Abstract
Manganese (Mn) is widely used in industry. However, its extensive applications have generated a great amount of manganese waste, which has become an ecological problem and has led to a decrease in natural resources. The use of microorganisms capable of accumulating Mn ions from contaminated ecosystems offers a potential alternative for the removal and recovery of this metal. The main aim of this work was an investigation of removal potential of Mn from soil by isolated bacterial. For this purpose, eleven bacterial strains were isolated from the soil from metallurgical waste heap in Upper Silesia, Poland. Strain named 2De with the highest Mn removal potential was selected and characterized taking into account its ability for Mn sorption and bioaccumulation from soil and medium containing manganese dioxide. Moreover, the protein profile of 2De strain before and after exposition to Mn was analyzed using SDS/PAGE technique. The 2De strain was identified as a Pseudomonas sp. The results revealed that this strain has an ability to grow at high Mn concentration and possesses an enhanced ability to remove it from the solution enriched with the soil or manganese dioxide via a biosorption mechanism. Moreover, changes in cellular protein expression of the isolated strain were observed. This study demonstrated that autochthonous 2De strain can be an effective tool to remove and recover Mn from contaminated soil.
1. Introduction
Manganese (Mn) is principally found in its oxide form and is available as a major constituent in many naturally occurring minerals [1]. It is also an important enzyme cofactor and provides protection to bacteria cells against reactive oxygen species. Many bacteria require it to form symbiotic or pathogenic interactions with eukaryotic host cells [2]. Moreover, Mn is a trace element essential for proper metabolic activity of humans and other mammals [3]. This metal is also important to a variety of industries. Mn is present in chemical and metallurgic products, alloys of iron and steel, ceramic fertilizers, varnishes, and fungicide products in livestock feeding supplements [4]. The global utilization of Mn is above 1.5 million tons per year, and it is intended to rise [5]. As a result of elevated Mn demand, high-grade reserved Mn ore is gradually depleting, which could lead to higher prices of both Mn ore and alloys [6].
The extensive use of Mn leads to the generation of a substantial amount of waste, among which a considerable quantity does not possess any market value [7]. Some of this waste is discharged into the environment leading to its contamination [1]. The excess Mn in terrestrial and aquatic ecosystems is known to have many toxic effects on organisms. In plants, Mn can lead to thickened palisade tissue, a high abscisic acid concentration, and growth retardation. In humans, exposure to high levels of Mn causes neurological changes and abnormalities of the immune and reproductive systems [4,8,9].
The improving awareness of both the health risks linked with Mn waste and increasing demand of this metal, necessitating its recovery, make it desirable to seek an environmentally friendly and economic process to recycle Mn from contaminated ecosystems. Chemical reagents used in traditional mineral processing, including foam flotation, which require the use of frothers, depressants, collectors, and other compounds, can be toxic and dangerous to the environment [10,11]. Due to the environmental risks posed by conventional ore enrichment methods, the use of biotechnological methods is gaining more interest. Great hopes are placed in the use of bacteria and their metabolic products for metal bioleaching, but also for bioremediation [12].
In recent years, bacteria that are able to recover Mn have been isolated, characterized, and used in biotechnology [5,6,13]. Depending on the microorganism, the mechanisms that they use for this process are biosorption and bioaccumulation [14]. Biosorption is usually not associated with cellular viability, whereas bioaccumulation is metabolically regulated. In the biosorption process, bacteria adsorb metals on the cellular surface through attachment/linkage onto many anionic functional groups [15]. During bioaccumulation, the intracellular uptake of metals takes place via precipitation linked with metabolic functions, chemical reactions, and/or ATP-driven active transport, after which metals are mainly accumulated inside the cells [16,17]. The accumulation of manganese by bacteria is possible through possession of complex protein systems to maintain proper Mn levels in the cells; thus, avoiding its toxicity. These systems are composed of manganese transporters, manganese-dependent transcription regulators, and manganese efflux pumps like MntE, MntP, MntX, CtpC, and P-type ATPase [18,19,20].
Many studies have demonstrated that biosorption of metals might be applicable for treatment of metallurgical and mining wastewater. Bacteria are potential organic sorbents, which show cell surface sorption or/and intracellular accumulation as well as extracellular accumulation [21,22]. The process can be metabolic-independent or metabolic-dependent. During the first process, a physical adsorption utilizes van der Waals’ forces and it is usually very fast and can be reversible. Organic compounds such as proteins, polysaccharides, lipids, and other components present on the surface of the microorganism cells, provide a rich variety of functional groups ready for exchange or binding of metal ions. In metabolic-dependent process, accumulation of metals in the cell involves transport of ions across the wall and usually requires more time [11].
The main aim of this work was to investigate the removal potential of Mn from soil by isolated bacteria. For this purpose, eleven bacterial strains were isolated from soil with a high Mn concentration collected from metallurgical waste heap in Upper Silesia, Poland. Among these strains, we selected and characterized the most Mn-tolerant bacterial strain and studied its potential for Mn removal in systems enriched in the soil or manganese dioxide (MnO2) solution taking into account biosorption and bioaccumulation. We assume that the metal accumulation observed by us can be an effect of biosorption (metabolic-dependent process) and 2De strain can be used for further tests, aimed at its use in in situ bioremediation of Mn-contaminated soil.
2. Materials and Methods
2.1. Sample Collection and Characterization
The soil samples from a metallurgical waste heap in Upper Silesia (Poland) was collected in a container and stored at 4 °C until further studies were carried out. The soil was mixed, ground in an agate mortar, and sieved in order to obtain a particle size of less than 200 μm.
The total Mn content in the soil sample was determined by acid digestion. The total Mn content in the soil sample was determined by microwave mineralization. Briefly, 10 mL of nitric acid (65% m/m) and 5 mL of hydrochloric acid (36% m/m) were added to the dry soil sample (0.5 g). The sample was decomposed in a microwave mineralizer (MARS 5, Digestion Microwave System, CEM Co., Matthews, NC, US) using the US EPA 3051 program (microwave power 1400 W, 175–180 °C, 10 min.) [23,24,25,26]. Mn concentration was measured by inductively coupled plasma optical emission spectrometry (ICP-OES, Varian 710 ES, Spectrometer, Mulgrave, Vic, Australia).
The soil was used for experiments described in Section 2.3, Section 2.6, Section 2.7 and Section 2.8. The soil was previously heat-sterilized (2 h at 180 °C, repeated 3 times with 24 h intervals).
2.2. Isolation of Bacteria
Bacteria able to accumulate Mn were isolated from the collected soil. Ten grams of soil sample was suspended in 90 mL sterile 0.85% NaCl, followed by preparing a series of dilutions. Then, 100 μL from 10−4, 10−5, and 10−6 dilutions were inoculated into nutrient agar plates supplemented with 50 mM of magnesium dioxide (MnO2). After incubation at 28 °C for 48 h, colonies showing visible growth were selected for further screening. The obtained isolates were assessed for Gram staining and tested for their ability for Mn adsorption according to the ISO PN-C-04590-03:1992 protocol.
