Abstract
Prior studies have yielded mixed findings concerning the association between apolipoprotein E(APOE)-ε4 and serum lipids in patients with Alzheimer’s disease (AD) and healthy individuals. Some studies suggested a relationship between APOEε4 and serum lipids in patients with AD and healthy individuals, whereas others proposed that the APOEε4 allele affects lipids only in patients with AD. Our study aimed to investigate whether APOE alleles have a distinct impact on lipids in AD. We conducted a comprehensive search of the PubMed and Embase databases for all related studies that investigate APOE and serum lipids of AD from the inception to 30 May 2022. Elevated total cholesterol (TC) and low-density lipoprotein (LDL) levels were found in APOEε4 allele carriers compared with non-carriers. No significant differences were found for high-density lipoprotein (HDL) and triglyceride (TG) levels in APOEε4 allele carriers compared to non-carriers. Notably, elevated TC and LDL levels showed considerable heterogeneity between patients with AD and healthy controls. A network meta-analysis did not find a distinct effect of carrying one or two APOEε4 alleles on lipid profiles. Higher TC and LDL levels were found in APOEε4 allele carriers compared with non-carriers, and the difference was more significant in patients with AD than in healthy controls.
1. Introduction
Alzheimer’s disease (AD) presents as a prevalent progressive neurodegenerative disease characterized by an insidious onset, progressive memory decline, cognitive impairment, and a spectrum of behavioral and psychological symptoms [1]. The development of AD appears to be a result of the complex interplay of genetic and environmental factors [2], hence rendering effective treatment of AD a formidable challenge [3]. The multifactorial etiology of this global health challenge has driven many research endeavors to unravel the complex web of causative elements of AD. Among these factors are apolipoprotein E (APOE) and its allelic variants, specifically the APOE ε4 allele, which have emerged as being noteworthy.
The human APOE gene is encoded on chromosome 19, and it has three allelic variants: ε2, ε3, and ε4 [4]. Notably, the individuals carrying the APOEε4 allele exhibit a high risk of sporadic AD [5]. Individuals with a single APOEε4 allele have a 3.2 times higher risk of developing AD, whereas, in those with two APOEε4 alleles, the risk of developing AD is increased by 8 to 10 folds [6]. This can be attributed to the influence of the APOEε4 allele on amyloid-β (Aβ), either by reducing its clearance or by increasing its production in the brain [7].
In neuroimaging investigations of APOE polymorphism in healthy individuals, there has been a predominant focus on examining gray matter alterations in middle or late life, particularly in brain regions associated with significant AD pathological findings. Even in individuals showing no clinical symptoms, documentation has shown a reduction in the gray matter within the hippocampal and frontotemporal regions in APOEε4 allele carriers compared with non-carriers [8].
Moreover, the human APOE allele encodes a polyclonal lipoprotein integral to metabolic processes, including cholesterol transport [9]. Although APOE alleles have a certain impact on lipid profiles [10,11,12,13,14], current research results are inconsistent [11,14]. Some studies have identified elevated levels of low-density lipoprotein (LDL) and total cholesterol (TC) in APOEε4 allele carriers (APOEε4 allele-C) compared with non-carriers (APOEε4 allele-N), whereas others [10,12] have reported the opposite. Furthermore, some studies [13,15] have reported that significant differences exist in high-density lipoprotein (HDL) levels between carriers and non-carriers of the APOEε4 allele. However, such distinctions were not observed in other studies [12,13]. Intriguingly, no systematic analyses have focused on the differences in lipid profiles between single APOEε4 allele carriers and APOEε4 homozygous individuals concerning lipid profiles.
Most researchers believe that lipid metabolism is very important in the pathophysiological mechanism of AD [16]. Notably, the latest meta-analysis summarized the disparities in lipid profiles between individuals with AD and healthy controls [17]. Since the APOEε4 allele affects both lipid metabolism and the pathophysiology of AD, it has been hypothesized that the special relationship between the APOEε4 allele and lipid metabolism is unique in AD. Some studies have found a relationship between the APOEε4 allele and lipid profiles in patients with AD and healthy control populations [18], whereas others have discerned this association exclusively within the AD population [13].
Additionally, most meta-analyses summarized the differences in lipids between patients with AD and healthy controls, but there has been no relevant summary analysis that has explored whether the unique relationship between the APOEε4 allele and lipids differs between patients with AD and healthy controls. Therefore, we systematically compared the lipid profiles between carriers and non-carriers of the APOE4 allele among patients with AD and healthy controls and investigated whether the effect of APOE on lipids is unique in AD. We hypothesized that the APOEε4 allele might cause the development of AD by influencing lipid metabolism.
2. Materials and Methods
2.1. Search Strategy
Two independent investigators searched the PubMed, Embase, Web of Science, and Chinese databases on 30 May 2022. The following medical subject heading (MeSH) terms and topic terms were used as the search terms: “Lipid”, “Cholesterol”, “Triglycerides”, “Alzheimer’s disease”, “Alzheimer Dementia”, “Apoprotein E”, and “APOE”.
2.2. Inclusion and Exclusion Criteria
The inclusion criteria were as follows: (1) all articles that reported the results of APOE alleles and were grouped participants according to whether they carried the APOEε4 allele and/or different APOE alleles; (2) articles reporting data as mean ± standard derivation (SD); (3) studies that analyzed patients with AD patients or healthy controls as the study populations; (4) studies that included patients diagnosed with AD; and (5) studies that included healthy controls with normal cognitive function and no neurological disease.
The exclusion criteria were as follows: reviews, conference papers, letters, comments, editorials, case reports, and abstracts without an available full text.
