Abstract
Securing medical imaging poses a significant challenge in preserving the confidentiality of healthcare data. Numerous research efforts have focused on fortifying these images, with encryption emerging as a primary solution for maintaining data integrity without compromising confidentiality. However, applying conventional encryption techniques directly to e-health data encounters hurdles, including limitations in data size, redundancy, and capacity, particularly in open-channel patient data transmissions. As a result, the unique characteristics of images, marked by their risk of data loss and the need for confidentiality, make preserving the privacy of data contents a complex task. This underscores the pressing need for innovative approaches to ensure the security and confidentiality of sensitive healthcare information within medical images. The proposed algorithm outperforms referenced algorithms in both image fidelity and steganographic capacity across diverse medical imaging modalities. It consistently achieves higher Peak Signal-to-Noise Ratio (PSNR) values, indicating superior image fidelity, reduced noise, and preserved signal quality in CT, MRI, ultrasound, and X-ray modalities. The experimental results demonstrate a considerable improvement in both the Peak Signal-to-Noise Ratio (PSNR) and maximum embedding capacity. Specifically, the average PSNR value for the X-ray modality reached a notable 73 dB, signifying superior image quality. Moreover, the CT modality exhibited the highest maximum embedding capacity, measured at 0.52, showcasing its efficiency in accommodating data within the images. Moreover, the algorithm consistently offers increased steganographic data hiding capacity in these images without perceptibly degrading their quality or integrity.
1. Introduction
Medical imaging is pivotal in modern healthcare, enabling clinicians to visualize and diagnose various ailments. However, the security and integrity of these images are increasingly threatened by unauthorized access, tampering, or data breaches. Incorporating robust watermarking techniques is imperative to mitigate these risks, ensuring medical imagery’s authenticity and confidentiality [1,2,3]. Watermarking has emerged as a critical mechanism in image security, embedding imperceptible information within images to authenticate and protect against unauthorized alterations. While various watermarking techniques exist, applying such methods to medical images necessitates unique considerations due to the sensitivity and criticality of the data [4,5,6].
The current landscape of medical image watermarking research is multifaceted, showcasing an array of approaches and methodologies [7,8]. Several published works have highlighted the importance of robustness, imperceptibility, and computational efficiency in watermarking algorithms [9,10]. However, these methods often grapple with trade-offs between robustness and imperceptibility, challenging maintaining image quality while ensuring robust security measures. Controversies and diverging hypotheses persist, particularly concerning developing watermarking techniques explicitly tailored for medical images. Some studies advocate for highly complex algorithms, prioritizing an intricate embedding process for enhanced security, while others underscore the significance of simplicity to ensure computational efficiency without compromising robustness [11].
Telemedicine applications face a substantial vulnerability to cyber threats that severely impact the confidentiality, integrity, and authentication of sensitive medical data. The surge in internet usage, coupled with the widespread adoption of smartphones, mobile healthcare devices, and wearable health technology, has significantly propelled the growth of telemedicine [12,13]. This growth has led to the vast exchange and storage of electronic health records among physicians, patients, and healthcare professionals, aiming to enhance healthcare services. Among these records, multimedia, especially images obtained from various medical imaging technologies like X-rays, ultrasound, digital mammography, CT scans, PET scans, and MRI scans, play a critical role [14,15].
However, these medical images, being highly sensitive, are managed within a resource-constrained environment characterized by limited bandwidth, processing power, and memory [16,17]. The extensive use of these images necessitates robust security measures that accommodate stringent privacy requirements while operating under these resource limitations [18,19].
The significance of safeguarding these images lies in the fact that they are crucial for accurate diagnoses and treatment decisions in healthcare [20]. Ensuring these images’ integrity, authenticity, and confidentiality is fundamental, considering the potential repercussions of unauthorized access, tampering, or data breaches. Incorporating robust security algorithms tailored explicitly for resource-constrained environments is imperative for mitigating these risks. These algorithms must strike a balance between providing robust protection for sensitive medical images and operating within the limitations of the systems used in telemedicine. Finding a way to ensure data security without compromising processing efficiency becomes essential for maintaining the trustworthiness and privacy of these medical records within the telemedicine ecosystem [21,22].
This paper aims to contribute to this evolving landscape by presenting an efficient watermarking algorithm customized for medical images. Our primary objective is to develop a technique that optimally balances robustness, imperceptibility, and computational efficiency. This algorithm intends to offer a pioneering solution that fortifies the security of medical images and maintains their diagnostic integrity.
The Fast Discrete Curvelet Transform (FDCuT) stands out in medical imaging due to its multifaceted advantages. One of its primary strengths lies in its remarkable ability to ensure consistent performance across diverse image types, rendering it a dependable choice for a broad spectrum of medical imaging applications. Moreover, FDCuT’s efficiency shines through its capability to swiftly extract various frequency coefficients from medical images, a crucial feature that facilitates comprehensive analysis. Notably, its resilience against geometric attacks and other manipulations further solidifies its reliability in preserving the integrity and accuracy of medical image data. Overall, FDCuT emerges as a robust and versatile tool, offering rapid coefficient extraction, steadfastness across different image types, and resilience against adversarial alterations in medical imaging.
In summary, this paper aims to fill a crucial gap in the existing research by introducing an efficient watermarking algorithm that addresses the unique requirements of medical image security. The findings are expected to contribute significantly to the field by paving the way for more secure and reliable utilization of medical imagery for diagnostic and research purposes.
