Abstract
Background: We conducted this review to summarize the relation between viral mutation and infectivity of SARS-CoV-2 and also the severity of COVID-19 in vivo and in vitro. Method: Articles were identified through a literature search until 31 May 2021, in PubMed, Web of Science and Google Scholar. Results: Sixty-three studies were included. To date, most studies showed that the viral mutations, especially the D614G variant, correlate with a higher infectivity than the wild-type virus. However, the evidence of the association between viral mutation and severity of the disease is scant. A SARS-CoV-2 variant with a 382-nucleotide deletion was associated with less severe infection in patients. The 11,083G > U mutation was significantly associated with asymptomatic patients. By contrast, ORF1ab 4715L and S protein 614G variants were significantly more frequent in patients from countries where high fatality rates were also reported. The current evidence showed that variants of concern have led to increased infectivity and deteriorating epidemiological situations. However, the relation between this variant and severity of COVID-19 infection was contradictory. Conclusion: The COVID-19 pandemic continues to spread worldwide. It is necessary to anticipate large clinical cohorts to evaluate the virulence and transmissibility of SARS-CoV-2 mutants.
1. Introduction
At the end of 2019, an epidemic of severe respiratory infections and pneumonia (named COVID-19) began in Wuhan, China. The cause of this outbreak is the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) virus. The disease is highly contagious, and the spread of COVID-19 has been taking place at varying rates globally. The World Health Organization (WHO) declared it a Public Health Emergency of International Concern on 30 January 2020 and then a global pandemic on 11 March 2020, less than three months after its appearance [1] (https://www.who.int/emergencies/diseases/novel-coronavirus-2019/events-as-they-happen (accessed on 26 April 2021)). This pandemic is the cause of an unprecedented health care crisis worldwide, with more than 110 million confirmed cases and more than 2,400,000 caused deaths to date [2].
Differences in severity have been observed with respiratory viruses, including influenza viruses, rhinoviruses, and coronaviruses [3,4,5]. SARS-CoV-2 primarily affects the respiratory system and the severity of the disease ranges from asymptomatic infection to severe acute respiratory distress [6]. Additionally, neurological symptoms including, notably, anosmia and ageusia are frequent [6], and some patients may also present with cutaneous [7] and gastrointestinal symptoms [7]. Finally, thrombotic, and thromboembolic diseases appeared to be frequent complications in COVID-19 patients [7]. As a consequence, the severity of the disease may greatly vary depending on the clinical presentation and the organs affected by the disease. In addition, the severity of the disease and the mortality rate are related to many host factors, including age, gender, chronic conditions, comorbidities, race, and ethnicity [6].
On the other hand, the virus mutation is also thought to affect the severity of the disease [8,9,10,11]. Since the emergence of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), genetic variants have been identified. In particular, the recent identification of new variants of concerns (VOC) in the UK (named 20I/501Y.V1 or B.1.1.7), South Africa (20H/501Y.V2 or B.1.351), Brazil (P1) and California, USA (B.1.427 and B.1.429), is causing concerns (Table 1) [12,13]. In a study conducted on more than 10,000 SARS-CoV-2 genomes from four databases from patients in 68 countries, 5775 distinct genomes were identified, including 2969 missense mutations and 36 stop-gained variants [14]. Investigation of a possible selective advantage or of an association with clinical severity of these variants is of paramount importance. Mutations in the gene encoding Spike protein of SARS-CoV-2 have been showed to affect both the virus infectivity and antigenicity in vitro [15]. In Marseille, France, a two-act pattern of incidence of COVID-19 cases occurred and significant differences in clinical outcomes were observed between patients seen in March–April 2020 and those seen in June–August [16,17].

Table 1.
Description of variants of concern (VOCs), up to 31 May 2021.
We aim to conduct this review to summarize the relation between viral mutation and infectivity of SARS-CoV-2 and viral mutation and severity of COVID-19 in vivo and in vitro.
2. Materials and Methods
2.1. Search Strategy and Selection Criteria
This review was conducted according to the Preferred Reporting Items for Systematic Reviews and MetaAnalyses (PRISMA) guidelines (http://www.prisma-statement.org (accessed on 31 May 2021)). The following databases were investigated in an attempt to identify all relevant studies published on: PubMed (http://www.ncbi.nlm.nih.gov/pubmed (accessed on 31 May 2021)), Web of Science (https://www.webofknowledge.com/) (accessed on 31 May 2021) and Google Scholar (http://scholar.google.fr/) (accessed on 31 May 2021). The most recent search was conducted on 31 May 2021. The topic search terms used for searching through the databases were the following:
#1: “variant” OR “variation” OR “mutation”;
#2: “SARS-CoV-2” OR “COVID-19”;
#3: “severity” OR infectivity”;
#4: #1 AND #2 AND #3.
Only articles published in English were included. For inclusion, articles had to fulfill two criteria: (1) be related to variants of SARS-CoV-2 virus and (2) describe the relation between viral mutation and infectivity of SARS-CoV-2 or severity of COVID-19. Reference lists of selected articles were screened to identify studies that might have been missing from the research.
After manually removing duplicates, three researchers (TLD, HVT and GP) independently performed the screening of the abstracts, applying the inclusion and exclusion criteria. In addition, articles without an abstract were included for full-text screening and assessed at this stage. Any discordant results were discussed in a consensus meeting. After screening the abstracts, the full texts of the articles were assessed for eligibility by the same three researchers and were selected or rejected for inclusion in the systematic review.
2.2. Data Collection Process
The following data (if available) were extracted from each article: country where patients were sampled, time period of the study, number of patients, type of clinical sample, genomic methods, characteristics of variants and outcome.
2.3. Data Synthesis and Analysis
As a result of the nature of the studies and the heterogeneity in patient populations, a formal meta-analysis was not possible. Therefore, the study results were summarized to describe the relation between genetic variation in SARS-CoV-2 and viral infectivity or severity of COVID-19. When possible, percentages not presented in the articles were calculated from the available data.
3. Results
3.1. Study Selection and Types of Studies
The study selection is presented in the flow diagram (Figure 1). The search algorithm produced 1642 articles from the PubMed, Web of Science and Google Scholar databases. After removing duplicates, 758 articles were scanned, based on their titles and abstracts. A total of 254 articles were processed for full-text screening. Sixty-three articles met the inclusion criteria and were included in the qualitative synthesis of the systematic review (Figure 1) [8,9,10,11,15,16,17,18,19,20,21,22,23,24,25,26,27,28,29,30,31,32,33,34,35,36,37,38,39,40,41,42,43,44,45,46,47,48,49,50,51,52,53,54,55,56,57,58,59,60,61,62,63,64,65,66,67,68,69,70,71,72,73].
