Abstract
We are currently experiencing a revolution in data production and artificial intelligence (AI) applications. Data are produced much faster than they can be consumed. Thus, there is an urgent need to develop AI algorithms for all aspects of modern life. Furthermore, the medical field is a fertile field in which to apply AI techniques. Breast cancer is one of the most common cancers and a leading cause of death around the world. Early detection is critical to treating the disease effectively. Breast density plays a significant role in determining the likelihood and risk of breast cancer. Breast density describes the amount of fibrous and glandular tissue compared with the amount of fatty tissue in the breast. Breast density is categorized using a system called the ACR BI-RADS. The ACR assigns breast density to one of four classes. In class A, breasts are almost entirely fatty. In class B, scattered areas of fibroglandular density appear in the breasts. In class C, the breasts are heterogeneously dense. In class D, the breasts are extremely dense. This paper applies pre-trained Convolutional Neural Network (CNN) on a local mammogram dataset to classify breast density. Several transfer learning models were tested on a dataset consisting of more than 800 mammogram screenings from King Abdulaziz Medical City (KAMC). Inception V3, EfficientNet 2B0, and Xception gave the highest accuracy for both four- and two-class classification. To enhance the accuracy of density classification, we applied weighted average ensembles, and performance was visibly improved. The overall accuracy of ACR classification with weighted average ensembles was 78.11%.
1. Introduction
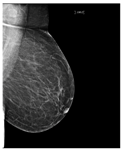
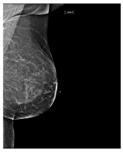
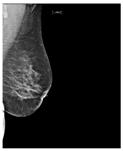
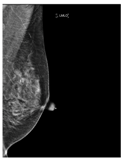
Artificial intelligence (AI) algorithms have improved and affected every aspect of modern life. In the last two decades, machine learning techniques and computer power have experienced momentous development, leading to an enormous increase in digital data in different fields. These advances have enabled researchers to solve real-world problems in many areas, including the medical image analysis field. Medical imaging was first used in 1895 [1]. Since that time, the devices and quality of medical imaging have continued to evolve. Today, medical imaging is an essential tool for diagnosing and detecting diseases such as cancer [2]. Cancer is a significant public health concern that affects people around the world, and it is the second leading cause of death in the US [3]. Breast cancer is one of the most prevalent cancers that affect women [4], but it can also arise in men [5]. In Saudi Arabia, in 2018, breast cancer was the most common type of cancer, with an incidence rate of 14.8% and a mortality rate of 8.5% [6]. In 2020, breast cancer remained the most common type of cancer in Saudi Arabia. Breast cancer is a disease caused by the abnormal growth of breast cells. Usually, somatic cells divide to produce new cells in an orderly manner. When the cells of a specific part of the body grow and divide out of control, a mass of tissue called a tumor is created. A tumor that consists of normal cells is benign. When the cells are abnormal and exhibit irregular behavior, it is a malignant or cancerous tumor [7]. Three distinct types of medical imaging are used to diagnose breast cancer. These are mammograms, ultrasound, and magnetic resonance imaging (MRI) [8]. Among the breast imaging techniques, the mammogram is considered the gold standard for detecting and diagnosis [9,10]. A mammogram is a type of medical imaging that uses a low-dose x-ray system to see inside the tissues of the breast [11]. There are two mammogram imaging modalities: digital and screen-film mammography. Screen-film mammography (SFM) consists of conventional analog mammographic films. SFM often contains various background objects, such as labels and markers, that need to be removed. Digital mammograms are also referred to as FFDM images; FFDM stands for full-field digital mammography. FFDM is the approved version in hospitals today [11]. Moreover, mammogram images can be found in several formats, including LJPG, DICOM, PGM, and TIFF. In the standard view for each breast, two X-ray images must be taken on both sides. Therefore, four images of both breasts must be examined. These four images are called: LEFT_CC, LEFT_MLO, RIGHT_CC, RIGHT_MLO [12,13]. The craniocaudal (CC) view is taken from above a horizontally compressed breast (head-to-foot picture). The CC view captures the medial portion and as much of the breast’s outer lateral region as possible. MedioLateralOblique (MLO) view captures the entire breast and often includes the lymph nodes and the pectoral muscle. Figure 1a,b show examples of the CC and MLO views, and Figure 2 illustrates the angle of each view [14]. Among the breast imaging techniques, the mammogram is considered the gold standard [9,10].
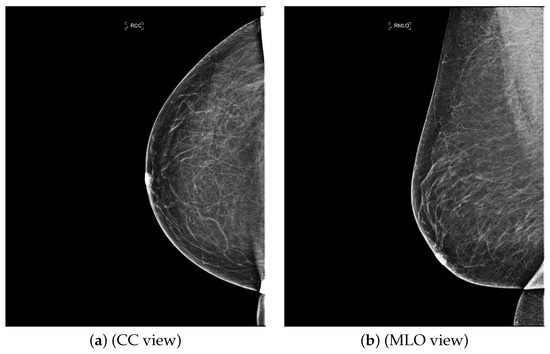
Figure 1.
FFDM view for right breast.
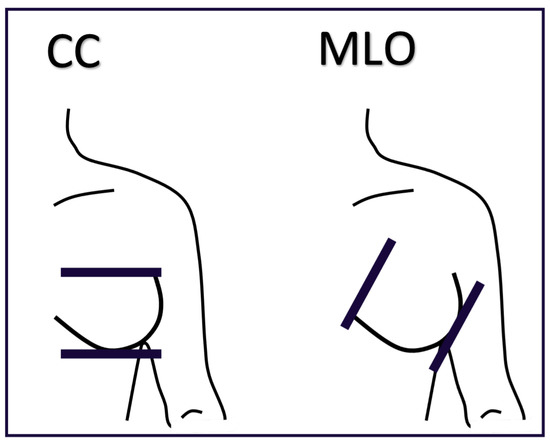
Figure 2.
The difference between CC and MLO views.
1.1. Breast Density
Breast density describes the amount of fibrous and glandular tissue in a breast compared with the amount of fatty tissue. The breast density is assigned to one of four classes in the mammogram report based on the ACR BI-RADS standard. In class A, the breasts are almost entirely fatty. A few areas of dense tissue scattered within the breasts indicate class B. In class C, the breasts are heterogeneously dense. Finally, in class D, breasts are extremely dense [8,15]. Breast density plays a significant role in detecting breast cancer and on the risk of developing breast cancer. The clinicians must identify breast density from a mammogram for each patient and write it in their reports. Usually, dense breasts—i.e., breasts categorized into class C or D—are more likely to be affected by breast cancer [16]. In [17], the authors studied the relationship between mammographic densities and breast cancer risk. The results showed that the ratio of positive cancer cases for the different ACR classes were as follows: D (13.7%), C (3.3%), B (2.7%), and A (2.2%). Table 1 illustrates the difference between ACR classes.