The main contributions of this work are as follows:
- We introduce a pioneering watermarking model designed explicitly for medical images, leveraging the Frequency Domain Curvelet Transform (FDCuT). This innovative approach dissects medical images into distinct frequency sub-bands, offering a unique and effective solution for watermark embedding in the medical imaging domain.
- Our paper conducts an extensive comparative analysis, evaluating the performance of the proposed algorithm against contemporary methodologies. Performance metrics, including Peak Signal-to-Noise Ratio (PSNR) and maximum embedding capacity, provide a thorough assessment, highlighting the strengths and advantages of our model in securing medical images.
- The algorithm presented in this paper is meticulously crafted to achieve an optimal balance between robustness, imperceptibility, and computational efficiency.
- Our approach incorporates RSA encryption to augment the security of medical images. By integrating public-key cryptography, we establish an additional layer of protection, safeguarding sensitive medical data against unauthorized access and ensuring the confidentiality of embedded information.
The rest of the paper is organized as follows: Section 2 provides a comprehensive overview of medical image encryption technology, detailing its principles, methodologies, and critical advancements. Section 3 introduces the proposed system and a detailed description of the novel approach or methodology proposed in this study within the context of medical image encryption. Moving forward, Section 4 delves into the presentation of results, discussing the outcomes derived from simulations and performance analysis. This section elaborates on the simulation results and performance metrics obtained from the proposed system. Following that, Section 5 engages in a detailed discussion concerning the performance evaluation of the proposed system in comparison to similar algorithms, elucidating the measures and metrics used for contrast and highlighting the system’s comparative strengths and weaknesses. Finally, Section 6 encapsulates the conclusions drawn from this study, summarizing key findings, insights, and potential implications for the field of medical image encryption.
2. Related Works
Hasan et al. [23] introduced a lightweight encryption technology to safeguard the privacy of patients’ medical images. The paper explores diverse security measures, components, and methods for encrypting medical images. It delves into analyzing various existing encryption techniques, assessing their encryption quality, memory requirements, and execution time. The investigation reveals that current methods involving key-based unsystematic sequence numbers result in extensive computational time. In contrast, the study’s results demonstrate that the proposed algorithm significantly reduces computational load. Thus, the meticulously devised algorithm aims to optimize security while ensuring minimal computational overhead to protect medical images. This encryption methodology operates through three stages, employing a 256-bit key value for logical operations on the images.
The work conducted by Araghi et al. [24] presented a novel watermarking scheme based on a Discrete Wavelet Transform (DWT) and 2-D Singular Value Decomposition (SVD). The innovation in this scheme primarily revolves around employing a two-level SVD, thereby enhancing its efficiency to be independent of the host image’s size. Comparative analysis was carried out between the proposed scheme and the authors’ prior DWT and 2-D SVD scheme, utilizing the same host and watermark images, attack parameters, and conditions. The experimental findings demonstrated that the new scheme achieved greater imperceptibility and robustness than the authors’ previous scheme and exhibited superior performance when contrasted with conventional DWT + SVD schemes. Moreover, a two-level authentication system was incorporated to ensure security, effectively identifying false positive and negative outcomes. Additionally, the proposed scheme addressed the limitations of the authors’ prior DWT and 2-D SVD schemes through image blocking and introduced a formula to optimize efficiency for increased capacity.
The method proposed by Khare et al. in [25] introduces an innovative approach for medical image watermarking (MIW) that integrates key features of Hough Transform (HT), Redundant Discrete Wavelet Transform (RDWT), and Singular Value Decomposition (SVD) transformations. The watermark embedding within the reflectance component ensures improved robustness and perceptual invisibility. To fortify resistance against attacks, a combination of RDWT and SVD is utilized. The technique incorporates a dual-layer security mechanism for the watermark image by employing chaotic mapping to safeguard critical diagnostic medical data against manipulation or unethical activities. Furthermore, the performance of the proposed technique is evaluated across several wavelet families. The experimental results notably demonstrate the superior robustness and perceptibility achieved by the proposed scheme, as various performance metrics exhibit enhanced values.
The novel strategy developed by Zermi et al. in [26] involves meticulously integrating hospital signature information and patient data within medical images. The primary aim of this endeavor is to seamlessly embed the watermark with minimal distortion, ensuring the preservation of essential medical information within the image. Employing a flexible approach, the initial step involves applying Discrete Wavelet Transform (DWT) decomposition to the image, enabling a highly adaptable adjustment during insertion. Subsequently, an SVD is employed on the three sub-bands, LL (Low-Low), LH (Low-High), and HL (High-Low), allowing for the preservation of maximum image energy using the essential minimum of singular values.
The research conducted by Alshanbari Hanan S. highlighted the effectiveness of multiple watermark insertions in addressing various security aspects of images, including ownership verification, tamper detection, and Region of Interest (ROI) recovery. The insertion of robust and fragile watermarks in succession was deemed essential to ensure the flawless operation of the proposed algorithm. Utilizing the principal components of the watermark for embedding purposes provided the proposed scheme’s resilience against security errors. Additionally, the application of DWT played a pivotal role in enhancing the watermarking scheme by improving imperceptibility and robustness [27].
In this investigation, Aparna et al. in [28] detailed a medical image watermarking technique employing a variety of algorithms. The utilization of biometric fingerprint technology notably enhances the security of the watermarking system. The study delved into analyzing various methods, including the RG algorithm, SHA-256, minutiae point extraction, elliptical curve cryptography, arithmetic encoding, and the embedding and extraction processes. The assessment focused on medical image integrity, authentication, and confidentiality.
