Abstract
Data science and machine learning are buzzwords of the early 21st century. Now pervasive through human civilization, how do these concepts translate to use by researchers and clinicians in the life-science and medical field? Here, we describe a software toolkit, just large enough in scale, so that it can be maintained and extended by a small team, optimised for problems that arise in small/medium laboratories. In particular, this system may be managed from data ingestion statistics preparation predictions by a single person. At the system’s core is a graph type database, so that it is flexible in terms of irregular, constantly changing data types, as such data types are common during explorative research. At the system’s outermost shell, the concept of ’user stories’ is introduced to help the end-user researchers perform various tasks separated by their expertise: these range from simple data input, data curation, statistics, and finally to predictions via machine learning algorithms. We compiled a sizable list of already existing, modular Python platform libraries usable for data analysis that may be used as a reference in the field and may be incorporated into this software. We also provide an insight into basic concepts, such as labelled-unlabelled data, supervised vs. unsupervised learning, regression vs. classification, evaluation by different error metrics, and an advanced concept of cross-validation. Finally, we show some examples from our laboratory using our blood sample and blood clot data from thrombosis patients (sufferers from stroke, heart and peripheral thrombosis disease) and how such tools can help to set up realistic expectations and show caveats.
Keywords:
thrombosis; stroke; data science; machine learning; graph database; python; PyTanito; user story; classification; cross-validation 1. Introduction
Coronary artery disease (CAD), acute ischemic stroke (AIS), peripheral artery disease (PAD) are cardiovascular diseases and represent the leading morbidity and mortality causes globally [1]. The acute tissue damage is mostly due to thrombi occluding the supplying arteries [2]. The lysis susceptibility and stability of these thrombi ultimately determines the fate of the patient [3]. Can we tell more from their structure and from common blood test data of patients? Can we predict the diseases from this data? Even better, can we predict it before disease onset? These are questions that we address with the help of the data analysis approach described in this paper.
2. Data Science Project Organisation—Industry ’Best Practices’
As with any project, the question arises: where to begin? This is especially so with complex, data-driven projects, such as the ‘bench-to-bedside’ projects often seen in the medical and life science field. Fortunately, this problem has been seen before and some solutions, or rather, guidelines were devised. Industrial Micro Machines (IBM) researchers and automotive engineers (Daimler-Chrysler) did indeed face the same problem in the 1990s. The resultant guidelines were standardized in the CRISP-DM standard which stands for Cross Industry Standard Process for Data Mining [4]. The model, illustrated in Figure 1, consists of six steps [5].
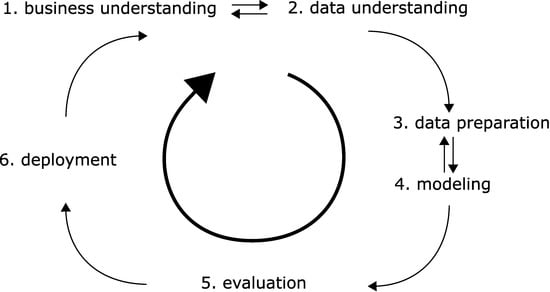
Figure 1.
The six elementary steps in a data science project as outlined in the CRISP-DM standard.
- Business understanding. In the medical field, we would need to determine if the project is for research purposes only. Do we have obvious clinical implications (with rigorous safety, regulatory and legal requirements)? Do we eye business potential—which in turn may need strict documentation and reporting?
- Data understanding. In the medical and life science field, we often have to gather data via experiments ourselves, and the quality of this data will be crucial. (In the era of big data, many projects may rely on published open data sources.) What data can we gather and in what amount? Can we collect enough in a realistic timeframe? Will the data be sufficient for the questions asked? Do we have good recipes so that the data is reproducible over large timespans, with changing personnel?
- Data preparation. We must gather data from handwritten notes or other sources, collect them in machine-readable form, and normalize or otherwise standardize them. Watch out for reproducibility issues often encountered with difficult experiments. Can our data be quantified and compared across different investigators and with human subjects involved? Even standard laboratory tests may differ in testing methodology. With human subjects, legal issues also arise privacy rights need to be respected, and data anonymized.