Table 1.
Breast density classes.
Breast density is important and must be specified in breast screening medical reports. Determining breast density can be challenging and mostly subjective, especially if they have been affected by treatment such as chemotherapy [18].
1.2. Convolutional Neural Network (CNN)
CNNs are a type of artificial neural network usually used for classification and computer vision tasks. Therefore, CNNs are considered efficient tools for medical imaging classification. In addition to input and output layers, CNNs include three main types of layers: convolutional, pooling, and fully connected. The convolutional layer is the main part of the CNN; it incorporates input data, a filter, and a feature map. The pooling layer or down-sampling layer seeks to reduce the number of parameters in the input. In the fully connected layer, a neuron applies a linear transformation to the input vector through a weight matrix [19]. Figure 3 illustrates the general structure of the CNN, assuming an input image size is 28 by 28, and a target task is classifying images into one of 10 classes [20].
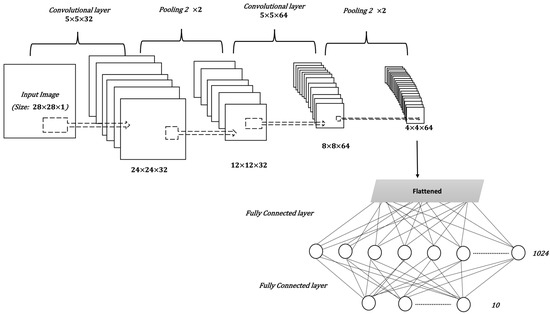
Figure 3.
The general structure of a CNN.
The CNN model is affected by many factors, including the number of layers, layer parameters, and other hyperparameters of the model, such as the optimizer and loss function. The loss function is used to calculate the difference between the predicted value and the actual value. An optimizer is a function that modifies the weights and learning rate of the DL model to reduce the loss value and increase accuracy. With classification problems, especially when there are more than two classes, the categorical_crossentropy function is the best choice to calculate loss value [21]. Root mean square propagation (RMSProp), and adaptive moment estimation(Adam), are the most commonly used optimizers [22,23].
1.3. Transfer Learning and Pre-Trained CNNs
Transfer learning reuses the knowledge gained from a previous task in a new deep learning model. Usually, deep learning requires a large amount of data to achieve good results. However, it is often difficult to gather enough data, especially in the medical field. Therefore, transfer learning enhances the learning process when the dataset has limited samples [24]. A pre-trained model is a model that was created and trained to solve a problem that is related to the target task. For example, in image classification tasks, such as flower image classification, we can use VGG19, which is a pre-trained CNN used to classify images that was trained on a huge image dataset called ImageNet [25]. Table 2, presents the main information about the pre-trained CNN that was used in this paper [26,27]. This information includes the model name, the number of layers, the top-1 accuracy of the model in classifying ImageNet data, and the year the model was established. The top-1 accuracy checks if the class with the highest probability is the same as the target label [28]. All of the models were trained on ImageNet. The ImageNet is a large dataset of 14,197,122 annotated images belonging to more than 1000 categories [29].

Table 2.
Pre-trained CNNs.
Many studies on applying machine learning and deep learning to classify breast density from mammogram have been conducted. However, we could not find a study that used local data from Saudi Arabia. Moreover, according to [30,31], breast density and ethnicity are associated. Therefore, an automated model to classify breast density trained on local mammogram datasets would be valuable. The main contribution of this paper is developing a breast density classification model that is suited for the local population in Saudi Arabia. The model is based on pre-trained CNNs and weighted average ensembles and applied to the KAMC dataset. The rest of this paper is divided as following, Section 2 explores recent related works that classify breast density from mammogram using deep learning techniques. Section 3 outlines our methodology. Section 4 presents and discusses our results. Finally, a conclusion is offered in Section 5.
2. Related Works
Recently, many studies have sought to develop deep learning models to assess breast density. Some of these studies target two classes of density (fatty or dense), while other studies classify the breast as fatty, glandular, or dense. However, most studies classify breast density into four classes according to the BI-RADS system. Here we mention only recent works related to ACR classification, as it is a standard in medical reporting. In [32], the authors proposed a breast density classification model based on convolutional neural networks (CNNs). They applied two techniques to 200,000 breast screenings. They called the first technique a baseline and the second a deep convolutional neural network (deep CNN). In the baseline, they used pixel intensity histograms of screening as input features. Then, softmax regression was used as a classifier. In deep CNN, the inputs were the four screening views, while the fully connected layer consisted of 1024 hidden units and the output layer used the softmax activation function. Additionally, they used the weights of the previously trained model of breast cancer detection to initialize the parameters of their network. Both techniques were measured by computing the area under the ROC curve (AUC), the accuracy of super-classes (dense or non-dense), and the ACR accuracy. For the baseline with 20 bins, AUC = 0.832, ACR accuracy = 67.9%, and super-classes accuracy = 81.1%. For deep CNN, AUC = 0.916, ACR accuracy = 76.7%, and super-classes accuracy = 86.5%. Another CNN was then applied to the MAIS dataset to classify breast density in [33]. Different preprocessing techniques were used, including pectoral muscle segmentation, image augmentation, and image resizing. The CNN consists of three convolutional layers, followed by two fully connected layers, and, finally, the output layer. The dataset was divided, with 20% used for testing and 80% for training, before five-fold cross-validation was applied. The overall accuracy of ACR classification was 83.6%. Moreover, in [34] the CNNs were applied with a squeeze-and-excitation network (SE-Net) mechanism to classify breast density from mammograms. The three CNN models used with SE-Net were Inception-V4, ResNeXt, and DenseNet. A 10-fold cross-validation was used to obtain better results. The dataset consisted of 18,157 images. The preprocessing entailed removing the background, grayscale transformation, augmentation by cropping and rotating images, and normalizing the images into a normal distribution. The classification accuracy was measured for each model with and without SE-Attention. The accuracy of Inception-V4 and Inception-V4-SE-Attention was 89.97% and 92.17%, respectively, while for ResNeXt50 and ResNeXt50-SE-Attention, the accuracy was 89.64% and 91.57%, respectively. Finally, the accuracy of DenseNet121 was 89.20%, and for DenseNet121-SE-Attention it was 91.79%. Furthermore, in [35], a fine-tuned model based on InceptionV3 was used to classify breast density. The dataset consists of 3813 mammogram screenings. The accuracy obtained by the model for the BI-RADS classification based on 150 screenings was 83.33%. Meanwhile, in [36], a deep learning model based on vgg16 was proposed to predict breast density class. The central idea of this work is to compute the amount of fibroglandular tissue in each image. The dataset consists of 1602 images, 70% of which were used for training, and 30% for testing. The accuracy of the model 79.6%. In [37], a deep CNN based on ResNet-18 was applied. The experiment was performed on a dataset with 41,479 digital screening mammograms for training and 8677 mammograms for testing. The accuracy of dense or non-dense classification was 86.88%. On the other hand, the accuracy of classification into the four BI-RADS categories was 76.78%. The authors in [38] used another deep learning-based approach for fully automated breast density classification. This approach comprised three main stages. In the first stage, the breast area is isolated from the mammogram by removing the background and pectoral muscles. In the second stage, a binary mask containing the dense tissue is created by a generative adversarial network (cGAN). Then, in the third stage, breast density is classified by feeding the binary mask into a multi-class CNN. The INbreast dataset was used for training and testing. The overall accuracy of density classification was 98.75%. In [39], a range of deep CNN architectures with different numbers of filters, layers, dropout rates, and epochs were evaluated. The database used included 20,578 images, which were then reduced to 12,932 images to avoid over-representing ACR densities B and C, before being divided into 70% for training and 30% for testing. The chosen CNN architecture consists of 13 convolutional layers followed by max-pooling and dropout at a rate of 50%. The number of epochs was 120 and the patch size was 40. The performance was measured for MLO and CC views separately and the ACR classification accuracy was 90.9% and 90.1%, respectively. Authors in [40], meanwhile, proposed a multi-path deep convolutional neural network (multi-path DCNN) in order to classify breast density. The proposed DCNN takes four inputs including subsamples of digital mammograms, largest square region of interest, a mask of dense area, and the percentage of breast density. They used ten-fold cross validation, resulting in an overall accuracy of 80.7%. In [41,42], a residual neural network was used to classify breast density according to two classes (fatty and dense), and BI-RADS classification. The collected dataset in this work consists of 7848 images. After excluding badly exposed images and cases involving one breast, the total number of images was reduced to 1962. The proposed model consists of 41 convolutional layers. The model was tested with different image sizes and the size gave the highest accuracy with BI-RADS classification. The obtained accuracy for two classes classification and BI-RADS classification was 86.3% and 76.0%, respectively. In [43], the author proposed an artificial neural net- work called DualViewNet as a means to classifying breast density. The structure of this model was based on MobileNetV2. In this model, a joint classification on MLO and CC mammograms corresponding to the same breast was performed. They used CBIS-DDSM and applied image enhancement techniques and image augmentation during the preprocessing stage. Moreover, they excluded the images with suspect labels. The performance measurement was carried out by computing AUC equal to 0.9882. In [44], the authors collected a dataset from 33 different clinics. The aim of this study was to test deep learning in classifying breast density from a large mammogram dataset that was collected from a range of multi-institutions. The dataset consists of 108,230 images. They used VGG16, ResNet, InceptionV3, and DenseNet121. The overall accuracy of the model was 66.7%. Authors in [45], employed federated learning (FL) to classify breast density across seven clinical institutions. They applied a pre-trained model based on DensNet12, achieving an overall accuracy of 77%. Several residual nets were used in [46] to classify breast density, including ResNet43, ResNet50, and ResNet101. The models applied on a clinical dataset consisted of 1985 mammograms, in addition to the INbreast dataset. During the preprocessing stage, images were resized to , the background was removed, and augmentation was applied. During the training stage, 5-fold cross validation was applied. The best performance was obtained from ResNet50, with an overall accuracy of 87.1% for the clinical dataset and 70% for INbreast. In [47], bilateral adaptive spatial and channel attention network (BASCNet) was used to classify breast density. The backbone of this model was ResNet. The training dataset that was used is a subset of DDSM consisting of 250 cases of each ACR class to handle the class imbalance problem. Various preprocessing stages were applied, including removing the background, image resizing to , and augmentation. In addition, 5-fold cross validation was applied during the training process. The testing was performed on both the DDSM and INbreast datasets, and the overall accuracy was 85.10% and 90.51%, respectively. Table 3, summarizes the deep learning classification techniques mentioned above.