2.3. Data Extraction and Quality Evaluation
FJJ and YJ conducted the preliminary screening of titles and abstracts and then screened potentially relevant full texts according to the inclusion criteria. A third investigator verified all the data. From each study, we collected the following data: the sample size, publication year, and participant characteristics (age, number of participants, sex ratio, country, and Mini-Mental State Examination scores). Relevant information was extracted independently by two investigators and verified by a third investigator. The Newcastle–Ottawa Quality Assessment Scale (NOS Scale) was used to assess the quality of the included studies [19]. The total score on this scale is 9, and a score of ≥6 is acceptable.
2.4. Statistical Analysis
We performed a meta-analysis using Stata, version 15.0 software (StataCorp LLC., College Station, TX, USA) and used the standardized mean difference (SMD) to obtain aggregate effects. The random-effects model was used if there was significant heterogeneity between the included studies (the Cochrane Q test result and I2 statistic: I2 > 50% or p < 0.1). The z-test was used to determine the overall effect. We assessed heterogeneity using sensitivity, meta-regression, and subgroup analyses and evaluated the publication bias using Begg’s and Egger’s tests. The standardized effect size was compared between multiple groups using network meta-analysis, and related indicators of each group were compared using the cumulative ranking curve (SUCRA).
3. Results
3.1. Study Selection and Characteristics
The flow chart illustrates the systematic search and selection process (Figure 1); 17 studies were included in the final analysis [10,12,13,14,15,18,20,21,22,23,24,25,26,27,28,29,30] (Table 1). These selected studies, which were carefully evaluated for their relevance and contribution to our research objectives, are shown in Table 1. Eight of these studies grouped the participants on the basis of their APOEε4 allele status. Among them, four studies exclusively focused on individuals with AD, whereas the other four studies examined both patients with AD and healthy controls. APOE allele classification was further extended in six studies, which divided participants into three specific groups: APOEε2 allele carriers, APOEε3/3 carriers, and APOEε4 allele carriers. Of these, two studies exclusively focused on the AD population, and the remaining four encompassed both AD and healthy control populations. The participants were divided into six subgroups based on their APOE alleles status across a total of five studies. Among them, one study was exclusively dedicated to the AD population, one study was exclusively dedicated to the control population, and three studies grouped both the AD and control populations. It is important to note that some studies did not analyze all pertinent variables, including but not limited to TC, triglycerides (TG), HDL, and LDL levels. These variances are essential to consider when interpreting the collective findings. The NOS Scale scores are shown in Table S1.
Figure 1.
The literature screening flow chart.

Table 1.
Details of the original studies included in the meta-analysis for APOE and AD.
3.2. Data Extraction and Study Population
We extracted data from eight articles focusing on TC and TG levels that included 652 individuals carrying APOEε4 allele-C and 1038 individuals with APOEε4 allele-N. Additionally, we collected information from seven articles regarding HDL and LDL levels that included 630 individuals carrying APOEε4 allele-C and 987 individuals with APOEε4 allele-N.
3.3. Overall Effect, Heterogeneity, Publication Bias, and Subgroup Analysis
To elucidate the overall effect across the studies, we used a random effect model to address the variances arising from differences present in the included studies. Noteworthy differences in TC and LDL levels were observed when comparing the APOEε4 allele-C and APOEε4 allele-N groups. Specifically, individuals in the APOEε4 allele-C group showed higher TC and LDL levels than those in the APOEε4 allele-N group (TC: SMD = 0.62 [0.2, 1.04], p = 0.004; LDL: SMD = 0.63 [0.9, 1.08], p = 0.005). However, studies indicated no difference in TG and HDL levels between those groups (TG: SMD = 0.08 [−0.19, 0.41], p = 0.108; HDL: SMD = −0.08 [−0.45, 0.28], p = 0.655) (Figure 2 and Figures S1–S4).
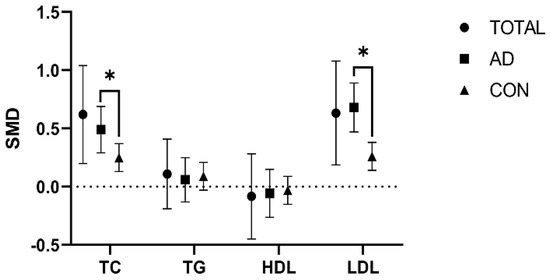
Figure 2.
Comparison of the effect of the APOEε4 allele on lipids in the AD and healthy control populations. The data are presented as the standardized mean difference (SMD) and 95% confidence interval. TOTAL: both the Alzheimer’s disease and healthy control populations; AD: Alzheimer’s disease population; CON: healthy control population; TC: total cholesterol; TG: triglycerides; HDL: high-density lipoprotein; LDL: low-density lipoprotein. *: p < 0.05.
Sensitivity analysis was conducted to identify the causes of heterogeneity and we were able to identify a clear cause of heterogeneity (Figures S5–S8). The funnel plot and bias test showed no significant publication bias (Figures S9–12, Tables S2–S5).
However, subgroup analysis showed great heterogeneity in TC and LDL levels between the APOEε4 allele-C and APOEε4 allele-N groups among the AD and healthy control populations (TC: p = 0.042; LDL: p = 0.001). However, no heterogeneity was shown in HDL and TG levels (TG: p = 0.794; HDL: p = 0.823) (Figure 2 and Figures S13–S16).
