Influence of Cholesterol on the Orientation of the Farnesylated GTP-Bound KRas-4B Binding with Anionic Model Membranes
Abstract
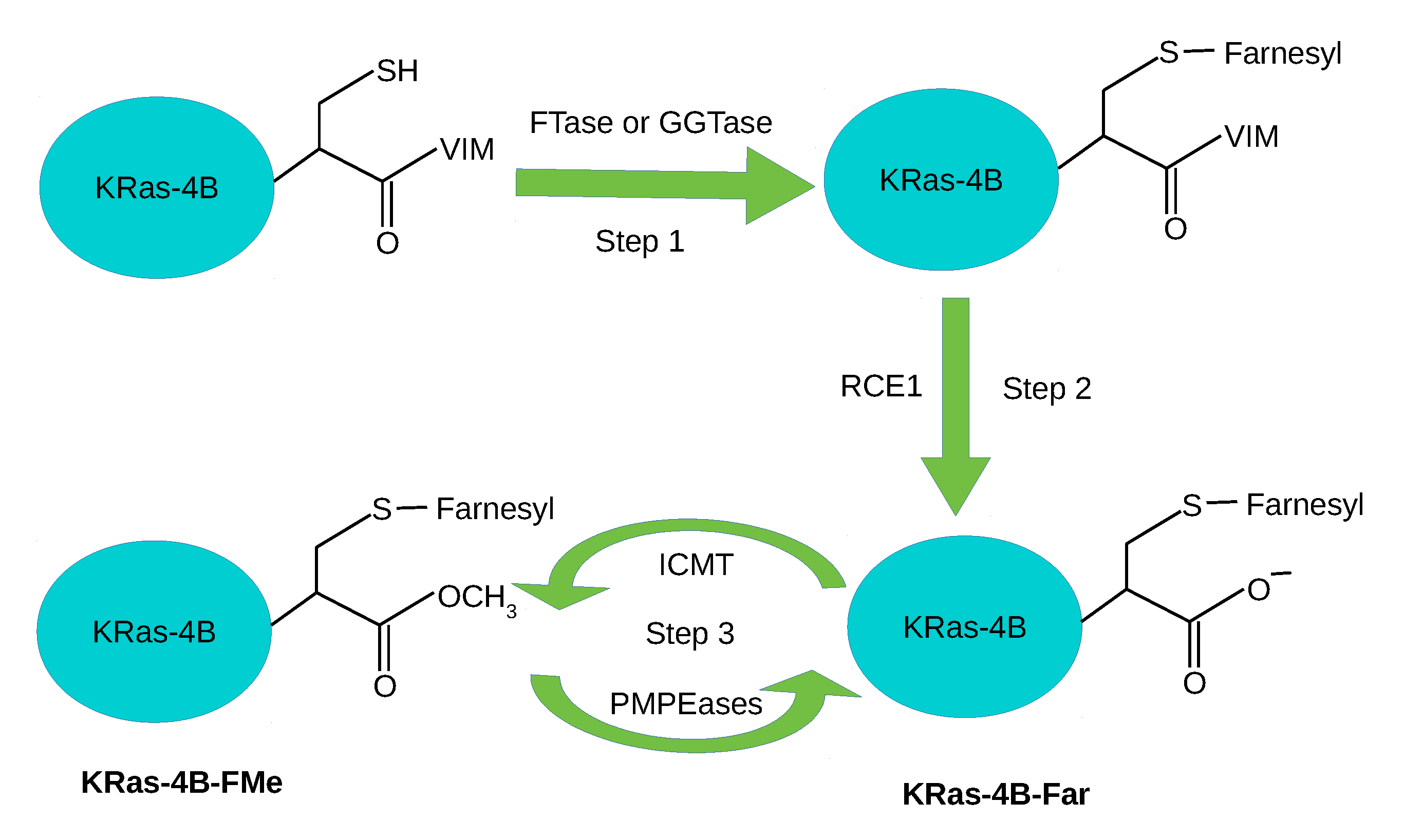
1. Introduction
2. Results and Discussion
2.1. Area Per Lipid
2.2. Preferential Localization of Kras-4b-Far on Membranes
2.3. Conformation of the 5-Aa-Sequence in The Hvr
2.4. Orientational Distributions of Kras-4b-Far on Different Anionic Membranes
2.5. Reorientation of Mutant Kras-4b-Far on the Anionic Membranes
3. Methods
4. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| MD | molecular dynamics |
| PM | plasma membrane |
| DOPC | 1,2-dioleoyl-sn-glycero-3-phosphocholine |
| DOPS | 1,2-dioleoyl-sn-glycero-3-phospho-L-serine |
| GDP | guanosine diphosphate |
| GTP | guanosine triphosphate |
| GEF | guanine-nucleotide-exchange factors |
| GAP | GTPase-activating proteins |
| COSMIC | catalog of somatic mutations in cancer |
| KRas-4B-Far | farnesylated and demethylated KRas-4B |
| KRas-4B-FMe | farnesylated and methylated KRas-4B |
| HVR | hypervariable region |
| FAR | farnesyl group |
| CD | catalytic domain |
| PHOS | phosphorylated serine |
| PTMs | post-translational modifications |
| OS | orientation state |
| RMSD | root mean square deviation |
| PDE | phosphodiest-erase |
| FTase | farnesyltrasferase |
| GGTase | geranylgeranyltransferase |
| RCE1 | Ras-converting enzyme 1 |
| ICMT | isoprenylcysteine carboxyl methyltransferase |
| PMPEases | prenylated/polyisoprenylated methylated protein methyl esterases |
| TSPO | translocator protein |
Appendix A. Supporting Information
Appendix A.1. Area Per Lipid
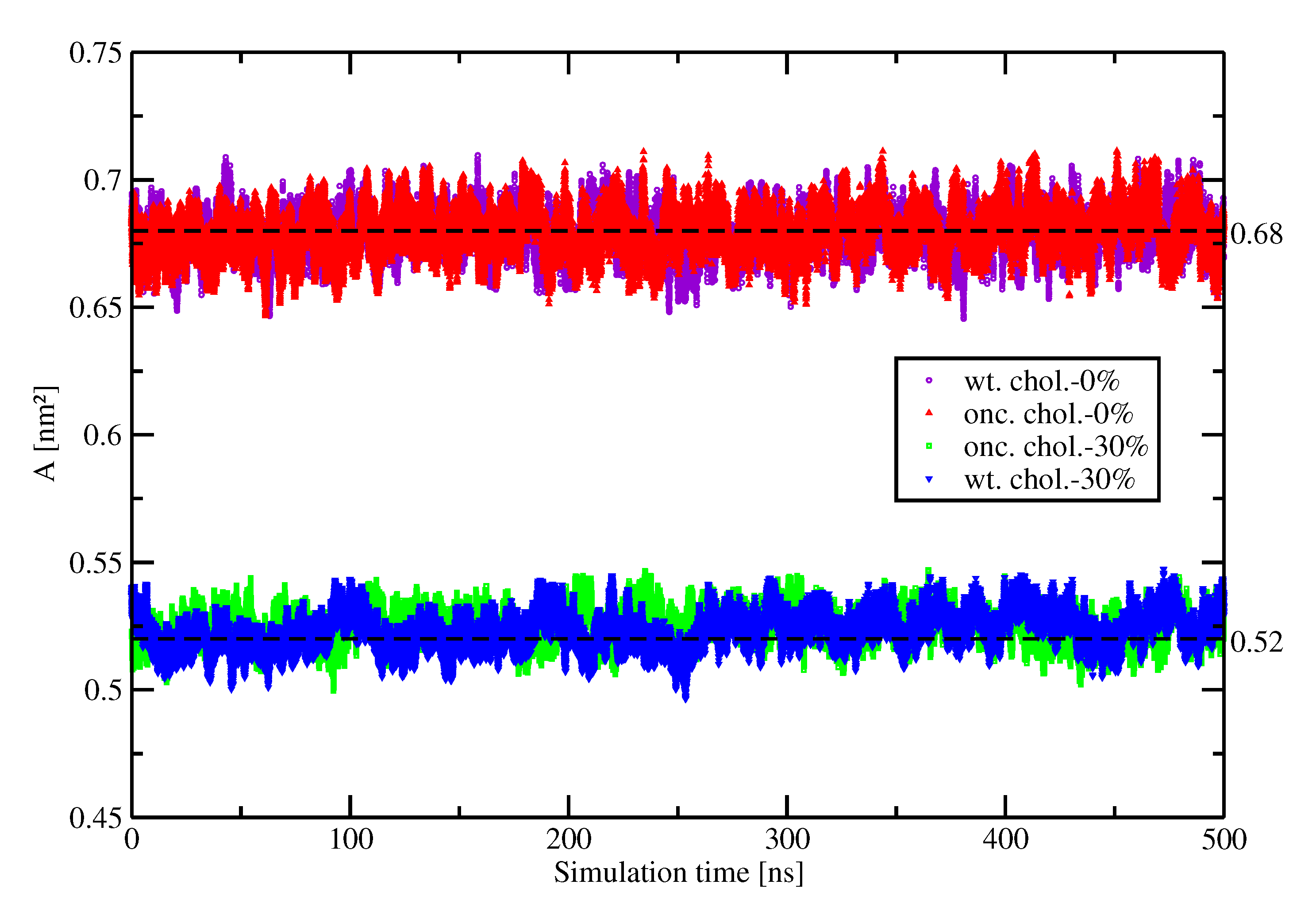
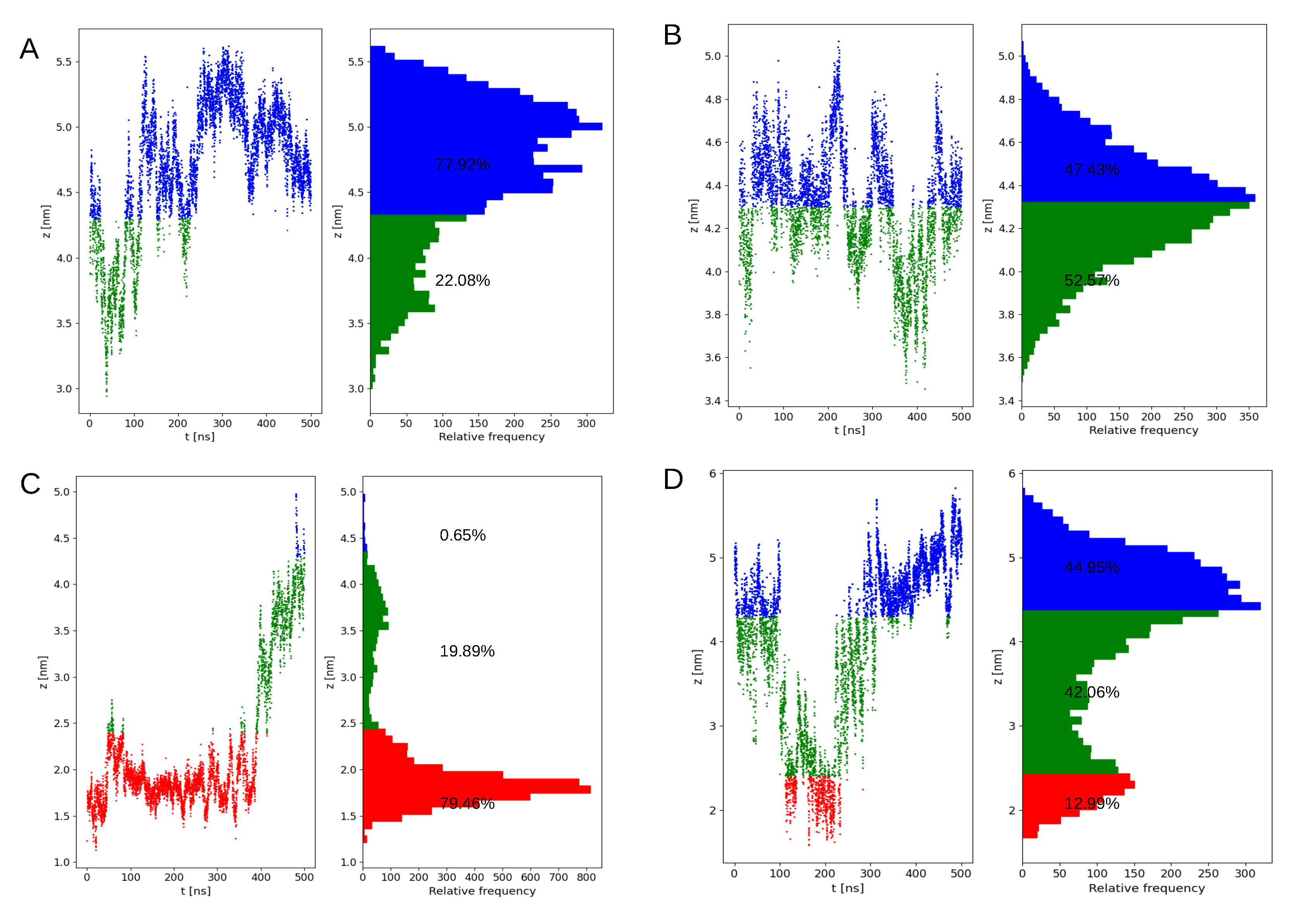
Appendix A.2. Orientation of Kras-4b on the Pm
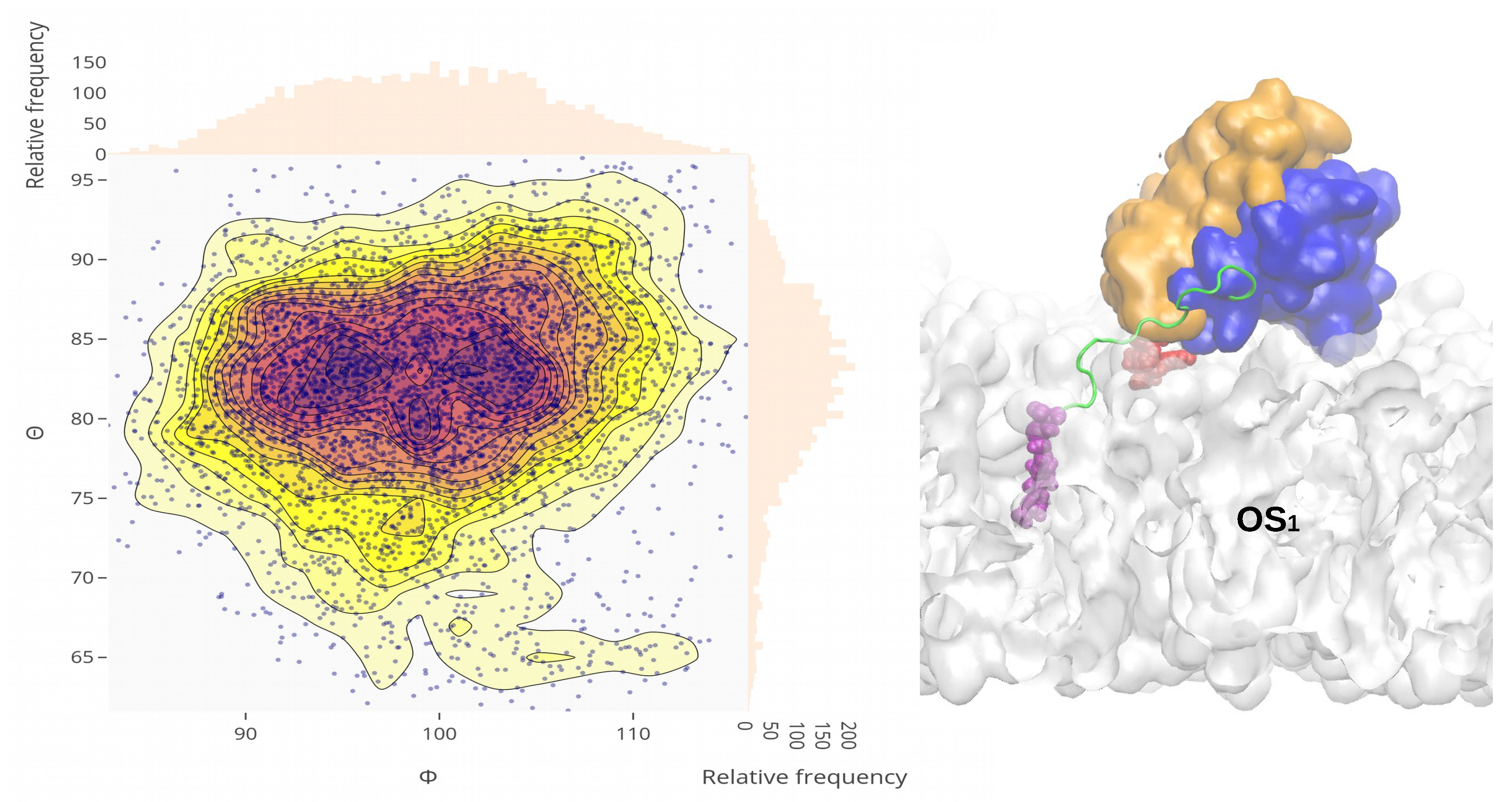
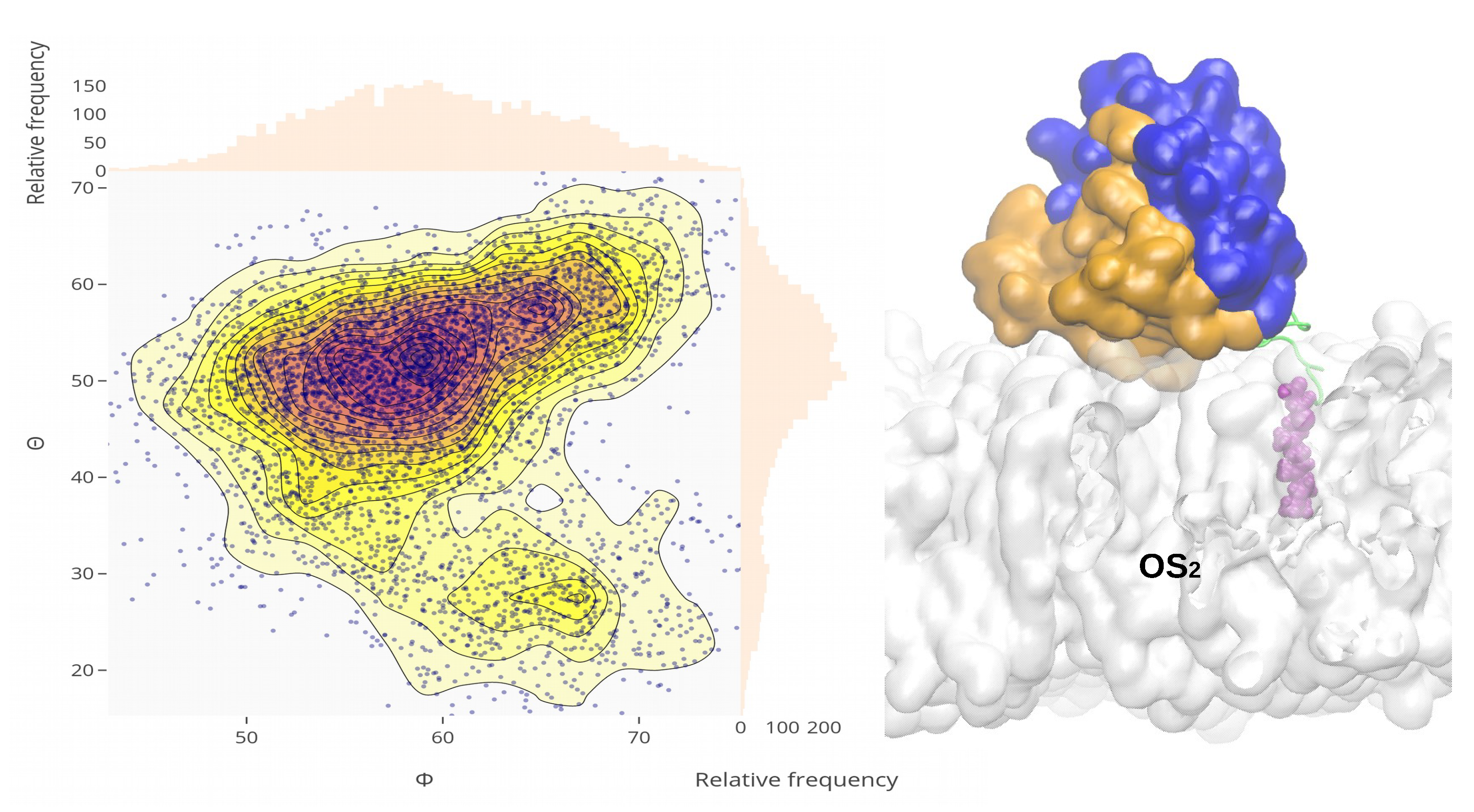

Appendix A.3. Sequences of Wild-Type and Mutant Kras-4b-Far Proteins

References
- Mouritsen, O.G. Life-as a Matter of Fat; Springer: Berlin/Heidelberg, Germany, 2005. [Google Scholar]
- Nagle, J.F.; Tristram-Nagle, S. Structure of lipid bilayers. Biochim. Biophys. Acta (BBA)-Rev. Biomembr. 2000, 1469, 159–195. [Google Scholar] [CrossRef]
- Zhou, Y.; Liang, H.; Rodkey, T.; Ariotti, N.; Parton, R.G.; Hancock, J.F. Signal integration by lipid-mediated spatial cross talk between Ras nanoclusters. Mol. Cell. Biol. 2014, 34, 862–876. [Google Scholar] [CrossRef] [PubMed]
- McLaughlin, S.; Murray, D. Plasma membrane phosphoinositide organization by protein electrostatics. Nature 2005, 438, 605–611. [Google Scholar] [CrossRef] [PubMed]
- Ingólfsson, H.I.; Melo, M.N.; Van Eerden, F.J.; Arnarez, C.; Lopez, C.A.; Wassenaar, T.A.; Periole, X.; De Vries, A.H.; Tieleman, D.P.; Marrink, S.J. Lipid organization of the plasma membrane. J. Am. Chem. Soc. 2014, 136, 14554–14559. [Google Scholar] [CrossRef]
