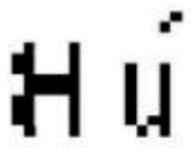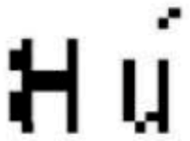Abstract
A hybrid watermarking approach for DICOM medical image protection is proposed in this paper. The medical image is first separated into two parts: the region used for the diagnosis, called Region Of Interest (ROI), and the remaining, called Region Of Non-Interest (RONI). The patient’s name is then extracted from the header of the DICOM image and pertinent features from the ROI. These data are therefore used to construct a watermark based on a Jacobian model. On the one hand, this watermark is used for the zero watermarking of the ROI. On the other hand, it is divided into blocks that are embedded in the RONI using a linear interpolation technique. Experiment results show a good performance of the proposed approach. The average values of SSIM, PSNR, NC, and BER are respectively 0.98, 71 dB, 0.98, and 0.0054, and the watermark embedding and extraction times do not exceed 12 s.
Keywords:
medical images; DICOM; ROI; RONI; watermarking; zero-watermarking; authentication; Jacobian model 1. Introduction
The work presented in this paper extends and enhances the work originally presented in [1]. Telemedicine uses telecommunications and information technology to offer health services in distant places. It adds access to medical services that are not available in distant rural areas. When using telecommunications technology, patients who live in distant communities can benefit from the medical analysis of remote professionals, so the patient does not have to travel to visit a doctor for consultation. Telemedicine facilitates the transmission of medical images and information to doctors and radiologists for diagnosis and treatment. It offers doctors the opportunity to send medical data and images from one hospital to another. So, doctors can help other doctors to find out adequate analytical methods to treat patients [2].
During transmission or storage, medical images may be intentionally or accidentally modified. Therefore, the authenticity of medical images needs to be controlled by the specialist to avoid a wrong diagnosis that may be caused by their modification [3].
Digital watermarking is a promising method to secure medical images and patient data. Indeed, it provides a solution making it possible to authenticate images at different steps of their use: during their storage, during their transmission over the network, and after their reception. There are two main watermarking principles. The first one consists in embedding a watermark in the image and extracting it later for image authentication. We will refer to watermarking approaches based on this principle as non-zero-watermarking approaches. The second principle is to generate a watermark from the image and to use the watermark as a key that will be used later to authenticate the original image by regenerating the key and comparing it to the original one. Watermarking approaches based on this second principle are called zero-watermarking approaches.
Medical images contain two disjoint areas respectively called Region Of Interest (ROI) and Region Of Non-Interest (RONI). The ROI is part of the image that is useful for the diagnosis and the RONI is part of the image that has no impact on the diagnosis.
There are four main categories of medical image watermarking methods [4]:
- Watermarking in RONI [5,6,7]
- Reversible watermarking [8,9,10]
- Zero-watermarking [11,12,13]
- Watermarking with image change not affecting diagnosis [14].
In this paper, we propose a watermarking approach that provides strong security for DICOM medical images by combining a zero-watermarking in ROI and a non-zero-watermarking in RONI, by using and ensuring the confidentiality of patient data available in the header of the DICOM image. DICOM is the format commonly used for the transfer of medical images. By embedding the watermark only in RONI, the diagnosis is not affected and the sensitive information of the patient is secured and not modified. Furthermore, by securing the ROI part with a zero-watermarking method, the ROI part is not modified because in zero-watermarking the watermark is not inserted directly into the original image. The proposed watermarking approach also takes into account the real-time constraints of medical applications.
The remainder of the paper is organized as follows. The second section is dedicated to related works. The proposed watermarking approach is described in section three. Experimentations performed for the evaluation of the proposed solution as well as the results obtained and their analysis are presented in section four. The paper ends with a conclusion and future works in section five.
2. Related Works
Various image watermarking methods have been proposed in the literature to achieve image authentication. In this section, we present some of them.
The authors in [4] (Ales Rocek et al.) proposed a fully reversible watermarking method combining the advantages of three approaches: reversible, zero, and RONI watermarking. The method is based on the separation of the original image into ROI and RONI parts. The ROI of the original image and the watermark are combined to generate a ‘secret share’ using a zero-watermarking approach. The ‘secret share’ is embedded as a watermark into the RONI part of the original image using a reversible watermarking method called reversible contrast mapping (RCM). Further, the original ROI and the watermarked RONI are concatenated to generate a watermarked image. The imperceptibility ratio in terms of SSIM reached 0.99, and the PSNR was around 81.