2.3. Manganese Determination Using the Formaldoxime Method
The soil sample at a concentration of 2% (w/v) was added to 250-mL Erlenmeyer flasks containing 100 mL of K medium (0.7 g L−1 yeast extract, 2 g L−1 dextrose, 2.2 g L−1 peptone, pH 6.5). In order to verify soil sterility, the mixtures were incubated in a shaker at 28 °C at 120 rpm. After 24 h, 1 mL of the log-phase cells of isolated strains was introduced to the mixtures. Each set containing soil and a specific strain was prepared in triplicate. Uninoculated medium with the 2% soil (w/v) in triplicate was used as the control to determine the abiotic influences on Mn removal. The flasks were incubated at 28 °C for 14 days with shaking at 120 rpm. Mn concentration in the medium was determined using the formaldoxime method according to the ISO PN-C-04590-03:1992 protocol. Briefly, to 50 mL of each sample, 1 mL of ammonium iron(II) sulphate hexahydrate, and 2 mL of EDTA (Sigma-Aldrich) were added. After mixing the samples, 1 mL of formaldoxime (prepared by dissolving 10 g of hydroxylamine hydrochloride solid in 50 mL of water and 5 mL of 37% formaldehyde solution was then added, and the volume was increased to 100 mL with water) and 1 mL of sodium hydroxide were added followed by incubation for 10 min. Subsequently, 3 mL of ammonia solution of hydrogen chloride was added. The samples were incubated 2 h at 22 °C. The manganese–formaldoxime complex produced in the colorimetric reaction was detected at λ = 450 nm, and the Mn concentration in the media was calculated based on a standard curve. The standard curve was prepared by adding to 0, 0.5, 1, 2.5, 5, 10, and 20 mL of standard solution (287 mg potassium permanganate in 3 mL sulphuric acid, some drops of 5% sodium sulphite was then added until the pink colour disappeared and the volume was increased to 1000 mL with water; 1 mL of this solution contained 0.01 mg of Mn) to the same compounds as for the tested samples. The analyses were performed before and after the 14 days incubation period. The Mn concentration (mg L−1) was calculated based on the following formula:
where A was Mn content in the sample calculated from the standard curve while V was the volume of analyzed sample.
The 2De strain showing the highest ability for Mn adsorption was chosen for further analysis.
2.4. Identification of Selected Strain
The selected isolate was identified using 16S rRNA analysis. Genomic DNA of the 2De strain was extracted from pure cultures grown from a single colony using a DNA extraction kit (Bacterial and Yeast Genomic DNA Purification Kit, EURx, Gdańsk, Poland) according to the manufacturer’s instructions. The DNA was used as the template with the universal bacterial primers 8F (5′ AGTTTGATCATCGCTCAG 3′) and 1492R (5′ GGTTACCTTGTTACGACTT 3′) targeting a fragment size of 1484 bp [27]. The identity of the obtained sequence was performed using the ClustalW alignment and compared with published sequences using the NCBI BLAST program in the GenBank database. The Average Nucleotide Identity (ANI) calculator [28] was used to determine the similarity of 16S rRNA sequences between tested and typed strains [29]. In order to elucidate the phylogenetic position of the strain, a phylogenetic tree was constructed based on the longest common fragment of the 16S rRNA gene sequence (1372 bp) selected from ClustalW alignment of the 2De strain and the 10 closest type strains of other Pseudomonas species obtained from GenBank. The analysis was performed using Mega X software with the maximum likelihood method and Tamura-3-parameter model, assuming that a certain fraction of sites are evolutionary invariable (þI) and 1000 bootstrap replicates [30,31].
2.5. Determination of Minimal Inhibitory Concentration (MIC) of Selected Strains
In order to evaluate a level of a resistance of the isolated strain to Mn, the minimum inhibitory concentration (MIC) against increasing concentrations of Mn was determined by the agar dilution method. Bacterial cells were cultured to log phase in medium K at 28 ℃, and 100 μL of cell suspension (OD600 = 0.1) was spread onto medium K plates containing MnCl2 in concentrations increasing in arithmetic progression from 200 to 1600 mM.
2.6. Whole Cell Protein Extraction and Profiling by SDS/PAGE
The 2De strain was cultivated in medium K with 0.5 mM MnO2 or with the sterile soil at a concentration of 4% (w/v) at 28 °C. Untreated strain cultivated in medium K was used as a control. After 18 days of incubation, equal amounts of proteins were normalized to the optical density of cultures at 600 nm. For this purpose, 1 mL of each culture was centrifuged (15 min, 4 °C, 13000 rpm), and the cell pellets were washed with phosphate buffer (pH 7.4) followed by re-centrifugation (15 min, 4 °C, 13000 rpm). The pellets were suspended in sample buffer (0.5 M Tris-HCl 12.5 mL; 10% SDS 20 mL; glycerol 10 mL, 2-mercaptoethanol 5 mL; bromophenol blue 0.01 g; final volume of 100 mL with Millipore H2O) at a ratio of 100 μL/OD600 = 1, boiled (100 °C, 30 min), and centrifuged for 15 s in a microcentrifuge. The obtained samples were used for protein profiling using SDS/PAGE (Mini Protean, Bio-Rad, Hercules, CA, US). Then, 20 μL of the protein sample was separated using a 5% stacking gel and a 12% separating gel. After electrophoresis for 3 h at 100 V, the gel was stained with Coomassie brilliant blue in 10% (v/v) acetic acid and 45% (v/v) methanol for one hour. Destaining was carried out using 10% (v/v) acetic acid and 20% (v/v) methanol for 10 min. In order to obtain a clear background, the gel was rinsed in distilled water.
The relative migration distance (Rf) of the protein standard and the unknown proteins was determined. For this aim, the Rf was calculated by dividing the migration distance of the analyzed protein by the migration distance of the dye front. A calibration curve was created based on the logarithmic molecular mass of proteins relative to the Rf factor. Using the resulting equation of the calibration curve, the molecular sizes of analyzed proteins were determined.
Moreover, the quantification of the protein content of the 2De strain from each of the above-mentioned cultures was performed. A 5 mL portion of the culture was centrifuged (15 min, 4 °C, 4700 rpm). The pellet was re-suspended in 1 mL of lysis buffer (50 mM Tris-HCl; 100 mM NaCl; 1 mM Tween 20, 5% glycerol, 1 mM EDTA). After centrifugation (4 min, 4 °C, 4700 rpm), 1600 µL chilled acetone was added to 200 µL of supernatant and incubated for 30 min at 4 °C. Then, acetone was evaporated under N2, and the pellet was suspended in 100 µL of phosphate-buffered saline (PBS). Protein estimation was carried out using the Bradford Protein Assay (Bio-Rad, Hercules, CA, US) using bovine serum albumin (BSA) as a standard, with a concentration ranging from 0 to 80 mg mL−1.