- Zhang, Y.; Chen, X.; Gueydan, C.; Han, J. Plasma membrane changes during programmed cell deaths. Cell Res. 2018, 28, 9–21. [Google Scholar] [CrossRef]
- Krapf, D. Compartmentalization of the plasma membrane. Curr. Opin. Cell Biol. 2018, 53, 15–21. [Google Scholar] [CrossRef]
- Zhang, J.; Jin, R.; Jiang, D.; Chen, H.Y. Electrochemiluminescence-based capacitance microscopy for label-free imaging of antigens on the cellular plasma membrane. J. Am. Chem. Soc. 2019, 141, 10294–10299. [Google Scholar] [CrossRef]
- Nussinov, R.; Tsai, C.J.; Jang, H. Oncogenic Ras isoforms signaling specificity at the membrane. Cancer Res. 2018, 78, 593–602. [Google Scholar] [CrossRef]
- Bernards, A.; Settleman, J. GAP control: Regulating the regulators of small GTPases. Trends Cell Biol. 2004, 14, 377–385. [Google Scholar] [CrossRef]
- Wennerberg, K.; Rossman, K.L.; Der, C.J. The Ras superfamily at a glance. J. Cell Sci. 2005, 118, 843–846. [Google Scholar] [CrossRef]
- Schmidt, A.; Hall, A. Guanine nucleotide exchange factors for Rho GTPases: Turning on the switch. Genes Dev. 2002, 16, 1587–1609. [Google Scholar] [CrossRef] [PubMed]
- Stephen, A.G.; Esposito, D.; Bagni, R.K.; McCormick, F. Dragging ras back in the ring. Cancer Cell 2014, 25, 272–281. [Google Scholar] [CrossRef] [PubMed]
- Forbes, S.A.; Bindal, N.; Bamford, S.; Cole, C.; Kok, C.Y.; Beare, D.; Jia, M.; Shepherd, R.; Leung, K.; Menzies, A.; et al. COSMIC: Mining complete cancer genomes in the Catalogue of Somatic Mutations in Cancer. Nucleic Acids Res. 2010, 39, D945–D950. [Google Scholar] [CrossRef] [PubMed]
- Hobbs, G.A.; Der, C.J.; Rossman, K.L. RAS isoforms and mutations in cancer at a glance. J. Cell Sci. 2016, 129, 1287–1292. [Google Scholar] [CrossRef] [PubMed]
- Gelabert-Baldrich, M.; Soriano-Castell, D.; Calvo, M.; Lu, A.; Viña-Vilaseca, A.; Rentero, C.; Pol, A.; Grinstein, S.; Enrich, C.; Tebar, F. Dynamics of KRas on endosomes: Involvement of acidic phospholipids in its association. FASEB J. 2014, 28, 3023–3037. [Google Scholar] [CrossRef]
- Cho, K.J.; van der Hoeven, D.; Zhou, Y.; Maekawa, M.; Ma, X.; Chen, W.; Fairn, G.D.; Hancock, J.F. Inhibition of acid sphingomyelinase depletes cellular phosphatidylserine and mislocalizes K-Ras from the plasma membrane. Mol. Cell. Biol. 2016, 36, 363–374. [Google Scholar] [CrossRef]
- Gregory, M.C.; McLean, M.A.; Sligar, S.G. Interaction of KRas4b with anionic membranes: A special role for PIP2. Biochem. Biophys. Res. Commun. 2017, 487, 351–355. [Google Scholar] [CrossRef]
- Zhou, Y.; Wong, C.O.; Cho, K.J.; Van Der Hoeven, D.; Liang, H.; Thakur, D.P.; Luo, J.; Babic, M.; Zinsmaier, K.E.; Zhu, M.X.; et al. Membrane potential modulates plasma membrane phospholipid dynamics and K-Ras signaling. Science 2015, 349, 873–876. [Google Scholar] [CrossRef]
