Current Methods for Identifying Plasma Membrane Proteins as Cancer Biomarkers
Abstract
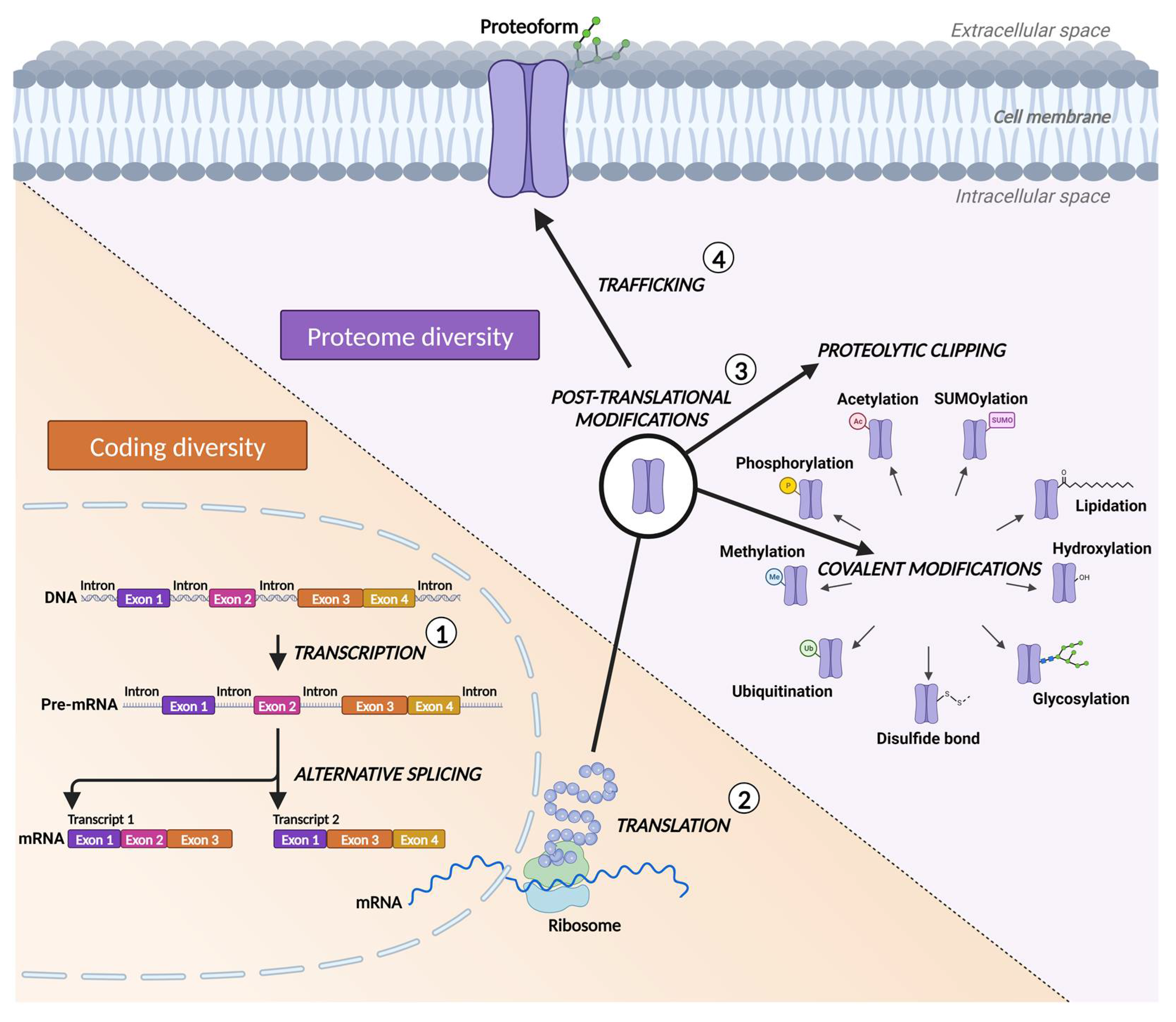
1. Introduction
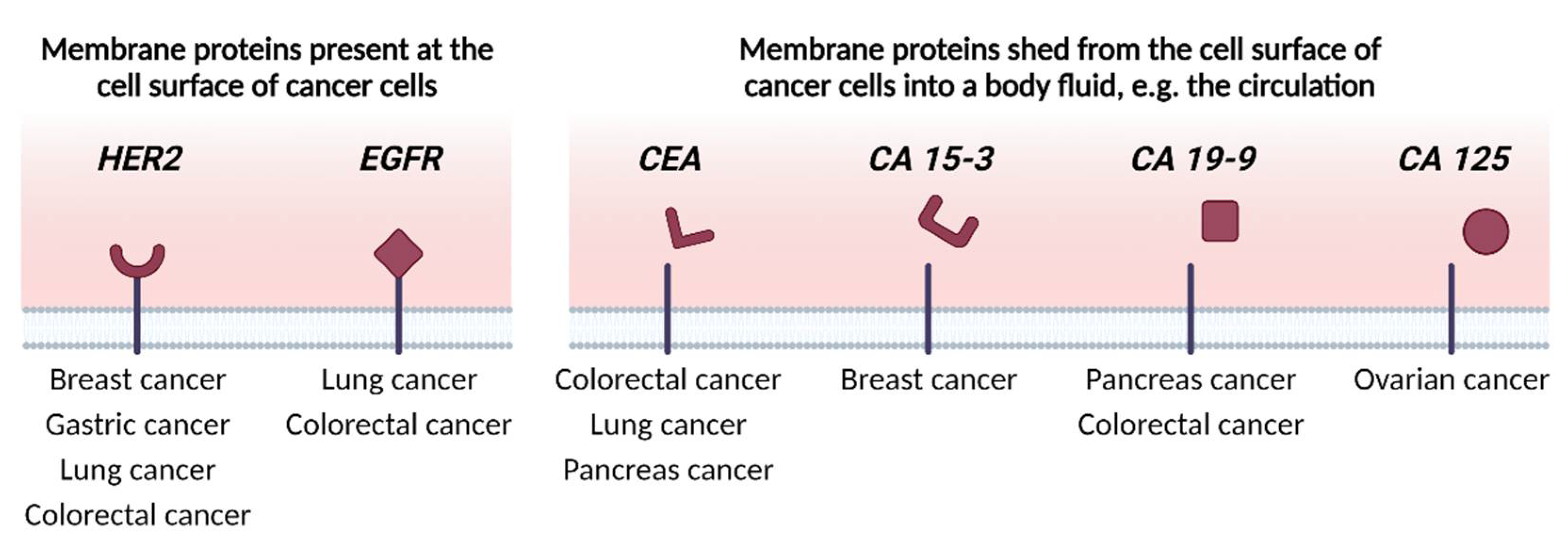
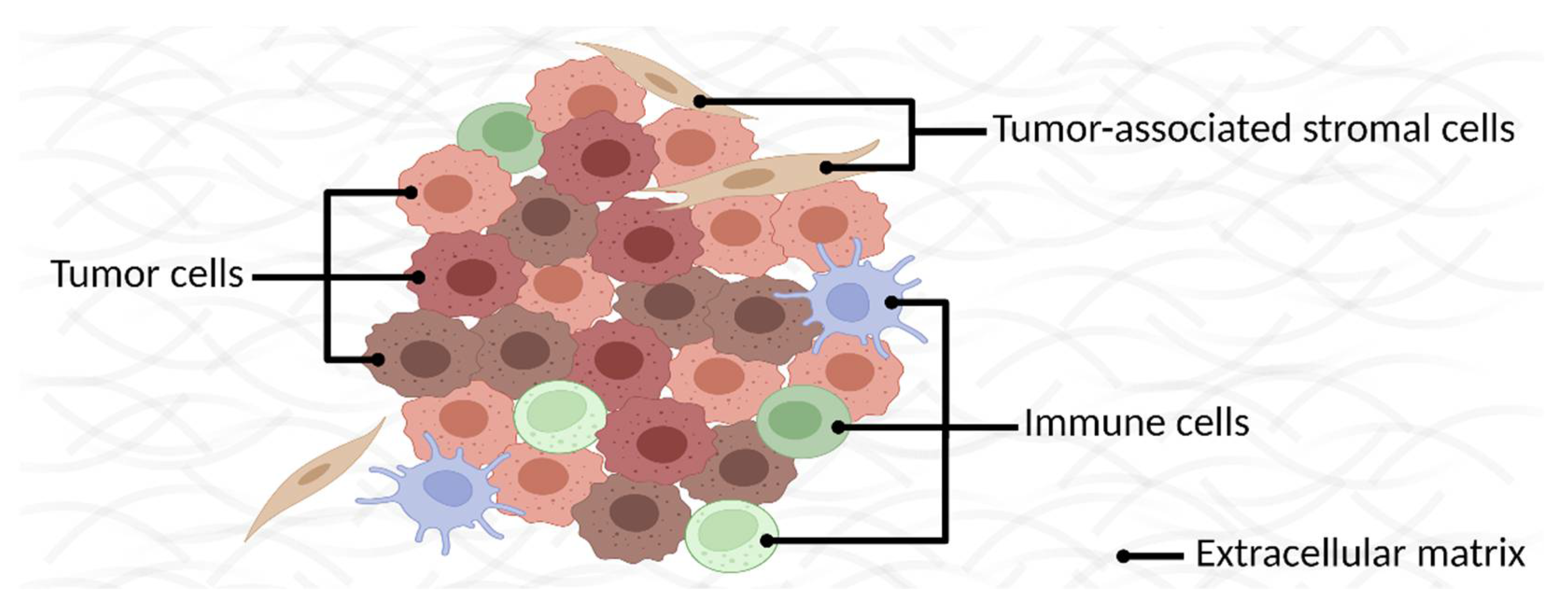
2. Identifying Membrane Proteins as Cancer Biomarkers
2.1. Indirect Discovery Methods
2.2. Direct, Biased Discovery Methods
2.2.1. Multiplexed Immunohistochemistry/Immunofluorescence
2.2.2. High-Throughput Flow Cytometry
2.3. Direct, Unbiased Discovery Methods
2.3.1. Bottom-Up Mass Spectrometry
Enrichment
Solubility
2.3.2. Top-Down Mass Spectrometry
2.4. Contemporary Methods
2.4.1. Mass Cytometry
2.4.2. Cell-SELEX
3. Conclusions and Future Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hübner, C.A.; Jentsch, T.J. Ion Channel Diseases. Hum. Mol. Genet 2002, 11, 2435–2445. [Google Scholar] [CrossRef] [PubMed]
- Rebhan, M.; Chalifa-Caspi, V.; Prilusky, J.; Lancet, D. GeneCards: Integrating Information about Genes, Proteins and Diseases. Trends Genet 1997, 13, 163. [Google Scholar] [CrossRef] [PubMed]
- Lehmann-Horn, F.; Jurkat-Rott, K. Voltage-Gated Ion Channels and Hereditary Disease. Physiol. Rev. 1999, 79, 1317–1372. [Google Scholar] [CrossRef] [PubMed]
- Leth-Larsen, R.; Lund, R.R.; Ditzel, H.J. Plasma Membrane Proteomics and Its Application in Clinical Cancer Biomarker Discovery. Mol. Cell. Proteom. 2010, 9, 1369. [Google Scholar] [CrossRef]
- Dobson, L.; Reményi, I.; Tusnády, G.E. The Human Transmembrane Proteome. Biol. Direct 2015, 10, 1–18. [Google Scholar] [CrossRef]
- Jentsch, T.J.; Hübner, C.A.; Fuhrmann, J.C. Ion Channels: Function Unravelled by Dysfunction. Nat. Cell Biol. 2004, 6, 1039–1047. [Google Scholar] [CrossRef]
- Kampen, K.R. Membrane Proteins: The Key Players of a Cancer Cell. J. Membr. Biol. 2011, 242, 69–74. [Google Scholar] [CrossRef]
- Imai, K.; Takaoka, A. Comparing Antibody and Small-Molecule Therapies for Cancer. Nat. Rev. Cancer 2006, 6, 714–727. [Google Scholar] [CrossRef]
- Yildirim, M.A.; Goh, K.I.; Cusick, M.E.; Barabási, A.L.; Vidal, M. Drug—Target Network. Nat. Biotechnol. 2007, 25, 1119–1126. [Google Scholar] [CrossRef]
- Ye, X.; Kaczmarczyk, J.A.; Luke, B.; Saul, R.G.; Whiteley, G.R.; Nissley, D.V.; Blonder, J. Cell Surface Protein Enrichment for Biomarker and Drug Target Discovery Using Mass Spectrometry-Based Proteomics. In Proteomic and Metabolomic Approaches to Biomarker Discovery; Academic Press: Cambridge, MA, USA, 2020; pp. 409–420. [Google Scholar] [CrossRef]
- Davey, G.C.; Currie, G.A.; Alexander, P. Spontaneous Shedding and Antibody Induced Modulation of Histocompatibility Antigens on Murine Lymphomata: Correlation with Metastatic Capacity. Br. J. Cancer 1976, 33, 9–14. [Google Scholar] [CrossRef]
- Prevarskaya, N.; Skryma, R.; Shuba, Y. Ion Channels in Cancer: Are Cancer Hallmarks Oncochannelopathies? Physiol. Rev. 2018, 98, 559–621. [Google Scholar] [CrossRef]
- Lund, R.; Leth-Larsen, R.; Jensen, O.N.; Ditzel, H.J. Efficient Isolation and Quantitative Proteomic Analysis of Cancer Cell Plasma Membrane Proteins for Identification of Metastasis-Associated Cell Surface Markers. J. Proteome Res. 2009, 8, 3078–3090. [Google Scholar] [CrossRef]
- Kuhlmann, L.; Cummins, E.; Samudio, I.; Kislinger, T. Cell-Surface Proteomics for the Identification of Novel Therapeutic Targets in Cancer. Expert Rev. Proteom. 2018, 15, 259–275. [Google Scholar] [CrossRef]
- Cordwell, S.J.; Thingholm, T.E. Technologies for Plasma Membrane Proteomics. Proteomics 2010, 10, 611–627. [Google Scholar] [CrossRef]
- Smolders, K.; Lombaert, N.; Valkenborg, D.; Baggerman, G.; Arckens, L. An Effective Plasma Membrane Proteomics Approach for Small Tissue Samples. Sci. Rep. 2015, 5, 1–18. [Google Scholar] [CrossRef]
- Karhemo, P.R.; Hyvönen, M.; Laakkonen, P. Metastasis-Associated Cell Surface Oncoproteomics. Front. Pharmacol. 2012, 3, 192. [Google Scholar] [CrossRef]
- Yang, J.; Griffin, A.; Qiang, Z.; Ren, J. Organelle-Targeted Therapies: A Comprehensive Review on System Design for Enabling Precision Oncology. Signal Transduct. Target. Ther. 2022, 7. [Google Scholar] [CrossRef]
- Díaz, P.; Sandoval-Bórquez, A.; Bravo-Sagua, R.; Quest, A.F.G.; Lavandero, S. Perspectives on Organelle Interaction, Protein Dysregulation, and Cancer Disease. Front. Cell Dev. Biol. 2021, 9, 1–10. [Google Scholar] [CrossRef]
