Featured Application
Model-informed drug discovery and development (MID3) is proposed to be applied throughout the preclinical to clinical phases to provide an informative prediction of drug exposure and efficacy in humans in order to select novel anti-tuberculosis drug combinations for the treatment of tuberculosis.
Abstract
The increasing emergence of drug-resistant tuberculosis requires new effective and safe drug regimens. However, drug discovery and development are challenging, lengthy and costly. The framework of model-informed drug discovery and development (MID3) is proposed to be applied throughout the preclinical to clinical phases to provide an informative prediction of drug exposure and efficacy in humans in order to select novel anti-tuberculosis drug combinations. The MID3 includes pharmacokinetic-pharmacodynamic and quantitative systems pharmacology models, machine learning and artificial intelligence, which integrates all the available knowledge related to disease and the compounds. A translational in vitro-in vivo link throughout modeling and simulation is crucial to optimize the selection of regimens with the highest probability of receiving approval from regulatory authorities. In vitro-in vivo correlation (IVIVC) and physiologically-based pharmacokinetic modeling provide powerful tools to predict pharmacokinetic drug-drug interactions based on preclinical information. Mechanistic or semi-mechanistic pharmacokinetic-pharmacodynamic models have been successfully applied to predict the clinical exposure-response profile for anti-tuberculosis drugs using preclinical data. Potential pharmacodynamic drug-drug interactions can be predicted from in vitro data through IVIVC and pharmacokinetic-pharmacodynamic modeling accounting for translational factors. It is essential for academic and industrial drug developers to collaborate across disciplines to realize the huge potential of MID3.
1. Introduction
Drug discovery and development is a challenging, lengthy, and costly process. The costs of a novel drug reaching the market can be as much as 2–3 billion dollars [1]. In the early discovery phase, libraries consisting of thousands of compounds can be synthesized chemically and tested for efficacy in vitro at a relatively low cost. The largest expenditures are in the late preclinical and clinical phases of drug development, where the efficacy and safety of treatment are assessed. Smart decisions need to be made regarding which compounds and regimens should progress through the preclinical phase and subsequently into clinical trials. Early characterization of each compound’s exposure-response relationship, i.e., pharmacokinetic (PK)-pharmacodynamic (PD) relationship and potential interactions within regimens and with commonly co-administered drugs, can allow for informative decision making throughout preclinical development and into clinical development [2].
Tuberculosis (TB) is the leading cause of adult mortality through infectious diseases and 10 million new cases are reported globally every year [3]. Sensitive TB is currently treated with a six-month regimen of antibiotics, consisting of isoniazid, pyrazinamide, rifampicin and ethambutol, which was developed in the mid-twentieth century. This therapy is believed to be suboptimal and was not developed using modern approaches for drug development, thereby lacking important information on the PK-PD relationship. Therefore, clinical trials have recently been conducted in order to define the relationship between exposure and efficacy, as well as safety, where statistically significant exposure-response relationships for rifampicin have been identified, in order to support a higher dose of rifampicin [4,5,6,7]. Almost one in five patients will acquire multidrug-resistant tuberculosis (MDR-TB) or rifampicin-resistant tuberculosis (RR-TB) [3]. Recently, the new anti-TB drugs bedaquiline, delamanid and pretomanid were conditionally approved against MDR-TB, which led to updates to the World Health Organization (WHO) treatment guideline for MDR-TB [8]. Bedaquiline is a diarylquinoline, a new class of antibiotics. It is an inhibitor of the membrane-bound adenosine triphosphate (ATP)-synthase enzyme, therefore blocking mycobacterial ATP formation and energy metabolism. Bedaquiline is therefore bactericidal for dormant mycobacteria as well, a preferable feature for the shortening of treatment duration and prevention of relapse [9]. Delamanid is a nitroimidazole and affects the mycobacterial cell wall, thereby also improving drug penetration into the mycobacterium. It is the most potent TB drug and is active against replicating and dormant mycobacteria as well [9]. The combination of delamanid with bedaquiline is, however, not recommended, due to QT-prolongation-related cardiotoxicity [10]. Pretomanid belongs to the same class of antibiotics as delamanid [9]. Pretomanid was developed as part of a drug combination together with bedaquiline and linezolid, an oxazolidinone-class otherwise used for the treatment of pneumonia and skin infection. There is a clear need for the additional development of new effective drug combinations. The European Medical Agency (EMA) drug development guideline for TB specifies that efforts should be made to develop entirely new regimens to treat TB, rather than focusing on single drugs [11]. Due to the burden of polypharmacy for the patients and the increased risk of side effects, the focus should be on developing new regimens instead of the development of single agents as an add-on to a current regimen which was recommended in the earlier EMA TB drug development guideline [12]. Of the three new drugs against TB, only pretomanid is approved as a new combination regimen, while bedaquiline and delamanid were developed as add-ons to existing therapy [13]. The development of new combination regimens is the way forward, the acceleration of which is the objective of the new Innovative Medicines Initiative (IMI)-funded consortium European Regimen Accelerator for Tuberculosis (ERA4TB). It is important to assess drug-drug interactions (DDI), with respect to both PK and PD, to understand how the different drugs behave in certain combinations and doses in order to maximize the efficacy and potentially learn how the efficacy of the combination varies with time and concentration. The development of drug combinations is, however, challenging. It is difficult to demonstrate the contribution of an individual drug to a regimen regarding efficacy or safety [14]. The duration of treatment is lengthy, especially when considering follow-up to ensure no relapse. Moreover, the design and execution of preclinical experiments and clinical trials are complex, as the number of treatments to test grows exponentially with every added drug or dose, leading to longer development times and higher costs. Tuberculosis drug development, which focuses on regimens rather than unique drugs as an add-on treatment, thus challenges our methods to assess and identify optimal regimens. Therefore, smart experimental designs and optimized data analysis are essential. Data from larger scale in vitro preclinical experiments, with different drug regimens that explore the PD interaction space in order to investigate the synergism and/or antagonism of the interacting drugs, should be used to select the best regimens to determine the exposure range in vivo. Based on the exposure-response relationship in animals, and/or pure in vitro predictions, the first in-human (FIH) and early bactericidal activity (EBA) trials can be designed. These steps all require a mathematical translational approach, taking into account the PK-PD and translational factors to account for differences between preclinical species and patients [15,16].
The European Medicines Agency/European Federation of Pharmaceutical Industries and Associations (EFPIA) Modeling and Simulation joint workshop held in 2011 assembled scientists from the pharmaceutical industry, academia and regulatory authorities from across Europe, the USA and Japan to consider the future role of modeling and simulation in drug development and regulatory assessment. As a follow up to the workshop, one of the EFPIA groups’ commitment to EMA was to generate a “good practice” manuscript covering aspects of planning, conduct and documentation of a variety of quantitative approaches for modeling and simulation methods where the concept of Model-Informed Drug Discovery and Development (MID3) was defined [17]. The aim of MID3 is to enable more efficient and robust research and development and regulatory decisions using an integrated model-based drug development approach [17,18]. The MID3 strategy for the development of drugs in any therapeutic area is supported by the EMA [19]. The MID3 framework has been defined as a “quantitative framework for prediction and extrapolation, centered on knowledge and inference generated from integrated models of compound, mechanism and disease level data and aimed at improving the quality, efficiency and cost effectiveness of decision making” [17]. The MID3 framework should be applied in the development of new TB drug regimens and is necessary for the reliable prediction of the optimal selection of novel TB drug combination therapies based on pre-clinical information, and subsequent decisions on which combinations to evaluate in clinical trials in order to confirm their efficacy and safety. The framework integrates all available data and information on the disease and the compounds. In addition to PK and PD models, systems biology or systems pharmacology models [18] and machine learning based on, for example, imaging data [20] or even artificial intelligence (AI) [21,22] are important tools. Figure 1 shows the proposed MID3 strategy for the rapid development of anti-TB regimens through the prediction of human-concentration-time relationships (PK), exposure-response relationships (PK-PD) and DDIs to select FIH doses, as well as the prediction of Phase II and Phase III drug regimens. Initially in a drug development program, preclinical data is mostly available. The impact of modeling and simulation increases towards the prediction of human exposure-response. With this input efficient decision can be made about the optimal combination of different drugs, and the right dose for each drug in the combination. Currently, limited modeling and simulation are required for market approval, which relies more on statistical comparison between treatment groups after phase III [23]. However, modeling and simulation can have a role in the analysis of Phase III data in order to define the relationship between exposure and clinical endpoint, evaluate PK DDI and simulate alternative potential regimens in certain subgroups, for example, patients with renal impairment [24]. A key step for successful TB drug development is to use modeling and simulation to predict the efficacy of combinations, including DDIs, for, for example, synergy. We will review the necessary steps from this perspective for the successful MID3 application to the preclinical to clinical translation of efficacious TB drug combinations, regarding the optimal doses of drugs in complex regimens.
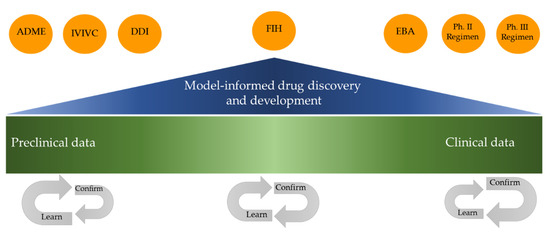
Figure 1.
Illustration of the role of Model-Informed Drug Discovery and Development (MID3) and the application across preclinical to clinical drug development.
MID3, with modeling and simulation as key tools, is suggested to be applied throughout the pre-clinical to clinical drug development phases in order to optimize and inform decision making with respect to clinical trial design and the selection of drugs and doses to be carried forward from the preclinical phase and into clinical trial programs. The prediction of human-concentration-time relationships (PK), exposure-response relationships (PK-PD) in monotherapy and combination therapy, as well as drug-drug interactions, (DDI), requires the application of MID3 techniques and integration of all available data. In early preclinical drug development, preclinical data is used for predictions using, for example, in vitro-in vivo correlation (IVIVC), physiology-based pharmacokinetics (PBPK) and a biopharmaceutics drug disposition classification system (BDDCS) in order to define absorption, distribution, metabolism, and excretion (ADME) properties. Further down the developmental process, MID3 becomes more important in order to define exposure-response relationships and pharmacodynamic (PD) interactions using preclinical data for optimal design of first-in-human (FIH) and early bactericidal activity (EBA) trials. The need to define the optimal combination regimen using preclinical information data is evident, as the necessary number of clinical trial arms/experimental groups grow exponentially with the number of drugs within a regimen. Techniques using optimal design and simulation studies are essential and part of the MID3 framework. Throughout the process, the precision of human predictions increases. Different important drug development decision steps (circles) are subject to learn-and-confirm cycles, for example, early EBA clinical studies where the earlier defined exposure-response relationship using pre-clinical data (learning phase) is confirmed (confirming phase).