- Prakash, P.; Litwin, D.; Liang, H.; Sarkar-Banerjee, S.; Dolino, D.; Zhou, Y.; Hancock, J.F.; Jayaraman, V.; Gorfe, A.A. Dynamics of membrane-bound G12V-KRAS from simulations and single-molecule FRET in native nanodiscs. Biophys. J. 2019, 116, 179–183. [Google Scholar] [CrossRef]
- Prakash, P.; Zhou, Y.; Liang, H.; Hancock, J.F.; Gorfe, A.A. Oncogenic K-Ras binds to an anionic membrane in two distinct orientations: A molecular dynamics analysis. Biophys. J. 2016, 110, 1125–1138. [Google Scholar] [CrossRef]
- Prakash, P.; Gorfe, A.A. Probing the conformational and energy landscapes of KRAS membrane orientation. J. Phys. Chem. B 2019, 123, 8644–8652. [Google Scholar] [CrossRef] [PubMed]
- Tsai, C.J.; Ma, B.; Sham, Y.Y.; Kumar, S.; Nussinov, R. Structured disorder and conformational selection. Proteins Struct. Funct. Bioinform. 2001, 44, 418–427. [Google Scholar] [CrossRef] [PubMed]
- Lu, H.; Martí, J. Long-lasting Salt Bridges Provide the Anchoring Mechanism of Oncogenic Kirsten Rat Sarcoma Proteins at Cell Membranes. J. Phys. Chem. Lett. 2020, 11, 9938–9945. [Google Scholar] [CrossRef] [PubMed]
- Cao, S.; Chung, S.; Kim, S.; Li, Z.; Manor, D.; Buck, M. K-Ras G-domain binding with signaling lipid phosphatidylinositol (4, 5)-phosphate (PIP2): Membrane association, protein orientation, and function. J. Biol. Chem. 2019, 294, 7068–7084. [Google Scholar] [CrossRef] [PubMed]
- Yang, M.H.; Laurent, G.; Bause, A.S.; Spang, R.; German, N.; Haigis, M.C.; Haigis, K.M. HDAC6 and SIRT2 regulate the acetylation state and oncogenic activity of mutant K-RAS. Mol. Cancer Res. 2013, 11, 1072–1077. [Google Scholar] [CrossRef]
- Lu, S.; Jang, H.; Gu, S.; Zhang, J.; Nussinov, R. Drugging Ras GTPase: A comprehensive mechanistic and signaling structural view. Chem. Soc. Rev. 2016, 45, 4929–4952. [Google Scholar] [CrossRef]
- Ahearn, I.M.; Haigis, K.; Bar-Sagi, D.; Philips, M.R. Regulating the regulator: Post-translational modification of RAS. Nat. Rev. Mol. Cell Biol. 2012, 13, 39–51. [Google Scholar] [CrossRef]
- Zhou, B.; Cox, A.D. Posttranslational Modifications of Small G Proteins. In Ras Superfamily Small G Proteins: Biology and Mechanisms 1; Springer: Vienna, Austria, 2014; pp. 99–131. [Google Scholar]
- Abdelkarim, H.; Banerjee, A.; Grudzien, P.; Leschinsky, N.; Abushaer, M.; Gaponenko, V. The Hypervariable Region of K-Ras4B Governs Molecular Recognition and Function. Int. J. Mol. Sci. 2019, 20, 5718. [Google Scholar] [CrossRef]
- Ahearn, I.; Zhou, M.; Philips, M.R. Posttranslational modifications of RAS proteins. Cold Spring Harb. Perspect. Med. 2018, a031484. [Google Scholar] [CrossRef]