- Modelling. We need to use our data—somehow. Do we have continuous variables or categorical ones? Do we have an initial hypothesis to test, or do we need methods that are suitable for ‘walking in the dark’?
- Evaluation. We need to use metrics by which we quantify ‘successes. Do we have concise research questions so that an answer to them can be made? Are we descriptive or do we have new hypotheses as a result? Without clear questions, no metrics will be satisfactory, as metrics are often meaningless themselves. Can we compare our work to that of others via the metrics?
- Deployment. Pure research may result in scientific outputs, such as presentations or scientific papers or openly published datasets. However, does our research yield tangible results such as new scientific hypotheses? Does our research have clinical implications?
This is the model we will follow and around which we build our homemade software, which will be described in more detail in the next sections.
2.1. Software Architecture—Incorporation of ‘Best Practices’ for User-Friendliness
The software we are developing is an aid for a small-scale medical research team. To borrow from the beforementioned concepts, we are developing software that helps with steps 2–5, with 3–4 essential parts of it. The focus is deliberately limited, it does not want to do more: it is simply a framework by which a team can gather all their research data, test various hypotheses and models with it, and report it automatically in a user-readable form. We feel that this scope fits with a few developers and new functions may be added as needed; in contrast with large software frameworks, where such incorporation may take a long time or workarounds.
However, the CRISP-DM standard deliberately does not provide a guideline, how to actually implement the functionality required. This problem is as old as software development itself. Fortunately, industry ‘best practices’ are available. One of these is the widely popular ‘Agile’ methodology [6]. The ‘Agile Manifesto’ of software developers describes essential concepts and introduced the concept of ‘user stories’ [7]. With Agile and with ‘user stories’, the system is approached from outside, from the perspective of the users. Everything else is developed around the needs of the users, every user has a ‘story’ with the software piece.
In a small research project, we usually have four types of participants:
- Assistants, who usually have a narrow, well-defined scope of work.
- Junior researchers (e.g., undergraduate students) and other personnel, who are strongly dependent on input in their work.
- Senior researchers (e.g., postdocs), who are able to work independently with minimal input.
- Principal investigator, who oversees the whole project and sets goals.
To meet the requirements of data processing in such teams, we have devised a software toolkit, a data learning framework we named ‘PyTanito’. The Py stands for ‘Python’, an increasingly popular, open-source, machine programming language [8], with a large following in the science field as of 2021. We have deliberately made this choice, so that our software may be extended by future researchers without ‘strings attached’ or without potential large costs that come with proprietary systems managed by large corporations. A secondary benefit of this adoption is that an increasingly large amount of ‘libraries’ are available for it. These ‘libraries’ are pieces of independent programs performing specific tasks so that one can rely on the work of others. We do not need to program every function, especially data analysis or machine learning tasks, which are usually difficult to code.
Illustrated in Figure 2, the system builds around the needs of four types of users. The respective four ‘user stories’ are as follows, roughly corresponding to the natural composition of a small medical research team:
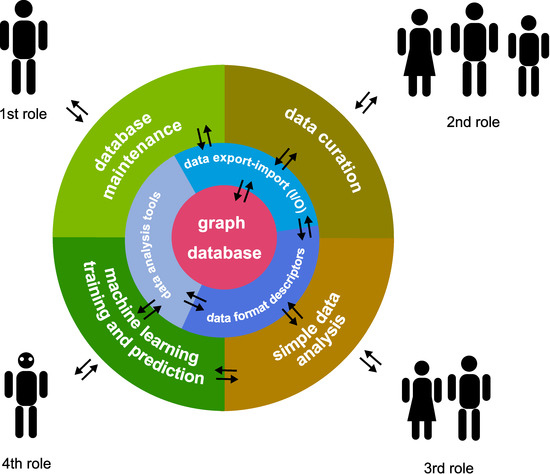
Figure 2.