2. Model-Informed Drug Discovery and Development
Model-informed drug discovery and development is given by a quantitative framework for prediction and extrapolation, aimed at improving the quality, efficiency and cost-effectiveness of decision making in drug development [17]. It can also be utilized in early drug discovery through target identification and validation, and in describing the PK-PD and toxicological properties of the candidate drug. In addition, it increases the efficiency of trials and reduces the cost through facilitating dose and sample size selection [17]. Because of the great potential of MID3, it has been received well and implemented by drug developers [18,25]. The EMA supports MID3 and has built competence to meet the increasing modeling and simulation work in the dossiers submitted to EMA through the implementation of the modeling and Simulation Working Group (MSWG). Further, the EMA stresses that, in order to benefit from the full potential of MID3, stand-alone applications of modeling and simulation, dissociated from clinical decisions with respect to the design and objectives of clinical trials, should be avoided [19]. This is also pointed out in the MID3 white paper [17], where the implementation process is described as very important, and where the modeling and simulation work should be clearly motivated in the analysis, with clear objectives that are relevant and understandable for the entire development team. To realize the full potential of MID3, it needs to be integrated into the development plan rather than being seen as an ad-hoc activity [23]. The FDA has implemented a new Model-Informed Drug Development Paired Meeting Pilot Program which refers to the application of a wide range of quantitative approaches in drug development to facilitate the decision-making process, such as dose optimization, supportive evidence for efficacy, clinical trial design, and informing policy [26]. Despite the recent efforts within academia, EFPIA and regulatory agencies, MID3 has not been utilized to its full potential within TB drug development, where the need is great due to the complex development of new drug regimens consisting of at least three drugs.
Model-informed drug discovery and development builds upon pharmacometrics, the discipline that applies mathematical and statistical methods to understand, quantify, translate, and predict PK and PD behavior, including uncertainty in that behavior [27,28]. Pharmacometric population PK and PK-PD modeling can quantify these processes to better predict the concentration-time and exposure-response relationships of anti-TB drugs as compared to non-modeling techniques, such as non-compartmental analyses (NCA) for PK or traditional statistical analysis of, for example, the relationship between dose and baseline-reduced response at the end of treatment [29] The advantage of pharmacometric modeling is that it takes the inter-individual and inter-occasion variabilities into account. Once a population model has been developed and evaluated, various simulation techniques can be used, e.g., Monte Carlo simulations where virtual patients are drawn from the earlier quantified variance of variability in the population. Pharmacokinetic models are usually nonlinear mixed-effects models with unique parameters for fixed effects and random effects. Pharmacodynamic models can consist of a statistical method suitable for the biomarker or endpoint where time-to-positivity and relapse would be described with a time-to-event model, while colony forming unit (CFU) is a continuous variable and, as such, can be described with similar nonlinear mixed effects modeling.
Model-informed drug discovery and development is likely most impactful in the translation from preclinical to clinical, where the understanding and extrapolation of the exposure-response from preclinical to clinical is crucial. Model-informed drug discovery and development is also very important in the early clinical phases of anti-TB drug development, specifically phase II EBA trials, as it is difficult to investigate all drug combinations and associated PD interactions in clinical trials. The majority of the knowledge about the potential PD interaction space needs to come from preclinical information. Additionally, MID3 can be used to design the next preclinical or clinical study in order to optimize the likelihood of collecting informative data. A crucial step in drug development is the prediction of FIH design and associated doses. Model-informed drug discovery and development strategies and methods can be used to scale preclinical information to humans to design the FIH trial. Pharmacometric techniques have been shown to reduce the sample size needed in comparison to traditional statistical methods [29,30,31], while MID3 has been reported to save significant costs through its impact on decision making [17]. Preclinical experiments should be designed to be able to quantify the exposure-response relationship, including quantitative biomarkers relative to the interspecies’ translation thereof [32]. An MID3 framework integrates all relevant preclinical and clinical information, and can therefore be used to back-translate results from the clinic to improve the preclinical understanding of the pathophysiology and pharmacology [33]. Even failed translations to humans are valuable in correcting the preclinical methods used. An iterative forward- and reverse-translational cycle has the potential to continuously enhance confidence in preclinical models [34]. The availability of large clinical datasets from, for example, electronic medical records accelerates reverse translation and improves the preclinical modeling of clinical manifestations [35]. Additionally, data from veterinary medicine can be utilized to guide human medicine development [36]. For this framework to really have an impact, data repositories and common languages are essential for application across different disciplines, disease areas, or stages of development [23,37]. In addition, to ensure that modeling and simulation adds value through an MID3 approach, pharmacometricians must communicate with their project teams before any data analysis starts to understand the key strategic development questions, clinical context, available data, assumptions, and decision criteria [23].
The prediction of efficacy and safety in new drug combinations with new or unknown mechanisms of action will benefit from the next paradigm in drug development and MID3, namely quantitative systems pharmacology (QSP). This is the pharmacological perspective on a systems’ modeling, a body-system-wide characterization of the health and disease of an organism based on a mechanistic and molecular understanding of the individual components in the context of the holistic network [38]. QSP is the middle-out interface between systems biology and pharmacometrics, describing the pharmacological perturbation within the studied context [39]. It accounts for differences in (molecular) mechanisms of a disease [40], which is very relative for TB with its heterogeneous pathophysiology of acute, chronic, and latent infections. Because of its mathematical description of all the relevant elements of the pharmacological and pathophysiological pathways, and their differences between species, it becomes key to translational medicine [41]. Because of this quantitative understanding of the network, the prediction of the effects of drugs with new mechanisms of action improves significantly [42]. The development of QSP models in the preclinical phase is, however, uncommon, and the dedicated acquisition of experimental data like transcriptomics or metabolomics for the development of QSP models is rare [43,44]. Quantitative systems pharmacology models are intended to be applied to a wider scale than the individual questions or problems they were originally developed for [45]. For TB specifically, this could mean a systems model of the M. tuberculosis infection in the human context of macrophage infiltration, granuloma formation and pulmonary lesion development, with all relevant pathways and drug targets quantitatively described. The effect of new combinations, including drugs with novel mechanism of actions, can be predicted.
3. Prediction of Human Pharmacokinetics
In silico ADME-PK (absorption, distribution, metabolism, excretion, and pharmacokinetics) is the use of computer modeling to understand structure−property relationships and to predict DMPK (drug metabolism and pharmacokinetics) properties from compound structure. This is related to but distinct from physiologically based pharmacokinetic (PBPK) modeling, which strives to provide accurate predictions of the PK profile of drug candidates [46]. The focus of in silico ADME-PK is to guide the design of novel compounds with superior ADME properties. Most often a quantitative structure−property relationship (QSPR) approach is used to relate a compound’s structure to the chemical property in question (e.g., cell permeability or metabolic clearance) measured in an in vitro assay. Related terms are also quantitative-structure-activity relationships (QSAR), when a set of predictor variables is related to the potency of the drug.
Orally administered products are subject to a sequence of transport and enzymatic barriers in enterohepatic systems affecting bioavailability, including extraction in the intestinal and liver tissues, which could impact the fraction of the orally administered dose that reaches the systemic circulation and thereby the site of action. Bioavailability is mainly dependent on three general and rather complex serial processes: the fraction of the oral dose that is absorbed (Fabs), the first-pass extraction of the drug in the gut wall (EG), and the first-pass extraction of the drug in the liver (EH) [47]. In general, oral products with a low F (<25%–35%) have a higher inter- and intra-individual variability in plasma exposure (coefficient of variation >60%–120%) [48]. Drugs with high degree of Fabs show sufficiently high solubility of the active pharmaceutical ingredient (API), no luminal degradation, and absorption along the small and/or large intestine [49,50]. The regulatory framework Biopharmaceutics Classification System (BCS) of drugs provides information relevant to understanding and predicting GI drug absorption and bioavailability in general, which is relevant to the absorption potential in the small and large intestine [51]. After the drug is absorbed, it passes to the liver, which expresses a broader range of different enzymes compared to the intestine [52], such as the family of CYP enzymes [53]. Other enzymes such as microsomal uridine 5’-diphospho-glucuronosyltransferases (UGTs), sulfotransferases, and glutathione S-transferases are found in great amounts in the human liver [54]. Humans show large inter-individual variation in the amount of different enzymes, which accounts, in part, for the large inter-individual variation reported for EH and CL. A considerable large inter-individual variation in the expression of the different CYP isoforms has also been observed, ranging from 20-(CYP2E1 and CYP3A4) to >1000-fold (CYP2D6). The liver is also the major organ for glucuronidation in the body. Glucuronide conjugates have molecular characteristics that are associated with biliary excretion of a compound, i.e., a high molecular weight, ionized and the presence of polar groups [55].
The identification and quantification of the important PK processes described above can be investigated in relevant in vitro models, and predictions of PK properties like regional intestinal permeability can be made early [56]. Most importantly, an estimate of human drug clearance will determine how fast a drug is eliminated, and conversely define the dose range to study in FIH [57]. Several different models have been suggested for the prediction of oral absorption for the biopharmaceutical design of oral drug delivery systems [58,59]. The proposed BDDCS has been shown to be useful in predicting some crucial ADME parameters and especially the transport/absorption/elimination interplay [60]. Preclinical data can then be translated through in vitro-in vivo extrapolation, or even through PBPK modeling to generate basic PK parameters such as fraction of dose absorbed, bioavailability, clearance (CL), volume of distribution, and terminal half-life. Furthermore, the accuracy of the QSAR predictions of effective intestinal permeability (Peff), is significantly improved when based on a combination of molecular physicochemical descriptors and molecular dynamics simulations from in vitro data [61]. Molecular simulations have been successfully used to predict the effects of cholesterol in the lipid membrane fluidity [62]. Additionally, molecular simulations have been reported to be useful as they are comparable to experimental data [63]. Among the molecular descriptors evaluated by Lipinski (e.g., polar surface area, hydrogen bond donors (HBDs)/acceptors, Log D), the number of HBDs is the most restrictive when it comes to intestinal membrane transport/absorption [64,65]. Two drugs violating this rule (i.e., >5 HBDs and low intestinal Peff and Fabs), one of which is rifampicin, have been investigated thoroughly and offered a potential explanation for drug absorption beyond the Lipinski Rule-of-five [66]. Based on a liposomal permeation assay, it has been proposed that drug molecules with more than five HBD can be sufficiently absorbed in the intestine by passive lipoidal diffusion [66]. Some drugs are absorbed by passive lipoidal diffusion despite their unfavorable physicochemical properties. It is therefore necessary to find more complex descriptions of the molecular interaction by applying a combination of experimental data and molecular dynamic modeling and simulation to further improve the accuracy in predicting general membrane transport across the cellular membrane barrier and not only in the GI-tract [66,67,68,69].
Physiologically based pharmacokinetic modeling is considered to assist drug product development by providing quantitative predictions through a systems approach [70]. A mechanism-based model, like that of the PBPK approach, separates drug-specific from system-specific elements, which allows for the interspecies translation of the time course of the drug [41]. Physiologically based pharmacokinetic models divide the body into anatomically and physiologically meaningful compartments, including the gastrointestinal tract for absorption, the eliminating organs, and non-eliminating tissue compartments [71]. In addition, compound-specific parameters such as physicochemical and biochemical parameters (e.g., tissue/blood partitioning and metabolic CL) are incorporated into the model to predict the plasma and tissue concentration versus the time profiles of a compound in an in vivo system following intravenous or oral administration. Translation between species, special populations, or disease states, are the result of changing these physiological parameters accordingly. Several variations are in use, including a whole-body PBPK model describing the complete organism, and hybrid PBPK models, combining PBPK elements with empirical compartmental PK to simplify the model [72]. An important element in the physiologically based translation of PK is binding of the drug of interest to proteins in the plasma, mainly albumin, lipo- or glycoproteins, or globulins. Protein binding differs between experimental settings (in vitro) and species (in vivo) and should be taken into account because it influences tissue penetration and the free drug that can interact with its target [73]. Physiologically based pharmacokinetic modeling predictions are a valuable tool in the pharmaceutical industry due to the possibility of combining all the available and relevant information that is generated during the preclinical stage, which helps improve decision making during the selection process [74]. For instance, biorelevant dissolution-absorption PBPK modeling and simulation has been reported as applied in 88% of early drug development processes [75]. Moreover, PBPK modeling provides a powerful tool to study potential PK DDIs through incorporating the drug’s physicochemical properties, PK properties, human physiological variables, and inter-individual variability estimates.