- Konstantinopoulos, P.A.; Karamouzis, M.V.; Papavassiliou, A.G. Post-translational modifications and regulation of the RAS superfamily of GTPases as anticancer targets. Nat. Rev. Drug Discov. 2007, 6, 541–555. [Google Scholar] [CrossRef]
- Zhang, S.Y.; Sperlich, B.; Li, F.Y.; Al-Ayoubi, S.; Chen, H.X.; Zhao, Y.F.; Li, Y.M.; Weise, K.; Winter, R.; Chen, Y.X. Phosphorylation weakens but does not inhibit membrane binding and clustering of K-Ras4B. ACS Chem. Biol. 2017, 12, 1703–1710. [Google Scholar] [CrossRef] [PubMed]
- Alvarez-Moya, B.; Lopez-Alcala, C.; Drosten, M.; Bachs, O.; Agell, N. K-Ras4B phosphorylation at Ser181 is inhibited by calmodulin and modulates K-Ras activity and function. Oncogene 2010, 29, 5911–5922. [Google Scholar] [CrossRef] [PubMed]
- Bivona, T.G.; Quatela, S.E.; Bodemann, B.O.; Ahearn, I.M.; Soskis, M.J.; Mor, A.; Miura, J.; Wiener, H.H.; Wright, L.; Saba, S.G.; et al. PKC regulates a farnesyl-electrostatic switch on K-Ras that promotes its association with Bcl-XL on mitochondria and induces apoptosis. Mol. Cell 2006, 21, 481–493. [Google Scholar] [CrossRef] [PubMed]
- Kollár, P.; Rajchard, J.; Balounová, Z.; Pazourek, J. Marine natural products: Bryostatins in preclinical and clinical studies. Pharm. Biol. 2014, 52, 237–242. [Google Scholar] [CrossRef]
- Barceló, C.; Paco, N.; Morell, M.; Alvarez-Moya, B.; Bota-Rabassedas, N.; Jaumot, M.; Vilardell, F.; Capella, G.; Agell, N. Phosphorylation at Ser-181 of oncogenic KRAS is required for tumor growth. Cancer Res. 2014, 74, 1190–1199. [Google Scholar] [CrossRef]
- Jang, H.; Abraham, S.J.; Chavan, T.S.; Hitchinson, B.; Khavrutskii, L.; Tarasova, N.I.; Nussinov, R.; Gaponenko, V. Mechanisms of membrane binding of small GTPase K-Ras4B farnesylated hypervariable region. J. Biol. Chem. 2015, 290, 9465–9477. [Google Scholar] [CrossRef]
- Alvarez-Moya, B.; Barceló, C.; Tebar, F.; Jaumot, M.; Agell, N. CaM interaction and Ser181 phosphorylation as new K-Ras signaling modulators. Small GTPases 2011, 2, 5911–5922. [Google Scholar] [CrossRef][Green Version]
- Dharmaiah, S.; Bindu, L.; Tran, T.H.; Gillette, W.K.; Frank, P.H.; Ghirlando, R.; Nissley, D.V.; Esposito, D.; McCormick, F.; Stephen, A.G.; et al. Structural basis of recognition of farnesylated and methylated KRAS4b by PDEδ. Proc. Natl. Acad. Sci. USA 2016, 113, E6766–E6775. [Google Scholar] [CrossRef]
- Schmick, M.; Vartak, N.; Papke, B.; Kovacevic, M.; Truxius, D.C.; Rossmannek, L.; Bastiaens, P.I. KRas localizes to the plasma membrane by spatial cycles of solubilization, trapping and vesicular transport. Cell 2014, 157, 459–471. [Google Scholar] [CrossRef]
- Zimmermann, G.; Papke, B.; Ismail, S.; Vartak, N.; Chandra, A.; Hoffmann, M.; Hahn, S.A.; Triola, G.; Wittinghofer, A.; Bastiaens, P.I.; et al. Small molecule inhibition of the KRAS–PDEδ interaction impairs oncogenic KRAS signalling. Nature 2013, 497, 638–642. [Google Scholar] [CrossRef]