Notably, the AD and healthy control populations had elevated TC and LDL levels in the APOEε4 allele-C group compared with the APOEε4 allele-N group, but the degree of elevation was lower in the AD population than in the healthy control population (AD population: TC: SMD = 0.49 [0.29, 0.69], p = 0.000; LDL: SMD = 0.68 [0.47, 0.89], p = 0.000) (healthy control population: TC: SMD = 0.25 [0.13, 0.37], p = 0.000; LDL: SMD = 0.26 [0.14,0.38], p = 0.000) (Figure 2 and Figures S13–S16).
3.4. Comparison of the Lipids in APOEε3/ε3, APOEε2 Allele, and APOEε4 Allele Carriers
The network meta-analysis was performed to compare TC, TG, and LDL levels in individuals carrying different APOE alleles; APOEε4 allele carriers had the highest SUCRA value, followed by APOEε3/3 and APOEε2 allele carriers, respectively (Tables S6–S8). However, regarding HDL levels, APOEε4 allele carriers had the lowest SUCRA value, followed by APOEε2 allele carriers, whereas APOEε3/3 allele carriers had the highest SUCRA value (Table S9).
3.5. Comparison of the Lipids between Six Groups of APOE Alleles
The network meta-analysis of six distinct groups formed by APOE alleles showed variations in the SUCRA values of TC levels as follows: APOEε3/ε4 > APOEε4/ε4 > APOEε3/ε3 > APOEε2/ε2 > APOEε2/ε4 > APOEε2/ε3 (Table S10).
Similarly, SUCRA values of TG levels were as follows: APOEε2/ε2 > APOEε3/ε4 > APOEε3/ε3 > APOEε4/ε4 > APOEε2/ε3 > APOEε2/ε4 (Table S11).
Owing to the limited availability of multiple data sets, sequencing comparisons of HDL and LD levels between these six groups could not be performed.
4. Discussion
4.1. Main Findings
We hypothesized that the APOEε4 allele might cause the development of AD by influencing lipid metabolism. Studies have found that high levels of serum cholesterol are positively associated with an increased risk of dementia, and the prevalence of AD is reduced in patients taking cholesterol-lowering drugs [32]. A Mendelian randomization study of AD metabolism and risk confirmed the causal role of LDL, cholesterol, and serum total cholesterol in the high-risk of AD [33]. Some studies have found an association between blood lipids and Alzheimer’s disease, proving that blood lipids can be used as biomarkers for the early diagnosis of Alzheimer’s disease. It can also help predict the stage of prognosis and disease severity, and further studies are needed to find out the exact mechanisms behind these changes [34]. This study focused on the relationship between APOE alleles and serum lipid profiles, specifically TC, LDL, TG, and HDL levels in individuals with AD compared to healthy controls. Through our meta-analysis, we found that individuals carrying the APOEε4 allele showed increased TC and LDL levels compared with those without the APOEε4 allele. There was a statistically significant difference in TC levels between the APOEε4 allele-C and APOEε4 allele-N groups. The p-value indicated that the difference did not occur by chance and is, therefore, statistically significant. APOEε4 allele-C carriers had higher LDL levels than non-APOEε4 allele-C carriers.
Notably, no significant statistical differences were found in TG and HDL levels between these groups. These data reinforce the absence of statistically significant differences in TG and HDL levels between individuals with APOEε4 allele-C and those without. Further analysis showed differences in TC and LDL levels between APOEε4 allele-C and APOEε4 allele-N groups with significant heterogeneity when considering AD and healthy controlled populations separately. These data suggest that in AD populations, TC and LDL levels are higher in APOEε4 allele-C carriers than in APOEε4 allele-N carriers, but the degree of elevation is lower than that seen in the healthy control populations.
It is crucial to note that APOEε4 acts as a main genetic risk factor for AD. Genome-wide association studies have shown that APOEε4 is the strongest genetic risk factor for AD, irrespective of the age of onset [31].
4.2. APOE Functions in the Brain
The APOE gene encodes the APOE protein, which plays an important role in the transportation and metabolism of lipids [35]. APOE is responsible for the transportation of lipids and the maintenance of cholesterol homeostasis in the brain. It plays a crucial role in supplying neurons with cholesterol and facilitating the removal of excess cholesterol. It is also involved in other brain functions, such as promoting synaptic plasticity, transmitting signals, maintaining protein balance, modulating the immune system, and repairing after an injury [36].
4.3. APOE Isomers and Their Binding Specificity
Research has shown that the C-terminal domain of APOE is the key to lipoprotein binding and determines the specificity of APOE subtype lipidosis [37]. Specifically, APOEε4 shows distinct characteristics, including poor lipidation compared with APOEε2 and APOEε3 alleles [38]. The APOEε3 and APOEε2 alleles prefer to bind to HDL, whereas the APOEε4 allele prefers to bind to very low-density lipoprotein (VLDL) [39]. This variation in lipoprotein association is determined by differences in the interactions of the carboxyl-terminal domains among the isoforms, leading to APOEε2 and APOEε3 binding to smaller more phospholipid-enriched HDL, and APOEε4 binding to larger triglyceride-rich VLDL [40].
4.4. Lipid-Binding Effects of APOEε 4 and Cholesterol Efflux
The lipid-binding features of APOEε4 have substantial effects on the efflux of cholesterol and the metabolism of amyloid-beta (Aβ). The functional attributes of APOE, including receptor binding capabilities, molecular stability, and overall functionality, are conditional based on its lipidation status [41]. In vitro model studies have shown a pivotal role of lipidation in preventing self-aggregation of APOE [42]. Given the considerable influence of lipidation on many roles of APOE, it has been proposed as a potential therapeutic treatment for AD. Hence, there is the possibility to correct, as well as prevent, certain outcomes associated with neurodegeneration. The benefit of increasing lipidation and reducing lipid-free availability may offer greater advantages to the individuals who carry the APOEε4 allele, which accounts for a larger percentage of both AD populations and healthy control populations [43].