In [29], Amine, Khaldi, et al. presented a robust and imperceptible watermarking method to secure medical images utilized in telemedicine. This technique is tailored to ensure traceability and integrity, fortifying the security of crucial medical data within telemedicine. Their paper introduces a blind watermarking methodology designed to secure electronic patient records effectively. The process involves a meticulous amalgamation of successive values’ parity. This innovative method is implemented across three insertion domains: spatial, frequency, and multiresolution. The watermark is intricately integrated within the image’s colorimetric values for spatial insertion. In the frequency domain, the least significant bit of the Discrete Cosine Transform (DCT) coefficients is replaced with the watermark bits. The integration process employs the LL sub-band coefficients obtained post-Discrete Wavelet Transform (DWT) computation in the multiresolution domain. Upon comparison with recent works in these domains, their approach exhibits noteworthy imperceptibility, particularly in the frequency and spatial domains.
In [30], a study by an undisclosed author proposes applying crypto-watermarking for securing medical images in E-healthcare settings. The work presents an effective crypto-watermarking system integrating cryptographic algorithms and embedding processes. This method is adaptable to various modalities of medical images and is suitable for different image sizes, formats, and bit depths. Notably, including face images enhances the security of the crypto-watermarking system. The analysis includes examining the region-growing algorithm, SHA-256, AES, arithmetic encoding, and the embedding and extraction processes.
Chao et al. proposed an innovative data concealment technique to share digital medical information between different healthcare facilities securely. Their method involved merging various types of medical data into an encoded image, accessible only to authorized individuals during extraction. However, this approach has limitations, notably in its incapacity to detect tampering or perform self-recovery in the event of data corruption or manipulation [6].
On a different front, Guo and Zhuang introduced a watermarking system designed to authenticate and ensure the integrity of medical images. While this system allows for complete image recovery without loss, it operates on a non-blind framework, requiring authentication of the original watermark data, such as electronic patient records. This reliance on specific original data presents a weakness, as it could pose challenges if the original information is unavailable or compromised, potentially hindering the verification process. Additionally, the non-blind nature of this method could lead to security vulnerabilities if unauthorized individuals gain access to the original data, compromising the authentication process [31].
Moreover, both techniques might need help with scalability and compatibility in larger healthcare systems, where diverse data formats and multiple access points could pose challenges in implementing and maintaining these security measures across different platforms and systems within various healthcare facilities. Furthermore, the potential complexity of integrating these methods into existing hospital systems might create barriers to their widespread adoption. It could necessitate extensive training or system updates, making their implementation less straightforward.
Ultimately, while these methods offer advancements in securing medical data transmission and image integrity, their reliance on specific data, potential vulnerabilities due to non-blind operation, and potential scalability issues in complex healthcare systems suggest areas that require further refinement for comprehensive and robust data security in the medical domain.
3. Proposed System
The proposed system for securing medical images implements an organized sequence of processes to fortify the integrity and confidentiality of sensitive healthcare data, as shown in Figure 1.
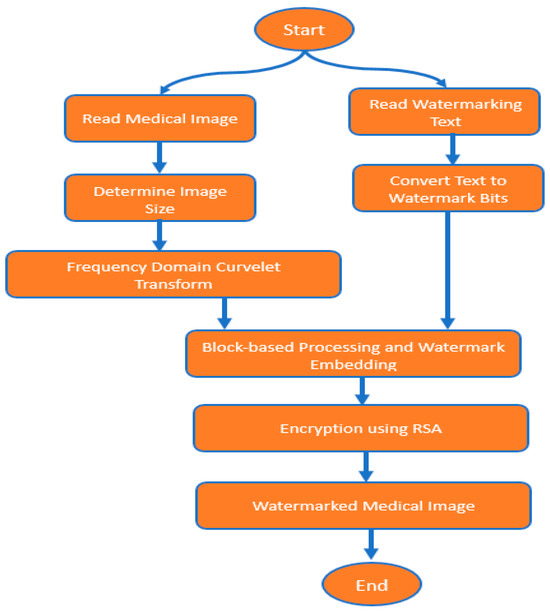
Figure 1.
SecureMediMark flowchart.
Initiating with medical images and the watermarking text, the algorithm systematically navigates through a series of strategic stages. These include reading the medical image and determining image dimensions, which are essential for watermark embedding. Leveraging specialized encoding schemes, textual information is converted into bits to serve as watermarks. The Algorithm 1, SecureMediMark algorithm then applies an advanced Frequency Domain Curvelet Transform (FDCuT), effectively dissecting the image into distinct frequency sub-bands crucial for subsequent watermark embedding and security measures. The process continues with block-based transformations, a Discrete Cosine Transform (DCT), and strategically embedding text watermark bits within transformed coefficients. Conclusively, encryption using the robust RSA algorithm fortifies the watermarked coefficients, ensuring the security and confidentiality of embedded information within the medical image. This amalgamation of diverse processes in the algorithm is designed to strengthen medical images against unauthorized access, ensuring the protection and privacy of the embedded information.
FDCuT, or Fast Discrete Curvelet Transform, presents notable advancements over prior methods like DCT and wavelet transforms. Its effectiveness lies in superior directional information capture and increased sparsity, particularly advantageous for representing signals with intricate directional features, curves, and edges. Unlike DCT, FDCuT offers improved sparsity, overcoming the limitations of traditional methods in handling sparse data efficiently. While it is more computationally intensive than DCT, FDCuT showcases competitive efficiency compared to certain wavelet transforms, making it ideal for applications reliant on capturing directional features such as medical imaging and edge detection in images. Overall, FDCuT’s ability to provide enhanced directional information, higher sparsity, and adaptability to specific application needs makes it a compelling choice for advanced signal processing tasks.