- Muratcioglu, S.; Jang, H.; Gursoy, A.; Keskin, O.; Nussinov, R. PDEδ binding to Ras isoforms provides a route to proper membrane localization. J. Phys. Chem. B 2017, 121, 5917–5927. [Google Scholar] [CrossRef] [PubMed]
- Murarka, S.; Martín-Gago, P.; Schultz-Fademrecht, C.; Al Saabi, A.; Baumann, M.; Fansa, E.K.; Ismail, S.; Nussbaumer, P.; Wittinghofer, A.; Waldmann, H. Development of pyridazinone chemotypes targeting the PDEδ prenyl binding site. Chem. Eur. J. 2017, 23, 6083–6093. [Google Scholar] [CrossRef] [PubMed]
- Ntai, I.; Fornelli, L.; DeHart, C.J.; Hutton, J.E.; Doubleday, P.F.; LeDuc, R.D.; van Nispen, A.J.; Fellers, R.T.; Whiteley, G.; Boja, E.S.; et al. Precise characterization of KRAS4b proteoforms in human colorectal cells and tumors reveals mutation/modification cross-talk. Proc. Natl. Acad. Sci. USA 2018, 115, 4140–4145. [Google Scholar] [CrossRef] [PubMed]
- McMullen, T.P.; Lewis, R.N.; McElhaney, R.N. Cholesterol–phospholipid interactions, the liquid-ordered phase and lipid rafts in model and biological membranes. Curr. Opin. Colloid Interface Sci. 2004, 8, 459–468. [Google Scholar] [CrossRef]
- Levitan, I.; Fang, Y.; Rosenhouse-Dantsker, A.; Romanenko, V. Cholesterol and ion channels. In Cholesterol Binding and Cholesterol Transport Proteins; Springer: Dordrecht, The Netherlands, 2010; pp. 509–549. [Google Scholar]
- Ding, X.; Zhang, W.; Li, S.; Yang, H. The role of cholesterol metabolism in cancer. Am. J. Cancer Res. 2019, 9, 219. [Google Scholar]
- Phillips, R. Membranes by the Numbers. In Physics of Biological Membranes; Springer: Cham, Switzerland, 2018; pp. 73–105. [Google Scholar]
- Liu, S.L.; Sheng, R.; Jung, J.H.; Wang, L.; Stec, E.; O’Connor, M.J.; Song, S.; Bikkavilli, R.K.; Winn, R.A.; Lee, D.; et al. Orthogonal lipid sensors identify transbilayer asymmetry of plasma membrane cholesterol. Nat. Chem. Biol. 2017, 13, 268–274. [Google Scholar] [CrossRef]
- Lange, Y.; Swaisgood, M.; Ramos, B.; Steck, T. Plasma membranes contain half the phospholipid and 90% of the cholesterol and sphingomyelin in cultured human fibroblasts. J. Biol. Chem. 1989, 264, 3786–3793. [Google Scholar]
- Das, A.; Brown, M.S.; Anderson, D.D.; Goldstein, J.L.; Radhakrishnan, A. Three pools of plasma membrane cholesterol and their relation to cholesterol homeostasis. Elife 2014, 3, e02882. [Google Scholar] [CrossRef]
- Lange, Y.; Slayton, J.M. Interaction of cholesterol and lysophosphatidylcholine in determining red cell shape. J. Lipid Res. 1982, 23, 1121–1127. [Google Scholar]
- Lu, H.; Martí, J. Effects of cholesterol on the binding of the precursor neurotransmitter tryptophan to zwitterionic membranes. J. Chem. Phys. 2018, 149, 164906. [Google Scholar] [CrossRef]