Predictions of PK parameters use information from preclinical studies in animals to transition to clinical trials, i.e., FIH studies, when no clinical information is available to guide the decision of the starting dose. Thus, the estimation of the starting dose in human subjects relies on the PK knowledge of the drug from different species. It is essential to have preclinical PK data based on blood, plasma, and/or tissue sampled longitudinally to optimally capture the complete drug profile during a dosing interval. A model-based approach to the starting dose often uses allometric scaling to predict human drug clearance and distribution volumes. Allometric scaling is based on the assumption that physiological similarities exist between different species arising from anatomical similarities, specifically similarities in body weights and body surface areas [76]. Historically, a maximum dose for FIH studies was based on the no observed adverse effect level (NOAEL) in preclinical experiments, an arbitrary safety factor, and allometric scaling. This approach is empirical by nature and therefore limited. When more mechanistic data and models are available, a minimal anticipated biological effect level (MABEL) can be estimated [42]. For example, preclinical PK-PD and interspecies differences in the target can be utilized to estimate the MABEL for a FIH trial [77]. This has the benefit of being driven by pharmacology, where FIH trials will answer pharmacological questions on PK and PD, rather than being driven by toxicology or tolerability. Taking into account MABEL and safety factors, a first study with single ascending doses (SAD study) will quantify the PK and ensure safety and tolerability. A second study with multiple administered doses (MAD) can subsequently be designed accordingly [78].
In addition to allometric scaling, the use of IVIVC has markedly increased [79,80,81,82]. It is suggested that animal PBPK models should be used as part of a stepwise approach, in which the first step uses animal data to understand the processes and verify the predictive power of in vitro systems, and the second step is about forecasting human PK from in vitro data and in silico methods (learn-and-confirm) [82]. The first step in predicting drug CL using IVIVC is to obtain intrinsic CL (CLint) from in vitro data [83,84]. In vitro CLint values determined from various systems including hepatocytes, cell transport models, liver or intestinal microsomes, or recombinant CYPs, either by substrate depletion or metabolite formation, are normalized for cell, microsomal protein or enzyme concentration. The next step consists of scaling the activity determined in vitro to the whole liver by the use of a scaling factor, to account for incomplete microsomal recovery from the tissue to obtain in vivo CLint. Finally, the third step involves the use of a liver model which incorporates the effects of hepatic blood flow, plasma protein-binding and blood cell partitioning to convert the estimated in vivo CLint into a hepatic CL (CLH). The well-stirred liver model is most commonly used, but the dispersion model or the parallel tube model is also available [84].
Using high doses of oral anti-TB drugs may result in high plasma concentrations, leading to an increased risk of adverse effects [85] while not ensuring adequate concentrations at the site of action [85,86]. This has prompted investigation into the use of the pulmonary route to deliver anti-TB drugs directly to the site of action in the lungs. Administering anti-TB drugs as inhaled formulations ensures the delivery of the drug directly to the target organ, avoiding any unwanted systemic side effects, thereby improving patient compliance [87]. Optimal pulmonary drug delivery for locally acting drugs includes a high local lung concentration, extended lung residence time and low systemic concentration [88]. A fundamental understanding of pulmonary dissolution, residence time, and lung absorption processes is key for the successful development of inhaled products [89,90]. However, inhaled formulations have many challenges, including formulation stability, pulmonary distribution, lung toxicity, and additives safety [91]. Furthermore, dosing of inhaled drugs is more complicated than other routes of administration as their absorption in the lung is highly variable [91]. Thus, many in vitro and in vivo models have been developed to study the PK of inhaled anti-TB drugs, specifically their absorption and distribution, in order to evaluate the efficacy and safety of anti-TB drugs in the lungs. These models, while allowing a reduction in biological complexity, still face many challenges, and can be demanding to build [92]. In vivo animal models are the gold standard regarding the assessment of drug clearance, systemic side effects and PK after pulmonary administration [92]. However, animal models are not always able to mimic human pulmonary anatomy and physiology, or TB disease progression in humans, and they do not exhibit extrapulmonary dissemination similar to humans [86]. Translation from animal data to the clinic has been recognized as challenging [93]. Understanding the pulmonary exposure is important, and animal data can contribute specific information about the lesion to plasma ratio [94], as similar lung distribution ratios can be obtained in human. However, little is known about the factors that influence drug distribution from plasma into the range of tissues, nodules and cavities that are inhabited by the TB pathogen. Pulmonary TB lesions consist of a diversity of cell types, tissue structures and vascular architectures which suggests that the distribution of the drug is not only governed by passive equilibration between unbound drug concentration in plasma and tissue [95]. MALDI mass spectrometry imaging (MALD-MSI) is a new technique to study the distribution of small molecules in the various compartments of pulmonary lesions [96]. Information from such studies not only provides knowledge of regional differences in drug exposure, but also confirms a high exposure in regions where a high density of persistent TB bacteria is found.
4. Prediction of Human Pharmacokinetic-Pharmacodynamic Relationship
In order to translate drug effects from preclinical information to the clinical phase of drug development, defining a drug exposure-response relationship using preclinical information is of importance. The PK-PD relationship is quantitative, predictive, and reproducible and is valid in all disease models [57,58]. Thus, characterizing this relationship is of great benefit in preclinical PK-PD studies to help guide dose selection and study design in humans. Exhaustive reviews of preclinical experimental methods that quantify exposure-response relationships have previously been performed [97,98]. These methods, such as classical time-kill experiments, hollow-fiber system (HF), different murine models, rabbits and guinea pig, all mimic elements of the human pathology to a certain extent, but all have their limitations. Here, the focus is on their informativeness of the exposure-response relationship for translation to human prediction.
In vitro determination of the minimum inhibitory concentration (MIC) is informative about the sensitivity of the bacterial strain to the compound. This is especially the case when the target sizes of the M. tuberculosis infection, macrophages, are utilized as environmental context [97]. The MIC is a measure of the net effect of the drug on bacterial growth and survival. However, it is very crude and undynamic as it is measured at a specific concentration and after a fixed time, which might cause it to deviate from the true MIC [99]. The MIC is also limited because the resolution is determined by the chosen dilution steps, and bigger dilution steps increase the risk of under- or overestimating the MIC. In addition, the determination of MIC is based on visual inspection which makes it prone to subjective error [99]. Mouton et al. have studied the variability between MIC measurements in Staphylococcus aureus treated with linezolid and have concluded that over half of the variability in the MIC measurements is either due to systemic and significant inter-laboratory differences or differences between strains [100]. The other half can be explained by assay variation and different environmental conditions, such as the media used and incubation temperature [100].
Several preclinical animal models for TB are in use. The advantage of an animal model over in vitro systems is the holistic environment of a whole organism, including a functioning immune system, physiological feedback systems and (drug) disposition. This results in more variability in the determination of the exposure-response relationship and requires more effort to elucidate drug effect from, for example, the immune system. The most emphasis is placed on murine models of TB [101], although there are arguments that the mouse is not a good model for TB in humans [102]. Mice can be housed in the required biosafety laboratories with ease, blood and tissue sampling is well established, and both chronic and latent infections have been successfully used [103]. However, the mice have a low susceptibility to M. tuberculosis and show only loosely organized granulomas, and are therefore limited when considering lesion-specific treatment. Granuloma formation in guinea pigs and rabbits is more representative of human granulomas, including caseous necrosis [102]. Guinea pigs are highly susceptible to M. tuberculosis which makes infection as straightforward as exposure to exhaustion from TB patients [103]. Rabbits are also utilized to study a slower response to treatment, disease relapse, and resistance development due to lung cavities, and their size makes studying drug distribution to TB lesions more feasible [98]. The experimental toolbox regarding immunologic reagents and genetic techniques is, however, more restricted in these animals, and both need more difficult and expensive husbandry. Granuloma formation can also be studied non-invasively in the zebrafish, a relatively new disease model organism in drug discovery and development [103]. Because of their transparency and easy genetic modification, fluorescence microscopy of pathogen and immune cells can be leveraged to follow infection and treatment [104]. With the small size and high fecundity of the zebrafish, high-throughput assays are available to test large numbers of compounds in short amounts of time with enough statistical power [105]. Methods to quantify internal drug exposure have also been established [106,107,108]. Recently, an exposure-response relationship has been developed for isoniazid in the zebrafish, which translated well to humans [109]. In general, non-invasive imaging of lesion pathology by computed tomography (CT) and positron emission tomography (PET) has the potential to improve the comparison between preclinical and clinical measurements of disease progression and treatment [102]. Ordonez et al have demonstrated this by using dynamic [11C]rifampicin PET-CT imaging in patients newly diagnosed with pulmonary TB and rabbits infected with cavitary TB to noninvasively measure intralesional drug concentration-time profiles and, consequently, time to bacterial extinction [110]. They also employed integrated modeling of the PET-captured concentration-time profiles in hollow-fiber bacterial kill curve experiments to predict the rifampicin dose required to achieve a cure in 4 months, which has a huge potential in antimicrobial drug development to shorten TB treatments [110]. It is clear that no single animal model represents a heterogeneous disease such as TB. A mechanistic understanding of TB in humans will identify which elements are characterized best by which animal model [103]. Independently of which preclinical experimental method is utilized, the sampling design of both PK (e.g., drug and/or metabolite concentration) and PD (e.g., infection, bacterial burden) biomarkers is of the utmost importance. The careful selection of datapoints over the duration of the experiment and at different drug concentrations is essential for a reliable quantification of the exposure-response relationship.
Regulatory agencies suggest determining PK/PD indices based on preclinical data for antibiotics, e.g., the area under the concentration curve over MIC (AUC/MIC), the maximum concentration (Cmax) over MIC (Cmax/MIC), and the percent of a 24-hour time period that the drug concentration is above MIC (T > MIC), for the establishment of the PK-PD profile of antimicrobials and for deciding the most optimal dosing regimens. PK/PD indices are based on preclinical studies that describe the PK-PD relationships of antimicrobials [111]. However, PK/PD indices suffer from several clear limitations, some of which are inherent to their use of MIC, the limitations of which are discussed above. Using PK/PD indices ignores information about the time-course of individual PK and PD processes [112]. As summary endpoints, they lack the ability to track the changes in the bacterial load over time [113]. Furthermore, when using AUC/MIC as a PK/PD index, the rate of drug administration is ignored, while, when using Cmax/MIC, bacterial killing is assumed to depend solely on the maximum drug concentration, ignoring drug half-life and infusion duration [99]. Using T > MIC assumes that the maximal drug effect has been reached when MIC is reached, regardless of whether higher concentrations were given [99]. Additionally, the colony-forming units (CFU) versus PK/PD indices profile shows great variability in the CFU observations for the same PK/PD indices value [99]. These PK/PD indices are selected and predicted as PD targets using HFS-TB to quantify a more realistic in vitro exposure-response relationship that is translatable to in vivo [98,99,114]. However, despite EMA’s qualification of the preclinical HFS-TB to be used to complement existing methodologies, it still suffers from a number of limitations. The EMA advises caution when interpreting HFS-TB results, as many instances of over- and under-estimates of the drug’s anti-TB activity have been reported [115]. In addition, HFS-TB cannot replace animal models or clinical studies [116], while the reproducibility of the method by other laboratories has not yet been assessed [115].