Another complementary study observed that pharmacologically promoting cholesterol efflux can increase myelination in vitro and in vivo and improve cognition in APOE4/e-TR mice. This finding indicates a link between cholesterol dysregulation and myelination in APOEε4 carriers, which may impact the onset and severity of cognitive decline in AD. Interventions such as pharmacological treatments, lifestyle, and dietary modifications aiming at restoring cholesterol equilibrium and myeline volume might help to increase the cognitive reserves in APOEε4 carriers [44].
This proposal to augment APOE lipidation as a therapeutic approach shows the increasing understanding of the complex connection between lipid metabolism, APOE genetics, and AD pathogenesis. Further investigations are required to determine the practicality and effectiveness of using this approach in the clinical setting as a means to develop successful therapeutic interventions for AD.
Furthermore, the APOEε4 allele has a strong lipid-binding affinity and a low recovery capacity, leading to impaired cholesterol efflux, culminating in an increased accumulation of cholesterol in cell membranes [45]. The distribution of elevated cholesterol levels on the plasma membrane of neurons correlated with increases in the metabolism of Aβ precursor protein (APP), which results in increased Aβ production [46]. In addition to neurons, astrocytes and microglia are also affected by impaired cholesterol efflux. In these cells, less cholesterol efflux reduces Aβ degradation, which may increase aggregation of Aβ into plaques [47].
4.5. HDL and Cholesterol Metabolism in APOE Non-Carriers
Longitudinal studies have shown that individuals with AD who are non-carriers of the APOEε4 allele have elevated HDL levels. This elevation is associated with impaired cholesterol metabolism and impaired function, possibly resulting from reduced lipid availability in neuronal membranes [48]. Furthermore, in APOEε4 allele non-carriers of AD-stratified populations, the enzyme 3-hydroxy-3-methylglutaryl-CoA synthetase was significantly associated with sporadic AD. This suggests potential cholesterol metabolic dysfunction in patients with AD who do not carry the APOEε4 allele [49].
4.6. Heterogeneity and Implications in Clinical Practice
Subgroup analysis based on different populations yielded findings showing significant inter-group heterogeneity in patients with AD and the healthy controls, especially since the influence of the APOEε4 allele on TC and LDL levels appears to be more pronounced in patients with AD than in the healthy control population. One important consideration is that TC and LDL in peripheral blood rarely enter the central nervous system (CNS). These lipids typically do not cross the blood–brain barrier in substantial amounts to cause harm to CNS function. Therefore, any effect of APOE alleles on peripheral TC and LDL levels may differ from their potential roles in the CNS. This raises an important question as to whether the influence of APOE alleles on peripheral lipid levels is related to the central pathological mechanism of AD. The exact nature of this relationship remains unclear, so it is an important area that warrants more comprehensive investigations. Therefore, more attention should be paid to AD in clinical practice and future studies, especially the lipid levels of patients with AD carrying the APOEε4 allele.
Given the high degree of heterogeneity in this meta-analysis, we acknowledge that the exact cause of this variability has not been definitively identified despite performing sensitivity meta-regression and other analyses. We tried to exclude influential studies and found that heterogeneity could not be significantly reduced after re-analysis. This heterogeneity could be due to a combination of various factors, including different study populations, methodologies, and patient characteristics, such as age, sex, genetic background, medication use, ethnicity, and race.
4.7. Sex-Based Analysis
Sex-based analysis can provide more insight into how sex-specific hormonal factors interact with APOE alleles to modulate lipid profiles and AD risk differently in men and women. There are significant differences between males and females in the regulation of fatty acid metabolism. Premenopausal women tend to have higher levels of polyunsaturated fatty acids than men [31], which may be due to higher estrogen levels affecting lipid metabolism in premenopausal women [50]. Additionally, women in premenopausal, menopausal transition states have alterations in various body fats, which are also related to changes in their estrogen concentrations [51]. Decreased estrogen levels in postmenopausal women can affect lipid metabolism, which increases the risk of cognitive decline [52].
Females with one copy of the APOEε4 allele had about four times the risk of AD, whereas males with one copy of the APOEε4 allele had only twice the risk [53]. It is unknown whether there are differences in lipid metabolism between different APOEε4 allele groups with different sexes. A study conducted in 2022 showed that within the AD population, both sexes showed high levels of TC and LDL compared with the control group. Notably, among female patients with AD, TC and LDL levels were significantly higher in APOEε4 allele carriers than in non-carriers. In contrast, the presence of the APOEε2 allele was linked to reduced TC levels in male patients with AD compared with non-carriers. This particular influence was not evident among male controls, female controls, or female AD populations. However, further prospective studies are required to confirm these findings [54].
In our study, owing to insufficient data, it was not possible to conduct subgroup analysis based on sex, age, and medication use to explore various causes of heterogeneity. It is worth further exploring the sex-based differences in lipid metabolism between different APOEε4 allele groups and how these differences can influence AD risk.
4.8. Dual APOE4 and Lipid Profiles
In addition, this study used network meta-analyses to explore the effect of both single and dual APOEε4 alleles on lipid profiles. Interestingly, the presence of dual APOEε4 alleles did not increase the degree of influence on lipid profiles compared with a single APOEε4 allele. This finding negates the notion that having a higher genetic predisposition (possessing two APOEε4 alleles) leads to more lipid-related impacts in AD.