- Boughter, C.T.; Monje-Galvan, V.; Im, W.; Klauda, J.B. Influence of cholesterol on phospholipid bilayer structure and dynamics. J. Phys. Chem. B 2016, 120, 11761–11772. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.W.; Wiers, C.E.; Tyler, R.; Shokri-Kojori, E.; Jang, Y.J.; Zehra, A.; Freeman, C.; Ramirez, V.; Lindgren, E.; Miller, G.; et al. Influence of alcoholism and cholesterol on TSPO binding in brain: PET [11 C] PBR28 studies in humans and rodents. Neuropsychopharmacology 2018, 43, 1832–1839. [Google Scholar] [CrossRef] [PubMed]
- Miller, K.D.; Siegel, R.L.; Lin, C.C.; Mariotto, A.B.; Kramer, J.L.; Rowland, J.H.; Stein, K.D.; Alteri, R.; Jemal, A. Cancer treatment and survivorship statistics, 2016. CA Cancer J. Clin. 2016, 66, 271–289. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Qin, S.; Wang, M.; Zhang, T.; Zhang, S. Association between cholesterol intake and pancreatic cancer risk: Evidence from a meta-analysis. Sci. Rep. 2015, 5, 8243. [Google Scholar] [CrossRef] [PubMed]
- Huang, B.Z.; Chang, J.I.; Li, E.; Xiang, A.H.; Wu, B.U. Influence of statins and cholesterol on mortality among patients with pancreatic cancer. JNCI J. Natl. Cancer Inst. 2017, 109. [Google Scholar] [CrossRef]
- Karplus, M.; Petsko, G. Molecular dynamics simulations in biology. Nature 1990, 347, 631–639. [Google Scholar] [CrossRef]
- Karplus, M.; McCammon, J. Molecular dynamics simulations of biomolecules. Nat. Struct. Biol. 2002, 9, 646–652. [Google Scholar] [CrossRef]
- Nagy, G.; Gordillo, M.; Guàrdia, E.; Martí, J. Liquid water confined in carbon nanochannels at high temperatures. J. Phys. Chem. B 2007, 111, 12524–12530. [Google Scholar] [CrossRef]
- Poger, D.; Mark, A.E. On the validation of molecular dynamics simulations of saturated and cis-monounsaturated phosphatidylcholine lipid bilayers: A comparison with experiment. J. Chem. Theory Comput. 2010, 6, 325–336. [Google Scholar] [CrossRef]
- Lütgebaucks, C.; Macias-Romero, C.; Roke, S. Characterization of the interface of binary mixed DOPC: DOPS liposomes in water: The impact of charge condensation. J. Chem. Phys. 2017, 146, 044701. [Google Scholar] [CrossRef]
- Novakova, E.; Giewekemeyer, K.; Salditt, T. Structure of two-component lipid membranes on solid support: An x-ray reflectivity study. Phys. Rev. E 2006, 74, 051911. [Google Scholar] [CrossRef] [PubMed]
- Chaban, V. Computationally efficient prediction of area per lipid. Chem. Phys. Lett. 2014, 616, 25–29. [Google Scholar] [CrossRef]
- Petrache, H.I.; Tristram-Nagle, S.; Gawrisch, K.; Harries, D.; Parsegian, V.A.; Nagle, J.F. Structure and fluctuations of charged phosphatidylserine bilayers in the absence of salt. Biophys. J. 2004, 86, 1574–1586. [Google Scholar] [CrossRef]