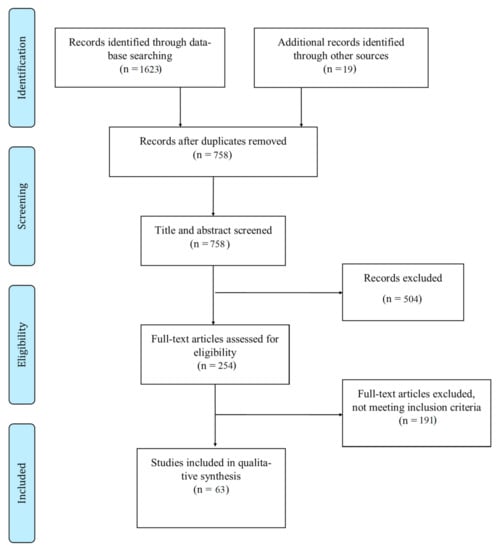
Figure 1.
Study flow chart.
Of the 63 publications included, five were non-peer reviewed preprints [16,17,52,54,66]. Eleven articles reported in vitro, in silico or animal model studies [15,18,19,22,23,24,25,26,27,28,29]; nineteen articles reported clinical studies [16,37,38,39,41,42,43,44,45,46,47,48,49,51,70,71,72,73]. The remaining 33 articles analyzed SARS-CoV-2 genomes downloaded from the GISAID or other available databases with patient status [8,9,10,11,20,21,30,31,32,33,34,35,36,40,50,52,53,54,55,56,57,58,59,60,61,62,63,64,65,66,67,68,69].
Most clinical studies were conducted before July 2020, corresponding to the first phase of the COVID-19 pandemic. Only four studies were conducted during two different episodes of the pandemic (from March to May and May to July 2020 in one study, from March to November in two studies and from March 2020 to January 2021 in one study) [41,46,70]. Most of the studies were conducted in the UK (9), followed by France (6), the USA (6), China (2), Singapore (2), Brazil (1), UAE (1), Vietnam (1) and Uruguay (1).
3.2. Relation between Viral Mutation and Infectivity of SARS-CoV-2
Twenty-five studies reported the relation between viral mutations and virus load or infectivity of SARS-CoV-2 (Table 2), of which five articles (two in vitro/animal model studies [18,19], two studies based on SARS-CoV-2 genomes downloaded from electronic databases or community-based testing datasets (bigdata analysis) [20,21] and one clinical study [16]) showed no impact of mutations with infectivity of SARS-CoV-2. The remaining 20 studies (nine in vitro/animal model or in silico studies [15,22,23,24,25,26,27,28,29], eight bigdata analyses [9,30,31,32,33,34,35,36] and three clinical studies [37,38,39]) showed that SARS-CoV-2 variants were significantly associated with increase in infectivity. All studies performed on the variants of concern (VOCs) have linked these mutations to increased viral transmissibility [23,27,28,29,34,35,36,38,39], except for one study by Li et al. [19]. In this in vitro study, the authors showed a non-significant increase in infectivity in cell lines for any of pseudotyped viruses with the B.1.351 variant compared to the D614G variant [19].

Table 2.
Viral mutation and infectivity of SARS-CoV-2.
3.3. Relation between Viral Mutations and Severity of COVID-19 Infection
A total of 45 articles addressed the effect of viral mutations on severity of COVID-19 in patients, of which fifteen studies (five bigdata analyses [21,30,33,34,40] and ten clinical studies [39,41,42,43,44,45,46,47,48,49]) showed no association between SARS-CoV-2 variants and severity of disease. Three studies (one bigdata analysis [50] and two clinical studies [17,51]) reported that the SARS-CoV-2 variants were significantly associated with decrease in severity of disease. The remaining 27 studies (one animal study [18], 22 bigdata analyses [8,9,10,11,52,53,54,55,56,57,58,59,60,61,62,63,64,65,66,67,68,69] and four clinical studies [70,71,72,73]) described the positive association between the different variants with severity of COVID-19 (Table 3).

Table 3.
Viral mutation and severity of COVID-19.
A clinical study by Long et al. showed that infection with SARS-CoV-2 variants harboring the D614G substitution was not associated with disease severity, overall mortality, transfer to ICU, mechanical ventilation and length of stay at hospital [41]. This result was supported by other research [33,45]. In a study conducted among 44 Vietnamese patients, 85 mutations covering 67 variant types were reported, of which P323L and D614G variants were the most frequent (present in 40/44 patients), followed by C241U (39/44) and GGG to AAC at 28881-3 variants (33/44). However, these mutations were not significantly associated with phenotype of illness [44]. Genomic investigation of 309 SARS-CoV-2 isolates obtained from patients seen in Marseille in March–April revealed specific mutations clustering in five main groups with no marked correlation with clinical severity of the disease [16]. Additionally, no significant difference in rate of transfer to ICU or mortality was identified between patients infected with the Marseille-1 or Marseille-4 variants (during the second episode of the pandemic) and those with clade 20A strains that predominantly circulated during the first phase of the pandemic in Marseille [46,47]. A study by Zhang et al. conducted in China showed no significant differences between two variants (clade I (ORF3a: p.251G > V, or S: p.614D > G (subclade G)); clade II (ORF8: p.84L > S (28,144U > C) and ORF1ab: p.2839S (8782C > U)) regarding disease severity and blood parameters indicative of severity [24]. No significant difference in clinical presentation was observed between hospitalized patients harboring or not harboring the D26 and D34 variants in the ORF6 protein [48]. A study conducted on isolates from patients in Washington, US, allowed the identification of two major clades distinguished by twelve polymorphisms in five genes. No significant difference regarding mortality was observed among patients infected with these two clades [43]. In a study conducted on 88 patients in the USA, most of the sequences (93%) were clustered in three main clades (clades 1, 2 and 3), defining mutations at the US level. The authors showed that the viral mutations have had no effect on time to symptom onset or disease severity [37]. Pawan et al. identified seven different variants from 3068 SARS-CoV-2 genomes obtained from GISAID, of which three clades (V, S and GH) were of no effect on the outcome of patients [54]. The new variant N501Y has been described as being more transmissible but is not associated with severity of COVID-19 infection [34].