Table 3.
Classifying breast density using deep learning.
3. Methodology
In this paper, we developed a model to classify breast density from mammograms based on pre-trained CNNs and weighted average ensembles. The key steps of our methodology are illustrate in Figure 4. The first stage entailed collecting the dataset from KAMC. Next, we prepared the data. Then, several pre-trained models were applied to classify breast density. Finally, a weighted average ensemble was employed to enhance the CNN performance. Below, we describe each of the steps in sequence.
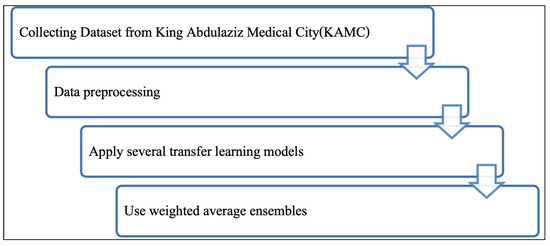
Figure 4.
Methodology.
3.1. Dataset
The dataset was collected from King Abdulaziz Medical City, and it consists of 879 mammogram screenings taken between 1 January 2020 and 12 January 2021. The data collection took around three months, from 3 December 2020 to 28 February 2021, because the system allows only one record to be downloaded at a time, and we could not access the system when the radiologists were using it. Each case is associated with a medical report written by a radiologist. Patients’ private information, such as their names and IDs, was hidden from the reports. Only data that relates to the research and statistics were included. These data include the diagnosis, BI-RADS stage, ACR class, age, and gender. Most of the screenings were diagnosed as BI-RADS 2, but the data include all BI-RADS categories. As each screening included two or more images, there were 3083 images in total. The majority of cases were for women, with just a few cases for men. Figure 5 shows the gender distribution.
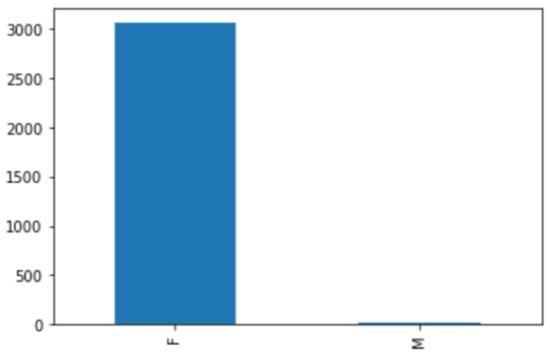
Figure 5.
Gender distribution.
The dataset includes people between the ages of 28 and 89. The majority were above 40 years. Patients younger than 40 years of age had their mammograms done for specified indications. Figure 6 shows the age distribution chart. The four density classes in the ACR standard are represented in this dataset: 36.9% of the cases are in class B, 34.9% in class C, 21.6% in class A, and 6.6% in class D. This follows the normal distribution of ACR classes in other mammogram datasets; where class B is the majority class, there are more samples in class C than in class A, and class D is the minority class [38,48]. The chart in Figure 7 illustrates this distribution.
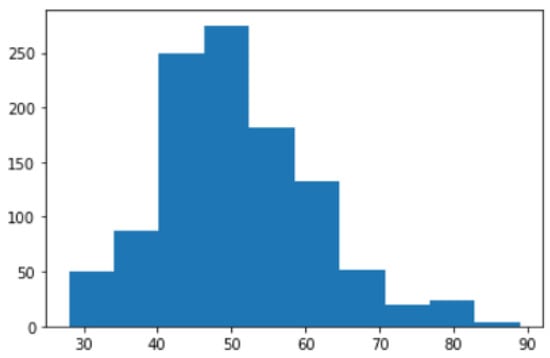
Figure 6.
Age distribution.
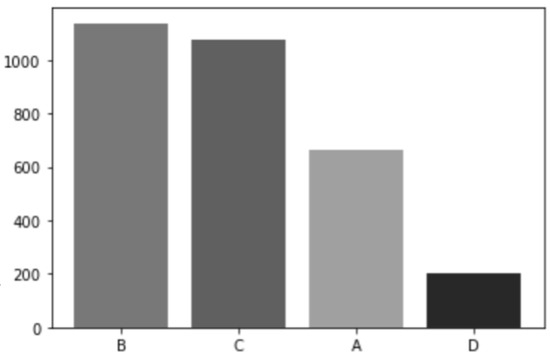
Figure 7.
ACR distribution.
3.2. Data Preparation
At this stage, we cleaned the data by removing mislabeled cases. The dataset was divided as follows: 80% was used for training and validation and 20% was used for testing with a fixed seed number. The images were converted into PNG format; the original image size was 4096 by 3328 pixels in RGB color mode. Converting images to PNG format does not compromise image quality through compression. Moreover, PNG format supports a wide color range. The images were then resized to 300 by 300 pixels, as their original size was too large to use during training. Finally, several augmentation techniques were applied, including horizontal flip, rotation, and translation, as in most of the related works.