- Lu, H.; Martí, J. Binding and dynamics of melatonin at the interface of phosphatidylcholine-cholesterol membranes. PLoS ONE 2019, 14, e0224624. [Google Scholar] [CrossRef]
- Litz, J.P.; Thakkar, N.; Portet, T.; Keller, S.L. Depletion with cyclodextrin reveals two populations of cholesterol in model lipid membranes. Biophys. J. 2016, 110, 635–645. [Google Scholar] [CrossRef]
- Kufareva, I.; Abagyan, R. Methods of protein structure comparison. In Homology Modeling; Springer: Berlin/Heidelberg, Germany; Humana Press: Totowa, NJ, USA, 2011; pp. 231–257. [Google Scholar]
- Abankwa, D.; Gorfe, A.A.; Inder, K.; Hancock, J.F. Ras membrane orientation and nanodomain localization generate isoform diversity. Proc. Natl. Acad. Sci. USA 2010, 107, 1130–1135. [Google Scholar] [CrossRef]
- Kapoor, S.; Triola, G.; Vetter, I.R.; Erlkamp, M.; Waldmann, H.; Winter, R. Revealing conformational substates of lipidated N-Ras protein by pressure modulation. Proc. Natl. Acad. Sci. USA 2012, 109, 460–465. [Google Scholar] [CrossRef]
- Mazhab-Jafari, M.T.; Marshall, C.B.; Smith, M.J.; Gasmi-Seabrook, G.M.; Stathopulos, P.B.; Inagaki, F.; Kay, L.E.; Neel, B.G.; Ikura, M. Oncogenic and RASopathy-associated K-RAS mutations relieve membrane-dependent occlusion of the effector-binding site. Proc. Natl. Acad. Sci. USA 2015, 112, 6625–6630. [Google Scholar] [CrossRef]
- Yan, S.; Qu, X.; Xu, L.; Che, X.; Ma, Y.; Zhang, L.; Teng, Y.; Zou, H.; Liu, Y. Bufalin enhances TRAIL-induced apoptosis by redistributing death receptors in lipid rafts in breast cancer cells. Anti-Cancer Drugs 2014, 25, 683–689. [Google Scholar] [CrossRef]
- Kuzu, O.F.; Noory, M.A.; Robertson, G.P. The role of cholesterol in cancer. Cancer Res. 2016, 76, 2063–2070. [Google Scholar] [CrossRef]
- Ravnskov, U.; Rosch, P.J.; McCully, K.S. Statins do not protect against cancer: Quite the opposite. J. Clin. Oncol. 2015, 33, 810–811. [Google Scholar] [CrossRef] [PubMed]
- Ravnskov, U.; McCully, K.; Rosch, P. The statin-low cholesterol-cancer conundrum. QJM Int. J. Med. 2012, 105, 383–388. [Google Scholar] [CrossRef] [PubMed]
- Nielsen, S.F.; Nordestgaard, B.G.; Bojesen, S.E. Statin use and reduced cancer-related mortality. N. Engl. J. Med. 2012, 367, 1792–1802. [Google Scholar] [CrossRef] [PubMed]
- Radišauskas, R.; Kuzmickienė, I.; Milinavičienė, E.; Everatt, R. Hypertension, serum lipids and cancer risk: A review of epidemiological evidence. Medicina 2016, 52, 89–98. [Google Scholar] [CrossRef]
- Murai, T. Cholesterol lowering: Role in cancer prevention and treatment. Biol. Chem. 2015, 396, 1–11. [Google Scholar] [CrossRef]
- Khan, I.; Spencer-Smith, R.; O’Bryan, J.P. Targeting the α4–α5 dimerization interface of K-RAS inhibits tumor formation in vivo. Oncogene 2019, 38, 2984–2993. [Google Scholar] [CrossRef]