- Gil-Hernández, A.; Arroyo-Campuzano, M.; Simoni-Nieves, A.; Zazueta, C.; Gomez-Quiroz, L.E.; Silva-Palacios, A. Relevance of Membrane Contact Sites in Cancer Progression. Front. Cell Dev. Biol. 2021, 8, 1–19. [Google Scholar] [CrossRef]
- Crick, F. Central Dogma of Molecular Biology. Nature 1970, 227, 561–563. [Google Scholar] [CrossRef]
- Da Cunha, J.P.C.; Galante, P.A.F.; De Souza, J.E.; De Souza, R.F.; Carvalho, P.M.; Ohara, D.T.; Moura, R.P.; Oba-Shinja, S.M.; Marie, S.K.N.; Silva, W.A.; et al. Bioinformatics Construction of the Human Cell Surfaceome. Proc. Natl. Acad. Sci. USA 2009, 106, 16752–16757. [Google Scholar] [CrossRef] [PubMed]
- Almén, M.S.; Nordström, K.J.; Fredriksson, R.; Schiöth, H.B. Mapping the Human Membrane Proteome: A Majority of the Human Membrane Proteins Can Be Classified According to Function and Evolutionary Origin. BMC Biol. 2009, 7, 50. [Google Scholar] [CrossRef]
- Babcock, J.J.; Li, M. Deorphanizing the Human Transmembrane Genome: A Landscape of Uncharacterized Membrane Proteins. Acta Pharmacol. Sin. 2014, 35, 11–23. [Google Scholar] [CrossRef] [PubMed]
- Carbonara, K.; Andonovski, M.; Coorssen, J.R. Proteomes Are of Proteoforms: Embracing the Complexity. Proteomes 2021, 9, 38. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.; Yu, X.; Xue, L.; Li, S.; Li, J.; Tong, D.; Du, Y. TP53-Associated Ion Channel Genes Serve as Prognostic Predictor and Therapeutic Targets in Head and Neck Squamous Cell Carcinoma. Technol. Cancer Res. Treat. 2020, 19, 1–13. [Google Scholar] [CrossRef]
- Zhao, Z.Y.; Liu, W. Pancreatic Cancer: A Review of Risk Factors, Diagnosis, and Treatment. Technol. Cancer Res. Treat. 2020, 19. [Google Scholar] [CrossRef]
- Diehn, M.; Eisen, M.B.; Botstein, D.; Brown, P.O. Large-Scale Identification of Secreted and Membrane-Associated Gene Products Using DNA Microarrays. Nat. Genet. 2000, 25, 58–62. [Google Scholar] [CrossRef]
- Diehn, M.; Bhattacharya, R.; Botstein, D.; Brown, P.O. Genome-Scale Identification of Membrane-Associated Human MRNAs. PLoS Genet 2006, 2, 39–50. [Google Scholar] [CrossRef]
- Walsh, C.T.; Garneau-Tsodikova, S.; Gatto, G.J. Protein Posttranslational Modifications: The Chemistry of Proteome Diversifications. Angew. Chem. Int. Ed. Engl. 2005, 44, 7342–7372. [Google Scholar] [CrossRef]
- Zhang, Y.; Qian, J.; Gu, C.; Yang, Y. Alternative Splicing and Cancer: A Systematic Review. Signal Transduct. Target. Ther. 2021, 6, 78. [Google Scholar] [CrossRef]
- Smith, L.M.; Kelleher, N.L.; Linial, M.; Goodlett, D.; Langridge-Smith, P.; Goo, Y.A.; Safford, G.; Bonilla, L.; Kruppa, G.; Zubarev, R.; et al. Proteoform: A Single Term Describing Protein Complexity. Nat. Methods 2013, 10, 186–187. [Google Scholar] [CrossRef]
- Tan, W.C.C.; Nerurkar, S.N.; Cai, H.Y.; Ng, H.H.M.; Wu, D.; Wee, Y.T.F.; Lim, J.C.T.; Yeong, J.; Lim, T.K.H. Overview of Multiplex Immunohistochemistry/Immunofluorescence Techniques in the Era of Cancer Immunotherapy. Cancer Commun. 2020, 40, 135–153. [Google Scholar] [CrossRef]
- Solier, C.; Langen, H. Antibody-Based Proteomics and Biomarker Research-Current Status and Limitations. Proteomics 2014, 14, 774–783. [Google Scholar] [CrossRef]
- Lopes, N.; Bergsland, C.H.; Bjørnslett, M.; Pellinen, T.; Svindland, A.; Nesbakken, A.; Almeida, R.; Lothe, R.A.; David, L.; Bruun, J. Digital Image Analysis of Multiplex Fluorescence IHC in Colorectal Cancer Recognizes the Prognostic Value of CDX2 and Its Negative Correlation with SOX2. Lab. Investig. 2019, 100, 120–134. [Google Scholar] [CrossRef]
- Toh, J.; Hoppe, M.M.; Thakur, T.; Yang, H.; Tan, K.T.; Pang, B.; Ho, S.; Roy, R.; Ho, K.Y.; Yeoh, K.G.; et al. Profiling of Gastric Cancer Cell-Surface Markers to Achieve Tumour-Normal Discrimination. BMJ Open Gastroenterol. 2020, 7, e000452. [Google Scholar] [CrossRef]
- Adan, A.; Alizada, G.; Kiraz, Y.; Baran, Y.; Nalbant, A. Flow Cytometry: Basic Principles and Applications. Crit. Rev. Biotechnol. 2017, 37, 163–176. [Google Scholar] [CrossRef]
- Gedye, C.A.; Hussain, A.; Paterson, J.; Smrke, A.; Saini, H.; Sirskyj, D.; Pereira, K.; Lobo, N.; Stewart, J.; Go, C.; et al. Cell Surface Profiling Using High-Throughput Flow Cytometry: A Platform for Biomarker Discovery and Analysis of Cellular Heterogeneity. PLoS ONE 2014, 9, e105602. [Google Scholar] [CrossRef]