3.3. Pre-Trained Model
We applied several pre-trained CNN models used in related works to classify breast density. These models were ResNet, Inception, DensNet, and VGG16 [34,36,37,44,47]. Moreover, we applied models based on VGG19, InceptionResNetV2, EfficientNetV2, and Xception. The models were applied to the entire dataset, without excluding cases that were affected by treatment. The structure of our proposed model is shown in Figure 8. First, the pre-trained CNN is loaded. Then a flatten layer is applied. After that, the model going through several dense and dropout layers. Finally, the output layer gives the density class.
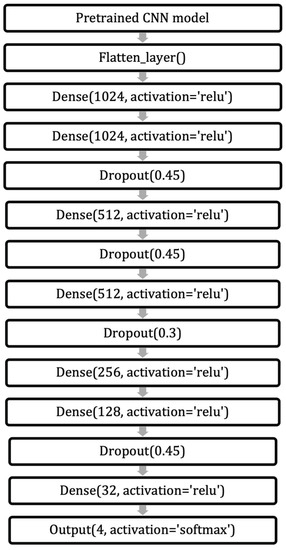
Figure 8.
Structure of the model.
3.4. Average Ensembles
Ensembles is a technique that applies machine learning and deep learning to enhance performance. Basically, ensembles combine multiple models instead of using just a single model. How these models are combined determines the type of ensembles technique used, which include stacking, bagging, boosting, and averaging. In stacking ensembles, a meta-learning model integrates the outputs of basic models that were trained on the entire training set. In contrast, the bagging technique trains many models separately on a subset of data before combining their predictions to generate an ensemble predictor. Boosting, meanwhile, uses sequential training, enabling subsequent models to focus on previously misclassified samples. Finally, in averaging ensembles, several models are trained on the training set, and during testing, the average of their predictions is taken. A subtype of averaging ensembles is weighted average ensembles. In a weighted average ensemble, each model is weighted according to certain criteria or based on a grid search. The final result is obtained through a dot product of the weight vector and the model predictions vector; after that, the sum or highest probable class is taking [49,50].
In our model, after we trained several pre-trained CNNs, we applied averaging ensembles to the CNNs. A grid search was applied to find the optimal weight for each model, and then weighted average ensembles were employed. Figure 9 and Figure 10 illustrate the idea of average and weighted average ensembles.
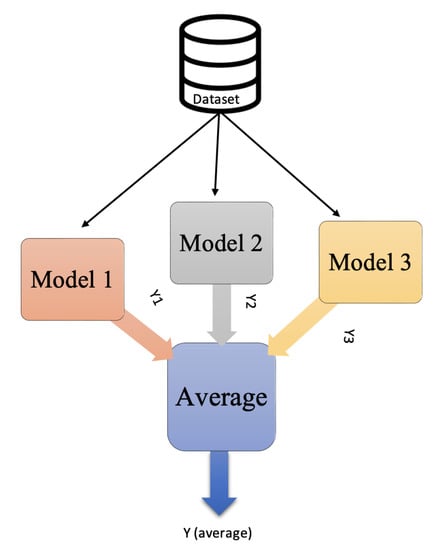
Figure 9.
Average ensembles.
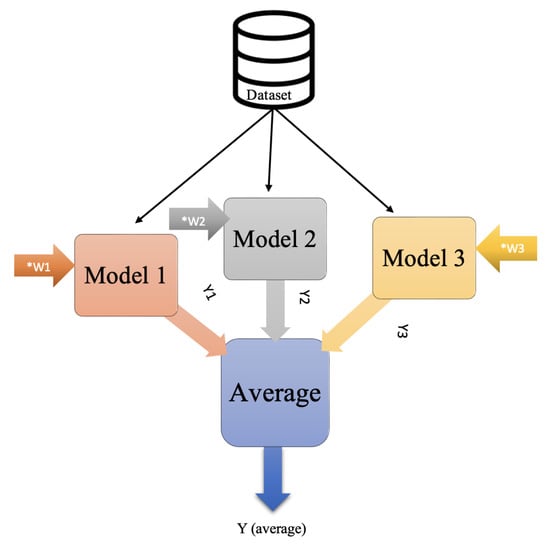
Figure 10.
Weighted average ensembles.
3.5. Experimental Tools
We used the Tensorflow Keras library and the training was conducted on Google Colab Pro+. The loss function was categorical cross-entropy loss, and the optimizer was RMSprop with a learning rate of 0.0001. We used 100 epochs. The hyperparameters were chosen based on related works and a wide range of experiments. The dataset was divided, with 80% for training and validation and 20% for testing. A fixed seed number was used during data splitting to ensure that the data were divided in the same way each time and that splitting would not affect the testing of other hyperparameters. The performance was measured with accuracy and F1 scores, in addition to monitoring the confusion matrix and F1 of each class.