The authors in [15] (Liew Siau-Chuin et al.) proposed a reversible tamper detection and recovery watermarking method. The watermarking method applied the concept of the separation of ROI and RONI. The separation of ROI and RONI is done manually by using a rectangle to define the ROI of the medical image. Four rectangles are used to form a pyramid shape for the ROI, and six rectangles are used to define areas in the RONI. The ROI is divided into blocks of 8 × 8 pixels and RONI into blocks of 6 × 6 pixels. A one-to-one mapping technique is used to assign a unique integer to each block in the ROI and RONI. Each block in RONI corresponds to a block in ROI. The number of blocks in the ROI must not exceed the number of blocks in the RONI to ensure that the original LSBs that will be removed from the ROI for watermark embedding can be stored in the RONI without any storage issue.
Jasni Mohamad Zain et al. [16] presented a reversible region of non-interest (RONI) watermarking to verify the authenticity and integrity of DICOM images. This method is based on the separation of ROI and RONI also. The watermark is generated from the ROI by creating a hash value of the whole image, using SHA-256. After that, the watermark is embedded in the least significant bits (LSB) of the RONI. This method is reversible. If the image has not been altered, the watermark will be extracted and the original image will be recovered and the SHA-256 of the recovered image will be compared with the extracted watermark for authentication purposes. Experimental results showed that the PSNR was equal to 32 dB.
In [17], Wei Pan et al. described a fragile reversible watermarking method of radiographic images for integrity and authenticity control for telemedicine applications. Quantum noise is computed using half of the wavelet coefficients of the anatomical object. The method uses a classification process to differentiate the image’s black background from the anatomical object which is the region of interest. From the image background, histogram shifting HS is applied to pixels; and from the anatomical object, HS is applied to wavelet detail coefficients, masking the watermark into the image quantum noise. The experimental results showed that the PSNR was equal to 76.5 dB and the SSIM was equal to 0.99.
Afaf Tareef et al. [18] described a recovery algorithm to confirm the integrity and authenticity of medical images. The whole image is divided into two regions, ROI and RONI. The sparse coding of the EPR data and the reshaped ROI are jointly embedded in the transform domain of the RONI using singular value decomposition (SVD). In the first part of the sparse coding, the patient information was saved along with the image, while the second part was used for verifying the authenticity. The hidden sparse coded ROI can be extracted to reconstruct the altered image. This technique can be used for several purposes such as EPR data hiding, authentication of the ROI, and retrieving the manipulated area. Experiment results showed that the PSNR was equal to 69.82 dB and the NC was equal to 0.80.
Rayachoti Eswaraiah et al. [19] proposed a novel medical image watermarking method based on integer wavelet transform (IWT). This proposal verifies the integrity of ROI, precisely identifies tampered blocks inside ROI, provides robustness to the data embedded inside the region of non-interest (RONI), and recovers the original ROI. In the proposed method, the medical image is segmented into ROI and RONI regions. The hash value of ROI, recovery data of ROI, and patient data are embedded into RONI using IWT. Experimental results show that the proposed method provides robustness to the watermark data embedded inside RONI and accurately detects and locates tampered areas inside ROI and recovers the original ROI. The PSNR was equal to 40 dB and the NC was equal to 0.9.
Baoru Han et al. [2] proposed a method combining a Hermite chaotic neural network and a zero-watermarking algorithm for the security of medical volume data. Firstly, it employs a novel Hermite chaotic neural network to generate the pseudo-random chaotic sequence for scrambling. Secondly, the three-dimensional medical image is transformed by a three-dimensional discrete wavelet transform and a three-dimensional discrete Fourier transform. Then, the transformed low and intermediate frequency coefficients are selected as medical volume data characteristics to structure zero-watermarking. The watermarking extraction does not need the original image, and its security depends on the chaotic sequence complexity and unpredictability, solving the watermark embedding, extraction safety, and efficiency. The simulation results show that the algorithm is simple to implement, with good robustness, security, and invisibility. The NC value was equal to 0.92.
Fares Kahlessenane et al. [20] present a watermarking method that consists in concatenating patient information and image acquisition data, computing the corresponding hash using the MD5 algorithm, and embedding the result in the original image. The proposed approach is applied in the frequency domain. Four transforms were used: discrete wavelets transform, non-subsampled contourlet transform, non-subsampled shearlet transform, and discreet cosine transform. All these transforms were combined with Schur decomposition, and the watermark bits were embedded in the upper triangular matrix. Experimental results show that the proposed method is robust against several conventional attacks. The PSNR value was equal to 54 dB, and the NC value to 0.99.
Chengshan Yang et al. [21] proposed a zero watermarking method for medical images based on Zernike-DCT. To encrypt the watermark, the authors used a chaotic logic sequence, then applied edge detection and Zernike moment processing on the original medical image to get the accurate edge points, and then discrete cosine transform (DCT) on them to get the feature vector. The perceptual hash and the zero-watermark are combined to generate the key (watermark) used in the embedding and extraction processes. Experimental results show that the NC value is 0.80, and the PSNR value is 16.3 dB.