2.7. Effects of Soil Concentration on Manganese Removal Efficiency
Erlenmeyer flasks containing 100 mL of medium K or 1%, 2%, 4%, or 6% (w/v) sieved and sterile soil were incubated at 28 °C to check soil sterility. After 24 h, each set was inoculated with 1 mL of the log-phase 2De cells. Controls were performed in the absence of bacteria using media with 1%, 2%, 4%, or 6% (w/v) soil. All the experiments, including the controls, were performed in triplicate. Cultures were incubated at 28 °C with shaking at 120 rpm. After 18 days, the cultures were centrifuged (4 °C, 4700 rpm), and supernatants were analyzed by atomic absorption spectrometry (AAS). The percentage of Mn removal efficiency (RE) was calculated by using the following equation:
where Co and Cf are the initial and final concentrations of Mn in the solution (mg L−1), respectively.
2.8. Bioaccumulation Experiments
The log-phase cells of the 2De strain (1 mL) were used as inoculum in 100 mL medium K in an Erlenmeyer flask containing 0.5 mM MnO2 or 4% soil and incubated at 28 °C with shaking at 120 rpm. The bacterial cells were harvested at 0, 3, 6, and 18 days after incubation by centrifugation (20 min, 4 °C, 4700 rpm) followed by washing with sterile deionized water to remove free metal ions. The concentration of Mn in cells was measured by AAS.
To examine Mn accumulation in the cell walls and intracellular spaces of the isolated strain, after washing with deionized water the pellet was treated with sterile 10 mM EDTA and incubated at 28 °C with agitation for 30 min [32]. The solution was centrifuged (20 min, 4 °C, 4700 rpm), and the resulting supernatant and pellet were separated and analyzed with AAS.
2.9. Statistical Analysis
Statistical analysis was performed using the STATISTICA 13.1 PL software (StatSoft, Tulsa, OK, USA). Analysis of variance was performed followed by a post hoc least significant difference test (ANOVA; p < 0.05) to identify any significant differences among the experimental groups. Accumulation of Mn by bacterial strain was expressed as mean ± SD.
3. Results and Discussion
3.1. Selection of Manganese-Accumulating Strain
In 1 g of D.W. of analyzed soil 43 ± 5.22 mg of Mn(II) and 0.819 ± 0.76 mg of Mn(IV) was present. Other metals identified in tested soil sample was shown in Table 1.

Table 1.
Metals other than manganese and their content in tested soil sample. “c” represents the range in which the content of the given metal was in the tested soil.
Eleven Mn-tolerant bacteria were isolated from Mn-contaminated waste soil from the heap in Upper Silesia using a spread plate procedure and were designated 2Ge, 2Dh, 2De, 2Dm, 2Dl, 2D10, 2Gh, 2Dj, 2Gm, 2Gi, and 2D12. In order to identify a strain with an ability to accumulate manganese, estimation of metal content in medium with the soil was performed. It was predicted that when isolated bacteria were able to accumulate Mn, the concentration of this metal in medium would decrease. The introduction of most of the isolated bacteria significantly (p < 0.05) reduced Mn content in the samples (Table 2). After 14 days, the best removal percentage was obtained for 2De, and it was 71.07%. This strain was chosen for further analysis.

Table 2.
Decrease in Mn concentration in eleven samples containing 2% (w/v) of the soil from metallurgical waste heap and a single, isolated strain. Error bars are ± standard deviation (n = 3). Values within a column followed by the same letter are not significantly different at p < 0.05 (LSD).
3.2. Phylogenic Analyses
The 16S rRNA sequence of the 2De strain has been submitted to GenBank and was assigned the accession number MK570454. The sequence was aligned and compared with known sequences in the GeneBank database. The analysis indicated that the closest related species was Pseudomonas baetica a390 with 99.80% similarity. The following closest related species were: Pseudomonas umsongensis Ps 3-10 (98.99%), Pseudomonas migulae NBRC 103157 (98.91%), Pseudomonas reinekei MT1 (98.91%), and Pseudomonas helmanticensis OHA11 (98.83%). The highest ANI for the 2De strain was observed for P. baetica a390 (100%) and P. helmanticensis OHA11 (99.40%). Figure 1 presents a phylogenetic tree of the 2De strain and the 10 closest type strains of other Pseudomonas species and shows the isolate cluster separately in a group as a sister taxon to Pseudomonas baetica a390.
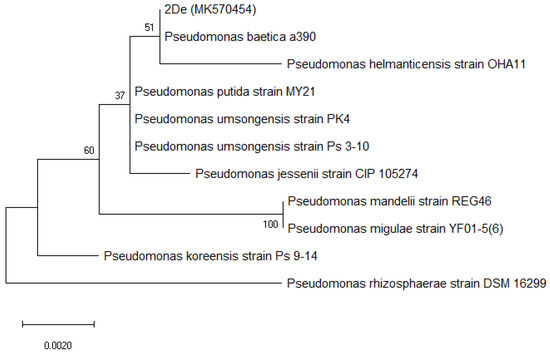
Figure 1.
Maximum likelihood-based phylogeny derived from 16S rRNA sequence data, representing the relative position of the 2De strain among other Pseudomonas. All positions containing gaps or missing data were deleted. Bootstrap values are represented at the branching points. The bar represents 0.002 substitutions per site, accession number from GenBank for 2De strain is shown in brackets.
3.3. Determination of MIC and Effects of Mn on Cellular Protein Expression
The MIC was defined as the lowest metal concentration at which bacterial growth was not observed. The isolated strain was found to be viable in medium with a MnCl2 concentration up to 800 mM. The MIC was therefore estimated to be 800 mM. The ability to tolerate a high concentration of Mn was established for strains that were isolated from Mn-rich environments. For example, Marinomonas sp. S11-S-4 isolated from sediment collected from the Arctic Ocean was able to grow when the Mn2+ concentration was up to 100 mM [33], while strains isolated from Sanindipur mines (Odisha, India) showed visible growth at a concentration up to 500 mM of Mn [1,6]. Interestingly, Acinetobacter sp. MSB 5 isolated from Sanindipur mines was also viable at even a 1000 mM concentration of MnCl2 [5]. Meanwhile, Bacillus thuringiensis HM7 isolated from Xiangtan Mn ore in China was able to grow at 31.78 mM [34].