A negative relation between severity of COVID-19 infection and virus mutations was described in several studies. In Singapore, a SARS-CoV-2 variant with a 382-nucleotide deletion (Δ382) that eliminates open-reading frame (ORF) 8 transcription was detected in a cluster of cases in January and February, 2020, and was associated with less severe infection in patients, in terms of hypoxia requiring supplemental oxygen [51]. A French study conducted during the second phase of the epidemic in the country showed that SARS-CoV-2 mutation rate was negatively associated with mortality rate [17]. Additionally, the Marseille-1 variant was associated with lower frequency of hospitalization than clade 20A strains [46]. In addition, the 11,083G > U mutation was significantly associated with asymptomatic patients [50].
By contrast, ORF1ab 4715L and S protein 614G variants were significantly more frequent in patients from countries where high fatality rates were also reported [8,10,11,57,59,60]. Patients infected with virus clades L, G and O were also exposed to a higher risk of severe infection than the base level [54]. Mutations at NSP6 and S proteins show a tendency to increase the death rate [56]. Majumda et al. analyzed 218 viral strains obtained from 15 countries. Their result showed that mutation in ORF3 protein increased the mortality of COVID-19 infection [9]. Among 1096 SARS-CoV-2 complete sequences downloaded from UK Biobank, 216 different verified super-variants were identified with eight predominant generic variants (chr6_148, chr7_23, chr2_197, chr2_221, chr8_99, chr10_57, chr16_4 and chr17_26). These variants were significantly associated with an increase in COVID-19 mortality [55]. By analyzing the data from datasets provided by Public Health England, Davies et al. showed that the 501Y variant was associated with a 12–64% higher risk of death [63].
Stauft et al. conducted an animal model study to evaluate the pathogenicity of D614 and G614 variants in the Spike protein. The authors showed that there was no difference in pathology in lung tissues of hamsters infected with these variants [18].
A total of ten studies reported the relation between new VOCs and severity of COVID-19, of which three studies (two bigdata analyses and one clinical study) showed negative associations. Most studies (6/7) describing a positive association between these variants and severity of infection used bigdata analyses. Only one clinical study [73] conducted using 68 patients showed a higher hospitalization rate in patients infected with the P.1 variant.
4. Discussion
The SARS-CoV-2 virus, due to the lack of proofreading activity of the RNA-dependent RNA polymerase, has high mutation rates that may have important effects on the pathogenicity and transmissibility of the virus [16]. The identification of genome variations of SARS-CoV-2 and their relationships with viral infectivity or severity of COVID-19 is therefore important for controlling and surveying the evolution of the pandemic [14,74]. In addition, mutation rate of SARS-CoV-2 determines the evolution of this virus and the risk of emergent infectious diseases [74]. In a study by Koyama et al., the median mutation rate of SARS-CoV-2 was estimated at 1.12.10−3 mutations per site/year (95%CI = [9.86.10−4–1.85.10−4]) [74]. A high mutation rate of around 30% was observed among 95 full-length genomic sequences [75]. An analysis of 48,635 samples showed an average of 7.23 mutations per sample [76]. To date, 46,251 SARS-CoV-2 mutations were documented in the public databases (http://cov-glue.cvr.gla.ac.uk/#/replacement (accessed on 31 May 2021)). Numbers of variations are the highest in the NSP3 protein, followed by S protein, NSP12 protein, NSP2 protein, NSP 13 protein, NSP14, and NSP4 protein. By contrast, very little divergence was documented in NSP11, ORF10, ORF7b and E proteins [74] (http://cov-glue.cvr.gla.ac.uk/#/replacement (accessed on 31 May 2021)). Figure 2 shows the positions of the mutations and deletions in the genome and of amino acid substitutions in the virion.
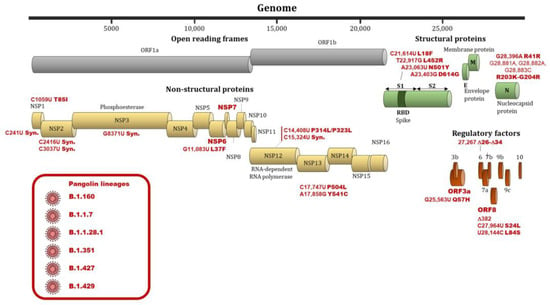
Figure 2.
Nucleotide and amino substitutions or deletions in the SARS-CoV-2 genome and viral clades associated with differences regarding infectivity and clinical severity. Footnote: Substitutions, deletions and genes involved in differences regarding infectivity and clinical severity are indicated by a red bold font. Viral clades involved in differences regarding infectivity and clinical severity are indicated by a red font and are shown in a red frame.
A key element of the coronavirus host range is determined by the binding affinity between the spike S protein and the cellular receptor. All mutations in the S protein could influence host range and transmissibility of the virus [16]. The SARS-CoV-2 Spike protein is a class I fusion protein that forms trimers on the viral surface: it is heavily glycosylated, which enables entry into host cells [77]. Angiotensin-converting enzyme 2 (ACE2) is the target receptor of the SARS-CoV-2 virus for entry into the host cell [77]. The main effect of the D614G mutation is to increase the availability of spike trimer components in the conformation and permits enhancing the binding of the virus spike to the ACE2 receptor. The in vitro and in vivo studies to date showed that the mutation D614G in Spike protein was associated with higher viral loads and probably with enhanced transmissibility of the virus [22,25,30]. Therefore, this mutation emerged and has become the dominant form in the global pandemic worldwide within a matter of months. It suggests that G614 may have a fitness advantage [78]. The frequency of S protein 614G was significantly associated with high fatality rates in several countries, as reported in studies which analyzed SARS-CoV-2 sequences from GISAID database [8,10,11]. However, clinical studies showed that this mutation did not correlate with severity of COVID-19, including mortality, transfer to ICU, mechanic ventilation, or length of stay at hospital [30,37,41,42,44,45].
In addition, clinical studies have shown that other viral mutations were not related to severity of COVID-19 infection or were associated with less severe infection in patients. Young et al. showed that the patients infected with Δ382 had lower concentrations of inflammatory biomarkers. Furthermore, these patients had a higher concentration of SDF-1 α, which is low in patients with hypoxemia [51]. Interestingly, the replication capacity of ∆382 variant in vitro is similar to that of wild-type SARS-CoV-2. It suggests that this mutation does not reduce replicative fitness [51]. In a study by Colson et al., the authors demonstrated seven new mutations of SARS-CoV-2, named “Marseille-1” to “Marseille-7”. Moreover, heterogeneity of the sequences produced from June to August 2020 (second outbreak) was higher than in sequences produced from February to May 2020 (first outbreak) (7.6.10−4 ± 3.8.10−4 versus 2.3.10−4 ± 1.1.10−4). This result indicates that the rate of virus mutation has increased rapidly. By contrast, the mortality of COVID-19 patients during the second outbreak was lower than that of those in the first one [17].