Analysis of the above methods shows that only one deals with the DICOM format commonly used for medical image transfer. Most of the proposed methods (Ales Rocek et al. [4], Afaf Tareef et al. [18], Rayachoti Eswaraiah et al. [19], Baoru Han et al. [2], Fares Kahlessenane et al. [20], Chengshan Yang et al. [21]) are implemented in the transform domain which generally leads to higher time complexity than implementations in the spatial domain and may cause difficulties in meeting time constraints related to medical diagnosis. Some of the proposed methods (Wei Pan et al. [17]) also embed the watermark in ROI and this may cause wrong diagnosis. Although the approach proposed by Ales Rocek et al. [4] provides strong medical images security by combining zero-watermarking in ROI and watermarking in RONI, the extraction of the secret share is expected to be done from the RONI and only a single copy of the secret share is inserted and uses all the RONI. Since images are subject to attacks during their transfer, an attack of the RONI that does not affect the ROI may lead to the impossibility to recover the secret share that is required for the authentication process.
The watermarking approach proposed in this paper aims at filling some of the gaps in the existing solutions. It takes into account the DICOM format and is based on the spatial domain. To enforce the security of medical image, it extracts patient information from the header of the DICOM image and combines it with specific image features to generate a strong authentication key that is only inserted in the RONI of the medical image, leaving the ROI unmodified to ensure the safety of the medical diagnosis. The proposed approach also integrates the insertion of multiple copies of the authentication key to cope with attacks that may modify parts of the RONI.
The proposed approach is detailed in the next section.
3. Proposed Method
3.1. General Description
The proposed method combines a zero-watermarking in the anatomical region of the image (ROI) and a non-zero-watermarking (insertion of a watermark) in the black background region of the image (RONI).
The medical image is initially divided into two parts: the anatomical part (ROI) and the black background part (RONI). Then from the header of the DICOM image, the first letter of the given name and the first letter of the family name of the patient are extracted, and pertinent features are extracted from the anatomical part. These pertinent features are skewness, entropy, and median. The patient name initials and pertinent features are then used as input to a Jacobian Model to build the zero-watermarking secret share as well as an encoded watermark. Several copies of the watermark are then embedded in the RONI, using a linear interpolation technique [22]. In this embedding process, the watermark and the RONI are split into several parts with a mapping between the watermark parts and RONI parts, and each watermark part is inserted in the corresponding RONI part. Finally, the anatomical region and the watermarked black background region (RONI) are merged to obtain the watermarked image.
On the receiver side, the process is similar to that used on the sender’s side. The image is divided into two parts (the anatomical part and the black background part), the watermark is extracted from the black background region using the extraction procedure of the linear interpolation technique, and the original watermark is compared to the extracted watermark. On the other hand, the zero-watermarking secret share is rebuilt from the ROI of the received image and compared to the original secret share.
Figure 1 shows the framework of the proposed watermarking method.
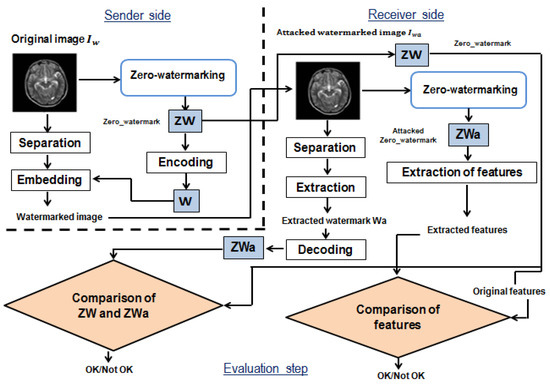
Figure 1.
The framework of the proposed watermarking method.
The proposed model is based on three phases on both the sender and receiver sides: the anatomical region and the black background region separation, zero-watermarking secret share generation from the anatomical region, and watermark embedding/extraction in/from the black background region. These three phases are followed by a last step on the receiver’s side, consisting of the comparison of the original and recovered secret shares and watermarks, to authenticate the medical image.
3.2. Separation of the Medical Image
The proposed method is based on the separation of the medical image into two regions: the anatomical region and the black background region. The separation can be done automatically by using an algorithm [4] or manually by using a rectangle to form the ROI [15,19]. In our method, we have used an automatic separation based on a threshold. The threshold was determined starting from the theoretical value 0 corresponding to the black pixel and iteratively increasing it by 1 until reaching a value making it possible to obtain a good separation between the anatomical region (ROI) and the black background region (RONI) for the images of the medical images database. A gray value greater than the threshold belongs to the anatomical object, and a gray value below or equal to the threshold belongs to the black background region of the image. Figure 2 shows an example of the anatomical region and the black background region separation.