The total protein content of the 2De strain was found to be 29.97 mg mL−1 for cells cultivated in the presence of 0.5 mM MnO2, 28.30 mg mL−1 when bacteria were cultivated in the presence of the soil, and 31.34 mg mL−1 for control cells. To analyze the effect of a high level of Mn on the protein profiles of the isolated bacterial strain, SDS/PAGE of the whole cell lysate was carried out (Figure 2).
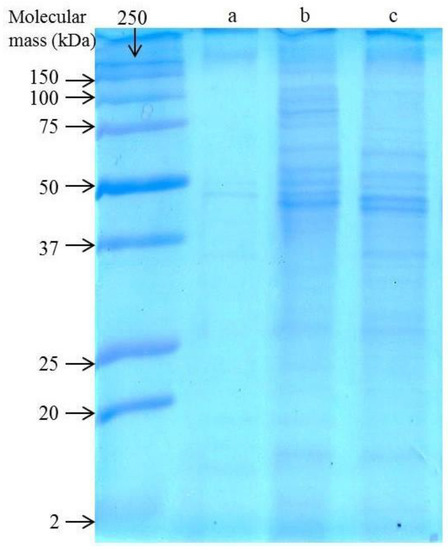
Figure 2.
SDS/PAGE of whole cell lysates of the untreated 2De strain (a) in comparison to the strain cultivated with 0.5 M MnO2 (b) or 4% soil from metallurgical waste heap (c).
When the 2De strain was cultivated in the absence of Mn, weakly visible bands in the range of 12 to 250 kDa were obtained (Figure 2a), while exposure of the strain both to the soil or MnO2 influenced most of the protein synthesis, indicated by more intense staining (Figure 2b,c). Mn serves as a cofactor for bacterial enzymes involved in the removal of harmful free radicals produced during metabolism. However, high Mn accumulation exerts toxic effects on cells. Therefore, it is necessary for bacteria to regulate the Mn acquisition system [35]. They adjust to changing environmental conditions due to utilizing numerous transcriptional and biochemical regulators for uptake and export of Mn. These include intracellular accumulation, enhanced activity of efflux pumps, detoxifying enzymes, intracellular, and extracellular sequestration, and Mn exporter protein like MntE, MntP, MntX, CtpC, and P-type ATPase [2,4,18,19,20]. These exporters are critical for proper Mn homeostasis and cell function for maintaining sufficient Mn levels. Their metal-binding capability may lead to its preferential association with Mn(II) or Mn(IV) particles [36]. On the other hand, the protein Alx, which has a mechanism of action that is not primarily through Mn export, can increase Mn levels and contribute to cell growth under high exposure to Mn [2]. The presence of Mn might also induce synthesis of oxide-associated proteins including metalloproteases. This phenomenon was also observed in two bacterial cultures of Mn(II)-oxidizers: Roseobacter sp. AzwK- 3b and Erythrobacter sp. SD- 21 [36]. In turn, Robinson-Lora and Brennan discovered that chitin-assosiated proteins are involved in the removal of manganese from mine impacted waters through adsorption. These authors showed that the presence of proteins significantly increases the sorption capacity of the crab-shield particles [37]. Considering the increased intensity of protein bands of the 2De strain after treatment with metallurgical waste or MnO2, it seems very likely that some of these mechanisms are present in its cells. For Acinetobacter sp. MSB 5 and Chromohalobacter beijerinckii, the presence of Mn was accompanied by the upregulation of proteins in the bacterial cells as well [5,38]. In summary, it is predicted that the observed changes in the 2De protein profile before and after Mn stress resulted from the mineral–microbe interaction, which finally leads to metal bioaccumulation or biosorption.
3.4. Removal Efficiency
The potential of the 2De strain to reduce the Mn concentration was investigated. During this process, bacteria can remove metals from soil by both bioaccumulation and biosorption [39]. It was reported that living cells have a greater capacity for metal removal than non-living cells [40]; therefore, live bacterial cells were used in the experiment. The pH value and surface tension were carefully controlled over the entire period of bacteria cell and soil contact. When soil concentration increased from 1% to 6%, Mn removal efficiency increased from 46% to 81% after three days of incubation (Figure 3). After six days, the Mn removal efficiencies in all research samples were comparable and were within the range of 85–89%. At the end of the experiment, the highest Mn removal (88%) was recorded in the set containing soil at a concentration (w/v) of 4%, but it was not significantly higher compared to the sets enriched in 2% and 6% (w/v) soil.
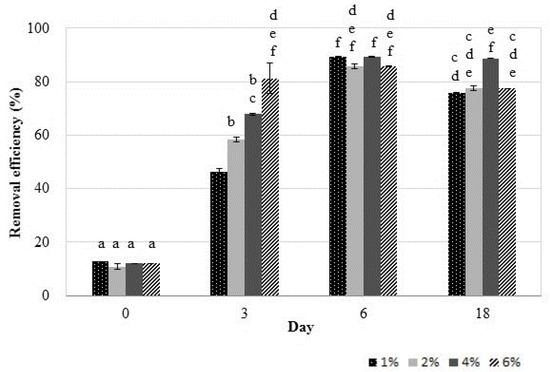
Figure 3.
Manganese removal efficiencies of the 2De strain after incubation with different concentrations of soil at 28 °C for 18 days. The same letter indicates no significant differences among groups (LSD, p < 0.05). Error bars are the standard deviation (n = 3).
The high percentage of Mn accumulation can be attributed to the fact that the 2De strain has been isolated from the soil from metallurgical waste heap. In such an environment, autochthonous strains demonstrate high tolerance to excess heavy metal concentration due to the stress coping mechanisms. Some of these mechanisms, especially utilizing proteins, are certainly present in 2De cells, as shown in the SDS/PAGE analysis (Figure 2). The results obtained in this study showed that throughout the duration of the experiment the pH increased (Figure 4a), and this certainly affects the increase in Mn removal efficiency (Figure 3).
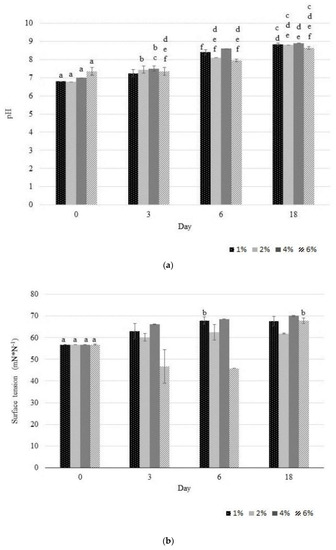
Figure 4.
Variation in pH (a) and in surface tension (b) leachate observed during the accumulation study of Mn by the 2De strain using different soil concentration from the metallurgical waste. The same letter indicates no significant differences among groups (LSD, p < 0.05). Error bars are standard deviation (n = 3).