4.9. Comparisons with Other Studies and What This Study Added to the Existing Knowledge
In contrast to previous meta-analyses that primarily examined the differences in lipids between AD and healthy controls, this study took a more focused approach. We investigated the difference in lipid levels between those carrying APOEε4 allele-C and APOEε4 allele-N within the context of AD. Thus, we were able to evaluate the specific influence of the APOEε4 allele on lipids in AD, which adds novel knowledge to improve understanding of the complex interplay between genetics and lipid metabolism in AD pathogenesis.
4.10. Study Strengths and Limitations
This study is the first comprehensive analysis of the distinctive relationship between the APOEε4 allele and lipids in patients with AD and healthy controls. The influence of the presence of the APOEε4 allele on blood lipids, and the differences between single and dual APOEε4 allele lipids, were analyzed using MeSH terms in meta-analysis, which is the strength of this study. However, this study has some limitations. First, since the data on age and the sex ratio of the APOEε4 allele carriers and the non-carriers were insufficient, we could not conduct a deeper subgroup analysis stratified by age and sex. Second, despite our best efforts to contact the respective authors, some articles had incomplete data.
5. Conclusions
This meta-analysis showed that APOEε4 allele-C carriers had higher TC and LDL levels than APOEε4 allele-N carriers, and the difference was significant between patients with AD and healthy participants. The dual APOEε4 allele may not have an increased effect on the lipid profiles. The effect of dyslipidemia and interventions on lipids levels in AD, especially in APOEε4 allele carriers, should be extensively studied in the future. Currently, there are no therapies targeting APOE for AD treatment. These studies offer new insights for potential future AD treatments and provide a basis for precision medicine.
Supplementary Materials
The following supporting information can be downloaded: https://www.mdpi.com/article/10.3390/brainsci13111554/s1, Figure S1: random effect forest map of TC; Figure S2: random effect forest map of LDL; Figure S3: random effect forest map of TG; Figure S4: random effect forest map of HDL; Figure S5: sensitivity analysis for TC; Figure S6: sensitivity analysis for LDL; Figure S7: sensitivity analysis for TG; Figure S8: sensitivity analysis for HDL; Figure S9: funnel plot of TC; Figure S10: funnel plot of LDL; Figure S11: funnel plot of TG; Figure S12: funnel plot of HDL; Figure S13: fixed effect subgroup analysis of TC; Figure S14: fixed effect subgroup analysis of LDL; Figure S15: fixed effect subgroup analysis of TG; Figure S16: fixed effect subgroup analysis of HDL; Table S1: quality assessment of NOS; Table S2: Begg bias test and Egger’s test of TC; Table S3: Begg bias test and Egger’s test of LDL; Table S4: Begg bias test and Egger’s test of TG; Table S5: Begg bias test and Egger’s test of HDL; Table S6: SUCRA ranking of ApoE4 allele carrying, ApoE3/3, and ApoE4 allele carrying (TC); Table S7: SUCRA ranking of ApoE4 allele carrying, ApoE3/3, and ApoE4 allele carrying (TG); Table S8: SUCRA ranking of ApoE4 allele carrying, ApoE3/3, and ApoE4 allele carrying (LDL); Table S9: SUCRA ranking of ApoE4 allele carrying, ApoE3/3, and ApoE4 allele carrying (HDL); Table S10: SUCRA ranking of 6 genotypes (TC); Table S11: SUCRA ranking of 6 genotypes (TG).
Author Contributions
H.X., J.F., R.B.M.N. and J.Y. contributed to the literature search, data extraction, and manuscript preparation. S.C., Y.H. and T.B. verified all the data. H.X., J.F. and X.C. conceived this study and revised this article. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by a grant from the Cadres Health Care Project in Sichuan Province (grant number: 2023–111).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The original material presented in this study is included as Supplementary Material.
Acknowledgments
The authors thank all those who contributed to the maintenance of the database.
Conflicts of Interest
The authors declare no conflict of interest.
Abbreviations
APOE: apolipoprotein E; AD: Alzheimer’s disease; TC: total cholesterol; LDL: low-density lipoprotein; TG: triglycerides; HDL: high-density lipoprotein; APOEε4 allele-C: APOEε4 allele carriers; APOEε4 allele-N: APOEε4 allele non-carriers; Aβ: amyloid-β; NOS Scale: the Newcastle–Ottawa Quality Assessment Scale; SMD: standardized mean difference; CI: confidence interval; SUCRA: the cumulative ranking curve; APP: Aβ precursor protein; VLDL: very low-density lipoprotein; CNS: the central nervous system.