Mechanistic, or semi-mechanistic, PK-PD models in TB based on preclinical data allow for the description of the multiple mycobacterial populations present. A mechanism-based PK-PD model by Hollow-fiber systems for TB has the advantage of being able to mimic dynamic PK in comparison to more traditional static time-kill experiments. A semi-mechanistic PK-PD model can be derived using HTS data [117,118,119]. Khan et al. describes susceptible, resting, and non-colony-forming bacterial populations [120]. The multistate tuberculosis pharmacometric (MTP) model is a semi-mechanistic mathematical model that can describe and identify the exposure-response profile of a drug towards three bacterial subpopulations: fast-, slow-, and non-multiplying bacteria. It has been successfully applied to describe in vitro [121], mouse [122], and clinical data [123]. In addition, the MTP model has been successfully used in an MID3 approach, to predict observations from early clinical studies using clinical dose-response forecasting from preclinical in vitro studies of rifampicin and in combination with isoniazid [15,16]. This model has been selected by The Impact and Influence Initiative of the Quantitative Pharmacology (QP) Network of the American society of Clinical Pharmacology and Therapeutics (ASCPT) to highlight the most impactful examples of QP applications where the role of quantitative translational pharmacology has bridged science and practice to make better, faster, and more efficient decisions in drug discovery and development [25]. Another mechanism-based model is the Magombedze et al. model that mimics the disease state in TB patients by describing the mycobacterial population as logarithmic growth-phase, semi-dormant, and persister bacilli [117]. In addition, a pulmonary PK-PD model of isoniazid has been developed to better characterize the relationship between its PK and its anti-TB effects in the lungs [124].
5. Prediction of Human Drug-Drug Interactions
Tuberculosis requires a combination therapy of three different antibiotics or more, which increases the risk of DDIs. Drug-drug interactions between drugs that are intended to be used in combination should be considered as early as possible. The prediction of DDIs from preclinical data will improve the ability to predict the total efficacy of the combination in relation to the drugs in monotherapy, as well as compared to expected additivity, i.e., the sum of all effects from the drugs when given alone. DDIs that result in less efficacy in the combination than in a combination with one less drug should be avoided. However, combinations that result in an efficacy less than the expected additivity, but still result in more efficacy than when one drug is omitted, can be considered. Drug-drug interactions can relate to both PK interactions, i.e., one drug (the perpetrator) impacting the absorption, distribution, metabolism, or excretion of another drug (the victim), or PD interactions, i.e., the perpetrator impacting the potency or efficacy of the victim drug.
Regulatory guidelines on the investigation of DDIs are brief about the use of in vitro data, while in an MID3 context, knowledge on the relevant mechanisms of, e.g., metabolism combined with in vitro data can be leveraged to decide on suitable combinations of drugs without extensive experimentation [125]. Both in vitro studies as well as animal experiments can be utilized to assess the potential for PK DDIs [126]. In vitro studies make use of metabolically active hepatocytes or cells overexpressing drug transporters to determine the PK interaction potential of a new drug [127]. When studying DDIs in preclinical species, the between-species differences in transporters or enzymes should be taken into account [128]. Pharmacokinetic DDIs mostly impact drug clearance by the induction or inhibition of metabolic enzymes like those from the CYP family and, to some extent, ABC and transport proteins. Such an interaction by the perpetrator drug will greatly enhance or reduce the exposure of the victim drug. For example, rifampicin induces bedaquiline clearance 5-fold, and should therefore not be combined for therapy [129]. Because bedaquiline has a very long terminal half-life, potential DDIs are difficult to identify using traditional methods, whereas properly designed experiments and quantitative modeling are necessary to elucidate such interactions [130]. Drug distribution can also be impacted because of the induction or inhibition of drug transporters like the permeability glycoprotein (P-gp), which is present on the canalicular membrane and blood-brain barrier, among others. Physiology based pharmacokinetic modeling can be very successful to predict metabolic DDIs, and specific DDI studies can be assisted by modeling and simulations [131]. Some anti-TB drugs are reported to be substrates for different hepatic enzymes or known to be inducers or inhibitors of metabolic enzymes. Rifampicin is well known as a CYP3A4 modulator [132,133], as well as an inducer of P-gp [134]. Additionally, even though the effect of clofazimine on CYP3A4 and P-gp is still unclear, clofazimine has been shown to delay the time taken to reach Cmax of rifampicin [135]. Horita et al. studied the effects of anti-TB and antiretroviral drugs on CYP3A4 and P-gp, and they found that clofazimine exhibits weak inductive effects on CYP3A4 [136]. Furthermore, the co-administration of bedaquiline and clofazimine has been reported to increase the risk of QT prolongation [137,138]. As described above, these potential DDIs can be predicted from in vitro data through, for example, in vitro-in vivo scaling [139] or PBPK [140]. A transcription/translation model and a PBPK model have been developed to predict rifampicin-induced DDIs with reasonable accuracy [141].
In contrast to PK interactions, due to clearly defined processes of absorption, distribution, metabolism, and excretion, PD interactions are harder to investigate and quantify. This is because, since a clinical DDI study has to study the drugs both alone and in combination, the number of arms in the study will substantially increase when studying three or more interacting drugs. The Greco model [142], which is derived from Loewe additivity, was developed to assess PD interactions. However, such a model suffers from being limited to interactions between only two drugs. On the other hand, the general pharmacodynamic interaction (GPDI) model overcomes this limitation, in addition to being flexible to different drug interaction data without requiring knowledge on the modes of action of the studied drugs [143]. The GPDI model-based approach proposes a PD interaction to be quantifiable, as multidirectional shifts in drug efficacy (Emax) or potency (EC50) and explicates the drugs’ role as victim, perpetrator or even both at the same time. The GPDI model has been utilized along with the MTP model [121] to develop a model-informed preclinical approach for the prediction of PD interactions [144]. The MTP-GPDI model has been further employed to successfully evaluate and quantify the PD interactions of anti-TB drug combinations in mice [145]. Furthermore, it has been demonstrated that the GPDI model outperforms conventional methods in the evaluation of PD interactions for TB drugs [146].
It is clear that the need for a combination therapy of TB could potentially result in DDIs in the clinic. It is therefore essential to quantitatively understand the DDIs, both PK- and PD-interactions, as early as possible in drug development. Utilizing data from in vitro combination experiments combined with preclinical in vivo data on the exposure-response relationships of the drugs in combination and early clinical data, will inform on which combinations of drugs at which doses are efficacious and safe for patients. This quantitative integration of data and translation to the clinic is possible through the MID3 model-informed framework.
6. Conclusions
The development of new combinations of anti-TB drugs is both promising and challenging. Novel drug combinations and drug delivery routes require novel and innovative techniques. Model-informed drug discovery and development is an integrated framework of preclinical and clinical data through translational models that show great promise in selecting and predicting which drug regimens to carry forward to be evaluated in clinical trials. The MID3 framework supports decision making in drug development in relation to the prediction of efficacious and safe combinations of new drugs and translates this to the clinic. It is essential for drug developers to collaborate across disciplines, and academic and industry borders and train a new type of scientist in experimental and computational innovation.
Author Contributions
All authors contributed to conceptualization of this work. All authors contributed to original draft preparation, review and editing. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Conflicts of Interest
The authors declare no conflict of interest.
References
- DiMasi, J.A.; Grabowski, H.G.; Hansen, R.W. Innovation in the Pharmaceutical Industry: New Estimates of R & D Costs. J. Health Econ. 2016, 47, 20–33. [Google Scholar] [CrossRef]
- Bellanti, F.; Van Wijk, R.C.; Danhof, M.; Della Pasqua, O. Integration of PKPD Relationships into Benefit-Risk Analysis. Br. J. Clin. Pharmacol. 2015, 80, 979–991. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. Global Tuberculosis Report 2019; World Health Organization: Geneva, Sweitzerland, 2019. [Google Scholar]
- Boeree, M.J.; Diacon, A.H.; Dawson, R.; Narunsky, K.; Du Bois, J.; Venter, A.; Phillips, P.P.J.; Gillespie, S.H.; McHugh, T.D.; Hoelscher, M.; et al. A Dose-Ranging Trial to Optimize the Dose of Rifampin in the Treatment of Tuberculosis. Am. J. Respir. Crit. Care Med. 2015, 191, 1058–1065. [Google Scholar] [CrossRef] [PubMed]
- Svensson, E.M.; Svensson, R.J.; Te Brake, L.H.M.; Boeree, M.J.; Heinrich, N.; Konsten, S.; Churchyard, G.; Dawson, R.; Diacon, A.H.; Kibiki, G.S.; et al. The Potential for Treatment Shortening with Higher Rifampicin Doses: Relating Drug Exposure to Treatment Response in Patients with Pulmonary Tuberculosis. Clin. Infect. Dis. 2018, 67, 34–41. [Google Scholar] [CrossRef] [PubMed]
- Svensson, R.J.; Svensson, E.M.; Aarnoutse, R.E.; Diacon, A.H.; Dawson, R.; Gillespie, S.H.; Moodley, M.; Boeree, M.J.; Simonsson, U.S.H. Greater Early Bactericidal Activity at Higher Rifampicin Doses Revealed by Modeling and Clinical Trial Simulations. J. Infect. Dis. 2018, 218, 991–999. [Google Scholar] [CrossRef]
- Susanto, B.O.; Svensson, R.J.; Svensson, E.M.; Aarnoutse, R.; Boeree, M.J.; Simonsson, U.S.H. Rifampicin Can Be given as Flat-Dosing Instead of Weight-Band Dosing. Clin. Infect. Dis. 2019. [Google Scholar] [CrossRef]
- WHO. Consolidated Guidelines on Drug-Resistant Tuberculosis Treatment; Licence: CC BY-NC-SA 3.0 IGO; World Health Organization: Geneva, Sweitzerland, 2019. [Google Scholar]
- Bahuguna, A.; Rawat, D.S. An Overview of New Antitubercular Drugs, Drug Candidates, and Their Targets. Med. Res. Rev. 2020, 40, 263–292. [Google Scholar] [CrossRef]
- Pontali, E.; Sotgiu, G.; Tiberi, S.; Tadolini, M.; Visca, D.; D’Ambrosio, L.; Centis, R.; Spanevello, A.; Migliori, G.B. Combined Treatment of Drug-Resistant Tuberculosis with Bedaquiline and Delamanid: A Systematic Review. Eur. Respir. J. 2018, 52. [Google Scholar] [CrossRef]
- European Medicines Agency. Addendum to the Guideline on the Evaluation of Medicinal Products Indicated for Treatment of Bacterial Infections to Address the Clinical Development of New Agents to Treat Pulmonary Disease Due to Mycobacterium Tuberculosis; European Medicines Agency: London, UK, 2017.