4. Results and Discussion
The experiments were started with applying pre-trained CNN models that used in the related works. Then, we investigated and tested recent and efficient pre-trained CNNs supported by Keras. These models included VGG19, InceptionResNetV2, EfficientNetV2, and Xception. Some models required a significant amount of memory, such as NasNet; therefore, we could not use them. The training/validation behavior based on accuracy and loss for each model is illustrated in Figure 11, Figure 12, Figure 13, Figure 14, Figure 15, Figure 16, Figure 17, Figure 18 and Figure 19. Moreover, Table 4, Table 5, Table 6, Table 7, Table 8, Table 9, Table 10, Table 11 and Table 12 report the performance of each model. Figure 11 explaining the behavior of the DenseNet based model. It can be notice that, the validation loss increase in oscillatory manner after epoch 40. The performance of DenseNet model, was low in terms of overall accuracy and F1 of each class, as shown in Table 4. Moreover, MobileNet model was the worst model in classifying breast density. We can notice this from training behavior in Figure 12, and testing results in Table 5. The loss value of ResNet50, increasing dramatically after epoch 80, as shown in Figure 13. Both ResNet50 and InceptionV3, have overall accuracy exceed 70%. However, the F1 of each class was better with ResNet50 model comparing with InceptionV3, as shown in Table 6 and Table 7. The training behaviors of VGG16 and VGG19, are very oscillate as illustrated in Figure 15 and Figure 16. Meanwhile, the performance of VGG16 and VGG19 shown in Table 8 and Table 9 were low, the overall accuracy and F1 score were under 65%. Comparing with ResNet50 and Inception V3, the InceptionResNetV2 model was worse, the behavior of the model and its performance present in Figure 17 and Table 10. The training behavior of the model based on Xception is shown in Figure 18, and the model’s performance illustrated in Table 11. Finally, the training behavior and testing performance of EfficientNetV2 is shown in Figure 19 and Table 12, respectively.
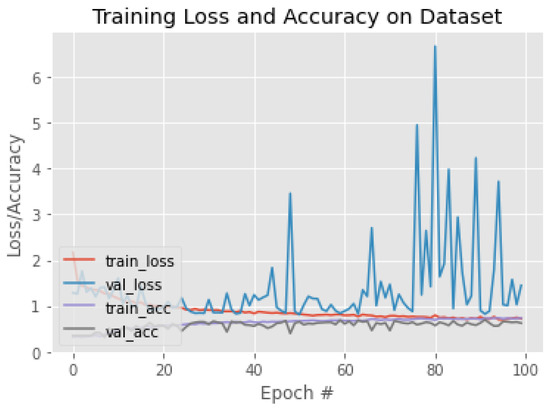
Figure 11.
The training behavior of the model based on DenseNet121.
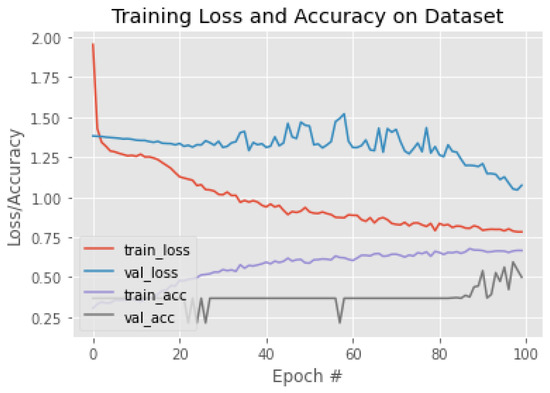
Figure 12.
The training behavior of the model based on MobileNet.
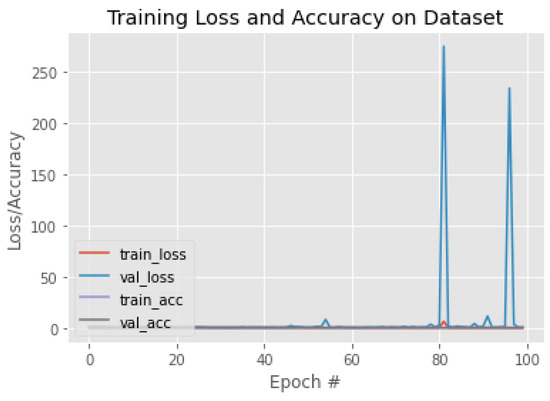
Figure 13.
The training behavior of the model based on ResNet50.
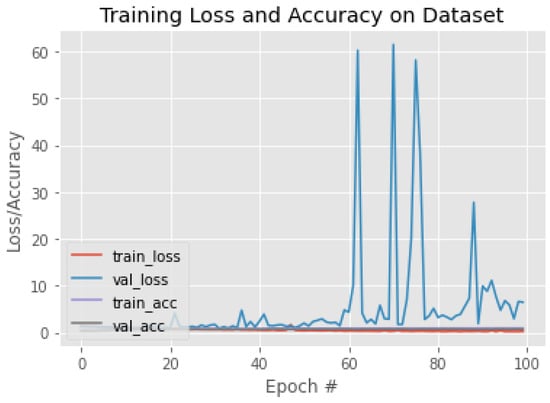
Figure 14.
The training behavior of the model based on InceptionV3.
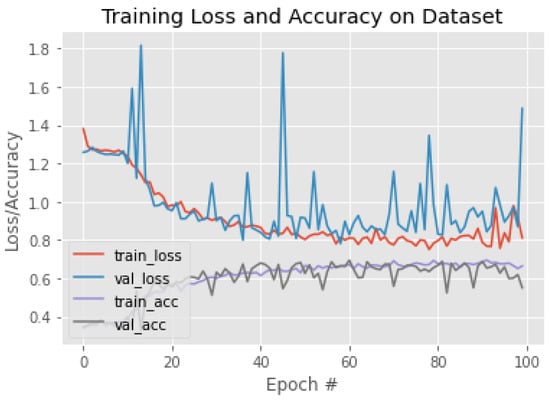
Figure 15.
The training behavior of the model based on VGG16.
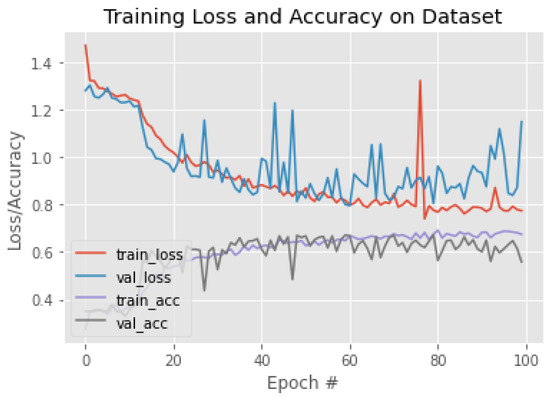
Figure 16.
The training behavior of the model based on VGG19.
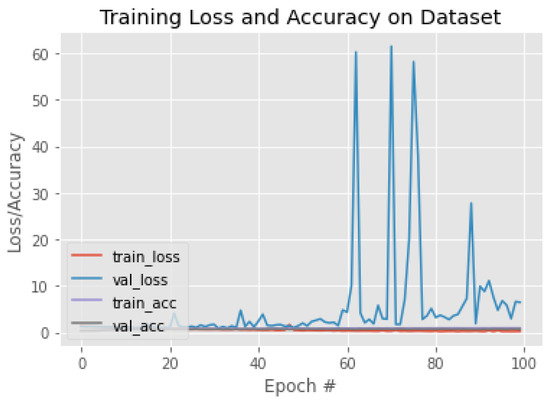
Figure 17.
The training behavior of the model based on InceptionResNetV2.
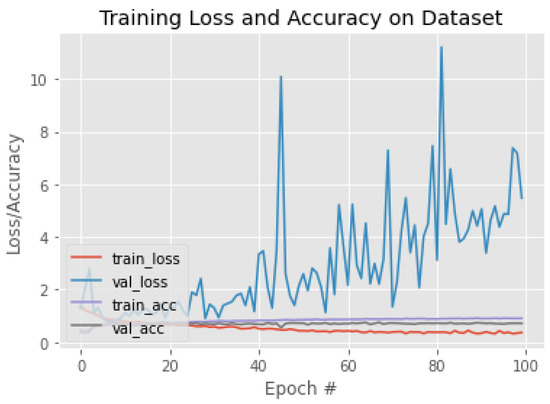
Figure 18.
The training behavior of the model based on Xception.
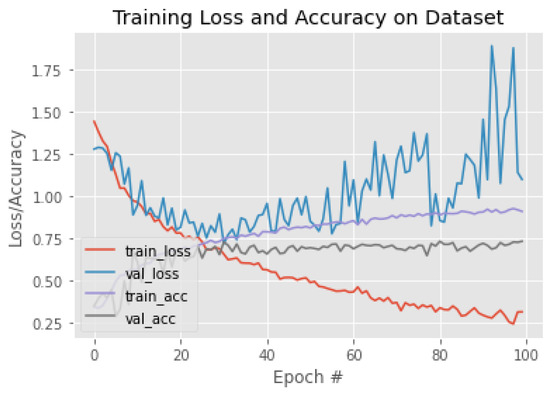
Figure 19.
The training behavior of the model based on EfficientNetV2B0.

