2019, 9, 15991. [Google Scholar] [CrossRef]
- Horn, L.N.; Ghebrehiwot, H.M.; Shimelis, H.A. Selection of novel cowpea genotypes derived through gamma irradiation. Front. Plant Sci. 2016, 7, 262. [Google Scholar] [CrossRef]
- Gerrano, A.S.; Adebola, O.P.; Jansen Van Rensburg, W.S.; Laurie, S.M. Genetic variability in cowpea (Vigna unguiculata (L.) Walp) genotypes. S. Afr. J. Plant Soil. 2015, 32, 165–174. [Google Scholar] [CrossRef]
- Atakora, K.; Essilfie, M.E.; Agyarko, K.; Dapaah, H.K.; Santo, K.G. Evaluation of yield and yield components of some cowpea (Vigna unguiculata (L.) Walp) genotypes in forest and transitional zones of Ghana. Agric. Sci. 2023, 14, 878–897. [Google Scholar] [CrossRef]
- Murdoch, G.M. Soils and Land Capability in Swaziland; Swaziland Ministry of Agriculture: Mbabane, Eswatini, 1968.
- Dlamini, W.M. Mapping Forest and woodland loss in Swaziland: 1990–2015. Remote Sens. Appl. Soc. Environ. 2017, 5, 45–53. [Google Scholar] [CrossRef]
- Ngwenya, Z.D.; Mohammed, M.; Dakora, F.D. Monocropping and Intercropping of Maize with Six Food Legumes at Malkerns in Eswatini: Their Effects on Plant Growth, Grain Yield and N2 Fixation, measured using the 15N Natural Abundance and Ureide Techniques. Symbiosis 2024, 92, 257–269. [Google Scholar] [CrossRef]
- IBPGR. Descriptors for Cowpea; International Board for Plant Genetic Resources: Rome, Italy, 1983. [Google Scholar]
- Parker, A.; Namuth-Covert, D. Guidelines for the Conduct of Tests for Distinctness, Uniformity and Stability; International Union for the Protection of New Varieties of Plants (UPOV): Geneva, Switzerland, 2017. [Google Scholar]
- SAS Institute Inc. The GLM Procedure—SAS/STAT User’s Guide; SAS Institute Inc.: Cary, NC, USA, 2019. [Google Scholar]
- Levene, H. Robust tests for equality of variances. In Contributions to Probability and Statistics; Olkin, I., Ed.; Stanford University Press: Redwood City, CA, USA, 1960; pp. 278–292. [Google Scholar]
- Rahimi, M.; Hernandez, M.V. A SAS code to estimate phenotypic-genotypic covariance and correlation matrices based on expected value of statistical designs to use in plant breeding. An. Acad. Bras. Ciênc. 2022, 94, e20200001. [Google Scholar] [CrossRef]
- Alvarado, G.; Rodríguez, F.M.; Pacheco, A.; Burgueño, J.; Crossa, J.; Vargas, M.; Prez-Rodríguez, P.; Lopez-Cruz, M.A. META-R: A software to analyze data from multi-environment plant breeding trials. Crop J. 2020, 8, 745–756. [Google Scholar] [CrossRef]
- Robinson, H.F.; Comstock, R.E.; Harvey, P.H. Estimates of heritability and the degree of dominance in corn. Agron. J. 1949, 41, 353–359. [Google Scholar] [CrossRef]
- Johnson, H.W.; Robinson, R.E. Comstock. Estimates of genetic and environmental variability in Soybeans. Agron. J. 1955, 47, 314. [Google Scholar] [CrossRef]
- Allard, R.W. Principles of Plant Breeding, 2nd ed.; John Wiley & Sons: Hoboken, NJ, USA, 1999. [Google Scholar]
- Singh, R.K.; Chaudhary, B.D. Biometrical Methods in Quantitative Genetic Analysis; Kalyani Publishers: New Delhi, India, 1985. [Google Scholar]
- Pallant, J. SPSS Survival Manual: A Step-by-Step Guide to Data Analysis Using IBM SPSS, 7th ed.; Routledge; Open University Press: London, UK, 2020. [Google Scholar]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2024. [Google Scholar]
- RStudio Team. RStudio: Integrated Development for R; RStudio, PBC: Boston, MA, USA, 2023; Available online: http://www.rstudio.com/ (accessed on 23 July 2025).
- SAS Institute Inc. JMP®, version 17; Computer software; SAS Institute Incorporated: Cary, NC, USA, 2021. [Google Scholar]
- FAO. FAOSTAT: Food and Agriculture Organization Statistical Databases; Food and Agriculture Organization of the United Nations: Rome, Italy, 2000; Available online: https://www.fao.org/faostat/ (accessed on 2 June 2025).