- European Medicines Agency. Addendum to the Note for Guidance on Evaluation of Medicinal Products Indicated for Treatment of Bacterial Infections to Specifically Address the Clinical Development of New Agents to Treat Disease Due to Mycobacterium Tuberculosis; European Medicines Agency: London, UK, 2010.
- Dheda, K.; Gumbo, T.; Maartens, G.; Dooley, K.E.; Murray, M.; Furin, J.; Nardell, E.A.; Warren, R.M.; Esmail, A.; Nardell, E.; et al. The Lancet Respiratory Medicine Commission: 2019 Update: Epidemiology, Pathogenesis, Transmission, Diagnosis, and Management of Multidrug-Resistant and Incurable Tuberculosis. Lancet Respir. Med. 2019, 7, 820–826. [Google Scholar] [CrossRef]
- European Medicines Agency. Guideline on Strategies to Identify and Mitigate Risks for First-in-Human and Early Clinical Trials with Investigational Medicinal Products; European Medicines Agency: London, UK, 2017.
- Wicha, S.G.; Clewe, O.; Svensson, R.J.; Gillespie, S.H.; Hu, Y.; Coates, A.R.M.; Simonsson, U.S.H. Forecasting Clinical Dose-Response From Preclinical Studies in Tuberculosis Research: Translational Predictions With Rifampicin. Clin. Pharmacol. Ther. 2018, 104, 1208–1218. [Google Scholar] [CrossRef]
- Susanto, B.O.; Wicha, S.G.; Hu, Y.; Coates, A.R.M.; Simonsson, U.S.H.; Biosciences, P.; Pharmacy, C.; Kingdom, U.; Simonsson, U.S.H.; Biosciences, P.; et al. Translational Model-Informed Approach for Selection of Tuberculosis Drug Combination Regimens in Early Clinical Development. Clin. Pharmacol. Ther. 2020. [Google Scholar] [CrossRef] [PubMed]
- Marshall, S.F.; Burghaus, R.; Cosson, V.; Cheung, S.; Chenel, M.; DellaPasqua, O.; Frey, N.; Hamrén, B.; Harnisch, L.; Ivanow, F.; et al. Good Practices in Model-Informed Drug Discovery and Development: Practice, Application, and Documentation. CPT Pharmacomet. Syst. Pharmacol. 2016, 5, 93–122. [Google Scholar] [CrossRef]
- Marshall, S.; Madabushi, R.; Manolis, E.; Krudys, K.; Staab, A.; Dykstra, K.; Visser, S.A.G. Model-Informed Drug Discovery and Development: Current Industry Good Practice and Regulatory Expectations and Future Perspectives. CPT Pharmacomet. Syst. Pharmacol. 2019, 8, 87–96. [Google Scholar] [CrossRef]
- Manolis, E.; Brogren, J.; Cole, S.; Hay, J.L.; Nordmark, A.; Karlsson, K.E.; Lentz, F.; Benda, N.; Wangorsch, G.; Pons, G.; et al. Commentary on the MID3 Good Practices Paper. CPT Pharmacomet. Syst. Pharmacol. 2017, 6, 416–417. [Google Scholar] [CrossRef]
- Goulooze, S.C.; Zwep, L.B.; Vogt, J.E.; Krekels, E.H.J.; Hankemeier, T.; Van den Anker, J.N.; Knibbe, C.A.J. Beyond the Randomized Clinical Trial: Innovative Data Science to Close the Pediatric Evidence Gap. Clin. Pharmacol. Ther. 2020. [Google Scholar] [CrossRef]
- Ribba, B. Model-Informed Artificial Intelligence: Reinforcement Learning for Precision Dosing. Clin. Pharmacol. Ther. 2020. [Google Scholar] [CrossRef]
- Harrer, S.; Shah, P.; Antony, B.; Hu, J. Artificial Intelligence for Clinical Trial Design. Trends Pharmacol. Sci. 2019, 40, 577–591. [Google Scholar] [CrossRef]
- Krishnaswami, S.; Austin, D.; Della Pasqua, O.; Gastonguay, M.R.; Gobburu, J.; van der Graaf, P.H.; Ouellet, D.; Tannenbaum, S.; Visser, S.A.G. MID3: Mission Impossible or Model-Informed Drug Discovery and Development? Point-Counterpoint Discussions on Key Challenges. Clin. Pharmacol. Ther. 2020. [Google Scholar] [CrossRef]
- Marshall, S.F.; Hemmings, R.; Josephson, F.; Karlsson, M.O.; Posch, M.; Steimer, J.L. Modeling and Simulation to Optimize the Design and Analysis of Confirmatory Trials, Characterize Risk-Benefit, and Support Label Claims. CPT Pharmacomet. Syst. Pharmacol. 2013, 2, 3–5. [Google Scholar] [CrossRef]
- Gupta, N.; Bottino, D.; Simonsson, U.S.H.; Musante, C.J.; Bueters, T.; Rieger, T.R.; Macha, S.; Chenel, M.; Fancourt, C.; Kanodia, J.; et al. Transforming Translation Through Quantitative Pharmacology for High-Impact Decision Making in Drug Discovery and Development. Clin. Pharmacol. Ther. 2019, 1–5. [Google Scholar] [CrossRef]
- Wang, Y.; Zhu, H.; Madabushi, R.; Liu, Q.; Huang, S.M.; Zineh, I. Model-Informed Drug Development: Current US Regulatory Practice and Future Considerations. Clin. Pharmacol. Ther. 2019, 105, 899–911. [Google Scholar] [CrossRef] [PubMed]
- Gieschke, R.; Steimer, J.L. Pharmacometrics: Modelling and Simulation Tools to Improve Decision Making in Clinical Drug Development. Eur. J. Drug Metab. Pharmacokinet. 2000, 25, 49–58. [Google Scholar] [CrossRef] [PubMed]
- Ette, E.I.; Williams, P.J. Pharmacometrics: The Science of Quantitative Pharmacology. John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2007; ISBN 978-0-471-67783-3. [Google Scholar]
- Svensson, R.J.; Gillespie, S.H.; Simonsson, U.S.H. Improved Power for TB Phase IIa Trials Using a Model-Based Pharmacokinetic-Pharmacodynamic Approach Compared with Commonly Used Analysis Methods. J. Antimicrob. Chemother. 2017, 72, 2311–2319. [Google Scholar] [CrossRef] [PubMed]
- Karlsson, K.E.; Vong, C.; Bergstrand, M.; Jonsson, E.N.; Karlsson, M.O. Comparisons of Analysis Methods for Proof-of-Concept Trials. CPT Pharmacomet. Syst. Pharmacol. 2013, 2. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Ortega, F.; Alameda, L.; Ferrer, S.; Simonsson, U.S.H. Population Pharmacokinetics, Optimised Design and Sample Size Determination for Rifampicin, Isoniazid, Ethambutol and Pyrazinamide in the Mouse. Eur. J. Pharm. Sci. 2016, 93, 319–333. [Google Scholar] [CrossRef]
- Muliaditan, M.; Davies, G.R.; Simonsson, U.S.H.; Gillespie, S.H.; Della Pasqua, O. The Implications of Model-Informed Drug Discovery and Development for Tuberculosis. Drug Discov. Today 2017, 22, 481–486. [Google Scholar] [CrossRef]
- Visser, S.A.G.; Aurell, M.; Jones, R.D.O.; Schuck, V.J.A.; Egnell, A.C.; Peters, S.A.; Brynne, L.; Yates, J.W.T.; Jansson-Löfmark, R.; Tan, B.; et al. Model-Based Drug Discovery: Implementation and Impact. Drug Discov. Today 2013, 18, 764–775. [Google Scholar] [CrossRef]
- Kasichayanula, S.; Venkatakrishnan, K. Reverse Translation: The Art of Cyclical Learning. Clin. Pharmacol. Ther. 2018, 103, 152–159. [Google Scholar] [CrossRef]
- Li, L. Reverse Translational Pharmacology Research Is Driven by Big Data. CPT Pharmacomet. Syst. Pharmacol. 2018, 7, 63–64. [Google Scholar] [CrossRef]
- Schneider, B.; Balbas-Martinez, V.; Jergens, A.E.; Troconiz, I.F.; Allenspach, K.; Mochel, J.P. Model-Based Reverse Translation Between Veterinary and Human Medicine: The One Health Initiative. CPT Pharmacometrics Syst. Pharmacol. 2017. [Google Scholar] [CrossRef]
- Van der Graaf, P.H.; Benson, N. The Role of Quantitative Systems Pharmacology in the Design of First-in-Human Trials. Clin. Pharmacol. Ther. 2018, 104, 797. [Google Scholar] [CrossRef] [PubMed]
- van der Greef, J.; McBurney, R.N. Rescuing Drug Discovery: In Vivo Systems Pathology and Systems Pharmacology. Nat. Rev. Drug Discov. 2005, 4, 961–967. [Google Scholar] [CrossRef] [PubMed]
- Vicini, P.; Van der Graaf, P.H. Systems Pharmacology for Drug Discovery and Development: Paradigm Shift or Flash in the Pan? Clin. Pharmacol. Ther. 2013, 93, 379–381. [Google Scholar] [CrossRef] [PubMed]
- Danhof, M.; Klein, K.; Stolk, P.; Aitken, M.; Leufkens, H. The Future of Drug Development: The Paradigm Shift towards Systems Therapeutics. Drug Discov. Today 2018, 23, 1990–1995. [Google Scholar] [CrossRef] [PubMed]
- Sorger, P.K.; Allerheiligen, S.R.B.; Abernethy, D.R.; Altmann, R.B.; Brouwer, K.L.R.; Califano, A.; D’Argenio, D.Z.; Iyengar, R.; Jusko, W.J.; Lalonde, R.; et al. Quantitative and Systems Pharmacology in the Post-Genomic Era: New Approaches to Discovering Drugs and Understanding Therapeutic Mechanisms (White Paper). In An NIH White Paper by the QSP Workshop Group; Bethesda: Rockville, MD, USA, 2011; Volume 48. [Google Scholar]
- Visser, S.A.G.; Manolis, E.; Danhof, M.; Kerbusch, T. Modeling and Simulation at the Interface of Nonclinical and Early Clinical Drug Development. CPT Pharmacomet. Syst. Pharmacol. 2013, 2, 8–10. [Google Scholar] [CrossRef]
- Nijsen, M.J.; Wu, F.; Bansal, L.; Bradshaw-Pierce, E.; Chan, J.R.; Liederer, B.M.; Mettetal, J.T.; Schroeder, P.; Schuck, E.; Tsai, A.; et al. Preclinical QSP Modeling in the Pharmaceutical Industry: An IQ Consortium Survey Examining the Current Landscape. CPT Pharmacomet. Syst. Pharmacol. 2018, 7, 135–146. [Google Scholar] [CrossRef]