ILF represents the reconstructed low-frequency sub-band.
IMF represents the reconstructed medium-frequency sub-band.
IHF_Reconstructed represents the previously reconstructed high-frequency sub-band after applying inverse DCT and block-based processing.
| Algorithm 1. SecureMediMark algorithm. |
| Step 1: Reading the Medical Image Input: Medical image file |
| Action: Read the medical image |
| Step 2: Determine Image Size and Convert Text to Watermark Bits |
| Input: The read medical image and the text watermark |
| Action: |
| Determine Image Size: Identify the size (dimensions) of the host (medical) image. |
| Ihost represent the host image with dimensions M*N. |
| Convert Text to Watermark Bits: Convert the text watermark into a sequence of bits using an encoding scheme. |
| Let Text_Watermark be the input text to be converted into bits. |
| Watermark_Bits = Text_To_Bits(Text_Watermark) |
| Step 3: Frequency Domain Curvelet Transform (FDCuT) |
| Input: Host (medical) image |
| Action: Apply FDCuT to the host image to obtain ILF, IMF, and IHF sub-bands. |
| ILF,IMF,IHF = FDCuT(Ihost) |
| Step 4: Block-based Processing and Watermark Embedding |
| Input: High-frequency (HF) sub-band, text watermark bits |
| Action: |
| Block-based processing: |
|
|
| IHF_block = Convert_to_Blocks(IHF,P,Q) |
| Watermark embedding: |
|
| IWatermarked_Coefficients = Embed_Text_Watermark(IHF_Blocks,Watermark_Bits) |
| Step 5: Encryption using RSA |
| Input: Watermarked coefficients |
| Action: Encrypt the watermarked coefficients using the RSA encryption algorithm. |
| Encrypted_Coefficients = RSA_Encrypt(IWatermarked_Coefficients, Public_Key) |
Following the securing process, the system’s decryption procedure employs the RSA decryption technique to retrieve and extract the watermark from the previously encrypted coefficients as shown in Algorithm 2 MediRestore decryption protocol. It proceeds by reverse block-based processing, applying inverse operations like the inverse DCT to reconstruct the original high-frequency (HF) sub-band, enabling the retrieval of the embedded watermark. Further, the algorithm employs the Frequency Domain Curvelet Transform (FDCuT) inverse to reconstruct the host (medical) image. Lastly, by utilizing specialized libraries or tools, the system visualizes and interprets the reconstructed medical image data, allowing for the restoration of the original medical image from its sub-bands. This reverse process effectively restores the original medical image, ensuring the reversal of the transformations previously applied and reconstituting the image to its pre-altered state, ensuring the preservation of the original information within [32].
| Algorithm 2. MediRestore decryption protocol. |
| Step 1: Decryption using RSA |
| Input: Encrypted watermarked coefficients |
| Action: Decrypt the encrypted coefficients using the RSA decryption method with the private key. |
| Decrypted_Coefficients = RSA_Decrypt(Encrypted_Coefficients,Private_Key) |
| Step 2: Extract Watermark from Watermarked Coefficients |
| Input: Decrypted coefficients |
| Action: |
|
|
|
| Step 3: Reverse Block-based Processing to Reconstruct HF Sub-band |
| Input: Extracted watermark and decrypted coefficients |
| Action: Apply the inverse of the transformations used during watermark embedding (e.g., inverse DCT, reverse block-based processing) to reconstruct the original HF sub-band. |
| IHF_Reconstructed_Blocks = Inverse_DCT(IWatermarked_Coefficients) |
| IHF_Reconstructed = Combine_Blocks(IHF_Reconstructed_Blocks) |
| Step 4: Inverse FDCuT to Reconstruct Host (Medical) Image |
| Input: Reconstructed HF sub-band and the rest of the FDCuT sub-bands (LF, MF) |
| Action: Apply the inverse of the Frequency Domain Curvelet Transform (FDCuT) to obtain the original host (medical) image. |
| Reconstructed_Image = Inverse_FDCuT(ILF,IMF,IHF_Reconstructed) |
| Step 5: Reading the Original Medical Image |
| Input: Reconstructed original medical image data |
| Action: Utilize specific libraries or tools to interpret the original image data. For instance, PyDICOM or related tools can be used to interpret and visualize the retrieved original medical image. |
| Visualization (IReconstructed_Image) |
4. Results
The suggested model is evaluated using a diverse range of medical images, encompassing X-ray, ultrasound (US), magnetic resonance imaging (MRI), and computed tomography (CT). These images, each with dimensions of 1024 × 1024 pixels, are sourced from the MedPixTM Medical Image Database [33]. All test medical images in the evaluation are grayscale images of 8-bit depth. The watermarking process in the algorithm is anticipated to be both efficient and robust, ensuring minimal distortion to the medical images while maintaining the watermark’s integrity against various image processing operations. This dual focus on efficiency and robustness is essential for preserving the diagnostic quality of the images while securing patient data.
4.1. Simulation
Python version 3.8 was employed to create the algorithms, and OpenCV served a pivotal role in image processing operations, such as reading and converting images to grayscale, and was crucial for feature detection and visualization in the realm of medical imaging. Numpy complemented these tasks by providing robust support for numerical and matrix operations, essential for the sophisticated data handling required in encryption algorithms. Together, these libraries formed the backbone of the script, enabling complex image manipulation and encryption processes critical for securing patient information in medical imaging.