These results suggested that strain 2De can self-mediate pH changes of medium K in whole cultivation period. This self-mediated pH of 2De strain is an interesting phenomenon, which is worthy further investigation to learn how this phenomenon affects biosorption and bioaccumulation of Mn by the growing 2De cells. Our results were well agreed with the findings of Li et al. [41] who revealed that Bacillus cereus strain Cd01-mediated pH decrease of the cultivation medium resulting in inhibition of Cd2+ biosorption of on Cd01 cells at the initial cultivation period. Along with the duration of the cultivation period an increased pH was observed, which was associated with facilitated biosorption of Cd2+ on Cd01 cells. Limcharoensuk et al. [42] noted that high pH (<7.5) promoted biosorption of metal on cell surface and its bioaccumulation inside cell due to the deprotonation of metal binding sites on cell surface. Other researches observed the low Mn2+ adsorption rate in an acidic condition (pH < 5), caused by the large amounts of protons like H3O+ and H+, which increased the difficulty in binding sites of Mn2+ to cell walls. When pH increased the higher removal rate was reached, which could be connected with more negatively charged cells available, promoting a greater metal uptake [34]. Another reason of this observation is that under acidic conditions, the carboxyl groups responsible for the retention of metals are protonated and restrict entry of metallic ions, whereas at a higher pH, these groups are negatively charged, facilitating the binding of the metals ions [43,44]. Other groups which were identified as functional groups involved in biosorption were hydroxyl, carboxyl, amino, ester, sulfhydryl, carbonyl, and phosphoryl groups [45]. Results of X-ray absorption fine structure (XAFS) spectroscopy analysis showed that below pH 4.4, metal binds mostly to phosphoryl ligands available in the Bacillus subtilis wall, whereas at more alkaline pH, adsorption to carboxyl groups becomes more important. At pH 7.8, activation of a different binding site was observed [46]. Presumably, the tested strain may also produce alkaline complexing agents as metabolites from the catabolism of proteins [47]. These metabolites may dissolve Mn by the formation of soluble metal complexion, which could easily bind to the bacteria cell surface. The increased Mn(II) as well as Cd(II) and Zn(II) adsorption onto bacteria with increasing pH has already been observed by other authors [42,43,48]. Most probably, the phenomena described above may be the cause of differences observed by us in the biosorption of Mn (Figure 4a), associated with a self-mediated change in pH.
Moreover, when soil at a concentration of 6% (w/v) was used, a decrease in surface tension (Figure 4b) was observed, which takes place when bacteria produce biosurfactants. It was speculated that this phenomenon was linked with high Mn extraction efficiency (81%) obtained on the third day of the experiment. Biosurfactants are metal-complexing agents that may increase the hydrophobicity of the bacterial cell surface, allowing hydrophobic substrates to associate more easily with bacterial cells [49,50]. According to the literature, biosurfactants are effective in remediation of metal-contaminated environments, especially under alkaline conditions [51]. This was confirmed by biosurfactant synthesizing Burkholderia sp. Z-90, which produces alkaline conditions during heavy metal (Zn, Pb, Mn, Cd, Cu, and As) removal from soil, indicating that this strain facilitated heavy metals removal via increasing pH. Nevertheless, apart from soil concentration and pH, the change of the leachate surface tension value was increased during the bioleaching process (Figure 4b), likely due to an interaction between Mn in the soil and the biosurfactant. The same results were obtained by Yang et al., [52] who concluded that it was caused by pre-modification the soil surface properties by the biosurfactant, due to the formation of a metal complex on the soil particles. Other authors also observed the significant contribution of biosurfactants in the removal of heavy metals-contaminated soils. For instance, Singh and Cameotra [53] reported that Bacillus subtilis producing lipopeptide biosurfactants removed a significant amount of metals: Zinc (32.07 %), Cadmium (44.2 %), Copper (26.2 %) Cobalt (35.4 %), Nickel (32.2 %), and Lead (40.3 %). Dahrazma and Mulligan [54], demonstrated that biosurfactants have the ability to remove the large amount of heavy metals from soil sediments. The removal percentage were up to 27% of Ni, 13% of Zn, and 37% of Cu, when the biosurfactants were used in a continuous flow configuration. Presence of rhamnolipids, supported recovery of Cd2+ from a soil component—kaolin [55]. In another study, dirhamnolipid selectively removed bioavailable and bound forms of Pb (88%) and Cd (92%) from contaminated soil [56].
3.5. Manganese Content in Bacterial Cells
Metal accumulation analysis revealed a significant increase in Mn content in 2De cells (Figure 5), which indicated that the strain was able to bind to Mn efficiently. Interestingly, in the duration of the experiment, the strain accumulated a higher level of Mn from the soil than from the manganese oxide. This result was confirmed when post-exposure Mn contents in cell walls and intracellular spaces of the isolate were determined. Likewise, when bacteria accumulated Mn from the soil, its level increased with incubation time opposite to the level of Mn adsorbed from MnO2 (Figure 6). In addition, the results suggested that a majority of Mn was cell-surface bound, as a high amount of this metal was present in supernatants after EDTA washing (Figure 6a). EDTA has the ability to porate the outer but not the inner membrane of gram negative bacteria; hence, it can chelate various cations from the exterior as well as from the envelope layer of the cells [57]. The ability of 2De strain to accumulate high concentrations of Mn, and the specificity of the accumulation process, suggest the existence of dedicated Mn storage mechanisms in cells of this strain.
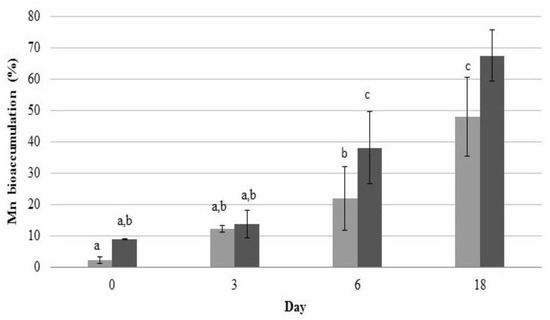
Figure 5.
Mn accumulation in cells of the 2De strain during accumulation studies with 0.5 M MnO2 (light grey column) and 4% soil from the metallurgical waste heap (dark grey column). Error bars are standard deviation (n = 3).
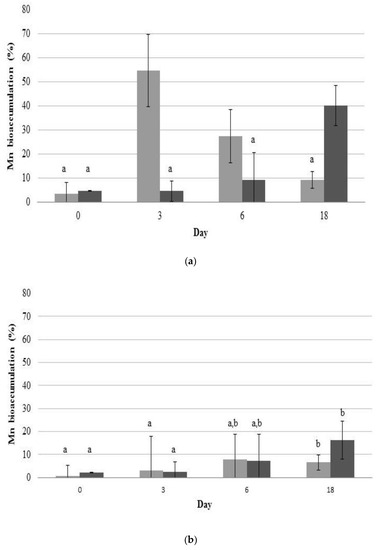
Figure 6.