Recently, 501Y1, 501Y2 and P1 variant emergence in UK, South Africa and Brazil and the successive spread beyond the country of origin [79] have led to significant concern by medical and political authorities in many countries with extensive media coverage. The current evidence may suggest that these variants have led to increased infectivity and deteriorating epidemiological situations [34,79,80]. However, the relation between these variants, particularly the 501Y variant, and severity of COVID-19 is contradictory [34,63,79,80]. In our analysis, 8/10 studies reported a positive association between these VOCs and severity of COVID-19. Almost all of these studies were bigdata analyses. In fact, in one clinical study conducted using 496 patients, with 341 having samples that could be sequenced, no evidence of an association between severity of disease and death and B.1.1.7 variant was observed [39]. In addition, Dao et al. showed a lower rate of hospitalization associated with N501Y variants as compared to Clade 20A and the Marseille-4 variant [70]. To date, only one clinical study reported a higher hospitalization rate in 68 patients infected with the P.1 variant [73]. These contradictory results can be explained by the much faster rate of spread of VOCs compared to that of the original virus strain or previous variants. This means that when the number of infected persons in the population increases, the absolute number of severe cases and deaths may also increase. Therefore, the studies based on the community-based testing dataset reported an increase in mortality in patients infected with VOCs. The absolute risk increase affecting individual patients is likely minimal.
Our study has some limitations. We screened papers published only in English and reported in PubMed, Web of Science and Google Scholar. Ongoing research projects have not been used. For example, in our University Hospital Institute, a large cohort study aiming at comparing the demographic and clinical characteristics of patients infected with several new variants of SARS-CoV-2 virus during July to September 2020 is ongoing. In addition, a new variant, L18F, has been recently reported with 1186 spike L18F VOC genomes in the UK. The weekly growth rate of the L18F increased 1.75-fold, compared with the VOC genomes non-mutated at residue 18 [81]. Additionally, the VOC P1 was significantly associated with lower Ct values and with an increase in positive samples from 0 to 87% in Manaus, Brazil, between 2 November 2020, and 4 January 2021, suggesting high transmissibility [38]. However, the number of infections with these variants remains too small to perform an analysis on their infectivity [38,81]. Nevertheless, our review gives an overview on the relation between SARS-CoV-2 genetic variations and viral infectivity or severity of COVID-19 infection. In conclusion, most studies showed that some genetic variants of the virus were associated with high virus load. However, to date, the evidence of the association between viral mutation and severity of the disease is scant. On the other hand, severity and outcome of COVID-19 infection depend on the host’s genetic factors, on the treatment and clinical management, which have been improved, and on increased hospital capacity and response speed. The COVID-19 pandemic continues to spread worldwide. It is necessary to anticipate large clinical cohorts to evaluate the virulence and infectivity of SARS-CoV-2 mutants.
Author Contributions
Conceptualization, P.G., and D.R.; methodology, T.L.D., V.T.H., P.C., J.C.L., M.M., D.R. and A.L.; formal analysis, T.L.D., V.T.H., P.C., A.L., and P.G.; investigation, T.L.D., V.T.H., P.C., A.L., and P.G.; data curation, T.L.D., V.T.H., and P.G.; writing—original draft preparation, T.L.D., V.T.H., and P.G.; writing—review and editing, T.L.D., V.T.H., P.C., J.C.L., M.M., D.R., A.L., and P.G.; supervision, P.G. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The datasets generated during and/or analyzed during the current study are available from the corresponding author, P.G., upon reasonable request.
Conflicts of Interest
The authors declare that they have no conflict of interest.
References
- WHO|Coronavirus. Available online: https://www.who.int/westernpacific/health-topics/coronavirus (accessed on 20 February 2021).
- Coronavirus Update (Live): 111,437,607 Cases and 2,467,436 Deaths from COVID-19 Virus Pandemic-Worldometer N.D. Available online: https://www.worldometers.info/coronavirus/ (accessed on 20 February 2021).
- Daoud, A.; Laktineh, A.; Macrander, C.; Mushtaq, A.; Soubani, A.O. Pulmonary Complications of Influenza Infection: A Targeted Narrative Review. Postgrad Med. 2019, 131, 299–308. [Google Scholar] [CrossRef]
- Hayden, F.G. Rhinovirus and the Lower Respiratory Tract. Rev. Med. Virol. 2004, 14, 17–31. [Google Scholar] [CrossRef]
- Su, S.; Wong, G.; Shi, W.; Liu, J.; Lai, A.C.K.; Zhou, J.; Liu, W.; Bi, Y.; Gao, G.F. Epidemiology, Genetic Recombination, and Pathogenesis of Coronaviruses. Trends Microbiol. 2016, 24, 490–502. [Google Scholar] [CrossRef]
- Chidambaram, V.; Tun, N.L.; Haque, W.Z.; Majella, M.G.; Sivakumar, R.K.; Kumar, A.; Hsu, A.T.-W.; Ishak, I.A.; Nur, A.A.; Ayeh, S.K.; et al. Factors Associated with Disease Severity and Mortality among Patients with COVID-19: A Systematic Review and Meta-Analysis. PLoS ONE 2020, 15, e0241541. [Google Scholar] [CrossRef] [PubMed]
- Lai, C.-C.; Ko, W.-C.; Lee, P.-I.; Jean, S.-S.; Hsueh, P.-R. Extra-Respiratory Manifestations of COVID-19. Int. J. Antimicrob. Agents 2020, 56, 106024. [Google Scholar] [CrossRef] [PubMed]
- Toyoshima, Y.; Nemoto, K.; Matsumoto, S.; Nakamura, Y.; Kiyotani, K. SARS-CoV-2 Genomic Variations Associated with Mortality Rate of COVID-19. J. Hum. Genet. 2020, 65, 1075–1082. [Google Scholar] [CrossRef] [PubMed]
- Majumdar, P.; Niyogi, S. ORF3a mutation associated higher mortality rate in SARS-CoV-2 infection. Epidemiol. Infect. 2020, 148, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Becerra-Flores, M.; Cardozo, T. SARS-CoV-2 Viral Spike G614 Mutation Exhibits Higher Case Fatality Rate. Int. J. Clin. Pr. 2020, 74, e13525. [Google Scholar] [CrossRef]
- Eaaswarkhanth, M.; Al Madhoun, A.; Al-Mulla, F. Could the D614G Substitution in the SARS-CoV-2 Spike (S) Protein Be Associated with Higher COVID-19 Mortality? Int. J. Infect. Dis. 2020, 96, 459–460. [Google Scholar] [CrossRef] [PubMed]
- WHO. WHO|SARS-CoV-2 Variants. Available online: http://www.who.int/csr/don/31-december-2020-sars-cov2-variants/en/ (accessed on 20 February 2021).