References
- Breijyeh, Z.; Karaman, R. Comprehensive review on Alzheimer’s disease: Causes and treatment. Molecules 2020, 25, 5789. [Google Scholar] [CrossRef]
- Poirier, J.; Miron, J.; Picard, C.; Gormley, P.; Théroux, L.; Breitner, J.; Dea, D. Apolipoprotein E and lipid homeostasis in the etiology and treatment of sporadic Alzheimer’s disease. Neurobiol. Aging 2014, 35 (Suppl. S2), S3–S10. [Google Scholar] [CrossRef]
- Gremer, L.; Schölzel, D.; Schenk, C.; Reinartz, E.; Labahn, J.; Ravelli, R.B.G.; Tusche, M.; Lopez-Iglesias, C.; Hoyer, W.; Heise, H.; et al. Fibril structure of amyloid-β(1–42) by cryo-electron microscopy. Science 2017, 358, 116–119. [Google Scholar] [CrossRef] [PubMed]
- Filippini, N.; MacIntosh, B.J.; Hough, M.G.; Goodwin, G.M.; Frisoni, G.B.; Smith, S.M.; Matthews, P.M.; Beckmann, C.F.; Mackay, C.E. Distinct patterns of brain activity in young carriers of the APOE-epsilon4 allele. Proc. Natl. Acad. Sci. USA 2009, 106, 7209–7214. [Google Scholar] [CrossRef]
- Corder, E.H.; Saunders, A.M.; Strittmatter, W.J.; Schmechel, D.E.; Gaskell, P.C.; Small, G.W.; Roses, A.D.; Haines, J.L.; Pericak-Vance, M.A. Gene dose of apolipoprotein E type 4 allele and the risk of Alzheimer’s disease in late onset families. Science 1993, 261, 921–923. [Google Scholar] [CrossRef] [PubMed]
- Farrer, L.A.; Cupples, L.A.; Haines, J.L.; Hyman, B.; Kukull, W.A.; Mayeux, R.; Myers, R.H.; Pericak-Vance, M.A.; Risch, N.; van Duijn, C.M. Effects of age, sex, and ethnicity on the association between apolipoprotein E genotype and Alzheimer disease. A meta-analysis. APOE and Alzheimer disease meta analysis consortium. JAMA 1997, 278, 1349–1356. [Google Scholar] [CrossRef]
- Stalmans, P.; Parys-Vanginderdeuren, R.; De Vos, R.; Feron, E.J. ICG staining of the inner limiting membrane facilitates its removal during surgery for macular holes and puckers. Bull. Soc. Belge Ophtalmol. 2001, 281, 21–26. [Google Scholar]
- Wishart, H.A.; Saykin, A.J.; McAllister, T.W.; Rabin, L.A.; McDonald, B.C.; Flashman, L.A.; Roth, R.M.; Mamourian, A.C.; Tsongalis, G.J.; Rhodes, C.H. Regional brain atrophy in cognitively intact adults with a single APOE epsilon4 allele. Neurology 2006, 67, 1221–1224. [Google Scholar] [CrossRef] [PubMed]
- Tarasoff-Conway, J.M.; Carare, R.O.; Osorio, R.S.; Glodzik, L.; Butler, T.; Fieremans, E.; Axel, L.; Rusinek, H.; Nicholson, C.; Zlokovic, B.V.; et al. Clearance systems in the brain-implications for Alzheimer disease. Nat. Rev. Neurol. 2015, 11, 457–470. [Google Scholar] [CrossRef]
- Hall, K.; Murrell, J.; Ogunniyi, A.; Deeg, M.; Baiyewu, O.; Gao, S.; Gureje, O.; Dickens, J.; Evans, R.; Smith-Gamble, V.; et al. Cholesterol, APOE genotype, and Alzheimer disease: An epidemiologic study of Nigerian Yoruba. Neurology 2006, 66, 223–227. [Google Scholar] [CrossRef] [PubMed]
- Hoshino, T.; Kamino, K.; Matsumoto, M. Gene dose effect of the APOE-epsilon4 allele on plasma HDL cholesterol level in patients with Alzheimer’s disease. Neurobiol. Aging 2002, 23, 41–45. [Google Scholar] [CrossRef]
- Isbir, T.; Agaçhan, B.; Yilmaz, H.; Aydin, M.; Kara, I.; Eker, E.; Eker, D. Apolipoprotein-E gene polymorphism and lipid profiles in Alzheimer’s disease. Am. J. Alzheimer’s Dis. Other Dement. 2001, 16, 77–81. [Google Scholar] [CrossRef] [PubMed]
- Raygani, A.V.; Rahimi, Z.; Kharazi, H.; Tavilani, H.; Pourmotabbed, T. Association between apolipoprotein E polymorphism and serum lipid and apolipoprotein levels with Alzheimer’s disease. Neurosci. Lett. 2006, 408, 68–72. [Google Scholar] [CrossRef]
- Shafagoj, Y.A.; Naffa, R.G.; El-Khateeb, M.S.; Abdulla, Y.L.; Al-Qaddoumi, A.A.; Khatib, F.A.; Al-Motassem, Y.F.; Al-Khateeb, E.M. APOE Gene polymorphism among Jordanian Alzheimer’s patients with relation to lipid profile. Neurosciences 2018, 23, 29–34. [Google Scholar] [CrossRef] [PubMed]
- Sabbagh, M.N.; Sandhu, S.; Kolody, H.; Lahti, T.; Silverberg, N.B.; Sparks, D.L. Studies on the effect of the apolipoprotein E genotype on the lipid profile in Alzheimer’s disease. Curr. Alzheimer Res. 2006, 3, 157–160. [Google Scholar] [CrossRef] [PubMed]
- Anstey, K.J.; Lipnicki, D.M.; Low, L.F. Cholesterol as a risk factor for dementia and cognitive decline: A systematic review of prospective studies with meta-analysis. Am. J. Geriatr. Psychiatry 2008, 16, 343–354. [Google Scholar] [CrossRef]
- Tang, Q.; Wang, F.; Yang, J.; Peng, H.; Li, Y.; Li, B.; Wang, S. Revealing a novel landscape of the association between blood lipid levels and Alzheimer’s disease: A meta-analysis of a case-control study. Front. Aging Neurosci. 2019, 11, 370. [Google Scholar] [CrossRef]
- Wang, P.; Zhang, H.; Wang, Y.; Zhang, M.; Zhou, Y. Plasma cholesterol in Alzheimer’s disease and frontotemporal dementia. Transl. Neurosci. 2020, 11, 116–123. [Google Scholar] [CrossRef]
- Tugwell, G.W.P.; Shea, B.; O’Connell, D.; Welch, J.P.V.; Losos, M. The Newcastle-Ottawa Scale (NOS) for Assessing the Quality of Nonrandomised Studies in Meta-Analyses. 2021. Available online: https://www.ohri.ca/programs/clinical_epidemiology/oxford.asp (accessed on 5 January 2022).