Mn contents accumulated in cell walls of 2De strain (a) and its intracellular spaces (b). Strain was cultivated in K medium containing 0.5 mM MnO2 (light grey column) or 4% soil from metallurgical waste heap (dark grey column) at 28 °C. Error bars are ± standard deviation (n = 3). Values within a column followed by the same letter are not significantly different at p < 0.05 (LSD).
Moreover, levels of intracellular and cell surface-bound Mn accumulated from MnO2 decreased with incubation time (Figure 6). It is believed that this difference resulted from the forms in which Mn occurred. When Mn was in the form of MnO2, it attached easily to the cell wall via physical adsorption or ion exchange, which is biosorption. The biosorption equilibrium occurs very quickly from the beginning of contact between the metal and microorganism as a result of rapid physical adsorption stage, followed by desorption of this metal into solution (Figure 6a) [58]. Mn from the metallurgical waste was mostly on the second oxidation stage and was sorbed onto bacterial cell walls as well but less rapidly than MnO2, presumably due to the presence of other metals (Pb, K, Zn, Si, Au, Sr, and S), which probably influences Mn sorption to bacterial cells. The change of metal concentration in 2De strain, that we observed in our experiment (Figure 5 and Figure 6), suggests the biosorption phenomenon, which involves mechanisms of binding metals to the cell surface, and not bioaccumulation, during which ion transport to the cell takes place. These results indicate that 2De strain might be a useful tool for bioremediation of Mn-contaminated soil, particularly when applying metal biosorption as a preliminary step to decrease metal concentrations prior to biological sulfate reduction to prevent microbial inhibition due to metal toxicity [59].
4. Conclusions
Undoubtedly, each soil deposit has a unique composition and properties that create special conditions for microorganisms. One universal soil bioremediation method is unlikely to be applied. Concerns about Mn contamination and its growing demand have created a need for reducing the content of this metal in the environment using biological techniques that allow for recovery of this metal. In this study, we isolated a Mn-tolerant 2De bacterial strain from metallurgical waste rich in Mn. This strain was found to have potential to biosorb this metal on its surface, and thus reduce the amount of Mn in the created systems. The manganese removal efficiency was correlated with soil concentration. The efficiency increased with soil concentration increasing. Moreover, during this process, the changes in pH of the K medium were observed resulting from pH self-mediating abilities of 2De strain. The tested strain was also able to biosurfactant production enhancing the efficiency of Mn removal, but the phenomenon was only observed when the soil was at concentration 6%. This suggests that organic and inorganic compounds that were present in the soil stimulate biosurfactant production. Furthermore, it seems that manganese both in the soil and in the form of MnO2 is the reason behind the induction of expression of cellular proteins. It is well known that proteins play a significant role in metal–microbe interphase with soil particles. It is imperative to study contact mechanisms in advance to identify specific proteins involved in the accumulation procedure so that information can pave the way for the better biological recovery of metals from soil.
The ability of the 2De isolate makes it a suitable candidate for the treatment of environments contaminated with Mn and its extraction from these ecosystems. However, further tests defining the best way to recover Mn utilizing biosorbents are still needed before using this strain in field-scale applications. In addition, detailed biosorption tests should always be performed before attempting to use microorganisms to enrich a given soil.
Author Contributions
K.N. and Z.P.-S. conceived the project, M.N. and K.Ł. executed the experiments, M.N. carried out the analyses. M.N. wrote the paper with input from other co-authors. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Conflicts of Interest
Authors declare no conflict of interest.
References
- Ghosh, S.; Mohanty, S.; Akcil, A.; Sukla, L.B.; Das, A.P. A greener approach for resource recycling: Manganese bioleaching. Chemosphere 2016, 154, 628–639. [Google Scholar] [CrossRef] [PubMed]
- Zeinert, R.; Martinez, E.; Schmitz, J.; Senn, K.; Usman, B.; Anantharaman, V.; Aravind, L.; Waters, L.S. Structure-function analysis of manganese exporter proteins across bacteria. J. Biol. Chem. 2018, 1–34. [Google Scholar] [CrossRef] [PubMed]
- Juttukonda, L.J.; Skaar, E.P. Manganese homeostasis and utilization in pathogenic bacteria. Mol. Microbiol. 2015, 97, 216–228. [Google Scholar] [CrossRef] [PubMed]
- Pereira, F. Bioremediation of Contaminated Estuarine Sites. In Marine Pollution and Microbial Remediation; Naik, M.M., Dubey, S.K., Eds.; Springer: Singapore, 2016; ISBN 9789811010422. [Google Scholar]
- Ghosh, S.; Das, A.P. Bioleaching of manganese from mining waste residues using Acinetobacter sp. Geol. Ecol. Landsc. 2017, 1, 77–83. [Google Scholar] [CrossRef]
- Ghosh, S.; Bal, B.; Das, A.P. Enhancing Manganese Recovery from Low-Grade Ores by Using Mixed Culture of Indigenously Isolated Bacterial Strains. Geomicrobiol. J. 2018, 35, 242–246. [Google Scholar] [CrossRef]
- Venugopal, R.; Engineering, M. Solid Waste Processing for Industrial Utilization—A Few Case Studies. In Proceedings of the XI International Seminar on Mineral Processing Technology (MPT-2010), Abhilash, India, 15–17 December 2010; pp. 1075–1083. [Google Scholar]