- Schulthess, P.; Van Wijk, R.C.; Krekels, E.H.J.; Yates, J.W.T.; Spaink, H.P.; Van der Graaf, P.H. Outside-in Systems Pharmacology Combines Innovative Computational Methods with High-Throughput Whole Vertebrate Studies. CPT Pharmacomet. Syst. Pharmacol. 2018, 7, 285–287. [Google Scholar] [CrossRef]
- Van der Graaf, P.H. Pharmacometrics and/or Systems Pharmacology. CPT Pharmacomet. Syst. Pharmacol. 2019, 8, 331–332. [Google Scholar] [CrossRef]
- Lombardo, F.; Desai, P.V.; Arimoto, R.; Desino, K.E.; Fischer, H.; Keefer, C.E.; Petersson, C.; Winiwarter, S.; Broccatelli, F. In Silico Absorption, Distribution, Metabolism, Excretion, and Pharmacokinetics (ADME-PK): Utility and Best Practices. An Industry Perspective from the International Consortium for Innovation through Quality in Pharmaceutical Development. J. Med. Chem. 2017, 60, 9097–9113. [Google Scholar] [CrossRef]
- Wu, C.Y.; Benet, L.Z.; Hebert, M.F.; Gupta, S.K.; Rowland, M.; Gomez, D.Y.; Wacher, V.J. Differentiation of Absorption and First-Pass Gut and Hepatic Metabolism in Humans: Studies with Cyclosporine. Clin. Pharmacol. Ther. 1995, 58, 492–497. [Google Scholar] [CrossRef]
- Hellriegel, E.T.; Bjornsson, T.D.; Hauck, W.W. Interpatient Variability in Bioavailability Is Related to the Extent of Absorption: Implications for Bioavailability and Bioequivalence Studies. Clin. Pharmacol. Ther. 1996, 60, 601–607. [Google Scholar] [CrossRef]
- Amidon, G.L.; Lennernäs, H.; Shah, V.P.; Crison, J.R. A Theoretical Basis for a Biopharmaceutic Drug Classification: The Correlation of in Vitro Drug Product Dissolution and in Vivo Bioavailability. Pharm. Res. 1995, 12, 413–420. [Google Scholar] [CrossRef] [PubMed]
- Dahlgren, D.; Roos, C.; Lundqvist, A.; Abrahamsson, B.; Tannergren, C.; Hellström, P.M.; Sjögren, E.; Lennernäs, H. Regional Intestinal Permeability of Three Model Drugs in Human. Mol. Pharm. 2016, 13, 3013–3021. [Google Scholar] [CrossRef] [PubMed]
- Tannergren, C.; Bergendal, A.; Lennernäs, H.; Abrahamsson, B. Toward an Increased Understanding of the Barriers to Colonic Drug Absorption in Humans: Implications for Early Controlled Release Candidate Assessment. Mol. Pharm. 2009, 6, 60–73. [Google Scholar] [CrossRef]
- Shimada, T.; Yamazaki, H.; Mimura, M.; Inui, Y.; Guengerich, F.P. Interindividual Variations in Human Liver Cytochrome P-450 Enzymes Involved in the Oxidation of Drugs, Carcinogens and Toxic Chemicals: Studies with Liver Microsomes of 30 Japanese and 30 Caucasians. J. Pharmacol. Exp. Ther. 1994, 270, 414–423. [Google Scholar]
- Rendic, S. Summary of Information on Human CYP Enzymes: Human P450 Metabolism Data. Drug Metab. Rev. 2002. [CrossRef]
- Zamek-Gliszczynski, M.J.; Hoffmaster, K.A.; Nezasa, K.; Tallman, M.N.; Brouwer, K.L.R. Integration of Hepatic Drug Transporters and Phase II Metabolizing Enzymes: Mechanisms of Hepatic Excretion of Sulfate, Glucuronide, and Glutathione Metabolites. Eur. J. Pharm. Sci. 2006, 27, 447–486. [Google Scholar] [CrossRef]
- Luo, G.; Johnson, S.; Hsueh, M.M.; Zheng, J.; Cai, H.; Xin, B.; Chong, S.; He, K.; Harper, T.W. In Silico Prediction of Biliary Excretion of Drugs in Rats Based on Physicochemical Properties. Drug Metab. Dispos. 2010, 38, 422–430. [Google Scholar] [CrossRef]
- Roos, C.; Dahlgren, D.; Tannergren, C.; Abrahamsson, B.; Sjögren, E.; Lennernas, H. Regional Intestinal Permeability in Rats: A Comparison of Methods. Mol. Pharm. 2017, 14, 4252–4261. [Google Scholar] [CrossRef]
- Gumbo, T.; Lenaerts, A.J.; Hanna, D.; Romero, K.; Nuermberger, E. Nonclinical Models for Antituberculosis Drug Development: A Landscape Analysis. J. Infect. Dis. 2015, 211, S83–S95. [Google Scholar] [CrossRef]
- Yu, L.X.; Lipka, E.; Crison, J.R.; Amidon, G.L. Transport Approaches to the Biopharmaceutical Design of Oral Drug Delivery Systems: Prediction of Intestinal Absorption. Adv. Drug Deliv. Rev. 1996, 19, 359–376. [Google Scholar] [CrossRef]
- Heimbach, T.; Suarez-Sharp, S.; Kakhi, M.; Holmstock, N.; Olivares-Morales, A.; Pepin, X.; Sjögren, E.; Tsakalozou, E.; Seo, P.; Li, M. Dissolution and Translational Modeling Strategies Toward Establishing an In Vitro-In Vivo Link—A Workshop Summary Report. AAPS J. 2019, 21, 29. [Google Scholar] [CrossRef]
- Wu, C.Y.; Benet, L.Z. Predicting Drug Disposition via Application of BCS: Transport/Absorption/ Elimination Interplay and Development of a Biopharmaceutics Drug Disposition Classification System. Pharm. Res. 2005, 22, 11–23. [Google Scholar] [CrossRef]
- Bennion, B.J.; Be, N.A.; McNerney, M.W.; Lao, V.; Carlson, E.M.; Valdez, C.A.; Malfatti, M.A.; Enright, H.A.; Nguyen, T.H.; Lightstone, F.C. Predicting a Drug’s Membrane Permeability: A Computational Model Validated with in Vitro Permeability Assay Data. J. Phys. Chem. B 2017, 121, 5228–5237. [Google Scholar] [CrossRef]
- Awoonor-Williams, E.; Rowley, C.N. Molecular Simulation of Nonfacilitated Membrane Permeation. Biochim. Biophys. Acta Biomembr. 2016, 1858, 1672–1687. [Google Scholar] [CrossRef]
- Orsi, M.; Essex, J.W. Permeability of Drugs and Hormones through a Lipid Bilayer: Insights from Dual-Resolution Molecular Dynamics. Soft Matter 2010, 6, 3797–3808. [Google Scholar] [CrossRef]
- Lipinski, C.A.; Lombardo, F.; Dominy, B.W.; Feeney, P.J. Experimental and Computational Approaches to Estimate Solubility and Permeability in Drug Discovery and Development Settings. Adv. Drug Deliv. Rev. 1997, 23, 3–25. [Google Scholar] [CrossRef]
- Bickerton, G.R.; Paolini, G.V.; Besnard, J.; Muresan, S.; Hopkins, A.L. Quantifying the Chemical Beauty of Drugs. Nat. Chem. 2012, 4, 90. [Google Scholar] [CrossRef]
- Krämer, S.D.; Aschmann, H.E.; Hatibovic, M.; Hermann, K.F.; Neuhaus, C.S.; Brunner, C.; Belli, S. When Barriers Ignore the “Rule-of-Five”. Adv. Drug Deliv. Rev. 2016, 101, 62–74. [Google Scholar] [CrossRef]
- Bemporad, D.; Luttmann, C.; Essex, J.W. Behaviour of Small Solutes and Large Drugs in a Lipid Bilayer from Computer Simulations. Biochim. Biophys. Acta Biomembr. 2005, 1718, 1–21. [Google Scholar] [CrossRef]
- Kuhn, B.; Mohr, P.; Stahl, M. Intramolecular Hydrogen Bonding in Medicinal Chemistry. J. Med. Chem. 2010, 53, 2601–2611. [Google Scholar] [CrossRef] [PubMed]
- Alex, A.; Millan, D.S.; Perez, M.; Wakenhut, F.; Whitlock, G.A. Intramolecular Hydrogen Bonding to Improve Membrane Permeability and Absorption in beyond Rule of Five Chemical Space. MedChemComm 2011, 2, 669–674. [Google Scholar] [CrossRef]
- Agoram, B.; Woltosz, W.S.; Bolger, M.B. Predicting the Impact of Physiological and Biochemical Processes on Oral Drug Bioavailability. Adv. Drug Deliv. Rev. 2001, 50, S41–S67. [Google Scholar] [CrossRef]
- Rowland, M.; Peck, C.; Tucker, G. Physiologically-Based Pharmacokinetics in Drug Development and Regulatory Science. Annu. Rev. Pharmacol. Toxicol. 2011, 51, 45–73. [Google Scholar] [CrossRef]
- Zhou, Q.; Gallo, J.M. The Pharmacokinetic/Pharmacodynamic Pipeline: Translating Anticancer Drug Pharmacology to the Clinic. AAPS J. 2011, 13, 111–120. [Google Scholar] [CrossRef]
- Woo, J.; Cheung, W.; Chan, R.; Chan, H.S.; Cheng, A.; Chan, K. In Vitro Protein Binding Characteristics of Isoniazid, Rifampicin, and Pyrazinamide to Whole Plasma, Albumin, and α-1-Acid Glycoprotein. Clin. Biochem. 1996, 29, 175–177. [Google Scholar] [CrossRef]
- Lavé, T.; Parrott, N.; Grimm, H.P.; Fleury, A.; Reddy, M. Challenges and Opportunities with Modelling and Simulation in Drug Discovery and Drug Development. Xenobiotica 2007, 37, 1295–1310. [Google Scholar] [CrossRef]
- Flanagan, T.; Van Peer, A.; Lindahl, A. Use of Physiologically Relevant Biopharmaceutics Tools within the Pharmaceutical Industry and in Regulatory Sciences: Where Are We Now and What Are the Gaps? Eur. J. Pharm. Sci. 2016, 91, 84–90. [Google Scholar] [CrossRef]
- Mahmood, I. Application of Allometric Principles for the Prediction of Pharmacokinetics in Human and Veterinary Drug Development. Adv. Drug Deliv. Rev. 2007, 59, 1177–1192. [Google Scholar] [CrossRef]
- Teitelbaum, Z.; Lave, T.; Freijer, J.; Cohen, A.F. Risk Assessment in Extrapolation of Pharmacokinetics from Preclinical Data to Humans. Clin. Pharmacokinet. 2010, 49, 619–632. [Google Scholar] [CrossRef]
- Gumbo, T.; Angulo-Barturen, I.; Ferrer-Bazaga, S. Pharmacokinetic-Pharmacodynamic and Dose-Response Relationships of Antituberculosis Drugs: Recommendations and Standards for Industry and Academia. J. Infect. Dis. 2015, 211, S96–S106. [Google Scholar] [CrossRef] [PubMed]