- Fernandes, M.A.; Proença, M.T.; Nogueira, A.J.; Oliveira, L.M.; Santiago, B.; Santana, I.; Oliveira, C.R. Effects of apolipoprotein E genotype on blood lipid composition and membrane platelet fluidity in Alzheimer’s disease. Biochim. Biophys. Acta 1999, 1454, 89–96. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Wehr, H.; Parnowski, T.; Puzyński, S.; Bednarska-Makaruk, M.; Bisko, M.; Kotapka-Minc, S.; Rodo, M.; Wołkowska, M. Apolipoprotein E genotype and lipid and lipoprotein levels in dementia. Dement. Geriatr. Cogn. Disord. 2000, 11, 70–73. [Google Scholar] [CrossRef]
- Sheng, B. Preliminary Studies on Genes Associated with Alzheimer’s Disease; Jilin University: Jilin, China, 2000. [Google Scholar]
- Cui, J.; Wang, J.; Guo, L. Effects of apolipoprotein E genotype on plasma apolipoprotein E, total cholesterol and triglyceride levels in patients with Alzheimer’s disease. Shanghai Med. 2002, 25, 444–446. [Google Scholar] [CrossRef]
- Zeng, X.; Qin, B.; Guo, H. Effect of apolipoprotein E gene on lipid metabolism in patients with Alzheimer’s disease. Chin. J. Psychiatry 2002, 3. [Google Scholar] [CrossRef]
- Al-Shammari, S.; Fatania, H.; Al-Radwan, R.; Akanji, A.O. Apolipoprotein E polymorphism and lipoprotein levels in a Gulf Arab population in Kuwait: A pilot study. Ann. Saudi Med. 2004, 24, 361–364. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Wang, Y.; Yang, J. Study on the association between APOE gene polymorphism and Alzheimer’s disease. J. Mod. Lab. Med. 2008, 23, 20–23. [Google Scholar] [CrossRef]
- Singh, N.K.; Chhillar, N.; Banerjee, B.D.; Bala, K.; Mukherjee, A.K.; Mustafa, M.D.; Mitrabasu. Gene-environment interaction in Alzheimer’s disease. Am. J. Alzheimer’s Dis. Other Demen. 2012, 27, 496–503. [Google Scholar] [CrossRef]
- Dai, T. Relationship between APOE Gene Polymorphism and Cognitive Function in Sporadic Alzheimer’s Disease; Central South University: Shenzhen, China, 2012. [Google Scholar]
- Liang, J.; Kabinver; Ke, Y. Study on the relationship between lipid levels and apolipoprotein E gene polymorphism in patients with sporadic Alzheimer’s disease in Uygurs and Han nationalities. Chin. J. Clin. Health Care 2013, 16, 565–568. [Google Scholar]
- Bian, M. Study on Apolipoprotein E Gene Polymorphism and Lipid Level in Alzheimer’s Disease Patients. Master’s Thesis, Dalian Medical University, Dalian, China, 2018. [Google Scholar]
- Lim, W.L.F.; Huynh, K.; Chatterjee, P.; Martins, I.; Jayawardana, K.S.; Giles, C.; Mellett, N.A.; Laws, S.M.; Bush, A.I.; Rowe, C.C.; et al. Relationships between plasma lipids species, gender, risk factors, and Alzheimer’s disease. J. Alzheimer’s Dis. 2020, 76, 303–315. [Google Scholar] [CrossRef]
- Loera-Valencia, R.; Goikolea, J.; Parrado-Fernandez, C.; Merino-Serrais, P.; Maioli, S. Alterations in cholesterol metabolism as a risk factor for developing Alzheimer’s disease: Potential novel targets for treatment. J. Steroid Biochem. Mol. Biol. 2019, 190, 104–114. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.-Y.; Yang, Y.-X.; Zhang, Y.-R.; Kuo, K.; Li, H.-Q.; Shen, X.-N.; Chen, S.-D.; Chen, K.-L.; Dong, Q.; Tan, L.; et al. Investigating Causal Relations Between Circulating Metabolites and Alzheimer’s Disease: A Mendelian Randomization Study. J. Alzheimer’s Dis. 2022, 87, 463–477. [Google Scholar] [CrossRef]
- Agarwal, M.; Khan, S. Plasma Lipids as Biomarkers for Alzheimer’s Disease: A Systematic Review. Cureus 2020, 12, e12008. [Google Scholar] [CrossRef]
- Strittmatter, W.J.; Saunders, A.M.; Schmechel, D.; Pericak-Vance, M.; Enghild, J.; Salvesen, G.S.; Roses, A.D. Apolipoprotein E: High-avidity binding to beta-amyloid and increased frequency of type 4 allele in late-onset familial Alzheimer disease. Proc. Natl. Acad. Sci. USA 1993, 90, 1977–1981. [Google Scholar] [CrossRef]
- Liu, C.C.; Liu, C.C.; Kanekiyo, T.; Xu, H.; Bu, G. Apolipoprotein E and Alzheimer disease: Risk, mechanisms and therapy. Nat. Rev. Neurol. 2013, 9, 106–118. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.; Liu, C.C.; Chen, X.F.; Zhang, Y.W.; Xu, H.; Bu, G. Opposing effects of viral mediated brain expression of apolipoprotein E2 (apoE2) and apoE4 on apoE lipidation and Aβ metabolism in apoE4-targeted replacement mice. Mol. Neurodegener. 2015, 10, 6. [Google Scholar] [CrossRef]