Figure 2.
(a) the original image, (b) the anatomical region IAntomical, (c) the black background region IBlackbackground and (d) original image with separation between IAntomical and IBlackbackground.
3.3. Zero-Watermarking Secret Share and Watermark Generation
The generation of the zero-watermarking secret share and the watermark is based on the extraction of preselected pertinent statistical features (skewness, entropy, and median) from the anatomical part of the original image. The pertinent statistical features were chosen after applying a statistical analysis aiming at selecting the smallest set of statistical parameters making it possible to uniquely identify each medical image within a large database of medical images.
The initials of the patient name (first letter of the given name and first letter of the family name) are extracted from the header of the DICOM image and presented as a matrix of size 16 × 16 pixels. An additional matrix is also generated from the anatomical part. The three pertinent features, the patient initials matrix, and the additional matrix are used as input of the Jacobian model to construct the zero-watermarking secret share of size 16 × 16 pixels. After generation, the zero-watermark is encoded into a watermark of size 32 × 32 pixels before its embedding in the original image.
Figure 3 illustrates the process of watermark generation. Figure 4 shows an example of a watermark generated from the original DICOM image of Figure 3, using the proposed watermark generation process.
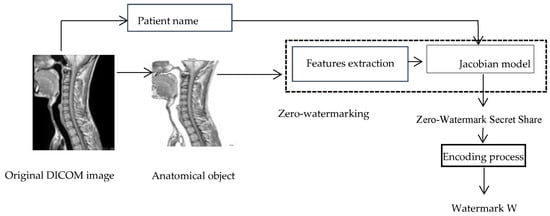
Figure 3.
Process of watermark generation.

Figure 4.
Example of zero-watermark and corresponding encoded Watermark.
3.4. Watermark Embedding Process
3.4.1. Black Background Region Verification
Before embedding the watermark in the black background region, the watermark is divided into blocks of size n x m. Its insertion depends on the availability of the blocks in the black background region. Therefore, the block size n × m should be chosen so as to ensure the availability of enough regions in the black background for the insertion of the watermark. In order to increase the robustness of the system, the targeted block size is that making it possible to ensure the insertion of two copies of the watermark.
In our model, an algorithm has been proposed for the search of the regions. The algorithm searches in the black background region the number of equal-sized regions R1, R2, R3, and Rn of size n × m pixels.
Figure 5 shows an example of the selection of the regions in the black background image to embed the watermark with n × m = 8 × 8. In this case, the block size chosen is 8 × 8. 8 regions have been selected in the RONI (black background) for the insertion of the copies of the watermark. The watermark will be divided into four blocks of 8 × 8 pixels that will be inserted in the RONI.
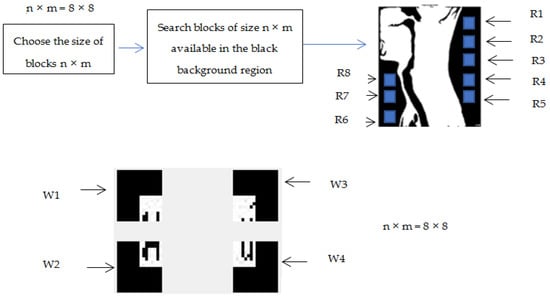
Figure 5.
Example of division of the black background region and the watermark with n × m = 8 × 8.
3.4.2. Embedding the Watermark in the Black Background Region
As mentioned earlier, the zero-watermark ZW of size 16 × 16 generated using the zero-watermarking approach is first encoded into a watermark W of size 32 × 32 before embedding into the original image. W is divided into K blocks of size n × m (, ), and the algorithm mentioned in the subsection above is used to find the regions (,…) of size n x m in the black background region for the insertion of the blocks of the watermark. The blocks are embedded into the regions selected using a linear interpolation technique [22]. In this technique, the embedding of a watermark W into a host image I uses the following equation (Equation (1)) to generate the watermarked image Iw:
Iw = (1 − t) W + t × I, where t is the interpolation factor and t ∈ ]0,1[
The imperceptibility of the embedded watermark is stronger when the value of t is close to 1. For this reason, a value of t close to 1 has been chosen for the embedding of the blocks of the watermark in the selected regions of the black background of the medical image.
After embedding the watermark blocks into the black background region, the anatomical object image and the watermarked black background image are merged to obtain the watermarked image.