- Jones, H.M.; Parrott, N.; Jorga, K.; Lavé, T. A Novel Strategy for Physiologically Based Predictions of Human Pharmacokinetics. Clin. Pharmacokinet. 2006, 45, 511–542. [Google Scholar] [CrossRef] [PubMed]
- Pelkonen, O.; Turpeinen, M. In Vitro-in Vivo Extrapolation of Hepatic Clearance: Biological Tools, Scaling Factors, Model Assumptions and Correct Concentrations. Xenobiotica 2007, 37, 1066–1089. [Google Scholar] [CrossRef] [PubMed]
- Houston, J.; Galetin, A. Methods for Predicting In Vivo Pharmacokinetics Using Data from In Vitro Assays. Curr. Drug Metab. 2008. [Google Scholar] [CrossRef] [PubMed]
- Lavé, T.; Chapman, K.; Goldsmith, P.; Rowland, M. Human Clearance Prediction: Shifting the Paradigm. Expert Opin. Drug Metab. Toxicol. 2009. [Google Scholar] [CrossRef]
- Ito, K.; Houston, J.B. Prediction of Human Drug Clearance from in Vitro and Preclinical Data Using Physiologically Based and Empirical Approaches. Pharm. Res. 2005, 22, 103–112. [Google Scholar] [CrossRef]
- Rostami-Hodjegan, A.; Tucker, G.T. Simulation and Prediction of in Vivo Drug Metabolism in Human Populations from in Vitro Data. Nat. Rev. Drug Discov. 2007, 6, 140–148. [Google Scholar] [CrossRef]
- Aït Moussa, L.; El Bouazzi, O.; Serragui, S.; Soussi Tanani, D.; Soulaymani, A.; Soulaymani, R. Rifampicin and Isoniazid Plasma Concentrations in Relation to Adverse Reactions in Tuberculosis Patients: A Retrospective Analysis. Ther. Adv. Drug Saf. 2016, 7, 239–247. [Google Scholar] [CrossRef]
- Muttil, P.; Wang, C.; Hickey, A.J. Inhaled Drug Delivery for Tuberculosis Therapy. Pharm. Res. 2009, 26, 2401–2416. [Google Scholar] [CrossRef]
- Pandey, R.; Khuller, G.K. Antitubercular Inhaled Therapy: Opportunities, Progress and Challenges. J. Antimicrob. Chemother. 2005, 55, 430–435. [Google Scholar] [CrossRef]
- Patton, J.S.; Byron, P.R. Inhaling Medicines: Delivering Drugs to the Body through the Lungs. Nat. Rev. Drug Discov. 2007, 6, 67–74. [Google Scholar] [CrossRef] [PubMed]
- Bäckman, P.; Arora, S.; Couet, W.; Forbes, B.; de Kruijf, W.; Paudel, A. Advances in Experimental and Mechanistic Computational Models to Understand Pulmonary Exposure to Inhaled Drugs. Eur. J. Pharm. Sci. 2018, 113, 41–52. [Google Scholar] [CrossRef] [PubMed]
- Eriksson, J.; Sjögren, E.; Thörn, H.; Rubin, K.; Bäckman, P.; Lennernäs, H. Pulmonary Absorption-Estimation of Effective Pulmonary Permeability and Tissue Retention of Ten Drugs Using an Ex Vivo Rat Model and Computational Analysis. Eur. J. Pharm. Biopharm. 2018, 124, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Scheuch, G.; Kohlhaeufl, M.J.; Brand, P.; Siekmeier, R. Clinical Perspectives on Pulmonary Systemic and Macromolecular Delivery. Adv. Drug Deliv. Rev. 2006, 58, 996–1008. [Google Scholar] [CrossRef]
- Hittinger, M.; Juntke, J.; Kletting, S.; Schneider-Daum, N.; de Souza Carvalho, C.; Lehr, C.-M. Preclinical Safety and Efficacy Models for Pulmonary Drug Delivery of Antimicrobials with Focus on in Vitro Models. Adv. Drug Deliv. Rev. 2015, 85, 44–56. [Google Scholar] [CrossRef]
- McGonigle, P.; Ruggeri, B. Animal Models of Human Disease: Challenges in Enabling Translation. Biochem. Pharmacol. 2014, 87, 162–171. [Google Scholar] [CrossRef]
- Kjellsson, M.C.; Via, L.E.; Goh, A.; Weiner, D.; Low, K.M.; Kern, S.; Pillai, G.; Barry, C.E.; Dartois, V. Pharmacokinetic Evaluation of the Penetration of Antituberculosis Agents in Rabbit Pulmonary Lesions. Antimicrob. Agents Chemother. 2012, 56, 446–457. [Google Scholar] [CrossRef]
- Dartois, V. The Path of Anti-Tuberculosis Drugs: From Blood to Lesions to Mycobacterial Cells. Nat. Rev. Microbiol. 2014, 12, 159–167. [Google Scholar] [CrossRef]
- Prideaux, B.; Dartois, V.; Staab, D.; Weiner, D.M.; Goh, A.; Via, L.E.; Barry III, C.E.; Stoeckli, M. High-Sensitivity MALDI-MRM-MS Imaging of Moxifloxacin Distribution in Tuberculosis-Infected Rabbit Lungs and Granulomatous Lesions. Anal. Chem. 2011, 83, 2112–2118. [Google Scholar] [CrossRef]
- Franzblau, S.G.; Degroote, M.A.; Cho, S.H.; Andries, K.; Nuermberger, E.; Orme, I.M.; Mdluli, K.; Angulo-Barturen, I.; Dick, T.; Dartois, V.; et al. Comprehensive Analysis of Methods Used for the Evaluation of Compounds against Mycobacterium Tuberculosis. Tuberculosis 2012, 92, 453–488. [Google Scholar] [CrossRef]
- Nuermberger, E.L. Preclinical Efficacy Testing of New Drug Candidates. Microbiol. Spectr. 2017, 5. [Google Scholar] [CrossRef]
- Nielsen, E.I.; Friberg, L.E. Pharmacokinetic-Pharmacodynamic Modeling of Antibacterial Drugs. Pharmacol. Rev. 2013, 65, 1053–1090. [Google Scholar] [CrossRef] [PubMed]
- Mouton, J.W.; Meletiadis, J.; Voss, A.; Turnidge, J. Variation of MIC Measurements: The Contribution of Strain and Laboratory Variability to Measurement Precision. J. Antimicrob. Chemother. 2018, 73, 2374–2379. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.K.; Gupta, U.D. Animal Models of Tuberculosis: Lesson Learnt. Indian J. Med. Res. 2018, 147, 456–463. [Google Scholar] [CrossRef] [PubMed]
- Young, D. Animal Models of Tuberculosis. Eur. J. Immunol. 2009, 39, 2011–2014. [Google Scholar] [CrossRef]
- Myllymäki, H.; Niskanen, M.; Oksanen, K.E.; Rämet, M. Animal Models in Tuberculosis Research-Where Is the Beef? Expert Opin. Drug Discov. 2015, 10, 871–883. [Google Scholar] [CrossRef]
- Meijer, A.H. Protection and Pathology in TB: Learning from the Zebrafish Model. Semin. Immunopathol. 2016, 38, 261–273. [Google Scholar] [CrossRef]
- Carvalho, R.; de Sonneville, J.; Stockhammer, O.W.; Savage, N.D.L.; Veneman, W.J.; Ottenhoff, T.H.M.; Dirks, R.P.; Meijer, A.H.; Spaink, H.P. A High-Throughput Screen for Tuberculosis Progression. PLoS ONE 2011, 6, e16779. [Google Scholar] [CrossRef]
- Kantae, V.; Krekels, E.H.; Ordas, A.; Gonzalez, O.; Van Wijk, R.C.; Harms, A.C.; Racz, P.I.; Van Der Graaf, P.H.; Spaink, H.P.; Hankemeier, T. Pharmacokinetic Modeling of Paracetamol Uptake and Clearance in Zebrafish Larvae: Expanding the Allometric Scale in Vertebrates with Five Orders of Magnitude. Zebrafish 2016, 13, 504–510. [Google Scholar] [CrossRef]
- Van Wijk, R.C.; Krekels, E.H.J.; Kantae, V.; Harms, A.C.; Hankemeier, T.; Van der Graaf, P.H.; Spaink, H.P. Impact of Post-Hatching Maturation on the Pharmacokinetics of Exogenous Compounds in Zebrafish Larvae. Sci. Rep. 2018, 9, 2149. [Google Scholar] [CrossRef]
- Van Wijk, R.C.; Krekels, E.H.J.; Kantae, V.; Ordas, A.; Kreling, T.; Harms, A.C.; Hankemeier, T.; Spaink, H.P.; van der Graaf, P.H. Mechanistic and Quantitative Understanding of Pharmacokinetics in Zebrafish Larvae through Nanoscale Blood Sampling and Metabolite Modelling of Paracetamol. J. Pharmacol. Exp. Ther. 2019, 371, 15–24. [Google Scholar] [CrossRef] [PubMed]
- Van Wijk, R.C.; Krekels, E.H.J.; Hu, W.; Van der Sar, A.M.; Dijkema, S.M.; Van den Berg, D.-J.; Bahi, R.; Liu, J.; Verboom, T.; Verbeek, F.J.; et al. Translational quantitative systems pharmacology; crossing borders between experimental and computational drug development using zebrafish as model organism. PAGE 2021, 29, 9455. [Google Scholar]
- Ordonez, A.A.; Wang, H.; Magombedze, G.; Ruiz-Bedoya, C.A.; Srivastava, S.; Chen, A.; Tucker, E.W.; Urbanowski, M.E.; Pieterse, L.; Fabian Cardozo, E.; et al. Dynamic Imaging in Patients with Tuberculosis Reveals Heterogeneous Drug Exposures in Pulmonary Lesions. Nat. Med. 2020, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Committee for Human Medicinal Products (CHMP). Guideline on the Use of Pharmacokinetics and Pharmacodynamics in the Development of Antibacterial Medicinal Products. (EMA/CHMP/594085/2015). Eur. Med. Agency 2016, 44, 1–21. [Google Scholar]
- Nielsen, E.I.; Cars, O.; Friberg, L.E. Pharmacokinetic/Pharmacodynamic (PK/PD) Indices of Antibiotics Predicted by a Semimechanistic PKPD Model: A Step toward Model-Based Dose Optimization. Antimicrob. Agents Chemother. 2011, 55, 4619–4630. [Google Scholar] [CrossRef]
- Khan, D.D.; Friberg, L.E.; Nielsen, E.I. A Pharmacokinetic-Pharmacodynamic (PKPD) Model Based on in Vitro Time-Kill Data Predicts the in Vivo PK/PD Index of Colistin. J. Antimicrob. Chemother. 2016, 71, 1881–1884. [Google Scholar] [CrossRef]
- Pasipanodya, J.G.; Nuermberger, E.; Romero, K.; Hanna, D.; Gumbo, T. Systematic Analysis of Hollow Fiber Model of Tuberculosis Experiments. Clin. Infect. Dis. 2015, 61 (Suppl. 1), S10–S17. [Google Scholar] [CrossRef]
- European Medicines Agency. Qualification Opinion In-Vitro Hollow Fiber System Model of Tuberculosis (HSF-TB); European Medicines Agency: London, UK, 2015.