- Kanekiyo, T.; Xu, H.; Bu, G. ApoE and Aβ in Alzheimer’s disease: Accidental encounters or partners? Neuron 2014, 81, 740–754. [Google Scholar] [CrossRef]
- Nguyen, D.; Dhanasekaran, P.; Nickel, M.; Nakatani, R.; Saito, H.; Phillips, M.C.; Lund-Katz, S. Molecular basis for the differences in lipid and lipoprotein binding properties of human apolipoproteins E3 and E4. Biochemistry 2010, 49, 10881–10889. [Google Scholar] [CrossRef] [PubMed]
- Morrow, J.A.; Segall, M.L.; Lund-Katz, S.; Phillips, M.C.; Knapp, M.; Rupp, B.; Weisgraber, K.H. Differences in stability among the human apolipoprotein E isoforms determined by the amino-terminal domain. Biochemistry 2000, 39, 11657–11666. [Google Scholar] [CrossRef] [PubMed]
- Mahley, R.W.; Rall, S.C. Apolipoprotein E: Far more than a lipid transport protein. Annu. Rev. Genomics Hum. Genet. 2000, 1, 507–537. [Google Scholar] [CrossRef] [PubMed]
- Hubin, E.; Verghese, P.B.; van Nuland, N.; Broersen, K. Apolipoprotein E associated with reconstituted high-density lipoprotein-like particles is protected from aggregation. FEBS Lett. 2019, 593, 1144–1153. [Google Scholar] [CrossRef]
- Fernández-Calle, R.; Konings, S.C.; Frontiñán-Rubio, J.; García-Revilla, J.; Camprubí-Ferrer, L.; Svensson, M.; Martinson, I.; Boza-Serrano, A.; Venero, J.L.; Nielsen, H.M.; et al. APOE in the bullseye of neurodegenerative diseases: Impact of the APOE genotype in Alzheimer’s disease pathology and brain diseases. Mol. Neurodegener. 2022, 17, 62. [Google Scholar] [CrossRef]
- Blanchard, J.W.; Akay, L.A.; Davila-Velderrain, J.; von Maydell, D.; Mathys, H.; Davidson, S.M.; Effenberger, A.; Chen, C.Y.; Maner-Smith, K.; Hajjar, I.; et al. APOE4 impairs myelination via cholesterol dysregulation in oligodendrocytes. Nature 2022, 611, 769–779. [Google Scholar] [CrossRef]
- Yassine, H.N.; Finch, C.E. APOE alleles and diet in brain aging and Alzheimer’s disease. Front. Aging Neurosci. 2020, 12, 150. [Google Scholar] [CrossRef] [PubMed]
- Cui, W.; Sun, Y.; Wang, Z.; Xu, C.; Xu, L.; Wang, F.; Chen, Z.; Peng, Y.; Li, R. Activation of liver X receptor decreases BACE1 expression and activity by reducing membrane cholesterol levels. Neurochem. Res. 2011, 36, 1910–1921. [Google Scholar] [CrossRef]
- Prasad, H.; Rao, R. Amyloid clearance defect in ApoE4 astrocytes is reversed by epigenetic correction of endosomal pH. Proc. Natl. Acad. Sci. USA 2018, 115, E6640–E6649. [Google Scholar] [CrossRef] [PubMed]
- Sacks, D.; Baxter, B.; Campbell, B.C.V.; Carpenter, J.S.; Cognard, C.; Dippel, D.; Eesa, M.; Fischer, U.; Hausegger, K.; Hirsch, J.A.; et al. Multisociety Consensus Quality Improvement Revised Consensus Statement for Endovascular Therapy of Acute Ischemic Stroke. Int. J. Stroke 2018, 13, 612–632. [Google Scholar] [CrossRef] [PubMed]
- Leduc, V.; De Beaumont, L.; Théroux, L.; Dea, D.; Aisen, P.; Petersen, R.C.; Alzheimer’s Disease Neuroimaging Initiative; Dufour, R.; Poirier, J. HMGCR is a genetic modifier for risk, age of onset and MCI conversion to Alzheimer’s disease in a three cohorts study. Mol. Psychiatry 2015, 20, 867–873. [Google Scholar] [CrossRef]
- Palmisano, B.T.; Zhu, L.; Eckel, R.H.; Stafford, J.M. Sex differences in lipid and lipoprotein metabolism. Mol. Metab. 2018, 15, 45–55. [Google Scholar] [CrossRef] [PubMed]
- Ko, S.H.; Kim, H.S. Menopause-associated lipid metabolic disorders and foods beneficial for postmenopausal women. Nutrients 2020, 12, 202. [Google Scholar] [CrossRef] [PubMed]
- Ancelin, M.L.; Ripoche, E.; Dupuy, A.M.; Samieri, C.; Rouaud, O.; Berr, C.; Carrière, I.; Ritchie, K. Gender-specific associations between lipids and cognitive decline in the elderly. Eur. Neuropsychopharmacol. 2014, 24, 1056–1066. [Google Scholar] [CrossRef]
- Altmann, A.; Tian, L.; Henderson, V.W.; Greicius, M.D.; Alzheimer’s Disease Neuroimaging Initiative Investigators. Sex modifies the APOE-related risk of developing Alzheimer disease. Ann. Neurol. 2014, 75, 563–573. [Google Scholar] [CrossRef]
- Fu, J.; Huang, Y.; Bao, T.; Ou, R.; Wei, Q.; Chen, Y.; Yang, J.; Chen, X.; Shang, H. Effects of sex on the relationship between apolipoprotein E gene and serum lipid profiles in Alzheimer’s disease. Front. Aging Neurosci. 2022, 14, 844066. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).