Figure 6 shows the watermark embedding process. Algorithm 1 presents the pseudo-code of the watermark embedding process.
| Algorithm 1: The pseudo code of the watermark embedding in the black background region of the image. |
| Input: host image I, the selected 2k blocks blocki (i = 1, … 2k) of size n × m in the black background region, original zero-watermark image ZW and interpolation factor t Output: watermarked image (Iw) Begin W ← Encode ZW Divide W into k blocks Wi (i = 1,…,k) of size nxm for i = 1 to k loop end loop Iw ← Merge Anatomical region of host image I and watermarked black background End |
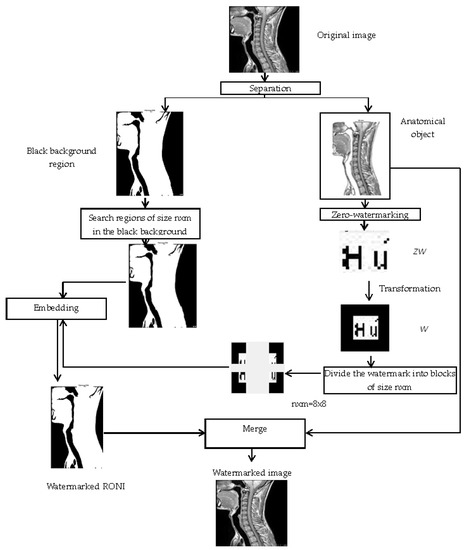
Figure 6.
The watermark embedding process.
3.5. Watermark Extraction Process
After the embedding process, the zero-watermark generated by the zero-watermarking process is sent as a key to the receiver through a secure channel. The obtained watermarked image will be sent to the receiver via public insecure networks and it will be exposed to different kinds of attacks.
Therefore, the received image is an attacked watermarked image and the extraction process must be applied to prove the origin of the image by extracting the set of attacked watermarks and from . The inverse form of the linear interpolation technique [22] is applied. The inverse form of the linear interpolation technique uses the following equation (Equation (2)) to extract the attacked watermark Wa from the attacked watermarked image Iwa:
Wa = (1/t) W + ((1−t)/t) × Iwa, where t is the interpolation factor and t ∈]0,1[
On the one hand, the extracted attacked watermarks are decoded to obtain corresponding zero-watermarks. These zero-watermarks are compared to the original ones using BER and NC metrics. If they are similar, the received image is authenticated and can be used for diagnosis. On the other hand, a comparison of the original features extracted from the original zero-watermark and the features extracted from the attacked zero-watermark is done for the authentication of the medical image. The image is considered authentic if the difference between the values of the initial and the extracted features is lower than a given threshold. Figure 7 illustrates the watermark extraction process.
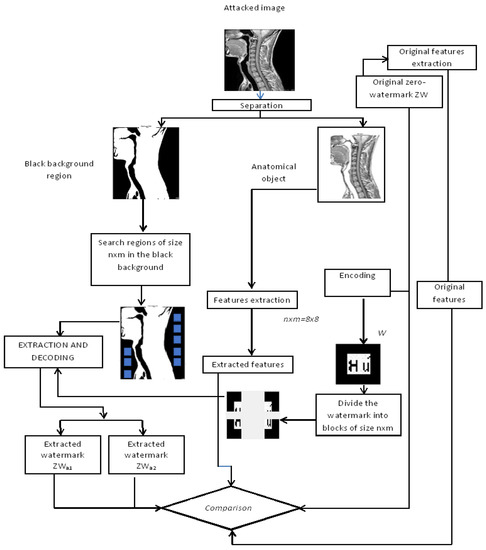
Figure 7.
Watermark extraction process.
The pseudo code of attacked watermark extraction process is illustrated in Algorithm 2.
| Algorithm 2: The pseudo code of extraction of the watermark from the black background region. |
| Input: Attacked watermarked image Iwa, the selected 2k attacked blocks blockai (i = 1, … 2k) of size nxm in the black background region, original zero-watermark image ZW and interpolation factor t Output: Set of attacked zero-watermarks (ZWa1,ZWa2) Begin W ← encode ZW Divide W into k blocks Wi (i = 1,…,k) of size nxm for i = 1 to k loop end loop Wa1 ← Merge Wai (i =1,…,k) (first copy of attacked watermark extracted) Wa2 ← Merge Wak+i, (i==1,…,k) (second copy of attacked watermark extracted) ZWa1← Decode Wa1 ZWa2← Decode Wa2 End |
4. Experiment Results
In this section, we present and analyze the results obtained with the proposed method, and compare it to methods proposed in similar works. The proposed watermarking system is implemented using MATLAB and executed on a Windows machine with the following characteristics: Intel R Core i5 processor, 4 GHz, 4 GB RAM, and Microsoft Windows 8 Professional operating system platform. In our experiments, we have used DICOM images of size 512 × 512 pixels as shown in Table 1.

Table 1.
Original images.