Architecture of ‘PyTanito’, organised into three layers. The outermost shell is the user-interactable components, corresponding to the four roles in a small research team. The middle layer is the part corresponding to data input-output, managing data formats, and the data analysis tools themselves. A database is placed at the core, serving as the ‘single point of truth’. The arrows denote flow of information.
- Software/database manager, who manages the software; may not need to be a research person; sets up environment and checks database and software function.
- Data curator, the role of assistants and junior researchers; collects and inputs data into the system.
- Simple data analyser, the role of more senior researchers; may perform simple analyses on certain selected data.
- Complex model builder and analysis, the role of the most senior researchers; sets up different machine learning models.
‘PyTanito’ incorporates another software development ‘best practice’: the layers only interact with the single layer below their level—or, at maximum, at their level. This key part is illustrated in Figure 2, which shows the direction of information flow. For example, it is not possible and not desirable for any user to interact directly with the database sitting in the centre. Simply use the 2nd role functions that read and input data into it. Another advantage is that 3rd and 4th role users do not need to know where the data comes from, as they work with the database, where all data is unified.
2.2. Data Storage and Organisation—An Up-to-Date Solution Using a Graph Database
At the core of the system is a database. It is a central, ‘single point of truth’ type organisation so that all data we work with will be consistent. We do not have to worry about differently formatted spreadsheet files or other nonstandard sources. We have chosen a graph-type database [9], instead of a traditional, relational type (such as SQL types, like Oracle SQL or MySQL [10]). Traditional relational type databases are organised into columns and rows, and usually need to have clearly defined data types/fields/tables [11]. That is, we need to have a clear concept beforehand of what we will store. Unfortunately, we found out that our exploratory research is not kind to this type of organisation. New variables emerge constantly as a result of research. In our case, not all parameters are measured for all patients, and new parameters are added routinely to certain patients. Patients do not have all their parameters measured—some patients have extra parameters. From a traditional relational database viewpoint, it is untidy and would require constantly changing its tables and columns, thus rebuilding the entire database structure.
A graph database does not rely on such assumptions. A graph, as a mathematical principle, consists of only ‘nodes’ and ‘links. The nodes may be connected with links to each other, describing their relation. We illustrate the use of this graph-type organisation on our dataset in Figure 3 [12], which consists of ~200 thrombosis patients along with their blood and blood clot parameters. Illustrated in Figure 3a, our minimal graph database consists of a ‘patient’ node and a ‘measurement’ node. That is, the minimal sensible unit is a single patient with a single laboratory parameter value.
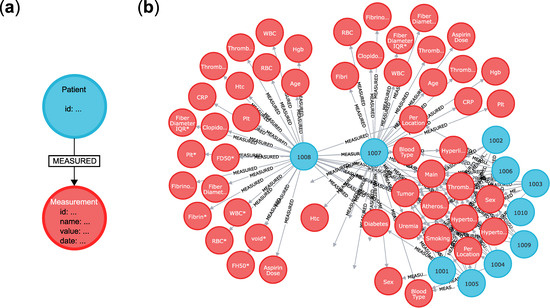
Figure 3.
The organisation of our graph-type database using Neo4j [13] on our actual blood and blood clot data [12]. (a) The minimal unit of the database consist of a ‘patient’ node (blue) and an attached ‘measurement’ node (red). The two elements are linked together with a link named ‘measured’. (b) A part of the actual database as viewed with the built-in Neo4j browser by opening the address http://localhost:7474 in any web browser. The patient nodes are connected to many red nodes. The patient IDs are shown (e.g., ‘1008′), just as the names of the measured parameters on the nodes (e.g., ‘Sex’ for sex, ‘Hgb’ for haemoglobin, ‘WBC*’ for white blood cell count, etc.).