- Chen, K.; Ding, A.; Ding, Y.; Ghanekar, A. High-Throughput Flow Cytometry Screening of Human Hepatocellular Carcinoma Reveals CD146 to Be a Novel Marker of Tumor-Initiating Cells. Biochem. Biophys. Rep. 2016, 8, 107. [Google Scholar] [CrossRef]
- Kelleher, N.L. Peer Reviewed: Top-Down Proteomics. Anal. Chem. 2004, 76, 196 A–203 A. [Google Scholar] [CrossRef]
- Cifani, P.; Kentsis, A. Towards Comprehensive and Quantitative Proteomics for Diagnosis and Therapy of Human Disease. Proteomics 2017, 17, 1600079. [Google Scholar] [CrossRef]
- Chait, B.T. Mass Spectrometry: Bottom-up or Top-Down? Science 2006, 314, 65–66. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Chen, X.; Fu, X.; Li, Z.; Huang, Y.; Liang, C. Advances in Aptamer-Based Biomarker Discovery. Front. Cell Dev. Biol. 2021, 9, 571. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Qin, H.; Ye, M. An Overview on Enrichment Methods for Cell Surface Proteome Profiling. J. Sep. Sci. 2020, 43, 292–312. [Google Scholar] [CrossRef] [PubMed]
- Mahmud, I.; Garrett, T.J. Mass Spectrometry Techniques in Emerging Pathogens Studies: COVID-19 Perspectives. J. Am. Soc. Mass Spectrom. 2020, 31, 2013–2024. [Google Scholar] [CrossRef] [PubMed]
- Vit, O.; Petrak, J. Integral Membrane Proteins in Proteomics. How to Break Open the Black Box? J. Proteom. 2017, 153, 8–20. [Google Scholar] [CrossRef]
- Wollscheid, B.; Bausch-Fluck, D.; Henderson, C.; O’Brien, R.; Bibel, M.; Schiess, R.; Aebersold, R.; Watts, J.D. Mass-Spectrometric Identification and Relative Quantification of N-Linked Cell Surface Glycoproteins. Nat. Biotechnol. 2009, 27, 378–386. [Google Scholar] [CrossRef]
- Elia, G. Biotinylation Reagents for the Study of Cell Surface Proteins. Proteomics 2008, 8, 4012–4024. [Google Scholar] [CrossRef]
- Gahmberg, C.G.; Andersson, L.C. Selective Radioactive Labeling of Cell Surface Sialoglycoproteins by Periodate-Tritiated Borohydride. J. Biol. Chem. 1977, 252, 5888–5894. [Google Scholar] [CrossRef]
- Bayer, E.A.; Ben-Hur, H.; Wilchek, M. Biocytin Hydrazide--a Selective Label for Sialic Acids, Galactose, and Other Sugars in Glycoconjugates Using Avidin-Biotin Technology. Anal. Biochem. 1988, 170, 271–281. [Google Scholar] [CrossRef]
- Gundry, R.L.; Riordon, D.R.; Tarasova, Y.; Chuppa, S.; Bhattacharya, S.; Juhasz, O.; Wiedemeier, O.; Milanovich, S.; Noto, F.K.; Tchernyshyov, I.; et al. A Cell Surfaceome Map for Immunophenotyping and Sorting Pluripotent Stem Cells. Mol. Cell. Proteom. 2012, 11, 303. [Google Scholar] [CrossRef]
- Kailemia, M.J.; Park, D.; Lebrilla, C.B. Glycans and Glycoproteins as Specific Biomarkers for Cancer. Anal. Bioanal. Chem. 2017, 409, 395–410. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Chen, J.; Sethi, A.; Li, Q.K.; Chen, L.; Collins, B.; Gillet, L.C.J.; Wollscheid, B.; Zhang, H.; Aebersold, R. Glycoproteomic Analysis of Prostate Cancer Tissues by SWATH Mass Spectrometry Discovers N-Acylethanolamine Acid Amidase and Protein Tyrosine Kinase 7 as Signatures for Tumor Aggressiveness. Mol. Cell. Proteom. 2014, 13, 1753. [Google Scholar] [CrossRef] [PubMed]
- Tian, Y.; Bova, G.S.; Zhang, H. Quantitative Glycoproteomic Analysis of Optimal Cutting Temperature-Embedded Frozen Tissues Identifying Glycoproteins Associated with Aggressive Prostate Cancer. Anal. Chem. 2011, 83, 7013–7019. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Xi, J.; Tian, Y.; Bova, G.S.; Zhang, H. Identification, Prioritization and Evaluation of Glycoproteins for Aggressive Prostate Cancer Using Quantitative Glycoproteomics and Antibody-Based Assays on Tissue Specimens. Proteomics 2013, 13, 2268. [Google Scholar] [CrossRef] [PubMed]
- van Oostrum, M.; Müller, M.; Klein, F.; Bruderer, R.; Zhang, H.; Pedrioli, P.G.A.; Reiter, L.; Tsapogas, P.; Rolink, A.; Wollscheid, B. Classification of Mouse B Cell Types Using Surfaceome Proteotype Maps. Nat. Commun. 2019, 10. [Google Scholar] [CrossRef]
- Bausch-Fluck, D.; Hofmann, A.; Bock, T.; Frei, A.P.; Cerciello, F.; Jacobs, A.; Moest, H.; Omasits, U.; Gundry, R.L.; Yoon, C.; et al. A Mass Spectrometric-Derived Cell Surface Protein Atlas. PLoS ONE 2015, 10, e0121314. [Google Scholar] [CrossRef]