- Spencer-Smith, R.; Koide, A.; Zhou, Y.; Eguchi, R.R.; Sha, F.; Gajwani, P.; Santana, D.; Gupta, A.; Jacobs, M.; Herrero-Garcia, E.; et al. Inhibition of RAS function through targeting an allosteric regulatory site. Nat. Chem. Biol. 2017, 13, 62–68. [Google Scholar] [CrossRef]
- Ambrogio, C.; Köhler, J.; Zhou, Z.W.; Wang, H.; Paranal, R.; Li, J.; Capelletti, M.; Caffarra, C.; Li, S.; Lv, Q.; et al. KRAS dimerization impacts MEK inhibitor sensitivity and oncogenic activity of mutant KRAS. Cell 2018, 172, 857–868. [Google Scholar] [CrossRef]
- Jo, S.; Kim, T.; Iyer, V.G.; Im, W. CHARMM-GUI: A web-based graphical user interface for CHARMM. J. Comput. Chem. 2008, 29, 1859–1865. [Google Scholar] [CrossRef]
- Huang, J.; Rauscher, S.; Nawrocki, G.; Ran, T.; Feig, M.; de Groot, B.L.; Grubmüller, H.; MacKerell, A.D. CHARMM36m: An improved force field for folded and intrinsically disordered proteins. Nat. Methods 2017, 14, 71–73. [Google Scholar] [CrossRef]
- Huang, J.; MacKerell, A.D., Jr. CHARMM36 all-atom additive protein force field: Validation based on comparison to NMR data. J. Comput. Chem. 2013, 34, 2135–2145. [Google Scholar] [CrossRef] [PubMed]
- Lemkul, J. From proteins to perturbed Hamiltonians: A suite of tutorials for the GROMACS-2018 molecular simulation package [article v1. 0]. Living J. Comput. Mol. Sci. 2018, 1, 5068. [Google Scholar] [CrossRef]
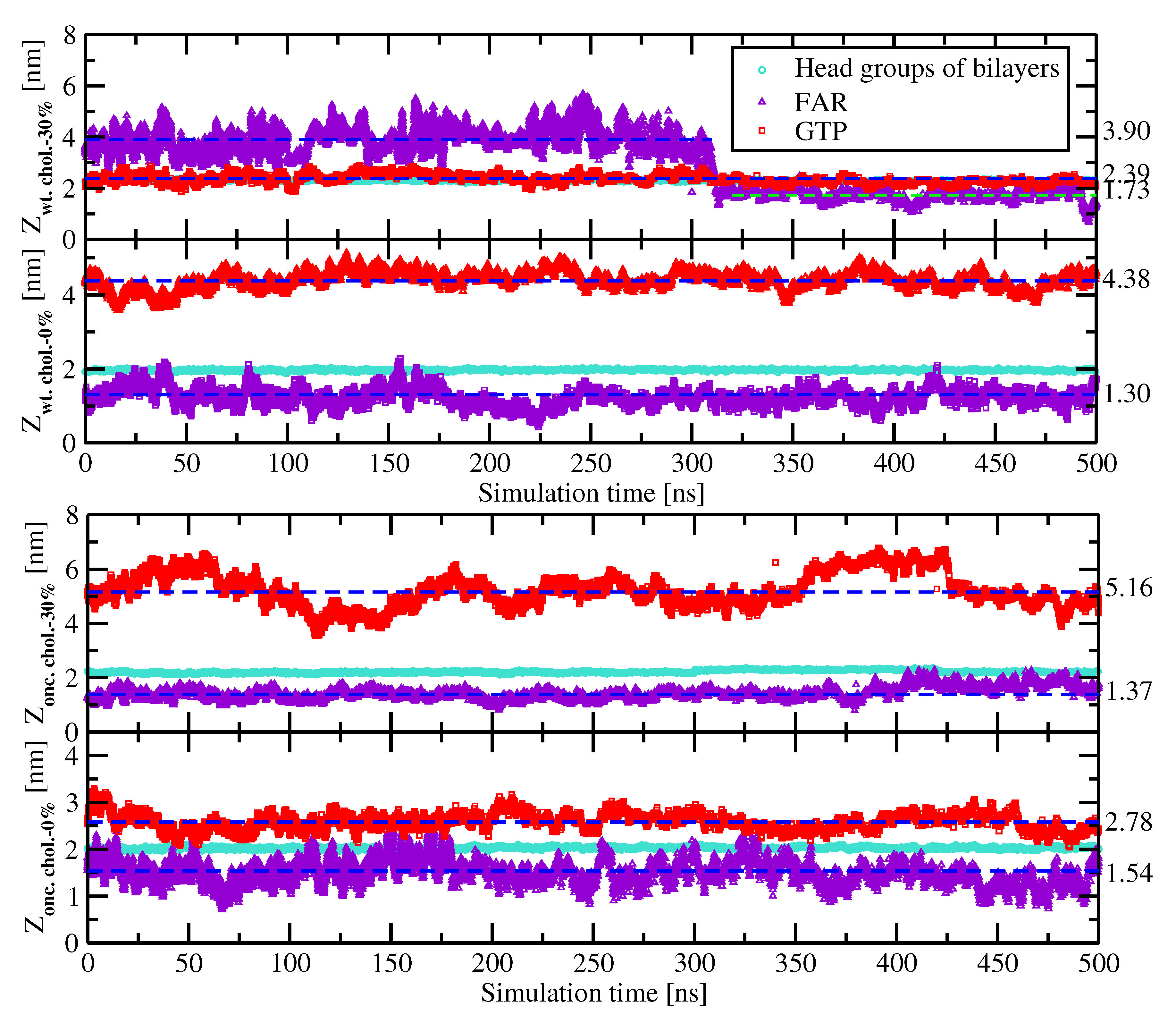

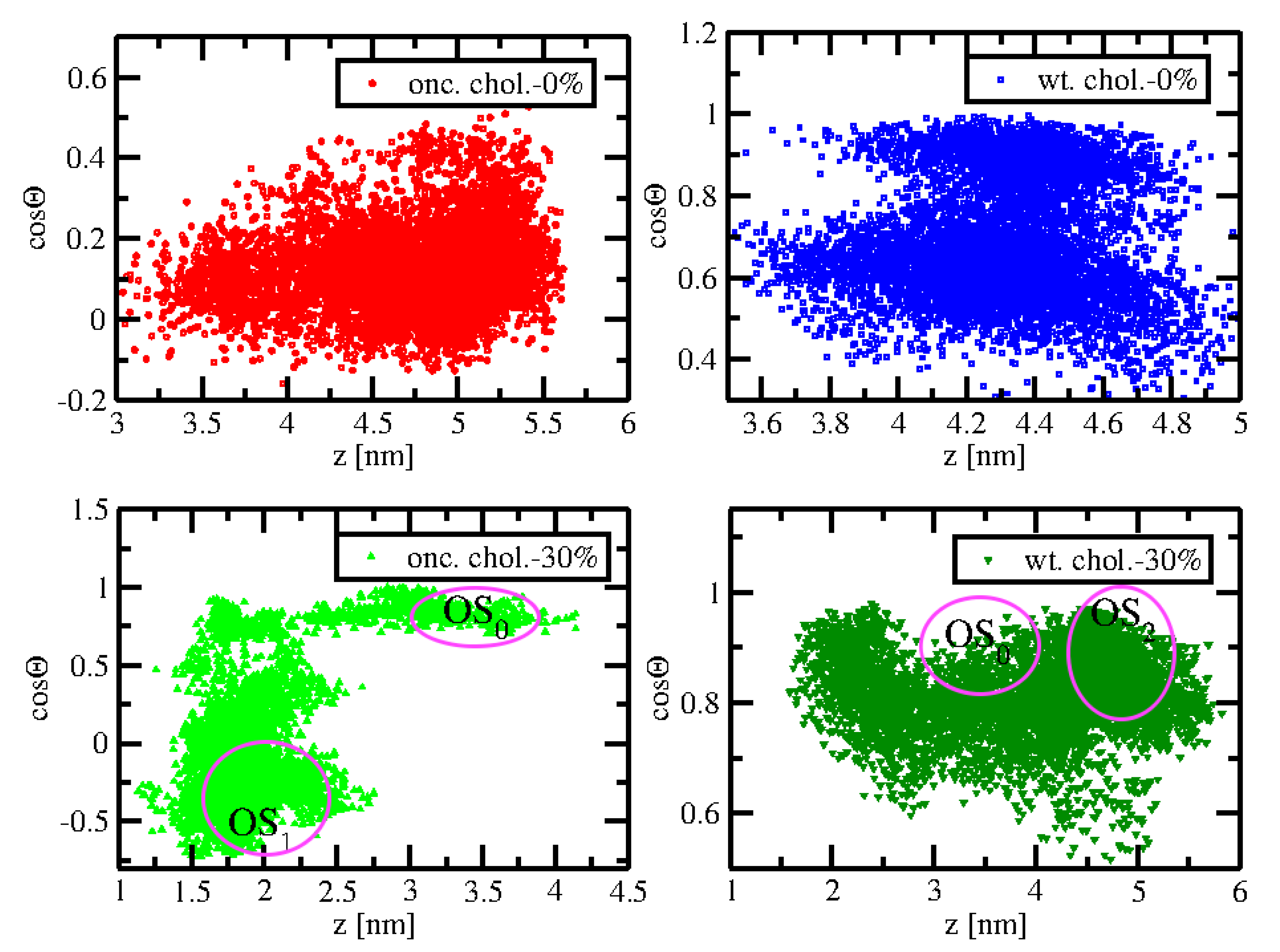
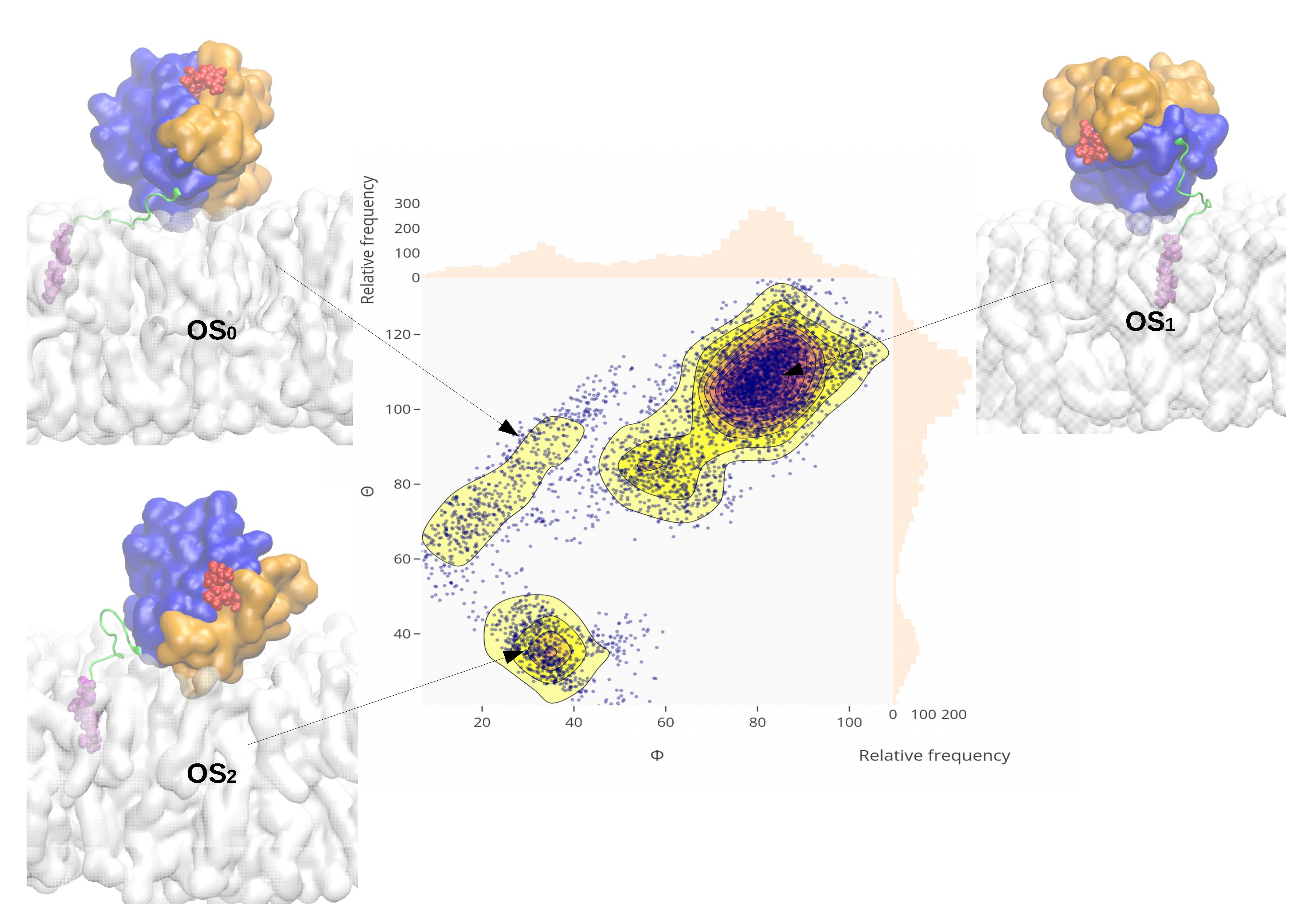
| System | A (nm) | z (nm) |
|---|---|---|
| wt. chol.-0% | 0.679 (0.008) | 3.84 (0.04) |
| wt. chol.-30% [24] | 0.523 (0.007) | 4.35 (0.05) |
| onc. chol.-0% | 0.679 (0.008) | 3.89 (0.04) |
| onc. chol.-30% [24] | 0.525 (0.006) | 4.23 (0.04) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lu, H.; Martí, J. Influence of Cholesterol on the Orientation of the Farnesylated GTP-Bound KRas-4B Binding with Anionic Model Membranes. Membranes 2020, 10, 364. https://doi.org/10.3390/membranes10110364
Lu H, Martí J. Influence of Cholesterol on the Orientation of the Farnesylated GTP-Bound KRas-4B Binding with Anionic Model Membranes. Membranes. 2020; 10(11):364. https://doi.org/10.3390/membranes10110364
Chicago/Turabian StyleLu, Huixia, and Jordi Martí. 2020. "Influence of Cholesterol on the Orientation of the Farnesylated GTP-Bound KRas-4B Binding with Anionic Model Membranes" Membranes 10, no. 11: 364. https://doi.org/10.3390/membranes10110364
APA StyleLu, H., & Martí, J. (2020). Influence of Cholesterol on the Orientation of the Farnesylated GTP-Bound KRas-4B Binding with Anionic Model Membranes. Membranes, 10(11), 364. https://doi.org/10.3390/membranes10110364