Patients have an ID as a property of the node. The ID we added is a four-letter number and is anonymized; it cannot be used to reveal the identity of the patients. The laboratory measurements are of course nodes themselves. Patients are linked to their laboratory parameter(s) with a ‘measured’ relation link. The ‘measurement’ nodes have the value of the measured parameter as property, but others may be added, such as the name of the parameter, the method by which it was measured, the date, etc.). This arrangement gives extreme flexibility: patients may be added without much data, new data types may be added as simple new nodes with different labels, and this does not need database rework, just an additional operation to an already existing database. The question arises, that with such advantages, why graph databases are not more widespread? The answer is that the background processes driving graph databases are more computationally intensive than those driving SQL databases, but this is not a concern when using relatively small datasets with contemporary hardware.
Our actual implementation uses the Neo4j Community Edition database, which is open source, and was chosen on grounds discussed before. We note that many more types of graphs exist, such as hypergraphs, where links (the edges) may link to any number of nodes. The more commonly encountered ‘property graphs’ described in this paper and software are a subset of such hypergraphs since a particular link ‘only’ connects two nodes. The universality of hypergraphs makes them more difficult to comprehend and use computationally, nevertheless, see [14].
2.3. Data Analysis—Typical Tasks, Terms and Algorithms
In the next sections, now that we have data on hand, we need to specify, what we do with it [15,16]. For the sake of simplicity, we will provide examples using our actual patient dataset with blood parameters. In broad terms, we usually have three different types of analysis. Let us also have two types of variables, X and Y.
- Regression/classification analysis. We would like to predict Y from given X. X is the independent variable and Y is the dependent variable (X → Y).
- Hypothesis testing. If, e.g., given two patient populations (YA and YB), is the so-called null hypothesis true? The ‘null hypothesis’ usually is that the two populations are different (YA → X is not equal to YB → X). The ‘alternative hypothesis’ is the opposite, exclusive hypothesis: the two populations are identical.
- Clustering/dimensionality reducing algorithms. Sometimes we do not even have labelled data (here denoted Y), only raw data X. We need algorithms that work when we want to reduce the complexity somehow. In effect, can we cluster/label/group/classify the data based on X alone?
To put these analysis possibilities in context, taken our clotting disorder data [12], usually the Y dependent variable is a fundamental, clinically relevant label: what disease did our patients suffer from? Stroke, myocardial infarction or peripheral thrombosis? The Xs, the independent variables are usually the measured blood/blood clot parameters, such as cell counts, molecular marker (e.g., CRP) levels, etc. Please note that in a ‘regression analysis’, the Y is a continuous, quantifiable variable, while in a ‘classification analysis’ the Ys are discrete, non-quantifiable labels.
It is also useful to know that when we talk about ‘labelled data’, we mean data, which was appropriately tagged with useful (in the medical field this means clinically relevant) properties using discrete, non-quantifiable variables. This is, e.g., the disease type mentioned before. Unlabelled data are usually less worthy, as the number of analysis tools available are reduced. Here, using our dataset as an example, raw data would be, e.g., blood test numbers. If we add disease type and other clinically relevant labels, our data becomes labelled. This labelling task can be a hard problem, and often requires human intervention.
Supervised vs. unsupervised learning are additional terms encountered. In supervised learning, we have some expectations about the data, we have labels on the data, and we direct the analysis using this expectation. The analysis tools mentioned before Types 1 and 2 are supervised types. Type 3 is of the unsupervised type.
We provide two tables: one lists the common machine learning algorithms Table 1. The second table lists the evaluation metrics that may be used to determine how successful we were by using the beforementioned algorithms Table 2.

Table 1.
Machine learning algorithms. This table provides an overview of algorithms that may be used in a Python language environment. The list may be extended significantly as new algorithms emerge and get incorporated into the Python environment. Of course, discussions on these methods fall beyond the scope of this paper, so the interested readers are directed to the excellent books of Hastie and Landau [15,16] for more information.

Table 2.
Metrics used for evaluation of machine learning models. In this table, we provide a list of useful metrics used when quantifying success in a data analysis experiment, be it a regression or classification problem. As with Table 1, the list is not exhaustive, and interested users are directed to [17,18,19] for more information.