- Garbis, S.; Lubec, G.; Fountoulakis, M. Limitations of Current Proteomics Technologies. J. Chromatogr. A 2005, 1077, 1–18. [Google Scholar] [CrossRef]
- Rabilloud, T.; Chevallet, M.; Luche, S.; Lelong, C. Fully Denaturing Two-Dimensional Electrophoresis of Membrane Proteins: A Critical Update. Proteomics 2008, 8, 3965–3973. [Google Scholar] [CrossRef]
- Olaya-Abril, A.; Jiménez-Munguía, I.; Gómez-Gascón, L.; Rodríguez-Ortega, M.J. Surfomics: Shaving Live Organisms for a Fast Proteomic Identification of Surface Proteins. J. Proteom. 2014, 97, 164–176. [Google Scholar] [CrossRef]
- Zhang, X. Less Is More: Membrane Protein Digestion Beyond Urea-Trypsin Solution for Next-Level Proteomics. Mol. Cell. Proteom. 2015, 14, 2441–2453. [Google Scholar] [CrossRef]
- Manza, L.L.; Stamer, S.L.; Ham, A.J.L.; Codreanu, S.G.; Liebler, D.C. Sample Preparation and Digestion for Proteomic Analyses Using Spin Filters. Proteomics 2005, 5, 1742–1745. [Google Scholar] [CrossRef]
- Wiśniewski, J.R.; Zougman, A.; Nagaraj, N.; Mann, M. Universal Sample Preparation Method for Proteome Analysis. Nat. Methods 2009, 6, 359–362. [Google Scholar] [CrossRef]
- Erde, J.; Loo, R.R.O.; Loo, J.A. Enhanced FASP (EFASP) to Increase Proteome Coverage and Sample Recovery for Quantitative Proteomic Experiments. J. Proteome Res. 2014, 13, 1885–1895. [Google Scholar] [CrossRef]
- Macklin, A.; Khan, S.; Kislinger, T. Recent Advances in Mass Spectrometry Based Clinical Proteomics: Applications to Cancer Research. Clin. Proteom. 2020, 17, 1–25. [Google Scholar] [CrossRef]
- Yu, Y.; Xie, L.; Gunawardena, H.P.; Khatun, J.; Maier, C.; Spitzer, W.; Leerkes, M.; Giddings, M.C.; Chen, X. GOFAST: An Integrated Approach for Efficient and Comprehensive Membrane Proteome Analysis. Anal. Chem. 2012, 84, 9008–9014. [Google Scholar] [CrossRef]
- Raimondo, F.; Corbetta, S.; Savoia, A.; Chinello, C.; Cazzaniga, M.; Rocco, F.; Bosari, S.; Grasso, M.; Bovo, G.; Magni, F.; et al. Comparative Membrane Proteomics: A Technical Advancement in the Search of Renal Cell Carcinoma Biomarkers. Mol. Biosyst. 2015, 11, 1708–1716. [Google Scholar] [CrossRef]
- Donnelly, D.P.; Rawlins, C.M.; DeHart, C.J.; Fornelli, L.; Schachner, L.F.; Lin, Z.; Lippens, J.L.; Aluri, K.C.; Sarin, R.; Chen, B.; et al. Best Practices and Benchmarks for Intact Protein Analysis for Top-down Mass Spectrometry. Nat. Methods 2019, 16, 587–594. [Google Scholar] [CrossRef] [PubMed]
- Konijnenberg, A.; Bannwarth, L.; Yilmaz, D.; Koçer, A.; Venien-Bryan, C.; Sobott, F. Top-down Mass Spectrometry of Intact Membrane Protein Complexes Reveals Oligomeric State and Sequence Information in a Single Experiment. Protein Sci. 2015, 24, 1292–1300. [Google Scholar] [CrossRef] [PubMed]
- Konijnenberg, A.; Yilmaz, D.; Ingólfsson, H.I.; Dimitrova, A.; Marrink, S.J.; Li, Z.; Vénien-Bryan, C.; Sobott, F.; Kocer, A. Global Structural Changes of an Ion Channel during Its Gating Are Followed by Ion Mobility Mass Spectrometry. Proc. Natl. Acad. Sci. USA 2014, 111, 17170–17175. [Google Scholar] [CrossRef] [PubMed]
- Laganowsky, A.; Reading, E.; Hopper, J.T.S.; Robinson, C.V. Mass Spectrometry of Intact Membrane Protein Complexes. Nat. Protoc. 2013, 8, 639–651. [Google Scholar] [CrossRef]
- Delcourt, V.; Franck, J.; Leblanc, E.; Narducci, F.; Robin, Y.M.; Gimeno, J.P.; Quanico, J.; Wisztorski, M.; Kobeissy, F.; Jacques, J.F.; et al. Combined Mass Spectrometry Imaging and Top-down Microproteomics Reveals Evidence of a Hidden Proteome in Ovarian Cancer. EBioMedicine 2017, 21, 55–64. [Google Scholar] [CrossRef] [PubMed]
- Erady, C.; Boxall, A.; Puntambekar, S.; Suhas Jagannathan, N.; Chauhan, R.; Chong, D.; Meena, N.; Kulkarni, A.; Kasabe, B.; Prathivadi Bhayankaram, K.; et al. Pan-Cancer Analysis of Transcripts Encoding Novel Open-Reading Frames (NORFs) and Their Potential Biological Functions. npj Genom. Med. 2021, 6, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Whitelegge, J.P.; Zhang, H.; Aguilera, R.; Taylor, R.M.; Cramer, W.A. Full Subunit Coverage Liquid Chromatography Electrospray Ionization Mass Spectrometry (LCMS+) of an Oligomeric Membrane Protein: Cytochrome B6f Complex From Spinach and the Cyanobacterium Mastigocladus Laminosus. Mol. Cell. Proteom. 2002, 1, 816–827. [Google Scholar] [CrossRef] [PubMed]