Table 4.
DensNet 121.

Table 5.
MobileNet.

Table 6.
ResNet50.

Table 7.
Inception V3.

Table 8.
VGG16.

Table 9.
VGG19.

Table 10.
InceptionResNetV2.

Table 11.
Xception.

Table 12.
EfficientNetV2.
Our target was to classify breast density according to ACR standards. However, fatty and dense (or superclass) classifications were also performed. Table 13 and Table 14 summarize the accuracy and F1 scores of each model for ACR and superclass classification, respectively.

Table 13.
ACR classification.

Table 14.
Superclass classification.
We noticed, from monitoring the performance of each model, that some models performed better with specific classes. For example, InceptionV3 gave the highest F1 for class A, whereas ResNet50 gave the highest F1 for class C. The model based on Xception achieved the overall highest F1 score for class D. Therefore, average and weighted average ensembles were applied to reap the benefits of all models. Table 15 shows the performance of the average ensembles of all models. The overall accuracy of the proposed model with average ensembles was 74.7%. Furthermore, weighted average ensembles with grid search were applied. These reference exampling the idea of grid search [51] and how to apply it with average ensembles [52]. First, we applied weighted averages using the best five models, achieving an accuracy of 77.9%. Next, we selected the best four models, InceptionV3, ResNet50, EfficientNet, and Xception, resulting in an overall accuracy of 78.11%. Finally, we tried weighted average ensembles with the three best models, achieving an accuracy of 75.8%. More details on the weighted average ensemble performance are provided in Table 16, Table 17 and Table 18. A summary of the average and weighted average of ACR and superclass classification is provided in Table 19 and Table 20, respectively.

Table 15.
Average ensembles.

Table 16.
Weighted average ensembles with best 5 models.

Table 17.
Weighted average ensembles with best 4 models.

Table 18.
Weighted average ensembles with best 3 models.

Table 19.
Ensembles for ACR classification.