2.4. A Machine Learning Workflow—Common Processes Explained with Examples
In a traditional scientific project, we are convinced of our models and hypotheses when these are repeated and verified by independent researchers. A single team collects data, and then ‘trains’ their model/algorithm on a part (or whole) of the available data [16]. Thus, this data is called ‘training data’. Training a model/algorithm means the setting of the model’s parameters so that the model from Table 1 describes the data ‘best’, according to selected metrics seen in Table 2. Training often involves other kinds of parameters (hidden variables) during the construction of the model (‘hyperparameters’) that refine/restrict the behaviour of an otherwise complex model. The model’s ordinary parameters are, e.g., the two coefficients of a simple linear regression, and in this case, the training may be called ‘linear fitting’. Linear regression is simple, and it has not hidden ‘hyperparameters’, but more complex models, such as a ‘decision tree’ may have more parameters and several hyperparameters that describe it.
However, it is possible that the model is not optimal for the problem, for example, a linear regression does not capture the phenomena studied, and so does not make sense from a medical/physical/chemical standpoint. To see that the fitted model is reasonable (and does not depend on cherry-picked ‘training’ datapoints), it is good practice to have a ‘validation set’ of data. This ‘validation dataset’ is data totally separated from the ‘training data’. It is not used for the training (e.g., linear regression fitting), and so is ‘unseen’ by the model-building procedure. The trained model’s error on this ‘validation dataset’ is more indicative of the model’s appropriateness.
However, even this train-validation procedure may be less than optimal in real life. For example, because of inherent systematic errors during the experiments or if it just happens that the particular picking of data into train/validation sets skew results. Figure 4 illustrates how independent observations by different researchers solve the problem. However, in the medical field, this is often not possible, because the experiments may be too costly or there is simply not enough interest or resources to reproduce results.
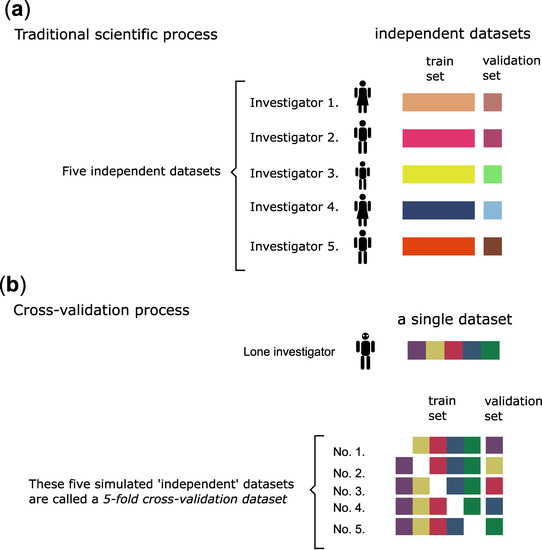
Figure 4.
The scientific process and the cross-validation process compared. (a) In an ideal world, experiments can be repeated many times, and the results are reproducible. (b) In the real world, relatively few experiments can be executed (e.g., expensive and time-consuming medical types with human subjects). In our case, ex vivo examination of blood clots from thrombosis patients is a costly and time-consuming endeavour. Therefore, a cross-validation type procedure is used to evaluate the data analysis results. A k-fold cross-validation procedure produces ‘k’ number of simulated ‘identical’ and ‘random’ datasets. The original dataset is split into ‘k’ number of equal parts, then each part is used for validation only once—while the rest is used for training.
A so-called cross-validation procedure Figure 4b may solve this problem with caveats [20,21]. In a cross-validation procedure, we produce simulated ‘independent’ datasets randomly. A k-fold cross-validation procedure produces ‘k’ number of simulated datasets. This procedure minimizes at least the ‘cherrypicking’ problem of the train-validation dataset split. Still, the problem of systematic errors remains, therefore, this procedure produces only a hint on the minimum error, and the real error is certainly larger than that. Still, the procedure will prevent over-optimistic error estimations by setting a minimum error expectation and is best used to compare the appropriateness of different machine learning model types (e.g., linear regression vs. polynomial regression, etc.). Another advantage is that this procedure may be used ‘on top’ of other error metrics, because any error metrics may be used with these ‘independent’ datasets.