- Kar, U.K.; Simonian, M.; Whitelegge, J.P. Integral Membrane Proteins: Bottom-up, Top-down and Structural Proteomics. Expert. Rev. Proteom. 2017, 14, 715. [Google Scholar] [CrossRef] [PubMed]
- Carroll, J.; Fearnley, I.M.; Walker, J.E. Definition of the Mitochondrial Proteome by Measurement of Molecular Masses of Membrane Proteins. Proc. Natl. Acad. Sci. USA 2006, 103, 16170. [Google Scholar] [CrossRef]
- Brown, K.A.; Tucholski, T.; Alpert, A.J.; Eken, C.; Wesemann, L.; Kyrvasilis, A.; Jin, S.; Ge, Y. Top-Down Proteomics of Endogenous Membrane Proteins Enabled by Cloud Point Enrichment and Multidimensional Liquid Chromatography-Mass Spectrometry. Anal. Chem. 2020, 92, 15726–15735. [Google Scholar] [CrossRef]
- Carroll, J.; Altman, M.C.; Fearnley, I.M.; Walker, J.E. Identification of Membrane Proteins by Tandem Mass Spectrometry of Protein Ions. Proc. Natl. Acad. Sci. USA 2007, 104, 14330–14335. [Google Scholar] [CrossRef]
- Chen, B.; Brown, K.A.; Lin, Z.; Ge, Y. Top-Down Proteomics: Ready for Prime Time? Anal. Chem. 2018, 90, 110–127. [Google Scholar] [CrossRef]
- Jeong, K.; Kim, J.; Gaikwad, M.; Hidayah, S.N.; Heikaus, L.; Schlüter, H.; Kohlbacher, O. FLASHDeconv: Ultrafast, High-Quality Feature Deconvolution for Top-Down Proteomics. Cell. Syst. 2020, 10, 213–218.e6. [Google Scholar] [CrossRef]
- Schaffer, L.V.; Millikin, R.J.; Miller, R.M.; Anderson, L.C.; Fellers, R.T.; Ge, Y.; Kelleher, N.L.; LeDuc, R.D.; Liu, X.; Payne, S.H.; et al. Identification and Quantification of Proteoforms by Mass Spectrometry. Proteomics 2019, 19, e1800361. [Google Scholar] [CrossRef]
- Wu, Z.; Roberts, D.S.; Melby, J.A.; Wenger, K.; Wetzel, M.; Gu, Y.; Ramanathan, S.G.; Bayne, E.F.; Liu, X.; Sun, R.; et al. MASH Explorer: A Universal Software Environment for Top-Down Proteomics. J. Proteome Res. 2020, 19, 3867–3876. [Google Scholar] [CrossRef]
- Brodin, P. The Biology of the Cell - Insights from Mass Cytometry. FEBS J. 2019, 286, 1514–1522. [Google Scholar] [CrossRef]
- Bandura, D.R.; Baranov, V.I.; Ornatsky, O.I.; Antonov, A.; Kinach, R.; Lou, X.; Pavlov, S.; Vorobiev, S.; Dick, J.E.; Tanner, S.D. Mass Cytometry: Technique for Real Time Single Cell Multitarget Immunoassay Based on Inductively Coupled Plasma Time-of-Flight Mass Spectrometry. Anal. Chem. 2009, 81, 6813–6822. [Google Scholar] [CrossRef]
- Zhang, T.; Warden, A.R.; Li, Y.; Ding, X. Progress and Applications of Mass Cytometry in Sketching Immune Landscapes. Clin. Transl. Med. 2020, 10. [Google Scholar] [CrossRef]
- Gadalla, R.; Noamani, B.; MacLeod, B.L.; Dickson, R.J.; Guo, M.; Xu, W.; Lukhele, S.; Elsaesser, H.J.; Razak, A.R.A.; Hirano, N.; et al. Validation of CyTOF against Flow Cytometry for Immunological Studies and Monitoring of Human Cancer Clinical Trials. Front. Oncol. 2019, 9, 415. [Google Scholar] [CrossRef]
- Li, L.; Yan, S.; Lin, B.; Shi, Q.; Lu, Y. Single-Cell Proteomics for Cancer Immunotherapy. Adv. Cancer Res. 2018, 139, 185–207. [Google Scholar] [CrossRef]
- Iyer, A.; Hamers, A.A.J.; Pillai, A.B. CyTOF® for the Masses. Front. Immunol. 2022, 13, 1636. [Google Scholar] [CrossRef]
- Spitzer, M.H.; Nolan, G.P. Mass Cytometry: Single Cells, Many Features. Cell 2016, 165, 780. [Google Scholar] [CrossRef]
- Sefah, K.; Shangguan, D.; Xiong, X.; O’Donoghue, M.B.; Tan, W. Development of DNA Aptamers Using Cell-SELEX. Nat. Protoc. 2010, 5, 1169–1185. [Google Scholar] [CrossRef]
- Shangguan, D.; Li, Y.; Tang, Z.; Cao, Z.C.; Chen, H.W.; Mallikaratchy, P.; Sefah, K.; Yang, C.J.; Tan, W. Aptamers Evolved from Live Cells as Effective Molecular Probes for Cancer Study. Proc. Natl. Acad. Sci. USA 2006, 103, 11838–11843. [Google Scholar] [CrossRef]
- Shigdar, S.; Agnello, L.; Fedele, M.; Camorani, S.; Cerchia, L. Profiling Cancer Cells by Cell-SELEX: Use of Aptamers for Discovery of Actionable Biomarkers and Therapeutic Applications Thereof. Pharmaceutics 2021, 14, 28. [Google Scholar] [CrossRef] [PubMed]
- Shangguan, D.; Cao, Z.C.; Li, Y.; Tan, W. Aptamers Evolved from Cultured Cancer Cells Reveal Molecular Differences of Cancer Cells in Patient Samples. Clin. Chem. 2007, 53, 1153–1155. [Google Scholar] [CrossRef] [PubMed]