The proposed methodology involves the evaluation of a medical image sized at 1024 × 1024 pixels as the host image, on which the FDCuT is executed to extract its curvelet coefficients. Specifically, the high-frequency curvelet coefficients, sized at 1024 × 1024, are selected to embed a watermark image. These high-frequency curvelet coefficients are transformed into 16,384 separate and non-overlapping blocks, each sized at 8 × 8. The Discrete Cosine Transform (DCT) coefficients for each block are then derived by applying block-wise DCT.
Figure 2 presents a comparative analysis of original and watermarked images and their respective histograms. A compelling advantage of our algorithm becomes evident when observing the histograms of these images. The generated histograms reveal a remarkably subtle difference, suggesting that the watermarking process minimally impacts the image’s overall distribution of pixel intensities. This slight dissimilarity implies that visually distinguishing between the original and watermarked images would be challenging for the human eye.
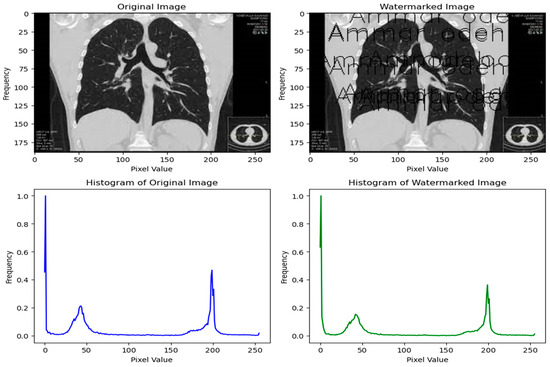
Figure 2.
Compare the original and watermarked image.
This inconspicuous alteration underscores the strength of our approach, ensuring that the embedded watermark has negligible visual impact on the image’s quality or appearance. By maintaining the integrity and aesthetic quality of the image, our algorithm offers a robust and inconspicuous method of safeguarding against unauthorized use or tampering.
Figure 3 showcases a compelling demonstration of our proposed algorithm’s advantage through a side-by-side comparison of original and watermarked images. The ‘Pixel Intensity Difference’ section shows the subtle yet significant difference. This visual representation highlights the altered pixel intensities resulting from the watermark embedding process, revealing the regions where changes have occurred. Despite the slight variations, the images maintain a strikingly similar appearance, affirming our algorithm’s effectiveness in preserving the original image’s visual integrity.
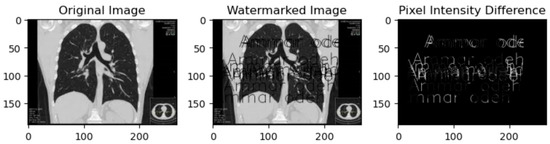
Figure 3.
Pixel intensity difference.
The pixel intensity differences portrayed in the ‘Pixel Intensity Difference’ panel, although existent, reinforce the robust nature of our algorithm. The essential advantage is the ability to embed a watermark into the image without drastically modifying the visual aspects. It ensures that while alterations are present, they need to be more inconspicuous, making it challenging for the human eye to discern any notable changes between the original and watermarked images. This subtle difference speaks to the algorithm’s ability to protect the image’s integrity while maintaining its overall visual appeal.
Figure 4 displays the outcome of applying RSA encryption to the watermarked image generated from the original image. The resulting image, termed the ‘Generated Encrypted Image’, is the product of our algorithm’s encryption process. Applying the robust RSA (Rivest–Shamir–Adleman) encryption technique, the watermarked image, containing the embedded watermark and subtle modifications, is further fortified and transformed into an encrypted form. This transformation ensures the secure concealment and protection of the embedded information within the image.
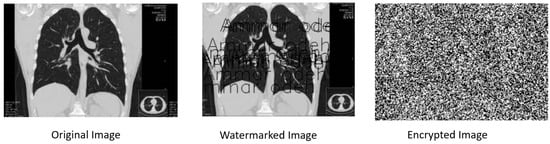
Figure 4.
The generated encrypted image after applying the RSA.
The ‘Generated Encrypted Image’ serves as a testament to the comprehensive security measures implemented by our proposed algorithm. It demonstrates the encryption of the watermark-embedded image, enhancing its resilience against unauthorized access and ensuring the confidentiality of the embedded data. This strategic utilization of encryption techniques strengthens the safeguarding of sensitive information within the image, further fortifying its integrity and confidentiality.
4.2. Performance Analysis
The proposed algorithm’s effectiveness is assessed using two performance measures. The initial metric gauges the distortion caused by the original image due to incorporating the spatial domain watermark quantified through the Peak Signal-to-Noise Ratio. The second metric assesses the maximum potential capacity for embedding within the domain, measured in bits per second (bps).
4.2.1. Peak Signal-to-Noise Ratio
The Peak Signal-to-Noise Ratio is a metric used to quantify the quality of a signal, such as an image or sound, by measuring the ratio of the maximum possible power of the signal to the power of its noise. It is commonly used to evaluate the quality of reconstructed or processed signals compared to their original unprocessed versions. PSNR is expressed in decibels (dB), and higher PSNR values typically indicate higher quality or fidelity in the signal.
where:
- MAXf is the maximum possible pixel value in the image (for an 8-bit image, it is typically 255).
- MSE represents the Mean Squared Error between the original and processed/reconstructed images.