In our method, several copies of the watermark are embedded in the RONI region, and the ROI region is kept intact. ROI and RONI are separated using the method presented in Section 3.2. The watermark is inserted in the RONI using a linear interpolation technique. From the results, one can see that there is no significant visual difference between the original and the watermarked images. Nevertheless, the difference appears through the values of SSIM and PSNR presented in next subsection.
Table 2 shows the attacked watermarked image and the corresponding extracted attacked zero-watermarks.

Table 2.
Attacked watermarked images and their corresponding extracted attacked watermarks Wa1 et Wa2.
From Table 2 we can see the effect of the attacks on the original image, along with the variation of the extracted attacked watermarks. It is worth indicating that in the case of rotation attacks, the proposed method applies the inverse rotation to the image before extracting the watermark. We notice that the cropping left top corner attack is the attack that affects images the most.
4.1. Evaluation Metrics
4.1.1. Perceptual Quality Analysis
The peak signal-to-noise ratio (PSNR) and the structural similarity index measurement (SSIM) are used to evaluate the perceptual quality of the DICOM images after embedding the watermark.
The peak signal-to-noise ratio (PSNR) measures the distortion between the original and the watermarked images. Higher values of PSNR indicate a lower distortion.
For grayscale images with 8-bit pixels, the PSNR is given by:
where MSE is the Mean Squared Error between the original and the watermarked images (Equation (4))
where M × N is the size of the images, is the original image, and is the watermarked image.
The Structural Similarity Index (SSIM) quantifies the similarity between the original and watermarked images. It compares the similarity of three parameters: luminance, contrast, and structure. The SSIM takes a value between −1 and 1. Higher values of SSIM (close to 1) indicate good imperceptibility.
where is the average of the original image , is the average of watermarked image , and are respectively the variance of and , cov is the covariance between and , and are variables to stabilize the division with weak denominator, and is the dynamic range of pixel values (). and are defined as follows:
Table 3 presents the values of the SSIM between the original image and the watermarked image under different attacks. The results in this table show that the proposed approach ensures a good level of imperceptibility. The SSIM values are over 0.9, which means good similarity between the original and watermarked images.

Table 3.
Values of SSIM between the original image and the attacked watermarked image.
Table 4 presents the values of the PSNR between the original and the watermarked medical image under different attacks. In this table, the PSNR values between the original and the watermarked images are greater than 71 dB. So, there is a very low distortion between the original image and the attacked watermarked images.

Table 4.
Values of PSNR between the original and attacked watermarked images.
4.1.2. Robustness Analysis
To evaluate the robustness of the embedded watermark against different attacks, Bit Error Rate (BER) and Normalized Correlation (NC) are used.
Bit Error Rate (BER) measures the percentage of erroneous bits between the extracted watermark and the original watermark. Lower BER values indicate higher robustness of the watermark against different attacks. The BER is given by Equation (6)
where represents the pixel in the original watermark , represents the pixel in the watermarked image , and is the watermark’s size.
Normalized Coefficient (NC) is a coefficient used to indicate the similarity between the original and the extracted watermark. Its value is between [–1, 1]. When the NC is 1 the original and extracted watermarks are absolutely identical. When NC = 0 the original and extracted watermarks are divergent. When NC = −1 the original and extracted watermarks are completely anti-similar.
NC is defined as follows:
where are the original and extracted watermark, is the () pixel in the original watermark is the pixel in the extracted watermark , is the mean of the original watermark , and is the mean of the extracted watermark , and is the dimension of the watermark.
Table 5 presents the values of NC and BER to prove the robustness of the watermark under different attacks. As mentioned in the previous section, two copies of the same watermark were embedded in the black background region of the original medical image. BER and NC values between the original and corresponding extracted watermarks were the same for the two copies of the watermark except in the case of the cropping left top corner attack. So, the values of BER and NC indicated in the table are common values for the two copies of the watermark, except for the Cropping left top corner attack where the values obtained for each copy of the watermark are mentioned.

Table 5.
Values of BER and NC between the original watermark 1 and the extracted attacked watermark 1.
The values of BER presented in Table 5 show the high robustness of the embedded watermark against various attacks. Indeed, on average, the BER values are equal to 0.5%. Furthermore, the NC values in Table 5 show that the embedded watermark withstood the different attacks. The NC values between the extracted attacked watermark and the original one are, on average, equal to 0.98.
4.1.3. Execution Time Analysis
Table 6 presents the processing time for embedding the watermark in the RONI region and the processing time for extraction of the watermark from the RONI region after the attacks.

Table 6.
Execution time for watermark embedding and extraction.
As shown in Table 6, the execution time for embedding the watermark is, on average, 10.8567 s, and the execution time of the extraction process of the watermark is, on average, 11.9031 s.