- Kim, S.I.; Jang, Y.S.; Han, S.H.; Choi, M.J.; Go, E.H.; Cheon, Y.P.; Lee, J.S.; Lee, S.H. Effect of Manganese Exposure on the Reproductive Organs in Immature Female Rats. Dev. Reprod. 2012, 295, 295–300. [Google Scholar] [CrossRef]
- Santamaria, A.B.; Sulsky, S.I. Risk assessment of an essential element: Manganese. J. Toxicol. Environ. Health Part A Curr. Issues 2010, 73, 128–155. [Google Scholar] [CrossRef]
- Sis, H.; Chander, S. Reagents used in the flotation of phosphate ores: A critical review. Miner. Eng. 2003, 16, 577–585. [Google Scholar] [CrossRef]
- Veglio’, F.; Beolchini, F. Removal of metals by biosorption: A review. Hydrometallurgy 1997, 44, 301–316. [Google Scholar] [CrossRef]
- Behera, S.K.; Mulaba-Bafubiandi, A.F. Advances in microbial leaching processes for nickel extraction from lateritic minerals—A review. Korean J. Chem. Eng. 2015, 32, 1447–1454. [Google Scholar] [CrossRef]
- Sadeghabad, M.S.; Bahaloo-Horeh, N.; Mousavi, S.M. Using bacterial culture supernatant for extraction of manganese and zinc from waste alkaline button-cell batteries. Hydrometallurgy 2019, 188, 81–91. [Google Scholar] [CrossRef]
- Sheng, X.F.; Xia, J.J.; Jiang, C.Y.; He, L.Y.; Qian, M. Characterization of heavy metal-resistant endophytic bacteria from rape (Brassica napus) roots and their potential in promoting the growth and lead accumulation of rape. Environ. Pollut. 2008, 156, 1164–1170. [Google Scholar] [CrossRef] [PubMed]
- Lo, Y.C.; Cheng, C.L.; Han, Y.L.; Chen, B.Y.; Chang, J.S. Recovery of high-value metals from geothermal sites by biosorption and bioaccumulation. Bioresour. Technol. 2014, 160, 182–190. [Google Scholar] [CrossRef] [PubMed]
- Dhankhar, R.; Hooda, A. Fungal biosorption-an alternative to meet the challenges of heavy metal pollution in aqueous solutions. Environ. Technol. 2011, 32, 467–491. [Google Scholar] [CrossRef]
- Yoo, K.; Shin, S.M.; Yang, D.H.; Sohn, J.S. Biological treatment of wastewater produced during recycling of spent lithium primary battery. Miner. Eng. 2010, 23, 219–224. [Google Scholar] [CrossRef]
- Rosch, J.W.; Gao, G.; Ridout, G.; Wang, Y.D.; Tuomanen, E.I. Role of the manganese efflux system mntE for signalling and pathogenesis in Streptococcus pneumoniae. Mol. Microbiol. 2009, 72, 12–25. [Google Scholar] [CrossRef]
- Veyrier, F.J.; Boneca, I.G.; Cellier, M.F.; Taha, M.K. A novel metal transporter mediating manganese export (mntx) regulates the mn to fe intracellular ratio and Neisseria meningitidis virulence. PLoS Pathog. 2011, 7. [Google Scholar] [CrossRef]
- Waters, L.S.; Sandoval, M.; Storz, G. The Escherichia coli MntR miniregulon includes genes encoding a small protein and an efflux pump required for manganese homeostasis. J. Bacteriol. 2011, 193, 5887–5897. [Google Scholar] [CrossRef]
- Yuan, W.; Cheng, J.; Huang, H.; Xiong, S.; Gao, J.; Zhang, J.; Feng, S. Optimization of cadmium biosorption by Shewanella putrefaciens using a Box-Behnken design. Ecotoxicol. Environ. Saf. 2019, 175, 138–147. [Google Scholar] [CrossRef]
- Endo, R.; Aoyagi, H. Adsorption preference for divalent metal ions by Lactobacillus casei JCM1134. Appl. Microbiol. Biotechnol. 2018, 102, 6155–6162. [Google Scholar] [CrossRef]
- Kluczka, J.; ZoŁotajkin, M.; Ciba, J. Speciation of aluminium in the water and bottom sediment of fish-breeding ponds. Arch. Environ. Prot. 2012, 38, 83–96. [Google Scholar] [CrossRef]
- Smoliński, A.; Zołotajkin, M.; Ciba, J.; Dydo, P.; Kluczka, J. PLS-EP algorithm to predict aluminum content in soils of Beskid Mountains region. Chemosphere 2009, 76, 565–571. [Google Scholar] [CrossRef] [PubMed]
- Kalembkiewicz, J.; Sitarz-Palczak, E.; Zapała, L. A study of the chemical forms or species of manganese found in coal fly ash and soil. Microchem. J. 2008, 90, 37–43. [Google Scholar] [CrossRef]
- Volkov, A.I.; Ossipov, K.B.; Seregin, A.N.; Zhdanov, P.A.; Seregina, I.F.; Bolshov, M.A. Determination of degree of oxidation and forms of manganese compounds in the Ulu-Telyak oxidized ore. Inorg. Mater. 2015, 51, 1394–1403. [Google Scholar] [CrossRef]
- Pacwa-Pł>ociniczak, M.; Płaza, G.A.; Paliwoda, A.; Piotrowska-Seget, Z. Characterization of hydrocarbon-degrading and biosurfactant-producing Pseudomonas sp. P-1 strain as a potential tool for bioremediation of petroleum-contaminated soil. Env. Sci. Pollut. Res. 2014, 9385–9395. [Google Scholar] [CrossRef]
- Yoon, S.H.; Ha, S.M.; Lim, J.M.; Kwon, S.J.; Chun, J. A large-scale evaluation of algorithms to calculate average nucleotide identity. Antonie Leeuwenhoek 2017, 110, 1281–1286. [Google Scholar] [CrossRef]
- Płociniczak, T.; Chodór, M.; Pacwa-Płociniczak, M.; Piotrowska-Seget, Z. Metal-tolerant endophytic bacteria associated with Silene vulgaris support the Cd and Zn phytoextraction in non-host plants. Chemosphere 2019, 219, 250–260. [Google Scholar] [CrossRef]
- Furmanczyk, E.M.; Kaminski, M.A.; Lipinski, L.; Dziembowski, A.; Sobczak, A. Pseudomonas laurylsulfatovorans sp. nov., sodium dodecyl sulfate degrading bacteria, isolated from the peaty soil of a wastewater treatment plant. Syst. Appl. Microbiol. 2018, 41, 348–354. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Tang, M.; Zhu, W.; Yu, J.; Fu, Y.; Fan, L.; Wei, G. Isolation and characterization of the heavy metal resistant bacteria CCNWRS33-2 isolated from root nodule of Lespedeza cuneata in gold mine tailings in China. J. Hazard. Mater. 2008, 162, 50–56. [Google Scholar] [CrossRef]
- Xuezheng, L.; Aiguo, G.; Haowen, C. Isolation and phylogenetic analysis of cultivable manganese bacteria in sediments from the Arctic Ocean. Acta Ecol. Sin. 2008, 28, 6364–6370. [Google Scholar] [CrossRef]
- Huang, H.; Zhao, Y.; Xu, Z.; Ding, Y.; Zhou, X.; Dong, M. A high Mn(II)-tolerance strain, Bacillus thuringiensis HM7, isolated from manganese ore and its biosorption characteristics. PeerJ 2020, 2020, 1–24. [Google Scholar] [CrossRef] [PubMed]