- Shangguan, D.; Cao, Z.; Meng, L.; Mallikaratchy, P.; Sefah, K.; Wang, H.; Li, Y.; Tan, W. Cell-Specific Aptamer Probes for Membrane Protein Elucidation in Cancer Cells. J. Proteome Res. 2008, 7, 2133–2139. [Google Scholar] [CrossRef] [PubMed]
- Sicco, E.; Baez, J.; Ibarra, M.; Fernández, M.; Cabral, P.; Moreno, M.; Cerecetto, H.; Calzada, V. Sgc8-c Aptamer as a Potential Theranostic Agent for Hemato-Oncological Malignancies. Cancer Biother Radiopharm 2020, 35, 262–270. [Google Scholar] [CrossRef]
- Jia, W.; Ren, C.; Wang, L.; Zhu, B.; Jia, W.; Gao, M.; Zeng, F.; Zeng, L.; Xia, X.; Zhang, X.; et al. CD109 Is Identified as a Potential Nasopharyngeal Carcinoma Biomarker Using Aptamer Selected by Cell-SELEX. Oncotarget 2016, 7, 55328–55342. [Google Scholar] [CrossRef]
- Yuan, B.; Jiang, X.; Chen, Y.; Guo, Q.; Wang, K.; Meng, X.; Huang, Z.; Wen, X. Metastatic Cancer Cell and Tissue-Specific Fluorescence Imaging Using a New DNA Aptamer Developed by Cell-SELEX. Talanta 2017, 170, 56–62. [Google Scholar] [CrossRef]
- Li, W.M.; Zhou, L.L.; Zheng, M.; Fang, J. Selection of Metastatic Breast Cancer Cell-Specific Aptamers for the Capture of CTCs with a Metastatic Phenotype by Cell-SELEX. Mol. Ther. Nucleic Acids 2018, 12, 707. [Google Scholar] [CrossRef]
- Li, X.; Zhang, W.; Liu, L.; Zhu, Z.; Ouyang, G.; An, Y.; Zhao, C.; Yang, C.J. In Vitro Selection of DNA Aptamers for Metastatic Breast Cancer Cell Recognition and Tissue Imaging. Anal. Chem. 2014, 86, 6596–6603. [Google Scholar] [CrossRef]
- Wang, L.; Li, P.; Xiao, X.; Li, J.; Li, J.; Yang, H.H.; Tan, W. Generating Lung-Metastatic Osteosarcoma Targeting Aptamers for in Vivo and Clinical Tissue Imaging. Talanta 2018, 188, 66–73. [Google Scholar] [CrossRef]
- Duan, M.; Long, Y.; Yang, C.; Wu, X.; Sun, Y.; Li, J.; Hu, X.; Lin, W.; Han, D.; Zhao, Y.; et al. Selection and Characterization of DNA Aptamer for Metastatic Prostate Cancer Recognition and Tissue Imaging. Oncotarget 2016, 7, 36436–36446. [Google Scholar] [CrossRef]
- Speransky, S.; Serafini, P.; Caroli, J.; Bicciato, S.; Lippman, M.E.; Bishopric, N.H. A Novel RNA Aptamer Identifies Plasma Membrane ATP Synthase Beta Subunit as an Early Marker and Therapeutic Target in Aggressive Cancer. Breast Cancer Res. Treat. 2019, 176, 271–289. [Google Scholar] [CrossRef]
- Wang, F.B.; Rong, Y.; Fang, M.; Yuan, J.P.; Peng, C.W.; Liu, S.P.; Li, Y. Recognition and Capture of Metastatic Hepatocellular Carcinoma Cells Using Aptamer-Conjugated Quantum Dots and Magnetic Particles. Biomaterials 2013, 34, 3816–3827. [Google Scholar] [CrossRef]
- Chen, H.; Yuan, C.H.; Yang, Y.F.; Yin, C.Q.; Guan, Q.; Wang, F.B.; Tu, J.C. Subtractive Cell-SELEX Selection of DNA Aptamers Binding Specifically and Selectively to Hepatocellular Carcinoma Cells with High Metastatic Potential. Biomed Res. Int. 2016, 2016, 5735869. [Google Scholar] [CrossRef]
- Rong, Y.; Chen, H.; Zhou, X.F.; Yin, C.Q.; Wang, B.C.; Peng, C.W.; Liu, S.P.; Wang, F.B. Identification of an Aptamer through Whole Cell-SELEX for Targeting High Metastatic Liver Cancers. Oncotarget 2016, 7, 8282–8294. [Google Scholar] [CrossRef]
- Li, X.; An, Y.; Jin, J.; Zhu, Z.; Hao, L.; Liu, L.; Shi, Y.; Fan, D.; Ji, T.; Yang, C.J. Evolution of DNA Aptamers through in Vitro Metastatic-Cell-Based Systematic Evolution of Ligands by Exponential Enrichment for Metastatic Cancer Recognition and Imaging. Anal. Chem. 2015, 87, 4941–4948. [Google Scholar] [CrossRef]
- Li, W.M.; Bing, T.; Wei, J.Y.; Chen, Z.Z.; Shangguan, D.H.; Fang, J. Cell-SELEX-Based Selection of Aptamers That Recognize Distinct Targets on Metastatic Colorectal Cancer Cells. Biomaterials 2014, 35, 6998–7007. [Google Scholar] [CrossRef]
- Ding, M.; Clark, R.; Bardelle, C.; Backmark, A.; Norris, T.; Williams, W.; Wigglesworth, M.; Howes, R. Application of High-Throughput Flow Cytometry in Early Drug Discovery: An AstraZeneca Perspective. SLAS Discov. 2018, 23, 719–731. [Google Scholar] [CrossRef]
- Yang, J.Y.; Herold, D.A. Evolving Platforms for Clinical Mass Spectrometry. In Mass Spectrometry for the Clinical Laboratory; Academic Press: Cambridge, MA, USA, 2017; pp. 261–276. [Google Scholar] [CrossRef]