The PSNR values measure the quality of the images produced by these modalities, where higher PSNR values generally indicate better image quality in terms of signal fidelity and lower noise.
According to Figure 5, the X-ray modality has the highest PSNR value of 73 dB. This indicates that X-ray images have the highest quality among the listed modalities in terms of the signal-to-noise ratio. The higher PSNR value suggests that the X-ray images have less noise and better fidelity in preserving the signal details than the other modalities listed.
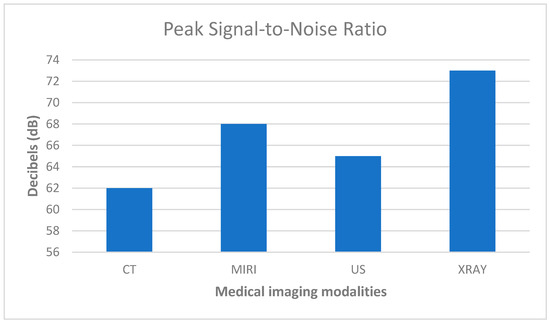
Figure 5.
Visual image quality measured in PSNR.
Conversely, the CT modality has the lowest PSNR value of 62 dB. This suggests that CT images have relatively lower quality in terms of signal fidelity and may have more noise or less preserved signal detail compared to the other modalities mentioned in the list.
4.2.2. Maximum Embedding Capacity
The maximum embedding capacity refers to the maximal amount of supplementary or hidden data that can be inconspicuously integrated into a carrier medium. It represents the upper limit of information that can be embedded within the carrier while maintaining a balance between the additional payload and the integrity of the original content.
Figure 6 shows that the CT modality exhibits the highest maximum embedding capacity of 0.52, indicating a more significant potential for concealing additional data within the images produced using CT scans without substantially compromising the original image quality or being easily detectable by observers or analysis tools. This suggests that CT images have a comparatively larger capacity for steganographic data hiding than the other modalities listed.
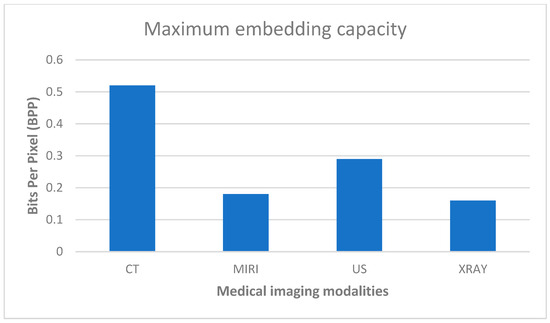
Figure 6.
Maximum embedding capacity.
Conversely, the X-ray modality demonstrates the lowest maximum embedding capacity of 0.16, indicating a smaller potential for hiding additional data within X-ray images. X-ray images have a more limited capacity for steganographic data embedding than the other modalities listed in the figures.
However, it is important to note that while the maximum embedding capacity quantifies the potential for data hiding within these modalities, the clinical relevance and ethical considerations must be carefully weighed, as altering medical images can have significant implications for diagnostic accuracy and patient care.
5. Discussion
The paper introduces an innovative watermarking model specifically designed for medical images, using the Frequency Domain Curvelet Transform (FDCuT). This method segments medical images into different frequency sub-bands, providing an effective way to embed watermarks. The paper also includes a comprehensive comparative analysis, using metrics like Peak Signal-to-Noise Ratio (PSNR) and maximum embedding capacity, to demonstrate the superiority of the proposed algorithm over existing methods in terms of securing medical images. The algorithm is carefully designed to balance robustness, imperceptibility, and computational efficiency. Additionally, the approach integrates RSA encryption, enhancing the security of medical images by adding a layer of public-key cryptography, which protects sensitive data from unauthorized access and maintains the confidentiality of embedded information.
Table 1 represents Peak Signal-to-Noise Ratio values for different medical imaging modalities, such as CT, MRI, ultrasound (US), and X-ray, obtained from various algorithms, including the proposed system and reference algorithms [34,35,36,37].

Table 1.
Comparison between the proposed algorithm and four other algorithms in terms of Peak Signal-to-Noise Ratio.
In the context of the PSNR values, which serve as a measure of image quality, the superiority of the proposed algorithm can be assessed based on the higher PSNR values it achieves compared to the reference algorithms across the different medical modalities.
Therefore, based on the PSNR values, the proposed algorithm demonstrates its superiority by consistently achieving higher PSNR values across all medical imaging modalities compared to the referenced algorithms. This suggests that the proposed system is more effective in maintaining image fidelity, preserving signal quality, and reducing noise in the resulting medical images for CT, MRI, ultrasound, and X-ray modalities.
Table 2 displays the maximum embedding capacity values for different medical imaging modalities, including CT, MRI, ultrasound (US), and X-ray, as achieved using various algorithms—the proposed system (Proposed System) and several reference algorithms [34,35,36,37].

Table 2.
Comparison between the proposed algorithm and four other algorithms in terms of maximum embedding capacity.
In evaluating the superiority of the proposed algorithm based on the maximum embedding capacity values, the proposed algorithm demonstrates its superiority by consistently providing higher capacities for steganographic data hiding across all medical imaging modalities (CT, MRI, ultrasound, and X-ray) compared to the reference algorithms. This suggests that the proposed system allows for a greater capacity to embed additional data within the images without significantly degrading the quality or perceptibility of the original content.