- Wakeman, C.A.; Skaar, E.P. Metalloregulation of Gram-positive pathogen physiology. Curr. Opin. Microbiol. 2012, 15, 169–174. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Estes, E.R.; Andeer, P.F.; Nordlund, D.; Wankel, S.D.; Hansel, C.M. Biogenic manganese oxides as reservoirs of organic carbon and proteins in terrestrial and marine environments. Geobiology 2017, 15, 158–172. [Google Scholar] [CrossRef]
- Robinson-Lora, M.A.; Brennan, R.A. Biosorption of manganese onto chitin and associated proteins during the treatment of mine impacted water. Chem. Eng. J. 2010, 162, 565–572. [Google Scholar] [CrossRef]
- Pereira, F.; Kerkar, S.; Krishnan, K.P. Bacterial response to dynamic metal concentrations in the surface sediments of a solar saltern (Goa, India). Environ. Monit. Assess. 2013, 3625–3636. [Google Scholar] [CrossRef]
- White, C.; Wilkinson, S.C.; Gadd, G.M. The role of microorganisms in biosorption of toxic metals and radionuclides. Int. Biodeterior. Biodegrad. 1995, 35, 17–40. [Google Scholar] [CrossRef]
- Zeng, X.; Tang, J.; Xue-duan, L.I.U.; Pei, J. Isolation, identification and characterization of cadmium-resistant Pseudomonas aeruginosa strain E 1. J. Cent. South Univ. Technol. 2009, 416–421. [Google Scholar] [CrossRef]
- Li, F.; Wang, W.; Li, C.; Zhu, R.; Ge, F.; Zheng, Y.; Tang, Y. Self-mediated pH changes in culture medium affecting biosorption and biomineralization of Cd2+ by Bacillus cereus Cd01. J. Hazard. Mater. 2018, 358, 178–186. [Google Scholar] [CrossRef]
- Limcharoensuk, T.; Sooksawat, N.; Sumarnrote, A.; Awutpet, T.; Kruatrachue, M.; Pokethitiyook, P.; Auesukaree, C. Bioaccumulation and biosorption of Cd2+ and Zn2+ by bacteria isolated from a zinc mine in Thailand. Ecotoxicol. Environ. Saf. 2015, 122, 322–330. [Google Scholar] [CrossRef]
- Calderón, M.E.; Elena, M.; Buitrón, G. Biosorption of Cd, Cr, Mn and Pb from aqueus solution by Bacillus sp. strains isolated from industrail waste activate sludge. TIP Rev.Esp.Cienc.Quím.Biol. 2016, 19, 5–14. [Google Scholar] [CrossRef]
- Pacheco, P.H.; Gil, R.A.; Cerutti, S.E.; Smichowski, P.; Martinez, L.D. Biosorption: A new rise for elemental solid phase extraction methods. Talanta 2011, 85, 2290–2300. [Google Scholar] [CrossRef] [PubMed]
- Javanbakht, V.; Alavi, S.A.; Zilouei, H. Mechanisms of heavy metal removal using microorganisms as biosorbent. Water Sci. Technol. 2014, 69, 1775–1787. [Google Scholar] [CrossRef] [PubMed]
- Boyanov, M.; Kelly, K.M.; Bunker, B.A.; Fein, J.B.; Fowle, D.A. Adsorption of cadmium to Bacillus subtilis bacterial cell walls: A pH-dependent X-ray absorption fine structure spectroscopy study. Geochim. Cosmochim. Acta 2003, 67, 3299–3311. [Google Scholar] [CrossRef]
- Anjum, F.; Shahid, M.; Akcil, A. Hydrometallurgy Biohydrometallurgy techniques of low grade ores: A review on black shale. Hydrometallurgy 2012, 117, 1–12. [Google Scholar] [CrossRef]
- Vázquez-Ortega, A.; Fein, J.B. Thermodynamic modeling of Mn (II) adsorption onto manganese oxidizing bacteria. Chem. Geol. 2017, 464, 147–154. [Google Scholar] [CrossRef]
- Das, P.; Mukherjee, S.; Sen, R. Bioresource Technology Biosurfactant of marine origin exhibiting heavy metal remediation properties. Bioresour. Technol. 2009, 100, 4887–4890. [Google Scholar] [CrossRef]
- Pacwa-Płociniczak, M.; Płaza, G.A.; Piotrowska-Seget, Z.; Cameotra, S.S. Environmental applications of biosurfactants: Recent advances. Int. J. Mol. Sci. 2011, 12, 633–654. [Google Scholar] [CrossRef]
- Reynier, N.; Blais, J.; Mercier, G. Optimization of arsenic and pentachlorophenol removal from soil using an experimental design methodology. J. Soils Sediments 2013, 13, 1189–1200. [Google Scholar] [CrossRef]
- Yang, L.; Cheng, Q.; Lin, L.; Su, W.; Luan, T. Contributions of Abiotic and Biotic Processes to the Aerobic Removal of Phenolic Endocrine-Disrupting Chemicals in a Simulated Estuarine Aquatic Environment. Environ. Sci. Technol. 2016, 50, 4324–4334. [Google Scholar] [CrossRef]
- Singh, A.K.; Cameotra, S.S. Efficiency of lipopeptide biosurfactants in removal of petroleum hydrocarbons and heavy metals from contaminated soil. Environ. Sci. Pollut. Res. 2013, 20, 7367–7376. [Google Scholar] [CrossRef]
- Dahrazma, B.; Mulligan, C.N. Investigation of the removal of heavy metals from sediments using rhamnolipid in a continuous flow configuration. Chemosphere 2007, 69, 705–711. [Google Scholar] [CrossRef] [PubMed]
- Aşçi, Y.; Nurbaş, M.; Saǧ Açikel, Y. A comparative study for the sorption of Cd(II) by K-feldspar and sepiolite as soil components, and the recovery of Cd(II) using rhamnolipid biosurfactant. J. Environ. Manage. 2008, 88, 383–392. [Google Scholar] [CrossRef] [PubMed]
- Juwarkar, A.A.; Nair, A.; Dubey, K.V.; Singh, S.K.; Devotta, S. Biosurfactant technology for remediation of cadmium and lead contaminated soils. Chemosphere 2007, 68, 1996–2002. [Google Scholar] [CrossRef] [PubMed]
- Keren, N.; Kidd, M.J.; Penner-Hahn, J.E.; Pakrasi, H.B. A light-dependent mechanism for massive accumulation of manganese in the photosynthetic bacterium Synechocystis sp. PCC 6803. Biochemistry 2002, 41, 15085–15092. [Google Scholar] [CrossRef] [PubMed]
- Fadel, M.; Hassanein, N.M.; Elshafei, M.M.; Mostafa, A.H.; Ahmed, M.A.; Khater, H.M. Biosorption of manganese from groundwater by biomass of Saccharomyces cerevisiae. HBRC J. 2017, 13, 106–113. [Google Scholar] [CrossRef]
- Utgikar, V.P.; Harmon, S.M.; Chaudhary, N.; Tabak, H.H.; Govind, R.; Haines, J.R. Inhibition of sulfate-reducing bacteria by metal sulfide formation in bioremediation of acid mine drainage. Environ. Toxicol. 2002, 17, 40–48. [Google Scholar] [CrossRef]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).