- Bendall, S.C.; Simonds, E.F.; Qiu, P.; Amir, E.A.D.; Krutzik, P.O.; Finck, R.; Bruggner, R.V.; Melamed, R.; Trejo, A.; Ornatsky, O.I.; et al. Single-Cell Mass Cytometry of Differential Immune and Drug Responses Across a Human Hematopoietic Continuum. Science 2011, 332, 687. [Google Scholar] [CrossRef]
- Meng, H.M.; Fu, T.; Zhang, X.B.; Tan, W. Cell-SELEX-Based Aptamer-Conjugated Nanomaterials for Cancer Diagnosis and Therapy. Natl. Sci. Rev. 2015, 2, 71–84. [Google Scholar] [CrossRef]
- Chen, M.; Yu, Y.; Jiang, F.; Zhou, J.; Li, Y.; Liang, C.; Dang, L.; Lu, A.; Zhang, G. Development of Cell-SELEX Technology and Its Application in Cancer Diagnosis and Therapy. Int. J. Mol. Sci. 2016, 17, 2079. [Google Scholar] [CrossRef]
- Dagogo-Jack, I.; Shaw, A.T. Tumour Heterogeneity and Resistance to Cancer Therapies. Nat. Rev. Clin. Oncol. 2018, 15, 81–94. [Google Scholar] [CrossRef] [PubMed]
- Crosby, D.; Bhatia, S.; Brindle, K.M.; Coussens, L.M.; Dive, C.; Emberton, M.; Esener, S.; Fitzgerald, R.C.; Gambhir, S.S.; Kuhn, P.; et al. Early Detection of Cancer. Science 2022, 375. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Sáez, O.; Prat, A. Current and Future Management of HER2-Positive Metastatic Breast Cancer. JCO Oncol. Pract. 2021, 17, 594–604. [Google Scholar] [CrossRef] [PubMed]
- Dormann, C. Metastatic Human Epidermal Growth Factor Receptor 2-Positive Breast Cancer: Current Treatment Standards and Future Perspectives. Breast Care 2020, 15, 570–578. [Google Scholar] [CrossRef] [PubMed]
- Zhao, D.; Klempner, S.J.; Chao, J. Progress and Challenges in HER2-Positive Gastroesophageal Adenocarcinoma. J. Hematol. Oncol. 2019, 12. [Google Scholar] [CrossRef]
- Van Cutsem, E.; Bang, Y.J.; Feng-yi, F.; Xu, J.M.; Lee, K.W.; Jiao, S.C.; Chong, J.L.; López-Sanchez, R.I.; Price, T.; Gladkov, O.; et al. HER2 Screening Data from ToGA: Targeting HER2 in Gastric and Gastroesophageal Junction Cancer. Gastric Cancer 2015, 18, 476–484. [Google Scholar] [CrossRef]
- Riudavets, M.; Sullivan, I.; Abdayem, P.; Planchard, D. Targeting HER2 in Non-Small-Cell Lung Cancer (NSCLC): A Glimpse of Hope? An Updated Review on Therapeutic Strategies in NSCLC Harbouring HER2 Alterations. ESMO Open 2021, 6. [Google Scholar] [CrossRef]
- Zhao, J.; Xia, Y. Targeting HER2 Alterations in Non-Small-Cell Lung Cancer: A Comprehensive Review. JCO Precis. Oncol. 2020, 4, 411–425. [Google Scholar] [CrossRef]
- La Salvia, A.; Lopez-Gomez, V.; Garcia-Carbonero, R. HER2-Targeted Therapy: An Emerging Strategy in Advanced Colorectal Cancer. Expert. Opin. Investig. Drugs 2019, 28, 29–38. [Google Scholar] [CrossRef]
- Greally, M.; Kelly, C.M.; Cercek, A. HER2: An Emerging Target in Colorectal Cancer. Curr. Probl. Cancer 2018, 42, 560–571. [Google Scholar] [CrossRef]
- Brückl, W.M.; Reck, M.; Griesinger, F.; Schäfer, H.; Kortsik, C.; Gaska, T.; Rawluk, J.; Krüger, S.; Kokowski, K.; Budweiser, S.; et al. Afatinib as First-Line Treatment in Patients with EGFR-Mutated Non-Small Cell Lung Cancer in Routine Clinical Practice. Ther. Adv. Med. Oncol. 2021, 13, 17588359211012361. [Google Scholar] [CrossRef]
- Harvey, R.D.; Adams, V.R.; Beardslee, T.; Medina, P. Afatinib for the Treatment of EGFR Mutation-Positive NSCLC: A Review of Clinical Findings. J. Oncol. Pharm. Pract. 2020, 26, 1461–1474. [Google Scholar] [CrossRef]
- Riedesser, J.E.; Ebert, M.P.; Betge, J. Precision Medicine for Metastatic Colorectal Cancer in Clinical Practice. Ther. Adv. Med. Oncol. 2022, 14, 175883592110727. [Google Scholar] [CrossRef]
- Gómez-España, M.A.; Gallego, J.; González-Flores, E.; Maurel, J.; Páez, D.; Sastre, J.; Aparicio, J.; Benavides, M.; Feliu, J.; Vera, R. SEOM Clinical Guidelines for Diagnosis and Treatment of Metastatic Colorectal Cancer (2018). Clin. Transl. Oncol. 2019, 21, 46–54. [Google Scholar] [CrossRef]
- Dervenis, C.; Xynos, E.; Sotiropoulos, G.; Gouvas, N.; Boukovinas, I.; Agalianos, C.; Androulakis, N.; Athanasiadis, A.; Christodoulou, C.; Chrysou, E.; et al. Clinical Practice Guidelines for the Management of Metastatic Colorectal Cancer: A Consensus Statement of the Hellenic Society of Medical Oncologists (HeSMO). Ann. Gastroenterol. 2016, 29, 390–416. [Google Scholar] [CrossRef]