In the last part of this paper, we show the actual outputs of the software. Figure 5 depicts the use of the software to check the data and available measurements for further analysis (a simple ‘data curator’ user role). Figure 5 displays the command-line interface which is always available for Python programming. The output is also always available as a command-line type. Although not as wieldy or ‘nice’ as graphical user interfaces (GUIs), command-line interfaces and outputs serve an important purpose: they are read and parsed easily by machines, e.g., by other parts of the same program or even third-party software. For this reason, efficient software implements both the command-line interface and the GUI.

Figure 5.
‘PyTanito’ software in actual use—example of a ‘data curation’ role use. (a) The input: these are code lines in Python for database import and for printing database statistics. (b) The output of the software in response to the commands in (a) lists the number of patients in our dataset 12 (209 total), as well as the number of measurements collected (6997 total), and as a bonus, it lists the type of measurements available for further analysis (43 different types). The asterisk (*) after a parameter name indicates that the parameter was measured using a pathological blood clot, unlike the rest, which are from ordinary blood samples.
In Figure 6, we show an actual example of decision making with the help of machine learning. The figure shows a pair of plots for two different, but related prediction problems using common ‘Decision Tree’ algorithm implementation. In each of the plots, we compare several ‘tree’ models and decide which ‘tree’ is the best if there is such a model. The provided example is realistic in the sense that it also displays commonly encountered, relatively unsuccessful models. A major takeaway is that relying on a single metric on a single training run is guaranteed to be flawed: even when it seems to be better, it is not when many random combinations of data are also validated. Extensive use of cross-validation helped in our case to set realistic expectations. For this particular example, the conclusion is that the decision trees are better than random guesses, but not by very much. It is an open question as to whether robust, clinically relevant predictions can be made. More data, more types of variables and more types of algorithms need to be tested. Thus, from this data only, the prediction of disease before onset is not practical.
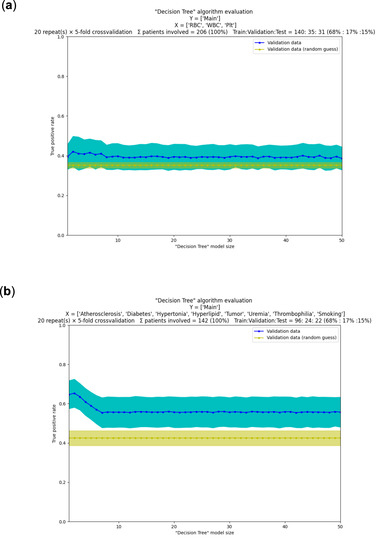
Figure 6.
‘PyTanito’ software in actual use—example use of the most advanced training and evaluation functions. (a) Decision tree models and their evaluation on our dataset [12]. Can we tell from mere blood cell counts (X independent variables are red ‘RBC’, white ‘WBC’ and platelet ‘Plt’ cell counts) what kind of thrombi were present (Y dependent variable is the ‘Main’ label, which has values ‘stroke’, ‘myocardial’ or ‘peripheral’)? We chose ‘true positive’ rate as error metrics (depicted on y scale) for a classification problem, 5-fold cross-validation was used with repeats. On the x scale are the different decision tree models, numbered 1–50. They are mostly identical in performance (on average they are ~40% correct, blue line) and not much better than random guessing (yellow line, 35% correct on average). (b) In a second problem, we used the same dataset and the same goal as before, but now X independent variables are clinically relevant labels (such as, do the patients have diabetes, atherosclerosis, etc.). Now, the results are better; all trained decision tree models (No. 1–50) seem better than random guessing. Models 1–8 appear better than the rest, with model 2 seemingly the best of all, with average success rate of ~65%, compared to the average success rate of ~45% of random guessing.
