Simple Summary
Currently, controversy about the intraspecific taxonomy of Amolops mantzorum remains unresolved. The Litang population was overlooked for several years in recent phylogenetic studies. We revisited this population and described the third subspecies of A. mantzorum, clearly dividing the four recognized lineages within A. mantzorum for the first time based on mitochondrial phylogeny. Our findings highlighted the importance of clear delimitation and naming of subspecies for biological conservation and taxonomic description, and provided insights for future investigations into the taxonomy of the Amolops mantzorum species group.
Abstract
Accurate species delimitation benefits detailed biological descriptions and conservation practices. In this study, we focus on a frog species distributed throughout the western edge of Sichuan Basin, China, the Sichuan torrent frog—Amolops mantzorum. Currently, A. mantzorum is recognized as comprising two subspecies—the nominate subspecies A. m. mantzorum and A. m. xinduqiao. We investigated a population from along the Yalong River, Sichuan Province, China which was previously overlooked in phylogenetic studies. Combining multiple mitochondrial genes (16S, CO1, cytb), we identified four distinct lineages within A. mantzorum using Bayesian inference and maximum likelihood phylogeny. Principal Component Analysis based on morphometrics revealed significant differences between the population from along the Yalong River and the other two subspecies for which morphological data were available. Combined with detailed morphological descriptions and comparisons, here we describe this population as the third subspecies of A. mantzorum—Amolops mantzorum feiye ssp. nov.
1. Introduction
The intraspecific taxonomy of Amolops mantzorum has been debated for several decades. Cai & Zhao [1] identified four genetic populations within the “A. mantzorum & A. kangtingensis complex” based on genetic distance. Two of these, exhibiting large morphometric divergence, are currently recognized as subspecies of A. mantzorum. Amolops m. xinduqiao (A. kangtingensis Kangding population sensu Cai & Zhao [1]) represents the lineage with smaller body size and a high-altitude distribution range (above 3000 m), distributed along the western slopes of Mt. Zheduo in the Yalong River Basin, whereas A. m. mantzorum (A. mantzorum Hongya and Dayi population sensu Cai & Zhao [1]) comprises larger-bodied, mid-to-high elevation (1200–2400 m) populations from the Dadu River Basin [2]. The two subspecies are also distinguished by different dorsal coloration patterns [2]. However, the coloration patterns can vary within a species and are thus insufficient for taxonomic diagnosis in the A. mantzorum group under current views. Given the small genetic divergence revealed by molecular phylogeny (i.e., Wu et al. [3]; Zeng et al. [4]), the subspecies-level classification proposed by Dufresnes & Litvinchuk [5] has been accepted in recent taxonomic systems (e.g., Tang et al. [6]; Frost [7]; Li et al. [8]).
Obviously, subspecies delimitation within A. mantzorum remains unresolved. Unfortunately, the CO1 and Cytb sequences used by Cai & Zhao [1] were not available in their original publication. However, some evidence could be traced. Lu et al. [9] recognized an undescribed lineage from Maoxian, Sichuan Province and Wenxian, Gansu Province, designating it as “A. mantzorum (northern lineage)”. This population is geographically close to the Lixian and Wolong populations of A. mantzorum in Cai & Zhao [1]. However, the Litang population—one of the four distinct genetic populations identified by Cai & Zhao [1]—was overlooked for several years in molecular phylogeny studies [3,4,5,10].
In this study, we revisited the Litang population of A. mantzorum, combining specimens collected from multiple localities along the Yalong River, Sichuan Province, China. Molecular phylogeny based on mitochondrial genes suggests that this population represents a monophyletic lineage. A comprehensive morphological examination was also conducted among the recognized subspecies of A. mantzorum. Based on this evidence, we describe this previously overlooked lineage as a new subspecies of A. mantzorum.
2. Material and Methods
2.1. Samples
From 2019 to 2021, 20 specimens and 2 tissue samples of the putative new subspecies were collected from Litang County, Yajiang County, Muli County, and Daocheng County in Sichuan Province (Figure 1). Additionally, 3 specimens of A. m. xinduqiao were collected from Yajiang County and Kangding City in Sichuan Province, and 14 specimens of A. m. mantzorum were collected from Baoxing County, Shimian County, Luding County and Kangding City in Sichuan Province. From the collected specimens and samples, we extracted tissues from 22 samples of the putative new subspecies, 2 of A. m. mantzorum, and 3 of A. m. xinduqiao. Muscle and/or liver tissues were preserved in 95% ethanol for molecular analyses. Specimens were fixed in formaldehyde solution and then transferred to 75% ethanol for long-term storage. All specimens were deposited in the Chengdu Institute of Biology (CIB) of Chinese Academy of Sciences (CAS). Museum specimens were also examined for morphological analyses (detailed in Appendix A).