4.2. Comparative Analysis with Other Related Works
A comparison of the proposed method with related methods separating ROI and RONI (methods of [4,17,18,23]) is presented in Table 7.

Table 7.
Comparison of the proposed method with other works in literature.
As far as robustness is concerned, our method is robust against the majority of attacks (median filtering, salt and pepper noise, average filtering, cropping left top corner, Gaussian filtering, histogram equalization, Gaussian noise, rotation, sharpening and translate attack), the method of Afaf Tareef et al. [18] is robust against fewer attacks (blurring, Gaussian noise, hard threshold and JPEG compression attacks), that of Wei Pan et al. [17] is only robust against compression attacks, and the method of NisarAhmed Memon et al. [23] against Gaussian noise, Median filtering, JPEG compressing, and copy attacks and not tested against other kinds of attacks.
By comparing the SSIM values, we can see that the methods proposed by Wei Pan et al. and Ales Rocek et al. achieve the same average value as our.
Regarding the PSNR, our method achieves a better value than that of Afaf Tareef et al. and Nisar Ahmed Memon et al., but less good than those achieved by Wei Pan et al. and Ales Rocek et al.
Regarding the BER, our approach reached 0.01. The other authors did not present their BER results.
By comparing the NC values, we can see that our method achieves a better result than that of Afaf et al.
5. Conclusions
Attacks on medical systems are increasing. It is essential to protect medical data exchanged or accessed to provide healthcare. Digital watermarking is a promising method to secure medical images and patient data. A watermarking method aiming at filling some gaps in existing watermarking solutions has been proposed in this paper. Unlike most existing solutions, it considers most domain-specific constraints of the medical domain (DICOM format commonly used for medical image transfer, timing constraints of medical diagnosis, no modification of the ROI, confidentiality of patient information). The proposed solution provides strong security of DICOM medical images by combining a zero-watermarking in ROI and a non-zero-watermarking in RONI with the insertion of several copies of the watermark for more robustness, by using and ensuring the confidentiality of patient data available in the header of the DICOM image. The proposed method has been applied to various DICOM images. Several performance parameters have been calculated (SSIM, PSNR, BER, NC) or measured (Time spent for watermarking embedding/extraction) to evaluate the proposed solution. The results analysis shows a good performance of the proposed approach. The average SSIM, PSNR, NC, and BER are respectively 0.98, 71 dB, 0.98, and 0.0054. Watermarking embedding/extraction time does not exceed 12s, which meets real-time requirements for medical diagnosis. Comparison of the proposed method to related methods separating ROI and RONI parts reveals its good results. Future works include investigating adaptive approaches to deal with the variable size of the RONI part of the medical image.
Author Contributions
Conceptualization, M.T., L.N., A.C.P. and F.B.; Methodology, M.T., L.N., A.C.P. and F.B.; Software, M.T.; Supervision, L.N., A.C.P. and F.B.; Validation, L.N. and A.C.P.; Formal analysis, M.T., L.N. and A.C.P.; Visualization, L.N. and A.C.P.; Writing—original draft preparation, M.T., L.N. and A.C.P.; Writing—review and editing, L.N. and A.C.P.; Project administration, L.N. and A.C.P. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by University of Brest in France and ENIT (National Engineering School of Tunis) in Tunisia. The APC was funded by University of Brest.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data presented in this study are available on request from the corresponding author. The data are not publicly available due to privacy issues.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Tayachi, M.; Nana, L.; Benzarti, F.; Pascu, A.C. A Dual Watermarking Approach of DICOM Images authentication. In Proceedings of the 2020 5th International Conference on Advanced Technologies for Signal and Image Processing (ATSIP), Sousse, Tunisia, 2–5 September 2020; pp. 1–6. [Google Scholar]
- Han, B.; Li, J.; Zong, L. A New Robust Zero-watermarking Algorithm for Medical Volume Data. Int. J. Signal Process. Image Process. Pattern Recognit. 2013, 6, 245–258. [Google Scholar] [CrossRef]