6. Conclusions
In health services information systems, the relevance of medical data in the diagnostic process is paramount. Most healthcare services rely on external systems to store patient information, making the security of these systems of utmost importance. With the advancements in communication and multimedia technologies, digital elements can be manipulated, copied, and replicated without leaving discernible traces, emphasizing the critical need for robust security measures. The effective transfer of medical information over public networks is significantly evolving, especially in telemedicine, telediagnosis, telesurgery, distance learning, and applications related to private database consultations. Transfer conditions mirror those encountered in electronic commerce, subjecting specific medical images to potential vulnerabilities such as communication errors and lossy compression. Preserving the integrity of medical information within the images is particularly crucial. To address these challenges, this paper introduces a novel blind Fast Discrete Curvelet Transform (FDCuT), Discrete Cosine Transform (DCT), and RSA-based medical image watermarking technique. The proposed algorithm demonstrates superior performance, specifically in terms of Peak Signal-to-Noise Ratio and maximum embedding capacity.
Author Contributions
Conceptualization, A.O. and A.A.T.; methodology, A.O. and T.A.; software, A.O.; validation, A.O., A.A.T. and F.N.; formal analysis, A.A.T.; investigation, T.A.; resources, A.O.; data curation, A.A.T.; writing—original draft preparation, A.O., A.A.T. and T.A.; writing—review and editing, F.N. and A.A.T.; visualization, T.A.; supervision, A.A.T.; project administration, A.O. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Publicly available datasets were analyzed in this study. This data can be found here: https://medpix.nlm.nih.gov/home accessed on 11 November 2023.
Acknowledgments
The authors sincerely acknowledge the Princess Sumaya University for Technology for supporting steps of this work.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Odeh, A.; Al-Haija, Q.A. Medical image encryption techniques: A technical survey and potential challenges. Int. J. Electr. Comput. Eng. (IJECE) 2023, 13, 3170–3177. [Google Scholar] [CrossRef]
- Ahmad, A.; AbuHour, Y.; Younisse, R.; Alslman, Y.; Alnagi, E.; Abu Al-Haija, Q. MID-Crypt: A cryptographic algorithm for advanced medical images protection. J. Sens. Actuator Netw. 2022, 11, 24. [Google Scholar] [CrossRef]
- Krichen, M.; Lahami, M.; Al–Haija, Q.A. Formal methods for the verification of smart contracts: A review. In Proceedings of the 2022 15th International Conference on Security of Information and Networks (SIN), Sousse, Tunisia, 11–13 November 2022; pp. 1–8. [Google Scholar]
- Odeh, A.; Keshta, I.; Al-Haija, Q.A. Analysis of Blockchain in the Healthcare Sector: Application and Issues. Symmetry 2022, 14, 1760. [Google Scholar] [CrossRef]
- Keshta, I.; Odeh, A. Security and privacy of electronic health records: Concerns and challenges. Egypt. Inform. J. 2021, 22, 177–183. [Google Scholar] [CrossRef]
- Chen, Y.-P.; Fan, T.-Y.; Chao, H.-C. Wmnet: A lossless watermarking technique using deep learning for medical image authentication. Electronics 2021, 10, 932. [Google Scholar] [CrossRef]
- Zhou, X.; Ma, Y.; Zhang, Q.; Mohammed, M.A.; Damaševičius, R. A reversible watermarking system for medical color images: Balancing capacity, imperceptibility, and robustness. Electronics 2021, 10, 1024. [Google Scholar] [CrossRef]
- Khafaga, D.S.; Karim, F.K.; Darwish, M.M.; Hosny, K.M. Robust Zero-Watermarking of Color Medical Images Using Multi-Channel Gaussian-Hermite Moments and 1D Chebyshev Chaotic Map. Sensors 2022, 22, 5612. [Google Scholar] [CrossRef]
- Tayachi, M.; Mulhem, S.; Adi, W.; Nana, L.; Pascu, A.; Benzarti, F. Tamper and clone-resistant authentication scheme for medical image systems. Cryptography 2020, 4, 19. [Google Scholar] [CrossRef]
- Salim, M.Z.; Abboud, A.J.; Yildirim, R. A visual cryptography-based watermarking approach for the detection and localization of image forgery. Electronics 2022, 11, 136. [Google Scholar] [CrossRef]
- Begum, M.; Uddin, M.S. Digital image watermarking techniques: A review. Information 2020, 11, 110. [Google Scholar] [CrossRef]
- Hafsa, A.; Gafsi, M.; Malek, J.; Machhout, M. FPGA implementation of improved security approach for medical image encryption and decryption. Sci. Program. 2021, 2021, 1–20. [Google Scholar] [CrossRef]
- Khare, P.; Srivastava, V.K. A secured and robust medical image watermarking approach for protecting integrity of medical images. Trans. Emerg. Telecommun. Technol. 2021, 32, e3918. [Google Scholar] [CrossRef]
- Singh, A.K. Fastmie: Faster medical image encryption without compromising security. Measurement 2022, 196, 111175. [Google Scholar]
- Thanki, R.; Kothari, A. Multi-level security of medical images based on encryption and watermarking for telemedicine applications. Multimed. Tools Appl. 2021, 80, 4307–4325. [Google Scholar] [CrossRef]