Figure 1.
(A) Geographical locations of the type localities of species within the A. mantzorum group. (B) Distribution of specimens (or sequences) used in this study of A. m. ssp., A. m. xinduqiao, A. m. mantzorum, and “A. m ssp. (northern lineage)”. The numbers (B) beside the distribution points correspond to the IDs in Table 1.

Table 1.
Sequences used in this study.
Table 1.
Sequences used in this study.
| ID | Species | Voucher Number | Locality | 16S | COI | Cytb |
|---|---|---|---|---|---|---|
| 1 | Amolops mantzorum ssp. | CIB QZ2021130 | Bajiaolou Village, Bajiaolou Town, Yajiang, Sichuan | PX599057 | PX599084 | PX620422 |
| 2 | A. m. ssp. | CIB QZ2021126 | Bajiaolou Village, Bajiaolou Town, Yajiang, Sichuan | PX599068 | PX599095 | PX620433 |
| 3 | A. m. ssp. | CIB QZ2021131 | Bajiaolou Village, Bajiaolou Town, Yajiang, Sichuan | PX599075 | PX599102 | PX620438 |
| 4 | A. m. ssp. | CIB QZ2021128 | Bajiaolou Village, Bajiaolou Town, Yajiang, Sichuan | PX599077 | PX599104 | PX620440 |
| 5 | A. m. ssp. | CIB QZ2021127 | Bajiaolou Village, Bajiaolou Town, Yajiang, Sichuan | PX599079 | PX599106 | PX620442 |
| 6 | A. m. ssp. | CIB YJ2019080602 | Bajiaolou Village, Bajiaolou Town, Yajiang, Sichuan | PX599080 | PX599107 | PX620443 |
| 7 | A. m. ssp. | CIB YJ2019080601 | Bajiaolou Village, Bajiaolou Town, Yajiang, Sichuan | PX599081 | PX599108 | PX620444 |
| 8 | A. m. ssp. | CIB YJ2019080603 | Bajiaolou Village, Bajiaolou Town, Yajiang, Sichuan | PX599082 | PX599109 | PX620445 |
| 9 | A. m. ssp. | CIB YJ202012 | Nimazong Village, Zhusang Town, Yajiang, Sichuan | PX599083 | PX599110 | PX620446 |
| 10 | A. m. ssp. | CIB YJ202011 | Nimazong Village, Zhusang Town, Yajiang, Sichuan | PX599058 | PX599085 | PX620423 |
| 11 | A. m. ssp. | CIB YJ202013 | Nimazong Village, Zhusang Town, Yajiang, Sichuan | PX599059 | PX599086 | PX620424 |
| 12 | A. m. ssp. | CIB LT20200712-4 | Dewu Town, Litang, Sichuan | PX599060 | PX599087 | PX620425 |
| 13 | A. m. ssp. | CIB LT20200712-5 | Dewu Town, Litang, Sichuan | PX599061 | PX599088 | PX620426 |
| 14 | A. m. ssp. | CIB LT20200712-6 | Dewu Town, Litang, Sichuan | PX599062 | PX599089 | PX620427 |
| 15 | A. m. ssp. | CIB LT20200712-8 | Dewu Town, Litang, Sichuan | PX599063 | PX599090 | PX620428 |
| 16 | A. m. ssp. | CIB DC20190905-89 | Rihuo Village, Shengmu Town, Daocheng, Sichuan | PX599064 | PX599091 | PX620429 |
| 17 | A. m. ssp. | CIB DC20190905-88 | Rihuo Village, Shengmu Town, Daocheng, Sichuan | PX599065 | PX599092 | PX620430 |
| 18 | A. m. ssp. | CIB DC20190905-91 | Rihuo Village, Shengmu Town, Daocheng, Sichuan | PX599066 | PX599093 | PX620431 |
| 19 | A. m. ssp. | CIB DC20190906-94 | Maize Village, Mula Town, Daocheng, Sichuan | PX599067 | PX599094 | PX620432 |
| 20 | A. m. ssp. | CIB DC20190906-95 | Maize Village, Mula Town, Daocheng, Sichuan | PX599069 | PX599096 | PX620434 |
| 21 | A. m. ssp. | CIB 2019ml0002 | Xianggelila Village, Shuiluo Town, Muli, Sichuan | PX599070 | PX599097 | PX620435 |
| 22 | A. m. ssp. | CIB 2019ml0001 | Xianggelila Village, Shuiluo Town, Muli, Sichuan | PX599071 | PX599098 | PX620436 |
| 23 | A. m. xinduqiao | CIB YJ202014 | Wangxia Village, Bajiaolou Town, Yajiang, Sichuan | PX599072 | PX599099 | PX620437 |