- Thabit, R. Review of medical image authentication techniques and their recent trends. Multimed. Tools Appl. 2021, 80, 13439–13473. [Google Scholar] [CrossRef]
- Roček, A.; Slavíček, K.; Dostál, O.; Javorník, M. A new approach to fully-reversible watermarking in medical imaging with breakthrough visibility parameters. Biomed. Signal Process. Control 2016, 29, 44–52. [Google Scholar] [CrossRef]
- Rahimi, F.; Rabbani, H. A dual adaptive watermarking scheme in contourlet domain for DICOM images. Biomed. Eng. Online 2011, 10, 53. [Google Scholar] [CrossRef] [PubMed]
- Al-Qershi, O.M.; Khoo, B.E. Authentication and data hiding using a hybrid ROI-based watermarking scheme for DICOM images. J. Digit. Imaging 2011, 24, 114–125. [Google Scholar] [CrossRef] [PubMed]
- Tan, C.K.; Ng, J.C.; Xu, X.; Poh, C.L.; Guan, Y.L.; Sheah, K. Security protection of DICOM medical images using dual-layer reversible watermarking with tamper detection capability. J. Digit. Imaging 2011, 24, 528–540. [Google Scholar] [CrossRef]
- Tsai, H.H.; Tseng, H.C.; Lai, Y.S. Robust lossless image watermarking based on a trimmed mean algorithm and support vector machine. J. Syst. Softw 2010, 83, 1015–1028. [Google Scholar] [CrossRef]
- Pan, W.; Coatrieux, G.; Montagner, J.; Cuppens, N.; Cuppens, F.; Roux, C. Comparison of some reversible watermarking methods in application to medical images. In Proceedings of the 31st Annual International Conference of the IEEE EMBS, Minneapolis, MN, USA, 2–6 September 2009; pp. 2172–2175. [Google Scholar]
- Kallel, I.F.; Bouhlel, M.S.; Lapayre, J.C. Improved Tian’s method for medical image reversible watermarking. Gvip J. 2007, 7, 1–5. [Google Scholar]
- Fu, R.; Jin, W. A Wavelet-Based Method of Zero-Watermark Utilizing Visual Cryptography. In Proceedings of the International Conference on Multimedia Technology (ICMT), Ningbo, China, 29–31 October 2010; pp. 1–4. [Google Scholar]
- Singh, A.; Dutta, M.K. A robust zero-watermarking scheme for tele-ophthalmological applications. J. King Saud Univ-Comput. Inf. Sci. 2017, 32, 895–908. [Google Scholar] [CrossRef]
- Ghadi, M.; Laouamer, L.; Nana, L.; Pascu, A. A novel zero-watermarking approach of medical images based on Jacobian matrix model. Secur. Commun. Netw. 2016, 9, 5203–5218. [Google Scholar] [CrossRef]
- Zain, J.M.; Fauzi, A.M.; Aziz, A.A. Clinical evaluation of watermarked medical images. In Proceedings of the 28th Annual International Conference of the IEEE EMBS, New York, NY, USA, 30 August 2006–3 September 2006; pp. 5459–5462. [Google Scholar]
- Liew, S.C.; Zain, J.M. Reversible Medical Image Watermarking For Tamper Detection And Recovery. In Proceedings of the 2010 3rd International Conference on Computer Science and Information Technology, Chengdu, China, 9–11 July 2010; Volume 5, pp. 417–420. [Google Scholar]
- Zain, J.M.; Clarke, M. Reversible region of non-interest (RONI) watermarking for authentication of DICOM images. arXiv 2011, arXiv:1101.1603. [Google Scholar]
- Pan, W.; Bouslimi, D.; Karasad, M.; Cozic, M.; Coatrieux, G. Imperceptible reversible watermarking of radiographic images based on quantum noise masking. Comput. Methods Programs Biomed. 2018, 160, 119–128. [Google Scholar] [CrossRef] [PubMed]
- Tareef, A.; Al-Ani, A.; Nguyen, H.; Chung, Y.Y. A Novel Tamper Detection-Recovery and Watermarking System for Medical Image Authentication and EPR Hiding 2014. In Proceedings of the 2014 36th Annual International Conference of the IEEE Engineering in Medicine and Biology Society, Chicago, IL, USA, 26–30 August 2014; pp. 5554–5557. [Google Scholar]
- Eswaraiah, R.; Reddy, E.S. Robust medical image watermarking technique for accurate detection of tampers inside region of interest and recovering original region of interest. IET Image Process. 2015, 9, 615–625. [Google Scholar] [CrossRef]
- Kahlessenane, F.; Khaldi, A.; Kafi, R.; Euschi, S. A robust blind medical image watermarking approach for telemedicine applications. Clust. Comput. 2021, 24, 2069–2082. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.; Li, J.; Bhatti, U.A.; Liu, J.; Ma, J.; Huang, M. Robust zero watermarking algorithm for medical images based on zernike-DCT. Secur. Commun. Netw. 2021, 2021, 4944797. [Google Scholar] [CrossRef]
- Benhocine, A.; Laoumer, L.; Nana, L.T.; Pascu, A.C. Improving extraction of watermarks in color attacked watermarked images. J. Communi. Comput. 2009, 6, 36–45. [Google Scholar]
- Memon, N.A.; Chaudhry, A.; Ahmad, M.; Keerio, Z.A. Hybrid watermarking of medical images for ROI authentication and recovery. Int. J. Comput. Math. 2011, 88, 2057–2071. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).