- CDC. SARS-CoV-2 Variant Classifications and Definitions. Available online: https://www.cdc.gov/coronavirus/2019-ncov/variants/variant-info.html (accessed on 31 May 2021).
- Koyama, T.; Platt, D.; Parida, L. Variant Analysis of SARS-CoV-2 Genomes. Bull. World Health Organ. 2020, 98, 495–504. [Google Scholar] [CrossRef]
- Li, Q.; Wu, J.; Nie, J.; Zhang, L.; Hao, H.; Liu, S.; Zhao, C.; Zhang, Q.; Liu, H.; Nie, L.; et al. The Impact of Mutations in SARS-CoV-2 Spike on Viral Infectivity and Antigenicity. Cell 2020, 182, 1284–1294.e9. [Google Scholar] [CrossRef]
- Levasseur, A.; Delerce, J.; Caputo, A.; Brechard, L.; Colson, P.; Lagier, J.-C.; Fournier, P.-E.; Raoult, D. Genomic Diversity and Evolution of Coronavirus (SARS-CoV-2) in France from 309 COVID-19-Infected Patients. bioRxiv 2020. [Google Scholar] [CrossRef]
- Colson, P.; Levasseur, A.; Delerce, J.; Chaudet, H.; Bossi, V.; Ben Khedher, M.; Fournier, P.E.; Lagier, J.C.; Raoult, D. Dramatic increase in the SARS-CoV-2 mutation rate and low mortality rate during the second epidemic in summer in Marseille. Preprints. Available online: https://doi.org/10.35088/68c3-ew82 (accessed on 31 May 2021).
- Stauft, C.B.; Lien, C.Z.; Selvaraj, P.; Liu, S.; Wang, T.T. The G614 Pandemic SARS-CoV-2 Variant Is Not More Pathogenic than the Original D614 Form in Adult Syrian Hamsters. Virology 2021, 556, 96–100. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Nie, J.; Wu, J.; Zhang, L.; Ding, R.; Wang, H.; Zhang, Y.; Li, T.; Liu, S.; Zhang, M.; et al. SARS-CoV-2 501Y.V2 Variants Lack Higher Infectivity but Do Have Immune Escape. Cell 2021, 184, 2362–2371.e9. [Google Scholar] [CrossRef]
- Van Dorp, L.; Richard, D.; Tan, C.C.S.; Shaw, L.P.; Acman, M.; Balloux, F. No Evidence for Increased Transmissibility from Recurrent Mutations in SARS-CoV-2. Nat. Commun. 2020, 11, 5986. [Google Scholar] [CrossRef] [PubMed]
- Young, B.E.; Wei, W.E.; Fong, S.-W.; Mak, T.-M.; Anderson, D.E.; Chan, Y.-H.; Pung, R.; Heng, C.S.; Ang, L.W.; Zheng, A.K.E.; et al. Association of SARS-CoV-2 Clades with Clinical, Inflammatory and Virologic Outcomes: An Observational Study. EBioMedicine 2021, 66, 103319. [Google Scholar] [CrossRef]
- Zhang, L.; Jackson, C.B.; Mou, H.; Ojha, A.; Peng, H.; Quinlan, B.D.; Rangarajan, E.S.; Pan, A.; Vanderheiden, A.; Suthar, M.S.; et al. SARS-CoV-2 Spike-Protein D614G Mutation Increases Virion Spike Density and Infectivity. Nat. Commun. 2020, 11, 6013. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Zhang, Q.; Wei, P.; Chen, Z.; Aviszus, K.; Yang, J.; Downing, W.; Jiang, C.; Liang, B.; Reynoso, L.; et al. The Basis of a More Contagious 501Y.V1 Variant of SARS-CoV-2. Cell Res. 2021. [Google Scholar] [CrossRef]
- Plante, J.A.; Liu, Y.; Liu, J.; Xia, H.; Johnson, B.A.; Lokugamage, K.G.; Zhang, X.; Muruato, A.E.; Zou, J.; Fontes-Garfias, C.R.; et al. Spike Mutation D614G Alters SARS-CoV-2 Fitness. Nature 2020, 1–9. [Google Scholar] [CrossRef]
- Hou, Y.J.; Chiba, S.; Halfmann, P.; Ehre, C.; Kuroda, M.; Dinnon, K.H.; Leist, S.R.; Schäfer, A.; Nakajima, N.; Takahashi, K.; et al. SARS-CoV-2 D614G Variant Exhibits Enhanced Replication Ex Vivo and Earlier Transmission in Vivo. bioRxiv 2020. [Google Scholar] [CrossRef]
- Yao, H.; Lu, X.; Chen, Q.; Xu, K.; Chen, Y.; Cheng, L.; Liu, F.; Wu, Z.; Wu, H.; Jin, C.; et al. Patient-derived SARS-CoV-2 mutations impact viral replication dynamics and infectivity in vitro and with clinical implications in vivo. Cell Discov. 2020, 182, 812–827.e19. [Google Scholar] [CrossRef]
- Deng, X.; Garcia-Knight, M.A.; Khalid, M.M.; Servellita, V.; Wang, C.; Morris, M.K.; Sotomayor-González, A.; Glasner, D.R.; Reyes, K.R.; Gliwa, A.S.; et al. Transmission, Infectivity, and Neutralization of a Spike L452R SARS-CoV-2 Variant. Cell 2021. [Google Scholar] [CrossRef]
- Pokhrel, S.; Kraemer, B.R.; Lee, L.; Samardzic, K.; Mochly-Rosen, D. Increased Elastase Sensitivity and Decreased Intramolecular Interactions in the More Transmissible 501Y.V1 and 501Y.V2 SARS-CoV-2 Variants’ Spike Protein–an in Silico Analysis. PLoS ONE 2021, 16, e0251426. [Google Scholar] [CrossRef]
- Kim, Y.J.; Jang, U.S.; Soh, S.M.; Lee, J.Y.; Lee, H.R. The Impact on Infectivity and Neutralization Efficiency of SARS-CoV-2 Lineage, B.1.351 Pseudovirus. Viruses 2021, 13, 633. [Google Scholar] [CrossRef] [PubMed]
- Korber, B.; Fischer, W.M.; Gnanakaran, S.; Yoon, H.; Theiler, J.; Abfalterer, W.; Hengartner, N.; Giorgi, E.E.; Bhattacharya, T.; Foley, B.; et al. Tracking Changes in SARS-CoV-2 Spike: Evidence That D614G Increases Infectivity of the COVID-19 Virus. Cell 2020, 182, 812–827.e19. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.; Chen, J.; Gao, K.; Hozumi, Y.; Yin, C.; Wei, G.-W. Analysis of SARS-CoV-2 Mutations in the United States Suggests Presence of Four Substrains and Novel Variants. Commun. Biol. 2021, 4, 1–14. [Google Scholar] [CrossRef]