- Mekonnen, T.W.; Mekbib, F.; Amsalu, B.; Gedil, M.; Labuschagne, M. Breeding implications of nodulation performance and root structure under natural inoculation for soil fertility enhancement and sustainable cowpea production. Front. Sustain. Food Syst. 2022, 6, 1076760. [Google Scholar] [CrossRef]
- Viswanatha, K.P.; Yogeesh, L.N. Genetic variation and morphological diversity in cowpea (Vigna unguiculata L. Walp). Arch. Agric. Environ. Sci. 2017, 2, 176–180. [Google Scholar]
- Pittelkow, C.M.; Liang, X.; Linquist, B.A.; van Groenigen, K.J.; Lee, J.; Lundy, M.E.; van Kessel, C. Productivity Limits and Potentials of the Principles of Conservation Agriculture. Nature 2015, 517, 365–368. [Google Scholar] [CrossRef]
- Sánchez, B.; Rasmussen, A.; Porter, J.R. Temperatures and the Growth and Development of Maize and Rice: A Review. Glob. Change Biol. 2014, 20, 408–417. [Google Scholar] [CrossRef]
- Nkhoma, N.; Shimelis, H.; Laing, M.D.; Shayanowako, A.; Mathew, I. Assessing the genetic diversity of cowpea [Vigna unguiculata (L.) Walp.] germplasm collections using phenotypic traits and SNP markers. BMC Genet. 2020, 21, 110. [Google Scholar] [CrossRef]
- Khadhem, F.; Baktash, F. AMMI Analysis of Adaptability and Yield Stability of Promising Lines of Bread Wheat (Triticum aestavum L.). Iraqi J. Agric. Sci. 2016, 47, 35–43. [Google Scholar]
- Darai, R.; Sarker, A.; Sah, R.P.; Pokhrel, K.; Chaudhary, R. AMMI Biplot Analysis for Genotype x Environment Interaction on Yield Trait of High Fe Content Lentil Genotypes in Terai and Mid-hill Environment of Nepal. Ann. Agric. Crop Sci. 2017, 2, 1028–1032. [Google Scholar]
- Angelova, S.; Stoilova, T. Maintenance, enrichment and utilization of grain legume collections in Bulgaria. In Proceedings of the IV Balkan Symposium on Vegetables and Potatoes, Plovdiv, Bulgaria, 9–12 September 2008; Volume 830, pp. 695–700. [Google Scholar]
- Hall, A. Phenotyping cowpeas for adaptation to drought. Front. Physiol. 2012, 3, 155. [Google Scholar] [CrossRef]
- Dugje, I.Y.; Omoigui, L.O.; Ekeleme, F.; Kamara, A.Y.; Ajeigbe, H. Farmers’ guide to cowpea production in West Africa. IITA Ib. Niger. 2009, 20, 12–14. [Google Scholar]
- Nkoana, D.K.; Gerrano, A.S.; Gwata, E.T. Agronomic Performance and Genetic Variability of Cowpea (Vigna unguiculata (L.) Walp) Accessions. Legume Res. 2019, 42, 757–762. [Google Scholar]
- Vidya, C.; Oommen, S.K.; Vijayaraghava, K. Genetic variability and heritability of yield and related characters in yard-long bean. J. Trop. Agric. 2002, 40, 11–13. [Google Scholar]
- Narayanankutty, C.; Mili, R.; Jaikumaran, U. Variability and genetic divergence in vegetable cowpea. J. Maharashtra Agric.Uni. 2003, 28, 26–29. [Google Scholar]
- Pal, A.K.; Maurya, A.N.; Singh, B.; Ram, D.; Kumar, S. Genetic variability, heritability and genetic advance in cowpea [Vigna unguiculata (L.) Walp]. Orissa J. Hortic. 2003, 31, 94–97. [Google Scholar]
- Girish, G.; Viswanatha, K.P.; Manjunath, A.; Yogeesh, L.N. Genetic variability, heritability and genetic advance analysis in cowpea [Vigna unguiculata L. (Walp.)]. Environ. Ecol. 2006, 24, 1172–1174. [Google Scholar]
- Owusu, E.Y.; Karikari, B.; Kusi, F.; Haruna, M.; Amoah, R.A.; Attamah, P.; Adazebra, G.; Sie, E.K.; Issahaku, M. Genetic variability, heritability and correlation analysis among maturity and yield traits in cowpea (Vigna unguiculata (L.) Walp) in Northern Ghana. Heliyon 2021, 7, e07890. [Google Scholar] [CrossRef]
- Mbuma, N.W.; Gerrano, A.S.; Lebaka, N.A.; Mofokeng, A.; Labuschagne, M. The evaluation of a southern African cowpea germplasm collection for seed yield and yield components. Crop Sci. 2021, 61, 466–489. [Google Scholar] [CrossRef]