- More, S.; Singla, J.; Verma, S.; Ghosh, U.; Rodrigues, J.J.; Hosen, A.S.; Ra, I.-H. Security assured CNN-based model for reconstruction of medical images on the internet of healthcare things. IEEE Access 2020, 8, 126333–126346. [Google Scholar] [CrossRef]
- Elhoseny, M.; Shankar, K. Optimal bilateral filter and convolutional neural network based denoising method of medical image measurements. Measurement 2019, 143, 125–135. [Google Scholar] [CrossRef]
- Singh, A.K. Robust and distortion control dual watermarking in LWT domain using DCT and error correction code for color medical image. Multimed. Tools Appl. 2019, 78, 30523–30533. [Google Scholar] [CrossRef]
- Belazi, A.; Talha, M.; Kharbech, S.; Xiang, W. Novel medical image encryption scheme based on chaos and DNA encoding. IEEE Access 2019, 7, 36667–36681. [Google Scholar] [CrossRef]
- Liu, X.; Lou, J.; Fang, H.; Chen, Y.; Ouyang, P.; Wang, Y.; Zou, B.; Wang, L. A novel robust reversible watermarking scheme for protecting authenticity and integrity of medical images. IEEE Access 2019, 7, 76580–76598. [Google Scholar] [CrossRef]
- Kaissis, G.; Ziller, A.; Passerat-Palmbach, J.; Ryffel, T.; Usynin, D.; Trask, A.; Lima Jr, I.; Mancuso, J.; Jungmann, F.; Steinborn, M.-M. End-to-end privacy preserving deep learning on multi-institutional medical imaging. Nat. Mach. Intell. 2021, 3, 473–484. [Google Scholar] [CrossRef]
- Singh, K.S.; Singh, H.V. Security of Medical Images Using DWT and SVD Watermarking Technique. In Proceedings of the Recent Trends in Electronics and Communication: Select Proceedings of VCAS 2020, Prayagraj, India, 14–16 October 2020; pp. 569–579. [Google Scholar]
- Hasan, M.K.; Islam, S.; Sulaiman, R.; Khan, S.; Hashim, A.-H.A.; Habib, S.; Islam, M.; Alyahya, S.; Ahmed, M.M.; Kamil, S. Lightweight encryption technique to enhance medical image security on internet of medical things applications. IEEE Access 2021, 9, 47731–47742. [Google Scholar] [CrossRef]
- Araghi, T.K.; Abd Manaf, A. An enhanced hybrid image watermarking scheme for security of medical and non-medical images based on DWT and 2-D SVD. Future Gener. Comput. Syst. 2019, 101, 1223–1246. [Google Scholar] [CrossRef]
- Awasthi, D.; Khare, P.; Srivastava, V.K. Multiple Image Watermarking in YCbCr Color Space Using Schur-SVD-DCT in Wavelet Domain and its authentication using SURF. In Proceedings of the 2023 10th International Conference on Signal Processing and Integrated Networks (SPIN), Delhi, India, 23–24 March 2023; pp. 192–197. [Google Scholar]
- Zermi, N.; Khaldi, A.; Kafi, M.R.; Kahlessenane, F.; Euschi, S. Robust SVD-based schemes for medical image watermarking. Microprocess. Microsyst. 2021, 84, 104134. [Google Scholar] [CrossRef]
- Alshanbari, H.S. Medical image watermarking for ownership & tamper detection. Multimed. Tools Appl. 2021, 80, 16549–16564. [Google Scholar]
- Aparna, P.; Kishore, P.V.V. Biometric-based efficient medical image watermarking in E-healthcare application. IET Image Process. 2019, 13, 421–428. [Google Scholar] [CrossRef]
- Amine, K.; Fares, K.; Redouane, K.M.; Salah, E. Medical image watermarking for telemedicine application security. J. Circuits Syst. Comput. 2022, 31, 2250097. [Google Scholar] [CrossRef]
- Khaldi, A.; Redouane, K.M.; Bilel, M. A Medical Image Watermarking System Based on Redundant Wavelets for Secure Transmission in Telemedicine Applications. Wirel. Pers. Commun. 2023, 132, 823–839. [Google Scholar] [CrossRef]
- Su, G.-D.; Chang, C.-C.; Lin, C.-C. Effective self-recovery and tampering localization fragile watermarking for medical images. IEEE Access 2020, 8, 160840–160857. [Google Scholar] [CrossRef]
- Zhang, X.; Zhang, X. Image Encryption Algorithm Based on the Matryoshka Transform and Modular-Inverse Matrix. 2023. Available online: https://www.researchsquare.com/article/rs-2663096/v1 (accessed on 11 November 2023).
- MedPixTM Medical Image Database MedPix isr Egistered Trademark of USUHS. 2020. Available online: https://medpix.nlm.nih.gov/home (accessed on 11 November 2023).
- Ke, G.; Wang, H.; Zhou, S.; Zhang, H. Encryption of medical image with most significant bit and high capacity in piecewise linear chaos graphics. Measurement 2019, 135, 385–391. [Google Scholar] [CrossRef]
- Liu, J.; Tang, S.; Lian, J.; Ma, Y.; Zhang, X. A novel fourth order chaotic system and its algorithm for medical image encryption. Multidimens. Syst. Signal Process. 2019, 30, 1637–1657. [Google Scholar] [CrossRef]
- Ravichandran, D.; Banu, S.A.; Murthy, B.; Balasubramanian, V.; Fathima, S.; Amirtharajan, R. An efficient medical image encryption using hybrid DNA computing and chaos in transform domain. Med. Biol. Eng. Comput. 2021, 59, 589–605. [Google Scholar] [CrossRef] [PubMed]
- Shafique, A.; Ahmed, J.; Rehman, M.U.; Hazzazi, M.M. Noise-resistant image encryption scheme for medical images in the chaos and wavelet domain. IEEE Access 2021, 9, 59108–59130. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).