- Chen, J.; Wang, R.; Wang, M.; Wei, G.-W. Mutations Strengthened SARS-CoV-2 Infectivity. J. Mol. Biol. 2020, 432, 5212–5226. [Google Scholar] [CrossRef] [PubMed]
- Volz, E.; Hill, V.; McCrone, J.T.; Price, A.; Jorgensen, D.; O’Toole, Á.; Southgate, J.; Johnson, R.; Jackson, B.; Nascimento, F.F.; et al. Evaluating the Effects of SARS-CoV-2 Spike Mutation D614G on Transmissibility and Pathogenicity. Cell 2021, 184, 64–75.e11. [Google Scholar] [CrossRef] [PubMed]
- Davies, N.G.; Abbott, S.; Barnard, R.C.; Jarvis, C.I.; Kucharski, A.J.; Munday, J.D.; Pearson, C.A.B.; Russell, T.W.; Tully, D.C.; Washburne, A.D.; et al. Estimated Transmissibility and Impact of SARS-CoV-2 Lineage, B.1.1.7 in England. Science 2021, 372. [Google Scholar] [CrossRef] [PubMed]
- Zhao, S.; Lou, J.; Cao, L.; Zheng, H.; Chong, M.K.C.; Chen, Z.; Chan, R.W.Y.; Zee, B.C.Y.; Chan, P.K.S.; Wang, M.H. Quantifying the Transmission Advantage Associated with N501Y Substitution of SARS-CoV-2 in the UK: An Early Data-Driven Analysis. J. Travel Med. 2021. [Google Scholar] [CrossRef] [PubMed]
- Krutikov, M.; Hayward, A.; Shallcross, L. Spread of a Variant SARS-CoV-2 in Long-Term Care Facilities in England. New Engl. J. Med. 2021, 384, 1671–1673. [Google Scholar] [CrossRef] [PubMed]
- Lorenzo-Redondo, R.; Nam, H.H.; Roberts, S.C.; Simons, L.M.; Jennings, L.J.; Qi, C.; Achenbach, C.J.; Hauser, A.R.; Ison, M.G.; Hultquist, J.F.; et al. A Clade of SARS-CoV-2 Viruses Associated with Lower Viral Loads in Patient Upper Airways. EBioMedicine 2020, 62, 103112. [Google Scholar] [CrossRef] [PubMed]
- Faria, N.R.; Mellan, T.A.; Whittaker, C.; Claro, I.M.; Candido, D.D.S.; Mishra, S.; Crispim, M.A.E.; Sales, F.C.; Hawryluk, I.; McCrone, J.T.; et al. Genomics and Epidemiology of the P.1 SARS-CoV-2 Lineage in Manaus, Brazil. Science 2021, 372, 815–821. [Google Scholar] [CrossRef]
- Frampton, D.; Rampling, T.; Cross, A.; Bailey, H.; Heaney, J.; Byott, M.; Scott, R.; Sconza, R.; Price, J.; Margaritis, M.; et al. Genomic Characteristics and Clinical Effect of the Emergent SARS-CoV-2 B.1.1.7 Lineage in London, UK: A Whole-Genome Sequencing and Hospital-Based Cohort Study. Lancet Infect. Dis. 2021, 0, S1473–S3099. [Google Scholar]
- Zhao, S.; Lou, J.; Chong, M.K.C.; Cao, L.; Zheng, H.; Chen, Z.; Chan, R.W.Y.; Zee, B.C.Y.; Chan, P.K.S.; Wang, M.H. Inferring the Association between the Risk of COVID-19 Case Fatality and N501Y Substitution in SARS-CoV-2. Viruses 2021, 13, 638. [Google Scholar] [CrossRef] [PubMed]
- Long, S.W.; Olsen, R.J.; Christensen, P.A.; Bernard, D.W.; Davis, J.J.; Shukla, M.; Nguyen, M.; Saavedra, M.O.; Yerramilli, P.; Pruitt, L.; et al. Molecular Architecture of Early Dissemination and Massive Second Wave of the SARS-CoV-2 Virus in a Major Metropolitan Area. mBio 2020, 11, e02707-20. [Google Scholar] [CrossRef]
- Zhang, X.; Tan, Y.; Ling, Y.; Lu, G.; Liu, F.; Yi, Z.; Jia, X.; Wu, M.; Shi, B.; Xu, S.; et al. Viral and Host Factors Related to the Clinical Outcome of COVID-19. Nature 2020, 583, 437. [Google Scholar] [CrossRef]
- Nakamichi, K.; Shen, J.Z.; Lee, C.S.; Lee, A.; Roberts, E.A.; Simonson, P.D.; Roychoudhury, P.; Andriesen, J.; Randhawa, A.K.; Mathias, P.C.; et al. Hospitalization and Mortality Associated with SARS-CoV-2 Viral Clades in COVID-19. Sci. Rep. 2021, 11, 4802. [Google Scholar] [CrossRef]
- Nguyen, T.T.; Pham, T.N.; Van, T.D.; Nguyen, T.T.; Nguyen, D.T.N.; Le, H.N.M.; Eden, J.-S.; Rockett, R.J.; Nguyen, T.T.H.; Vu, B.T.N.; et al. Genetic Diversity of SARS-CoV-2 and Clinical, Epidemiological Characteristics of COVID-19 Patients in Hanoi, Vietnam. PLoS ONE 2020, 15, e0242537. [Google Scholar] [CrossRef]
- Elizondo, V.; Harkins, G.W.; Mabvakure, B.; Smidt, S.; Zappile, P.; Marier, C.; Maurano, M.T.; Perez, V.; Mazza, N.; Beloso, C.; et al. SARS-CoV-2 Genomic Characterization and Clinical Manifestation of the COVID-19 Outbreak in Uruguay. Emerg. Microbes Infect. 2021, 10, 51–65. [Google Scholar] [CrossRef]
- Colson, P.; Levasseur, A.; Gautret, P.; Fenollar, F.; Thuan Hoang, V.; Delerce, J.; Bitam, I.; Saile, R.; Maaloum, M.; Padane, A.; et al. Introduction into the Marseille Geographical Area of a Mild SARS-CoV-2 Variant Originating from Sub-Saharan Africa: An Investigational Study. Travel Med. Infect. Dis. 2021, 40, 101980. [Google Scholar] [CrossRef] [PubMed]