- Chilukuri, D.; McMaster, O.; Bergman, K.; Colangelo, P.; Snow, K.; Toerner, J.G. The Hollow Fiber System Model in the Nonclinical Evaluation of Antituberculosis Drug Regimens. Clin. Infect. Dis. 2015, 61, S32–S33. [Google Scholar] [CrossRef][Green Version]
- Magombedze, G.; Pasipanodya, J.G.; Srivastava, S.; Deshpande, D.; Visser, M.E.; Chigutsa, E.; McIlleron, H.; Gumbo, T. Transformation Morphisms and Time-to-Extinction Analysis That Map Therapy Duration from Preclinical Models to Patients with Tuberculosis: Translating from Apples to Oranges. Clin. Infect. Dis. 2018, 67, S349–S358. [Google Scholar] [CrossRef]
- Deshpande, D.; Magombedze, G.; Srivastava, S.; Bendet, P.; Lee, P.S.; Cirrincione, K.N.; Martin, K.R.; Dheda, K.; Gumbo, T. Once-a-Week Tigecycline for the Treatment of Drug-Resistant TB. J. Antimicrob. Chemother. 2019, 74, 1607–1617. [Google Scholar] [CrossRef]
- Srivastava, S.; Deshpande, D.; Magombedze, G.; van Zyl, J.; Cirrincione, K.; Martin, K.; Bendet, P.; Berg, A.; Hanna, D.; Romero, K.; et al. Duration of Pretomanid/Moxifloxacin/Pyrazinamide Therapy Compared with Standard Therapy Based on Time-to-Extinction Mathematics. J. Antimicrob. Chemother. 2020, 75, 392–399. [Google Scholar] [CrossRef]
- Khan, D.D.; Lagerbäck, P.; Cao, S.; Lustig, U.; Nielsen, E.I.; Cars, O.; Hughes, D.; Andersson, D.I.; Friberg, L.E. A Mechanism-Based Pharmacokinetic/Pharmacodynamic Model Allows Prediction of Antibiotic Killing from MIC Values for WT and Mutants. J. Antimicrob. Chemother. 2015, 70, 3051–3060. [Google Scholar] [CrossRef] [PubMed]
- Clewe, O.; Aulin, L.; Hu, Y.; Coates, A.R.M.; Simonsson, U.S.H. A Multistate Tuberculosis Pharmacometric Model: A Framework for Studying Anti-Tubercular Drug Effects in Vitro. J. Antimicrob. Chemother. 2016, 71, 964–974. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Ortega, F.; Rullas, J.; Alameda, L.; Angulo-Barturen, I.; Ferrer, S.; Simonsson, U.S. The Multistate Tuberculosis Pharmacometric Model: A Semi-Mechanistic Pharmacokinetic-Pharmacodynamic Model for Studying Drug Effects in an Acute Tuberculosis Mouse Model. J. Pharmacokinet. Pharmacodyn. 2017, 44, 133–141. [Google Scholar] [CrossRef] [PubMed]
- Svensson, R.J.; Simonsson, U.S.H. Application of the Multistate Tuberculosis Pharmacometric Model in Patients with Rifampicin-Treated Pulmonary Tuberculosis. CPT Pharmacomet. Syst. Pharmacol. 2016, 5, 264–273. [Google Scholar] [CrossRef]
- Lalande, L.; Bourguignon, L.; Bihari, S.; Maire, P.; Neely, M.; Jelliffe, R.; Goutelle, S. Population Modeling and Simulation Study of the Pharmacokinetics and Antituberculosis Pharmacodynamics of Isoniazid in Lungs. Antimicrob. Agents Chemother. 2015, 59, 5181–5189. [Google Scholar] [CrossRef]
- European Medicines Agency. Guideline on the Investigation of Drug Interactions; European Medicines Agency: Lonon, UK, 2012. [CrossRef]
- Kuhlmann, J.; Mück, W. Clinical-Pharmacological Strategies to Assess Drug Interaction Potential during Drug Development. Drug Saf. 2001, 24, 715–725. [Google Scholar] [CrossRef]
- Barton, H.A.; Lai, Y.; Goosen, T.C.; Jones, H.M.; El-Kattan, A.F.; Gosset, J.R.; Lin, J.; Varma, M.V. Model-Based Approaches to Predict Drug-Drug Interactions Associated with Hepatic Uptake Transporters: Preclinical, Clinical and Beyond. Expert Opin. Drug Metab. Toxicol. 2013, 9, 459–472. [Google Scholar] [CrossRef]
- Chu, X.; Bleasby, K.; Evers, R. Species Differences in Drug Transporters and Implications for Translating Preclinical Findings to Humans. Expert Opin. Drug Metab. Toxicol. 2013, 9, 237–252. [Google Scholar] [CrossRef]
- McIlleron, H.; Chirehwa, M.T. Current Research toward Optimizing Dosing of First-Line Antituberculosis Treatment. Expert Rev. Anti. Infect. Ther. 2019, 17, 27–38. [Google Scholar] [CrossRef]
- Svensson, E.M.; Acharya, C.; Clauson, B.; Dooley, K.E.; Karlsson, M.O. Pharmacokinetic Interactions for Drugs with a Long Half-Life—Evidence for the Need of Model-Based Analysis. AAPS J. 2016, 18, 171–179. [Google Scholar] [CrossRef] [PubMed]
- Margolskee, A.; Darwich, A.S.; Pepin, X.; Pathak, S.M.; Bolger, M.B.; Aarons, L.; Rostami-Hodjegan, A.; Angstenberger, J.; Graf, F.; Laplanche, L.; et al. IMI-Oral Biopharmaceutics Tools Project-Evaluation of Bottom-up PBPK Prediction Success Part 1: Characterisation of the OrBiTo Database of Compounds. Eur. J. Pharm. Sci. 2017, 96, 598–609. [Google Scholar] [CrossRef]
- Chen, J.; Raymond, K. Roles of Rifampicin in Drug-Drug Interactions: Underlying Molecular Mechanisms Involving the Nuclear Pregnane X Receptor. Ann. Clin. Microbiol. Antimicrob. 2006, 5. [Google Scholar] [CrossRef] [PubMed]
- Chattopadhyay, N.; Kanacher, T.; Kanacher, T.; Frechen, S.; Ligges, S.; Zimmermann, T.; Rottmann, A.; Ploeger, B.; Höchel, J.; Schultze-Mosgau, M.H. CYP3A4-Mediated Effects of Rifampicin on the Pharmacokinetics of Vilaprisan and Its UGT1A1-Mediated Effects on Bilirubin Glucuronidation in Humans. Br. J. Clin. Pharmacol. 2018, 84, 2857–2866. [Google Scholar] [CrossRef] [PubMed]
- Greiner, B.; Eichelbaum, M.; Fritz, P.; Kreichgauer, H.P.; Von Richter, O.; Zundler, J.; Kroemer, H.K. The Role of Intestinal P-Glycoprotein in the Interaction of Digoxin and Rifampin. J. Clin. Invest. 1999, 104, 147–153. [Google Scholar] [CrossRef] [PubMed]
- Mehta, J.; Gandhit, I.S.; Sanet, S.B.; Wamburkart, M.N. Effect of Clofazimine and Dapsone on Rifampicin (Lositril) Pharmacokinetics in Multibacillary and Paucibacillary Leprosy Cases. Lepr. Rev. 1986, 57, 67–76. [Google Scholar] [CrossRef]
- Horita, Y.; Doi, N. Comparative Study of the Effects of Antituberculosis Drugs and Antiretroviral Drugs on Cytochrome P450 3a4 and P-Glycoprotein. Antimicrob. Agents Chemother. 2014, 58, 3168–3176. [Google Scholar] [CrossRef]
- Dannemann, B.; Bakare, N.; De Marez, T.; Lounis, N.; Van Heeswijk, R.P.G.; Meyvisch, P.; Haxaire-Theeuwes, M.; Andries, K.; Everitt, D.; Upton, A. Corrected QT Interval (QTcF) Prolongation in a Phase 2 Open-Label Trial of TMC207 plus Background Regimen as Treatment for MDR-TB: Effect of Co-Administration with Clofazimine. In Proceedings of the Abstract at 52nd Interscience Conference on Antimicrobial Agents and Chemotherapy, San Francisco, CA, USA, 9–12 September 2012. [Google Scholar]
- Pym, A.S.; Diacon, A.H.; Tang, S.J.; Conradie, F.; Danilovits, M.; Chuchottaworn, C.; Vasilyeva, I.; Andries, K.; Bakare, N.; De Marez, T.; et al. Bedaquiline in the Treatment of Multidrug- and Extensively Drugresistant Tuberculosis. Eur. Respir. J. 2016, 47, 564–574. [Google Scholar] [CrossRef]
- Iwatsubo, T.; Suzuki, H.; Shimada, N.; Chiba, K.; Ishizaki, T.; Green, C.E.; Tyson, C.A.; Yokoi, T.; Kamataki, T.; Sugiyama, Y. Prediction of in Vivo Hepatic Metabolic Clearance of YM796 from in Vitro Data by Use of Human Liver Microsomes and Recombinant P-450 Isozymes. J. Pharmacol. Exp. Ther. 1997, 282, 909–919. [Google Scholar]
- Zhao, P.; Zhang, L.; Grillo, J.A.; Liu, Q.; Bullock, J.M.; Moon, Y.J.; Song, P.; Brar, S.S.; Madabushi, R.; Wu, T.C.; et al. Applications of Physiologically Based Pharmacokinetic (PBPK) Modeling and Simulation during Regulatory Review. Clin. Pharmacol. Ther. 2011, 89, 259–267. [Google Scholar] [CrossRef]
- Yamashita, F.; Sasa, Y.; Yoshida, S.; Hisaka, A.; Asai, Y.; Kitano, H.; Hashida, M.; Suzuki, H. Modeling of Rifampicin-Induced CYP3A4 Activation Dynamics for the Prediction of Clinical Drug-Drug Interactions from In Vitro Data. PLoS ONE 2013, 8. [Google Scholar] [CrossRef] [PubMed]
- Greco, W.R.; Park, H.S.; Rustum, Y.M. Application of a New Approach for the Quantitation of Drug Synergism to the Combination of Cis-Diamminedichloroplatinum and 1-θ-d-Arabinofuranosylcytosine. Cancer Res. 1990, 50, 5318–5327. [Google Scholar] [PubMed]
- Wicha, S.G.; Chen, C.; Clewe, O.; Simonsson, U.S.H. A General Pharmacodynamic Interaction Model Identifies Perpetrators and Victims in Drug Interactions. Nat. Commun. 2017, 8. [Google Scholar] [CrossRef] [PubMed]
- Clewe, O.; Wicha, S.G.; de Vogel, C.P.; de Steenwinkel, J.E.M.; Simonsson, U.S.H. A Model-Informed Preclinical Approach for Prediction of Clinical Pharmacodynamic Interactions of Anti-TB Drug Combinations. J. Antimicrob. Chemother. 2018, 73, 437–447. [Google Scholar] [CrossRef]
- Chen, C.; Wicha, S.G.; De Knegt, G.J.; Ortega, F.; Alameda, L.; Sousa, V.; De Steenwinkel, J.E.M.; Simonsson, U.S.H. Assessing Pharmacodynamic Interactions in Mice Using the Multistate Tuberculosis Pharmacometric and General Pharmacodynamic Interaction Models. CPT Pharmacomet. Syst. Pharmacol. 2017, 6, 787–797. [Google Scholar] [CrossRef]
- Chen, C.; Wicha, S.G.; Nordgren, R.; Simonsson, U.S.H. Comparisons of Analysis Methods for Assessment of Pharmacodynamic Interactions Including Design Recommendations. AAPS J. 2018, 20, 1–12. [Google Scholar] [CrossRef]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).