Table 20.
Ensembles for superclass classification.
5. Conclusions
Breast density describes the amount of fibrous and glandular tissue in a breast compared with the amount of fatty tissue. Breast density plays a significant role in detecting breast cancer and on the risk of developing breast cancer. Dense breasts are more likely to be affected by breast cancer. The Breast Imaging-Reporting and Data System (BI-RADS) is an assessment standard for reporting the breast cancer risk based on a mammogram image. It sorts breast density into one of four ACR classes. In class A, breasts are almost entirely fatty. A few areas of dense tissue are scattered within the breasts in class B. In class C, the breasts are heterogeneously dense. Finally, in class D, the breasts are extremely dense. In this paper, a breast density classification model from mammograms was developed. The model is based on pre-trained CNNs and weighted average ensembles. The dataset was collected from King Abdulaziz Medical City under the supervision of the King Abdullah International Medical Research Center (KAIMRC). The dataset consists of 879 mammogram screenings. Several model pre-trained convolutional neural networks were applied, including ResNet, Inception, DensNet, vgg16, vgg19, InceptionResNetV2, EfficientNetV2, and Xception. Among these models, InceptionV3, EfficientNet 2B0, and Xception gave the highest accuracy in both four- and two-class classification. We noticed from monitoring the performance of each model that some models performed better with specific classes. Therefore, average and weighted average ensembles were applied to reap the benefits of all models. After applying weighted average ensembles, the performance was improved noticeably. The overall accuracy of ACR classification with weighted average ensembles was 78.11%. For future works we recommend the following: different ensembles techniques, such as boosting ensembles, could be investigated; increasing the size of the dataset; and applying unsupervised learning segmentation techniques to segment ROIs.
Author Contributions
Conceptualization, E.J., G.A., N.A. and A.A.; methodology, E.J. and A.A.; software, E.J.; validation, E.J.; formal analysis, E.J.; investigation, E.J.; resources, N.A.; data curation, E.J. and N.A.; writing—original draft preparation, E.J.; writing—review and editing, E.J. and N.A.; visualization, E.J.; supervision, G.A. and A.A. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
We got the ethical approval to collect this dataset and perform the study from King Abdullah International Medical Research Center (KAIMRC). The KAIMRC is the research center of King Abdulaziz medical city in Jeddah.
Informed Consent Statement
We got the ethical approval to collect this dataset and all private information such as patients names and IDs were removed.
Acknowledgments
We would like to acknowledge King Abdullah International Medical Research Center and King Abdulaziz Medical City radiology breast radiology section. They helped us collect the dataset and were very supportive.
Conflicts of Interest
The authors declare no conflict of interest.
Abbreviations
The following abbreviations are used in this manuscript:
| AI | Artificial Intelligence |
| BI-RADS | Breast imaging-reporting and data system |
| CNN | Convolutional neural network |
| DICOM | Digital image communication in medicine |
| KAMC | King Abdulaziz Medical City |
| RMS prop | Root mean square propagation |
References
- Bradley, W.G. History of medical imaging. Proc. Am. Philos. Soc. 2008, 152, 349–361. [Google Scholar] [PubMed]
- Giger, M.L. Machine learning in medical imaging. J. Am. Coll. Radiol. 2018, 15, 512–520. [Google Scholar] [CrossRef] [PubMed]
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Alotaibi, R.M.; Rezk, H.R.; Juliana, C.I.; Guure, C. Breast cancer mortality in Saudi Arabia: Modelling observed and unobserved factors. PLoS ONE 2018, 13, e0206148. [Google Scholar] [CrossRef] [PubMed]
- Reinisch, M.; Seiler, S.; Hauzenberger, T.; Kamischke, A.; Schmatloch, S.; Strittmatter, H.J.; Zahm, D.M.; Thode, C.; Furlanetto, J.; Strik, D.; et al. Efficacy of endocrine therapy for the treatment of breast cancer in men: Results from the MALE phase 2 randomized clinical trial. JAMA Oncol. 2021, 7, 565–572. [Google Scholar] [CrossRef]
- Alqahtani, W.S.; Almufareh, N.A.; Domiaty, D.M.; Albasher, G.; Alduwish, M.A.; Alkhalaf, H.; Almuzzaini, B.; Al-Marshidy, S.S.; Alfraihi, R.; Elasbali, A.M.; et al. Epidemiology of cancer in Saudi Arabia thru 2010–2019: A systematic review with constrained meta-analysis. AIMS Public Health 2020, 7, 679. [Google Scholar]
- Ministry of Health, Cancerous Diseases, Breast Cancer 2018. Available online: https://www.moh.gov.sa/en/HealthAwareness/EducationalContent/Diseases/Cancer/Pages/Cancer-2012-01-18-001.aspx (accessed on 12 September 2021).
- Kerlikowske, K.; Miglioretti, D.L.; Vachon, C.M. Discussions of Dense Breasts, Breast Cancer Risk, and Screening Choices in 2019. JAMA 2019, 322, 69–70. [Google Scholar] [CrossRef]
- Saha, D.; Vaishnav, N.; Jha, A.K. Imaging Techniques for Breast Cancer Diagnosis. In Biomedical Computing for Breast Cancer Detection and Diagnosis; IGI Global: Hershey, PA, USA, 2021; pp. 188–210. [Google Scholar]
- Jafari, S.H.; Saadatpour, Z.; Salmaninejad, A.; Momeni, F.; Mokhtari, M.; Nahand, J.S.; Rahmati, M.; Mirzaei, H.; Kianmehr, M. Breast cancer diagnosis: Imaging techniques and biochemical markers. J. Cell. Physiol. 2018, 233, 5200–5213. [Google Scholar] [CrossRef]
- Breast Cancer, R.T. Mammography 2019. Available online: https://www.radiologyinfo.org/en/info.cfm?pg=mammo#top/ (accessed on 4 April 2021).
- Komen, S.G. Imaging Methods Used to Find Breast Cancer; 2016; Available online: https://www.komen.org/wp-content/uploads/Imaging-Methods-used-to-Find-Breast-Cancer.pdf (accessed on 4 April 2021).
- Wolstencroft, R.T.W. Digital Database for Screening Mammography 2006, Computer Vision and Pattern Recognition. Available online: http://www.eng.usf.edu/cvprg/Mammography/Database.html (accessed on 30 September 2020).
- Moghbel, M.; Ooi, C.Y.; Ismail, N.; Hau, Y.W.; Memari, N. A review of breast boundary and pectoral muscle segmentation methods in computer-aided detection/diagnosis of breast mammography. Artif. Intell. Rev. 2019, 53, 1873–1918. [Google Scholar] [CrossRef]
- Center for Disease Control and Prevention. What Does It Mean to Have Dense Breasts? 2020. Available online: https://www.cdc.gov/cancer/breast/basic_info/dense-breasts.htm (accessed on 16 March 2021).
- Daniaux, M.; Gruber, L.; Santner, W.; Czech, T.; Knapp, R. Interval breast cancer: Analysis of occurrence, subtypes and implications for breast cancer screening in a model region. Eur. J. Radiol. 2021, 143, 109905. [Google Scholar] [CrossRef]
- Ali, E.A.; Raafat, M. Relationship of mammographic densities to breast cancer risk. Egypt. J. Radiol. Nucl. Med. 2021, 52, 1–5. [Google Scholar] [CrossRef]
- Mistry, K.A.; Thakur, M.H.; Kembhavi, S.A. The effect of chemotherapy on the mammographic appearance of breast cancer and correlation with histopathology. Br. J. Radiol. 2016, 89, 20150479. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Education, I.C. Convolutional Neural Networks 2020. Available online: https://www.ibm.com/cloud/learn/convolutional-neural-networks (accessed on 22 April 2022).
- Sebastian Raschka, V.M. Python Machine Learning; Packt: Birmingham, UK, 2017. [Google Scholar]
- Brownlee, J. How to Choose Loss Functions When Training Deep Learning Neural Networks. 2020. Available online: https://machinelearningmastery.com/how-to-choose-loss-functions-when-training-deep-learning-neural-networks/ (accessed on 12 January 2022).