3. Conclusions and Remarks
We have seen that modern workflow (CRISP-DM standard) with modern software ‘best practices’ (‘Agile’ and software layering) can produce useful tools in medical research. We also provide a glimpse into how data may be stored in a humanly reasonable way using a graph database. We also provided lists of the incredible amount of contemporary machine learning methods and metrics for their evaluation.
We consider two kinds of readers and have conclusions for each.
- First, from the perspective of the software developer involved in a ‘decision making’ project, it is notable that software specification is a bottleneck during the initial steps. This often stems from a lack of clarity and vision during the initial stages of the project. Going forward in the project, the tuning of the machine learning models (‘hyperparameter tuning’) becomes the bottleneck and this remains so until the end. Curiously, the choice of algorithms does not appear critical now. Computational capacities are larger than ever, and several highly advanced machine learning methods are available (in fact, often more than needed in a project). Projects, however, may fail easily because of data quality issues, and this brings us to the second set of conclusions below.
- Second, from the perspective of the medical professional, the bottleneck is usually a lack of understanding between them and software specialists during the initial stages of the project. Going forward, the single but often critical bottleneck is the quality of their data. Experiments may span years and may be performed by different personnel over time. This may lead to inconsistent procedures, even if for mundane reasons, e.g., because of no longer available reagents or altered cell lines. These can introduce large systematic errors that make even advanced machine learning algorithms ineffective. Other kinds of experiments are inherently difficult to perform consistently for physical, economical or legal reasons. For example, in our case, the blood clots of ex vivo human origin pose a significant problem, since the number of experiments is severely limited. In summary, medical professionals must do everything to ensure reproducibility and consistency of their experiments and data over long timespans in sufficient quantity.
Author Contributions
L.B. developed software, conceptualization, wrote paper; E.K. wrote paper; P.P. wrote paper; E.T. data collection; R.F. wrote paper; K.K. funding, conceptualization, wrote paper. All authors have read and agreed to the published version of the manuscript.
Funding
K.K. This work was supported by the Hungarian National Research, Development and Innovation Office (NKFIH) #137563. K.K., E.T. and L.B.—Thematic Institutional Excellence Programme of the Ministry of Human Capacities in Hungary for the Molecular Biology thematic programme of Semmelweis University (TKP2021-EGA-24).
Institutional Review Board Statement
The study was approved by the Semmelweis University and regional ethical board (Ref.#2014/18.09.2014). The research also conforms to the principles outlined in the Declaration of Helsinki.
Informed Consent Statement
Informed written consent was obtained from all participants or their legal guardians.
Data Availability Statement
The software described in this paper is under development and is in a pre-release state. Only for information purposes—it is not suitable for public use. Code reuse, distribution and third-party use are restricted. With these limitations, it is available upon request from L.B. (laszlo@beinrohr.com). The blood clot data may be provided by K.K. (Krasimir.Kolev@eok.sote.hu) upon reasonable request.
Conflicts of Interest
The authors declare no conflict of interest.
References
- The top 10 Causes of Death. Available online: https://www.who.int/news-room/fact-sheets/detail/the-top-10-causes-of-death (accessed on 11 November 2021).
- Santos-Gallego, C.G.; Bayón, J.; Badimón, J.J. Thrombi of Different Pathologies: Implications for Diagnosis and Treatment. Curr. Treat. Options Cardiovasc. Med. 2010, 12, 274–291. [Google Scholar] [CrossRef] [PubMed]
- Undas, A.; Ariëns, R.A.S. Fibrin Clot Structure and Function. Arterioscler. Thromb. Vasc. Biol. 2011, 31, e88–e99. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shearer, C. The CRISP-DM Model: The New Blueprint for Data Mining. J. Data Warehous. 2000, 5, 13–22. [Google Scholar]
- Chapman, P.; Clinton, J.; Kerber, R.; Khabaza, T.; Reinartz, T.; Shearer, C.; Wirth, R. Step-by-Step Data Mining Guide. Available online: https://the-modeling-agency.com/crisp-dm.pdf (accessed on 11 November 2021).