- Fournier, P.-E.; Colson, P.; Levasseur, A.; Devaux, C.A.; Gautret, P.; Bedotto, M.; Delerce, J.; Brechard, L.; Pinault, L.; Lagier, J.-C.; et al. Emergence and Outcomes of the SARS-CoV-2 “Marseille-4” Variant. Int J. Infect. Dis. 2021, 106, 228–236. [Google Scholar] [CrossRef] [PubMed]
- Quéromès, G.; Destras, G.; Bal, A.; Regue, H.; Burfin, G.; Brun, S.; Fanget, R.; Morfin, F.; Valette, M.; Trouillet-Assant, S.; et al. Characterization of SARS-CoV-2 ORF6 Deletion Variants Detected in a Nosocomial Cluster during Routine Genomic Surveillance, Lyon, France. Emerg. Microbes Infect. 2021, 10, 167–177. [Google Scholar] [CrossRef] [PubMed]
- Everett, J.; Hokama, P.; Roche, A.M.; Reddy, S.; Hwang, Y.; Kessler, L.; Glascock, A.; Li, Y.; Whelan, J.N.; Weiss, S.R.; et al. SARS-CoV-2 Genomic Variation in Space and Time in Hospitalized Patients in Philadelphia. mBio 2021, 12. [Google Scholar] [CrossRef]
- Wang, R.; Chen, J.; Hozumi, Y.; Yin, C.; Wei, G.-W. Decoding Asymptomatic COVID-19 Infection and Transmission. J. Phys. Chem. Lett. 2020, 11, 10007–10015. [Google Scholar] [CrossRef]
- Young, B.E.; Fong, S.-W.; Chan, Y.-H.; Mak, T.-M.; Ang, L.W.; Anderson, D.E.; Lee, C.Y.-P.; Amrun, S.N.; Lee, B.; Goh, Y.S.; et al. Effects of a Major Deletion in the SARS-CoV-2 Genome on the Severity of Infection and the Inflammatory Response: An Observational Cohort Study. The Lancet 2020, 396, 603–611. [Google Scholar] [CrossRef]
- Aiewsakun, P.; Wongtrakoongate, P.; Thawornwattana, Y.; Hongeng, S.; Thitithanyanont, A. SARS-CoV-2 Genetic Variations Associated with COVID-19 Severity. medRxiv 2020. [Google Scholar] [CrossRef]
- Nagy, Á.; Pongor, S.; Győrffy, B. Different mutations in SARS-CoV-2 associate with severe and mild outcome. Int J. Antimicrob Agents 2021, 57, 106272. [Google Scholar] [CrossRef]
- Verma, P.; Elkaffas, R.; Shodunke, O.C.; Hrab, P.; Adebayo, O.H.; Alam, H.M.; Gbedema, W.; Agbonyin, M.A.; Osman, A.M.; Igbokwe, J.O.; et al. COVID-19 Mortality Risk Assessment among Various Age Groups Using Phylogenetic Analysis. Preprints 2020. [Google Scholar] [CrossRef]
- Hu, J.; Li, C.; Wang, S.; Li, T.; Zhang, H. Genetic variants are identified to increase risk of COVID-19 related mortality from UK Biobank data. Hum. Genomics. 2021, 15, 10. [Google Scholar] [CrossRef]
- Bhadra, B. Significance of Mutation Rate of Structural and Non-Structural Proteins of SARS-Cov-2 Showed with Lower Death Rate of COVID-19. J. Biomed. Res. Environ. Sci. 2020, 6, 017–022. [Google Scholar]
- Biswas, S.K.; Mudi, S.R. Spike Protein D614G and RdRp P323L: The SARS-CoV-2 Mutations Associated with Severity of COVID-19. Genom. Inform. 2020, 18. [Google Scholar] [CrossRef]
- Dumonteil, E.; Fusco, D.; Drouin, A.; Herrera, C. Genomic Signatures of SARS-CoV-2 Associated with Patient Mortality. Viruses 2021, 13, 227. [Google Scholar] [CrossRef] [PubMed]
- Goyal, M.; De Bruyne, K.; van Belkum, A.; West, B. Different SARS-CoV-2 Haplotypes Associate with Geographic Origin and Case Fatality Rates of COVID-19 Patients. Infect. Genet. Evol. 2021, 90, 104730. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Chong, K.C.; Wong, M.C.S.; Boon, S.S.; Huang, J.; Wang, M.H.; Ng, R.W.Y.; Lai, C.K.C.; Chan, P.K.S. A Global Analysis on Replacement of Genetic Variants of SARS-CoV-2 in Associate with Containment Capacity and Changes in Disease Severity. Clin. Microbiol. Infect. 2021. [Google Scholar] [CrossRef] [PubMed]
- Challen, R.; Brooks-Pollock, E.; Read, J.M.; Dyson, L.; Tsaneva-Atanasova, K.; Danon, L. Risk of Mortality in Patients Infected with SARS-CoV-2 Variant of Concern 202012/1: Matched Cohort Study. BMJ 2021, 372. [Google Scholar] [CrossRef]
- Grint, D.J.; Wing, K.; Williamson, E.; McDonald, H.I.; Bhaskaran, K.; Evans, D.; Evans, S.J.; Walker, A.J.; Hickman, G.; Nightingale, E.; et al. Case Fatality Risk of the SARS-CoV-2 Variant of Concern B.1.1.7 in England, 16 November to 5 February. Eurosurveillance 2021, 26. [Google Scholar] [CrossRef] [PubMed]
- Davies, N.G.; Jarvis, C.I.; CMMID COVID-19 Working Group; Edmunds, W.J.; Jewell, N.P.; Diaz-Ordaz, K.; Keogh, R.H. Increased Mortality in Community-Tested Cases of SARS-CoV-2 Lineage B.1.1.7. Nature 2021, 593, 270–274. [Google Scholar] [CrossRef]
- Cao, C.; He, L.; Tian, Y.; Qin, Y.; Sun, H.; Ding, W.; Gui, L.; Wu, P. Molecular Epidemiology Analysis of Early Variants of SARS-CoV-2 Reveals the Potential Impact of Mutations P504L and Y541C (NSP13) in the Clinical COVID-19 Outcomes. Infect. Genet. Evol. 2021, 92, 104831. [Google Scholar] [CrossRef]
- Pandit, B.; Bhattacharjee, S.; Bhattacharjee, B. Association of Clade-G SARS-CoV-2 Viruses and Age with Increased Mortality Rates across 57 Countries and India. Infect. Genet. Evol. 2021, 90, 104734. [Google Scholar] [CrossRef]