- Gupta, A. A Comprehensive Guide on Deep Learning Optimizers. 2021. Available online: https://www.analyticsvidhya.com/blog/2021/10/a-comprehensive-guide-on-deep-learning-optimizers/ (accessed on 16 December 2021).
- Hamdia, K.M.; Ghasemi, H.; Bazi, Y.; AlHichri, H.; Alajlan, N.; Rabczuk, T. A novel deep learning based method for the computational material design of flexoelectric nanostructures with topology optimization. Finite Elem. Anal. Des. 2019, 165, 21–30. [Google Scholar] [CrossRef]
- Zhuang, F.; Qi, Z.; Duan, K.; Xi, D.; Zhu, Y.; Zhu, H.; Xiong, H.; He, Q. A comprehensive survey on transfer learning. Proc. IEEE 2020, 109, 43–76. [Google Scholar] [CrossRef]
- Wu, Y.; Qin, X.; Pan, Y.; Yuan, C. Convolution neural network based transfer learning for classification of flowers. In Proceedings of the 2018 IEEE 3rd international conference on signal and image processing (ICSIP), Shenzhen, China, 13–15 July 2018; pp. 562–566. [Google Scholar]
- Tsochatzidis, L.; Costaridou, L.; Pratikakis, I. Deep learning for breast cancer diagnosis from mammograms—A comparative study. J. Imaging 2019, 5, 37. [Google Scholar] [CrossRef] [Green Version]
- Keras Applications 2022. Available online: https://keras.io/api/applications/ (accessed on 16 November 2021).
- Dang, A.T. Accuracy and Loss: Things to Know about The Top 1 and Top 5 Accuracy. 2021. Available online: https://towardsdatascience.com/accuracy-and-loss-things-to-know-about-the-top-1-and-top-5-accuracy-1d6beb8f6df3 (accessed on 20 May 2021).
- Russakovsky, O.; Deng, J.; Su, H.; Krause, J.; Satheesh, S.; Ma, S.; Huang, Z.; Karpathy, A.; Khosla, A.; Bernstein, M.; et al. Imagenet large scale visual recognition challenge. Int. J. Comput. Vis. 2015, 115, 211–252. [Google Scholar] [CrossRef] [Green Version]
- Maskarinec, G.; Meng, L.; Ursin, G. Ethnic differences in mammographic densities. Int. J. Epidemiol. 2001, 30, 959–965. [Google Scholar] [CrossRef] [Green Version]
- Maskarinec, G.; Pagano, I.; Chen, Z.; Nagata, C.; Gram, I.T. Ethnic and geographic differences in mammographic density and their association with breast cancer incidence. Breast Cancer Res. Treat. 2007, 104, 47–56. [Google Scholar] [CrossRef]
- Wu, N.; Geras, K.J.; Shen, Y.; Su, J.; Kim, S.G.; Kim, E.; Wolfson, S.; Moy, L.; Cho, K. Breast density classification with deep convolutional neural networks. In Proceedings of the 2018 IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP), Calgary, AB, Canada, 15–20 April 2018; pp. 6682–6686. [Google Scholar]
- Shi, P.; Wu, C.; Zhong, J.; Wang, H. Deep Learning from Small Dataset for BI-RADS Density Classification of Mammography Images. In Proceedings of the 2019 10th International Conference on Information Technology in Medicine and Education (ITME), Qingdao, China, 23–25 August 2019; pp. 102–109. [Google Scholar]
- Deng, J.; Ma, Y.; Li, D.a.; Zhao, J.; Liu, Y.; Zhang, H. Classification of breast density categories based on SE-Attention neural networks. Comput. Methods Programs Biomed. 2020, 193, 105489. [Google Scholar] [CrossRef]
- Gandomkar, Z.; Suleiman, M.E.; Demchig, D.; Brennan, P.C.; McEntee, M.F. BI-RADS density categorization using deep neural networks. In Medical Imaging 2019: Image Perception, Observer Performance, and Technology Assessment; SPIE: San Diego, CA, USA, 2019; Volume 10952, pp. 149–155. [Google Scholar]
- Tardy, M.; Scheffer, B.; Mateus, D. Breast Density Quantification Using Weakly Annotated Dataset. In Proceedings of the 2019 IEEE 16th International Symposium on Biomedical Imaging (ISBI 2019), Venice, Italy, 8–11 April 2019; pp. 1087–1091. [Google Scholar]
- Lehman, C.D.; Yala, A.; Schuster, T.; Dontchos, B.; Bahl, M.; Swanson, K.; Barzilay, R. Mammographic breast density assessment using deep learning: Clinical implementation. Radiology 2019, 290, 52–58. [Google Scholar] [CrossRef] [Green Version]
- Saffari, N.; Rashwan, H.A.; Abdel-Nasser, M.; Kumar Singh, V.; Arenas, M.; Mangina, E.; Herrera, B.; Puig, D. Fully Automated Breast Density Segmentation and Classification Using Deep Learning. Diagnostics 2020, 10, 988. [Google Scholar] [CrossRef] [PubMed]
- Ciritsis, A.; Rossi, C.; Vittoria De Martini, I.; Eberhard, M.; Marcon, M.; Becker, A.S.; Berger, N.; Boss, A. Determination of mammographic breast density using a deep convolutional neural network. Br. J. Radiol. 2019, 92, 20180691. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.; Fisher, C.; Wei, J.; Helvie, M.A.; Chan, H.P.; Zhou, C.; Hadjiiski, L.; Lu, Y. Multi-path deep learning model for automated mammographic density categorization. In Medical Imaging 2019: Computer-Aided Diagnosis; SPIE: San Diego, CA, USA, 2019; Volume 10950, pp. 621–626. [Google Scholar]
- Lizzi, F.; Laruina, F.; Oliva, P.; Retico, A.; Fantacci, M.E. Residual convolutional neural networks to automatically extract significant breast density features. In International Conference on Computer Analysis of Images and Patterns; Springer: Berlin/Heidelberg, Germany, 2019; pp. 28–35. [Google Scholar]
- Lizzi, F.; Atzori, S.; Aringhieri, G.; Bosco, P.; Marini, C.; Retico, A.; Traino, A.C.; Caramella, D.; Fantacci, M.E. Residual Convolutional Neural Networks for Breast Density Classification. In BIOINFORMATICS; SciTePress: Setúbal, Portugal, 2019; pp. 258–263. [Google Scholar]
- Cogan, T.; Tamil, L. Deep Understanding of Breast Density Classification. In Proceedings of the 2020 42nd Annual International Conference of the IEEE Engineering in Medicine & Biology Society (EMBC), Montreal, QC, Canada, 20–24 July 2020; pp. 1140–1143. [Google Scholar]
- Chang, K.; Beers, A.L.; Brink, L.; Patel, J.B.; Singh, P.; Arun, N.T.; Hoebel, K.V.; Gaw, N.; Shah, M.; Pisano, E.D.; et al. Multi-institutional assessment and crowdsourcing evaluation of deep learning for automated classification of breast density. J. Am. Coll. Radiol. 2020, 17, 1653–1662. [Google Scholar] [CrossRef] [PubMed]
- Roth, H.R.; Chang, K.; Singh, P.; Neumark, N.; Li, W.; Gupta, V.; Gupta, S.; Qu, L.; Ihsani, A.; Bizzo, B.C.; et al. Federated learning for breast density classification: A real-world implementation. In Domain Adaptation and Representation Transfer, and Distributed and Collaborative Learning; Springer: Berlin/Heidelberg, Germany, 2020; pp. 181–191. [Google Scholar]
- Li, C.; Xu, J.; Liu, Q.; Zhou, Y.; Mou, L.; Pu, Z.; Xia, Y.; Zheng, H.; Wang, S. Multi-view mammographic density classification by dilated and attention-guided residual learning. IEEE/ACM Trans. Comput. Biol. Bioinform. 2020, 18, 1003–1013. [Google Scholar] [CrossRef] [PubMed]
- Zhao, W.; Wang, R.; Qi, Y.; Lou, M.; Wang, Y.; Yang, Y.; Deng, X.; Ma, Y. BASCNet: Bilateral adaptive spatial and channel attention network for breast density classification in the mammogram. Biomed. Signal Process. Control 2021, 70, 103073. [Google Scholar] [CrossRef]
- Tlusty, T.; Amit, G.; Ben-Ari, R. Unsupervised clustering of mammograms for outlier detection and breast density estimation. In Proceedings of the 2018 24th International Conference on Pattern Recognition (ICPR), Beijing, China, 20–24 August 2018; pp. 3808–3813. [Google Scholar]
- Cao, Y.; Geddes, T.A.; Yang, J.Y.H.; Yang, P. Ensemble deep learning in bioinformatics. Nat. Mach. Intell. 2020, 2, 500–508. [Google Scholar] [CrossRef]
- Ganaie, M.; Hu, M. Ensemble Deep Learning: A Review; Cornell University: Ithaca, NY, USA, 2021. [Google Scholar]
- Liashchynskyi, P.; Liashchynskyi, P. Grid search, random search, genetic algorithm: A big comparison for NAS. arXiv 2019, arXiv:1912.06059. [Google Scholar]
- Bhattiprolu, S. Ensemble Sign Language. 2021. Available online: https://github.com/bnsreenu/python_for_microscopists/blob/master/213-ensemble_sign_language.py (accessed on 4 January 2022).
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).