- Owusu, E.Y.; Mohammed, H.; Manigben, K.A.; Adjebeng-Danquah, J.; Kusi, F.; Karikari, B.; Sie, E.K. Diallel Analysis and Heritability of Grain Yield, Yield Components, and Maturity Traits in Cowpea (Vigna unguiculata (L.) Walp.). Sci. World J. 2020, 2020, 9390287. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Ajayi, A.T.; Gbadamosi, A.E. Genetic variability, character association and yield potentials of twenty-five accessions of cowpea (Vigna unguiculata L. Walp). J. Pure Appl. Agric. 2019, 5, 1–16. [Google Scholar]
- Manju Devi, S.; Jayamani, P. Genetic variability, heritability and genetic advance studies in cowpea (Vigna unguiculata [L.] Walp.) germplasm. Electron. J. Plant Breed. 2018, 9, 476–481. [Google Scholar] [CrossRef]
- Zaki, A.E.H.; Radwan, O. Estimates of genetic parameters and heterosis for horticultural traits in cowpea (Vigna unguiculata L. Walp). Front. Plant Sci. 2022, 13, 987985. [Google Scholar] [CrossRef]
- Meenatchi, T.; Thangaraj, K.; Gnanamalar, R.P.; Pushpam, K. Genetic variability and heritability study on yield and its component traits in segregating population of cowpea (Vigna unguiculata L. Walp). Electron. J. Plant Breed. 2019, 10, 736–741. [Google Scholar] [CrossRef]
- Bhasker, K.D.; Shashibhushan, K.; Murali Krishna, M. Genetic variability, heritability and genetic advance of grain yield in pearl millet [Pennisetum glaucum (L.) R. Br]. Int. J. Pure Appl. Biosci. 2017, 5, 1228–1231. [Google Scholar] [CrossRef]
- Kamara, A.Y.; Tefera, H.; Ewansiha, S.U.; Ajeigbe, H.A.; Okechukwu, R.; Boukar, O.; Omoigui, L.O. Genetic gain in yield and agronomic characteristics of cowpea cultivars developed in the Sudan Savannas of Nigeria over the past three decades. Crop Sci. 2011, 51, 1877–1886. [Google Scholar] [CrossRef]
- Arora, R.N.; Kumar, K.; Manav, K. Principal Component Analysis in Kabuli Chickpea (Cicer arietinum L.). Int. J. Chem. Stud. 2018, 6, 2767–2768. [Google Scholar]
- Walle, T.; Mekbib, F.; Amsalu, B.; Gedil, M. Genetic Diversity of Ethiopian Cowpea (Vigna unguiculata L. Walp) Genotypes using Multivariate Analysis. Ethiop. J. Agric. Sci. 2019, 29, 89–104. [Google Scholar]
- Mwadzingeni, L.; Shimelis, H.; Tesfay, S.; Tsilo, T.J. Screening of bread wheat genotypes for drought tolerance using phenotypic and proline analyses. Front. Plant Sci. 2016, 7, 1276. [Google Scholar] [CrossRef]
- Mbuma, N.W.; Gerrano, A.S.; Lebaka, N.; Labuschagne, M. Interrelationship between grain yield components and nutritional quality traits in cowpea genotypes. S. Afr. J. Bot. 2022, 150, 34–43. [Google Scholar] [CrossRef]
- Kutu, F.R. Agronomic evaluation and identification of potential cowpea (Vigna unguiculata L. Walp) genotypes in South Africa. Acta. Agric. Scand. B Soil Plant Sci. 2019, 69, 295–303. [Google Scholar]
- Gerrano, A.S.; van Rensburg, W.S.J.; Adebola, P. Nutritional Composition of Immature Pods in Selected Cowpea [Vigna unguiculata (L.) Walp.] Genotypes. South. Africa. Aust. J. Crop Sci. 2017, 11, 134–141. [Google Scholar] [CrossRef]
- Craufurd, P.Q.; Wheeler, T.R.; Ellis, R.H.; Summerfield, R.J.; Prasad, P.V. Escape and tolerance to high temperature at flowering in groundnut (Arachis hypogaea L.). J. Agric. Sci. 2000, 135, 371–378. [Google Scholar] [CrossRef]
- Pandey, I. Genetic diversity in grain cowpea (Vigna unguiculata [L.] Walp.). Legume Res. 2007, 30, 92–97. [Google Scholar]
- Chen, H.; Chen, H.; Hu, L.; Wang, L.; Wang, S.; Wang, M.; Chang, X. Genetic Diversity and Population Structure Analysis of Accessions in the Chinese Cowpea (Vigna unguiculata (L.) Walp) Germplasm Collection. Crop J. 2017, 5, 363–372. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).