- Manifesto for Agile Software Development. Available online: https://agilemanifesto.org/ (accessed on 11 November 2021).
- Dalpiaz, F.; Brinkkemper, S. Agile Requirements Engineering with User Stories. In Proceedings of the 2018 IEEE 26th International Requirements Engineering Conference (RE), Banff, AB, Canada, 20–24 August 2018; pp. 506–507. [Google Scholar] [CrossRef]
- Welcome to Python.org. Available online: https://www.python.org/ (accessed on 11 November 2021).
- Angles, R. A Comparison of Current Graph Database Models. In Proceedings of the 2012 IEEE 28th International Conference on Data Engineering Workshops, Arlington, VA, USA, 1–5 April 2012; pp. 171–177. [Google Scholar] [CrossRef]
- MySQL. Available online: https://www.mysql.com/ (accessed on 11 November 2021).
- Sahatqija, K.; Ajdari, J.; Zenuni, X.; Raufi, B.; Ismaili, F. Comparison between Relational and NOSQL Databases. In Proceedings of the 2018 41st International Convention on Information and Communication Technology, Electronics and Microelectronics (MIPRO), Opatija, Croatia, 21–25 May 2018; pp. 0216–0221. [Google Scholar] [CrossRef]
- Farkas, Á.Z.; Farkas, V.J.; Gubucz, I.; Szabó, L.; Bálint, K.; Tenekedjiev, K.; Nagy, A.I.; Sótonyi, P.; Hidi, L.; Nagy, Z.; et al. Neutrophil Extracellular Traps in Thrombi Retrieved during Interventional Treatment of Ischemic Arterial Diseases. Thromb. Res. 2019, 175, 46–52. [Google Scholar] [CrossRef] [PubMed]
- Neo4j Graph Data Platform—The Leader in Graph Databases. Available online: https://neo4j.com/ (accessed on 11 November 2021).
- Alam, M.T.; Ahmed, C.F.; Samiullah, M.; Leung, C.K. Mining Frequent Patterns from Hypergraph Databases. In Proceedings of the Advances in Knowledge Discovery and Data Mining; Karlapalem, k., Cheng, h., Ramakrishnan, N., Agrawal, R.K., Reddy, P.K., Srivastava, J., Chakrabortya, T., Eds.; Springer International Publishing: Cham, Switzerland, 2021; pp. 3–15. [Google Scholar]
- Landau, S.; Everitt, B. A Handbook of Statistical Analyses Using SPSS; Chapman & Hall/CRC: Boca Raton, FL, USA, 2004. [Google Scholar]
- Hastie, T.; Tibshirani, R.; Friedman, J. The Elements of Statistical Learning: Data Mining, Inference, and Prediction, Edition, 2nd ed.; Springer Series in Statistics; Springer: New York, NY, USA, 2009. [Google Scholar] [CrossRef]
- Hossin, M.; Sulaiman, M.N. A Review on Evaluation Metrics for Data Classification Evaluations. Int. J. Data Min. Knowl. Manag. Process 2015, 5, 1. [Google Scholar] [CrossRef]
- Fawcett, T. ROC graphs: Notes and practical considerations for researchers. Mach. Learn. 2004, 31, 1–38. [Google Scholar]
- Saito, T.; Rehmsmeier, M. The Precision-Recall Plot Is More Informative than the ROC Plot When Evaluating Binary Classifiers on Imbalanced Datasets. PLoS ONE 2015, 10, e0118432. [Google Scholar] [CrossRef] [Green Version]
- Yadav, S.; Shukla, S. Analysis of K-Fold Cross-Validation over Hold-Out Validation on Colossal Datasets for Quality Classification. In Proceedings of the 2016 IEEE 6th International Conference on Advanced Computing (IACC), Bhimavaram, India, 27–28 February 2016; pp. 78–83. [Google Scholar] [CrossRef]
- Wong, T.-T.; Yeh, P.-Y. Reliable Accuracy Estimates from K-Fold Cross Validation. IEEE Trans. Knowl. Data Eng. 2020, 32, 1586–1594. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).