- Patone, M.; Thomas, K.; Hatch, R.; Tan, P.S.; Coupland, C.; Liao, W.; Mouncey, P.; Harrison, D.; Rowan, K.; Horby, P.; et al. Analysis of Severe Outcomes Associated with the SARS-CoV-2 Variant of Concern 202012/01 in England Using ICNARC Case Mix Programme and QResearch Databases. medRxiv 2021. [Google Scholar] [CrossRef]
- Flores-Alanis, A.; Cruz-Rangel, A.; Rodríguez-Gómez, F.; González, J.; Torres-Guerrero, C.A.; Delgado, G.; Cravioto, A.; Morales-Espinosa, R. Molecular Epidemiology Surveillance of SARS-CoV-2: Mutations and Genetic Diversity One Year after Emerging. Pathogens 2021, 10, 184. [Google Scholar] [CrossRef] [PubMed]
- Thompson, C.N.; Hughes, S.; Ngai, S.; Baumgartner, J.; Wang, J.C.; McGibbon, E.; Devinney, K.; Luoma, E.; Bertolino, D.; Hwang, C.; et al. Rapid Emergence and Epidemiologic Characteristics of the SARS-CoV-2 B.1.526 Variant—New York City, New York, January 1–April 5, 2021. MMWR Morb. Mortal. Wkly. Rep. 2021, 70, 712–716. [Google Scholar] [CrossRef] [PubMed]
- Martin Webb, L.; Matzinger, S.; Grano, C.; Kawasaki, B.; Stringer, G.; Bankers, L.; Herlihy, R. Identification of and Surveillance for the SARS-CoV-2 Variants B.1.427 and B.1.429—Colorado, January–March 2021. MMWR Morb. Mortal. Wkly. Rep. 2021, 70, 717–718. [Google Scholar] [CrossRef] [PubMed]
- Dao, T.L.; Hoang, V.T.; Nguyen, N.N.; Delerce, J.; Chaudet, H.; Levasseur, A.; Lagier, J.C.; Raoult, D.; Colson, P.; Gautret, P. Clinical Outcomes in COVID-19 Patients Infected with Different SARS-Cov-2 Variants in Marseille, France. Clin. Microbiol. Infect. 2021. [Google Scholar] [CrossRef] [PubMed]
- Esper, F.P.; Cheng, Y.-W.; Adhikari, T.M.; Tu, Z.J.; Li, D.; Li, E.A.; Farkas, D.H.; Procop, G.W.; Ko, J.S.; Chan, T.A.; et al. Genomic Epidemiology of SARS-CoV-2 Infection During the Initial Pandemic Wave and Association with Disease Severity. JAMA Netw. Open 2021, 4, e217746. [Google Scholar] [CrossRef] [PubMed]
- Loney, T.; Khansaheb, H.; Ramaswamy, S.; Harilal, D.; Deesi, Z.O.; Varghese, R.M.; Ali, A.B.A.; Khadeeja, A.; Suwaidi, H.A.; Alkhajeh, A.; et al. Genotype-Phenotype Correlation Identified a Novel SARS-CoV-2 Variant Possibly Linked to Severe Disease. Transbound Emerg. Dis. 2021. [Google Scholar] [CrossRef]
- Martins, A.F.; Zavascki, A.P.; Wink, P.L.; Volpato, F.C.Z.; Monteiro, F.L.; Rosset, C.; De-Paris, F.; Ramos, Á.K.; Barth, A.L. Detection of SARS-CoV-2 Lineage P.1 in Patients from a Region with Exponentially Increasing Hospitalisation Rate, February 2021, Rio Grande Do Sul, Southern Brazil. Eurosurveillance 2021, 26. [Google Scholar] [CrossRef]
- Sardar, R.; Satish, D.; Birla, S.; Gupta, D. Integrative Analyses of SARS-CoV-2 Genomes from Different Geographical Locations Reveal Unique Features Potentially Consequential to Host-Virus Interaction, Pathogenesis and Clues for Novel Therapies. Heliyon 2020, 6, e04658. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Liu, Z.; Chen, Z.; Huang, X.; Xu, M.; He, T.; Zhang, Z. The Establishment of Reference Sequence for SARS-CoV-2 and Variation Analysis. J. Med. Virol. 2020, 92, 667–674. [Google Scholar] [CrossRef]
- Mercatelli, D.; Giorgi, F.M. Geographic and Genomic Distribution of SARS-CoV-2 Mutations. Front. Microbiol. 2020, 11, 1800. [Google Scholar] [CrossRef] [PubMed]
- Yurkovetskiy, L.; Wang, X.; Pascal, K.E.; Tomkins-Tinch, C.; Nyalile, T.P.; Wang, Y.; Baum, A.; Diehl, W.E.; Dauphin, A.; Carbone, C.; et al. Structural and Functional Analysis of the D614G SARS-CoV-2 Spike Protein Variant. Cell 2020, 183, 739–751.e8. [Google Scholar] [CrossRef] [PubMed]
- Groves, D.C.; Rowland-Jones, S.L.; Angyal, A. The D614G Mutations in the SARS-CoV-2 Spike Protein: Implications for Viral Infectivity, Disease Severity and Vaccine Design. Biochem Biophys Res. Commun. 2021, 538, 104–107. [Google Scholar] [CrossRef] [PubMed]
- Risk Assessment: Risk Related to the Spread of New SARS-CoV-2 Variants of Concern in the EU/EEA–First Update. Available online: https://www.ecdc.europa.eu/en/publications-data/covid-19-risk-assessment-spread-new-variants-concern-eueea-first-update (accessed on 2 June 2021).
- NERVTAG. Paper on COVID-19 Variant of Concern B.1.1.7. Available online: https://www.gov.uk/government/publications/nervtag-paper-on-covid-19-variant-of-concern-b117 (accessed on 2 June 2021).
- Grabowski, F.; Preibisch, G.; Giziński, S.; Kochańczyk, M.; Lipniacki, T. SARS-CoV-2 Variant of Concern 202012/01 Has about Twofold Replicative Advantage and Acquires Concerning Mutations. Viruses 2021, 13, 392. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).