- Labianca, R.; Nordlinger, B.; Beretta, G.D.; Mosconi, S.; Mandalà, M.; Cervantes, A.; Arnold, D. Early Colon Cancer: ESMO Clinical Practice Guidelines for Diagnosis, Treatment and Follow-Up. Ann. Oncol. 2013, 24 (Suppl. 6), vi64-72. [Google Scholar] [CrossRef]
- Jiao, Z.; Cao, S.; Li, J.; Hu, N.; Gong, Y.; Wang, L.; Jin, S. Clinical Associations of Preoperative and Postoperative Serum CEA and Lung Cancer Outcome. Front. Mol. Biosci. 2021, 8. [Google Scholar] [CrossRef]
- Arrieta, O.; Villarreal-Garza, C.; Martínez-Barrera, L.; Morales, M.; Dorantes-Gallareta, Y.; Peña-Curiel, O.; Contreras-Reyes, S.; Macedo-Pérez, E.O.; Alatorre-Alexander, J. Usefulness of Serum Carcinoembryonic Antigen (CEA) in Evaluating Response to Chemotherapy in Patients with Advanced Non Small-Cell Lung Cancer: A Prospective Cohort Study. BMC Cancer 2013, 13. [Google Scholar] [CrossRef]
- Xing, H.; Wang, J.; Wang, Y.; Tong, M.; Hu, H.; Huang, C.; Li, D. Diagnostic Value of CA 19-9 and Carcinoembryonic Antigen for Pancreatic Cancer: A Meta-Analysis. Gastroenterol. Res. Pract. 2018, 2018, 8704751. [Google Scholar] [CrossRef]
- Meng, Q.; Shi, S.; Liang, C.; Liang, D.; Xu, W.; Ji, S.; Zhang, B.; Ni, Q.; Xu, J.; Yu, X. Diagnostic and Prognostic Value of Carcinoembryonic Antigen in Pancreatic Cancer: A Systematic Review and Meta-Analysis. Onco. Targets Ther. 2017, 10, 4591–4598. [Google Scholar] [CrossRef]
- Ravelli, A.; Reuben, J.M.; Lanza, F.; Anfossi, S.; Cappelletti, M.R.; Zanotti, L.; Gobbi, A.; Senti, C.; Brambilla, P.; Milani, M.; et al. Breast Cancer Circulating Biomarkers: Advantages, Drawbacks, and New Insights. Tumour Biol. 2015, 36, 6653–6665. [Google Scholar] [CrossRef] [PubMed]
- Chourin, S.; Georgescu, D.; Gray, C.; Guillemet, C.; Loeb, A.; Veyret, C.; Basuyau, J.P. Value of CA 15-3 Determination in the Initial Management of Breast Cancer Patients. Ann. Oncol. 2009, 20, 962–964. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.J.; Koh, H.K.; Chie, E.K.; Oh, D.Y.; Bang, Y.J.; Nam, E.M.; Kim, K. Change in Carbohydrate Antigen 19-9 Level as a Prognostic Marker of Overall Survival in Locally Advanced Pancreatic Cancer Treated with Concurrent Chemoradiotherapy. Int. J. Clin. Oncol. 2017, 22, 1069–1075. [Google Scholar] [CrossRef] [PubMed]
- Stiksma, J.; Grootendorst, D.C.; Van Der Linden, P.W.G. CA 19-9 as a Marker in Addition to CEA to Monitor Colorectal Cancer. Clin. Colorectal Cancer 2014, 13, 239–244. [Google Scholar] [CrossRef]
- Charkhchi, P.; Cybulski, C.; Gronwald, J.; Wong, F.O.; Narod, S.A.; Akbari, M.R. CA125 and Ovarian Cancer: A Comprehensive Review. Cancers 2020, 12, 3730. [Google Scholar] [CrossRef]
- Funston, G.; Van Melle, M.; Baun, M.L.L.; Jensen, H.; Helsper, C.; Emery, J.; Crosbie, E.J.; Thompson, M.; Hamilton, W.; Walter, F.M. Variation in the Initial Assessment and Investigation for Ovarian Cancer in Symptomatic Women: A Systematic Review of International Guidelines. BMC Cancer 2019, 19. [Google Scholar] [CrossRef]
- Mckertish, C.M.; Kayser, V. Advances and Limitations of Antibody Drug Conjugates for Cancer. Biomedicines 2021, 9, 872. [Google Scholar] [CrossRef]
- Jin, S.; Sun, Y.; Liang, X.; Gu, X.; Ning, J.; Xu, Y.; Chen, S.; Pan, L. Emerging New Therapeutic Antibody Derivatives for Cancer Treatment. Signal Transduct. Target. Ther. 2022, 7, 1–28. [Google Scholar] [CrossRef]
- Karcini, A.; Lazar, I.M. The SKBR3 Cell-Membrane Proteome Reveals Telltales of Aberrant Cancer Cell Proliferation and Targets for Precision Medicine Applications. Sci. Rep. 2022, 12, 10847. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
de Jong, E.; Kocer, A. Current Methods for Identifying Plasma Membrane Proteins as Cancer Biomarkers. Membranes 2023, 13, 409. https://doi.org/10.3390/membranes13040409
de Jong E, Kocer A. Current Methods for Identifying Plasma Membrane Proteins as Cancer Biomarkers. Membranes. 2023; 13(4):409. https://doi.org/10.3390/membranes13040409
Chicago/Turabian Stylede Jong, Edwin, and Armagan Kocer. 2023. "Current Methods for Identifying Plasma Membrane Proteins as Cancer Biomarkers" Membranes 13, no. 4: 409. https://doi.org/10.3390/membranes13040409
APA Stylede Jong, E., & Kocer, A. (2023). Current Methods for Identifying Plasma Membrane Proteins as Cancer Biomarkers. Membranes, 13(4), 409. https://doi.org/10.3390/membranes13040409