| 24 | A. m. xinduqiao | KIZ 041127 | Xinduqiao Town, Kangding, Sichuan | MN953764 | MN961465 | - |
| 25 | A. m. xinduqiao | KIZ 041129 | Xinduqiao Town, Kangding, Sichuan | MN953765 | MN961466 | - |
| 26 | A. m. xinduqiao | CIB GGS-PBX4-2 | Geridi Village, Jiagenba Town, Kangding, Sichuan | PX599073 | PX599100 | - |
| 27 | A. m. xinduqiao | CIB GGS-SDX1-6 | Laha Village, Shade Town, Kangding, Sichuan | PX599074 | PX599101 | - |
| 28 | A. m. xinduqiao | 700307 | Yajiang, Sichuan | - | - | KJ008410 |
| 29 | A. m. xinduqiao | LCLH017 | Luhuo, Sichuan | - | - | KJ008392 |
| 30 | A. m. mantzorum | SYS a005360 | Kangding, Sichuan | MK573807 | MK568322 | - |
| 31 | A. m. mantzorum | SYS a005336 | Mt. Wawu, Hongya, Sichuan | MK573804 | MK568319 | - |
| 32 | A. m. mantzorum | SYS a005337 | Mt. Wawu, Hongya, Sichuan | MK604853 | MK605611 | - |
| 33 | A. m. mantzorum | 700114 | Longdong Town, Baoxing, Sichuan | - | - | KJ008297 |
| 34 | A. m. mantzorum | CIB 2020061501 | Dongsheng Village, Wulong Town, Baoxing, Sichuan | PX599076 | PX599103 | PX620439 |
| 35 | A. m. mantzorum | CIB 2020061502 | Dongsheng Village, Wulong Town, Baoxing, Sichuan | PX599078 | PX599105 | PX620441 |
| 36 | A. m. mantzorum | SYS a005370 | Fengtongzhai, Baoxing, Sichuan | MK604865 | MK605623 | - |
| 37 | A. m. mantzorum | SYS a005365 | Fengtongzhai, Baoxing, Sichuan | MK573808 | MK568323 | - |
| 38 | A. m. mantzorum | SCUM045825HX | Mt. Xiling Snow, Dayi, Sichuan | MN953707 | MN961409 | - |
| 39 | A. m. mantzorum | - | Mt. Xiling Snow, Dayi, Sichuan | KJ546429 | KJ546429 | KJ546429 |
| 40 | A. m. mantzorum | 700229 | Chongzhou, Sichuan | - | - | KJ008339 |
| 41 | A. m. mantzorum | SCUM045817HX | Wolong, Wenchuan, Sichuan | MN953706 | MN961408 | - |
| 42 | A. m. ssp. (northern lineage) | 700040 | Maoxian, Sichuan | - | - | KJ008277 |
| 43 | A. m. ssp. (northern lineage) | 700267 | Wenxian, Gansu | - | - | KJ008360 |
| 44 | A. tuberodepressus | SYS a003932 | Mt. Wuliang, Jingdong, Yunnan | MK573800 | MG991934 | - |
| 45 | A. tuberodepressus | CIB-XM3125 | Jingdong, Yunnan | KR559270 | KR559270 | KR559270 |
| 46 | A. ailao | GXNU YU000004 | Mt. Ailao, Xinping, Yunnan | MN650754 | MN650740 | MN650746 |
| 47 | A. shuichengicus | SYS a004956 | Shuicheng, Guizhou | MK604845 | MK605603 | - |
| 48 | A. granulosus | SCUM060911HX | Anxian, Sichuan | MN953681 | MN961381 | - |
| 49 | A. granulosus | 20130258 | Mt. Wawu, Sichuan, China | MH922934 | MH922934 | MH922934 |
| 50 | A. loloensis | SYS a005346 | Zhaojue, Sichuan | MK604854 | MK605612 | - |
| 51 | A. loloensis | SM-ZDTW-01 | Shimian, Sichuan | KT750963 | KT750963 | KT750963 |
| 52 | A. jinjiangensis | SYS a004571 | Mt. Gaoligong, Yunnan | MK573801 | MK568316 | - |
| 53 | A. jinjiangensis | SCUM050435CHX | Deqing, Yunnan | EF453741 | MN961403 | - |
| 54 | A. jinjiangensis | KIZ047095 | Chuxiong, Yunnan | MN953701 | MN961404 | - |
| 55 | A. sangzhiensis | CSUFT 907 | Mt. Doupeng, Sangzhi, Hunan | OQ079540 | OQ078905 | - |
| 56 | A. chayuensis | SYS a007509 | Baxoi, Xizang | MK573820 | MK568333 | - |
| 57 | A. lifanensis | SYS a005374 | Lixian, Sichuan | MK573809 | MK568324 | - |
| 58 | A. dafangensis dafangensis | MT DF20230601002 | Dafang, Guizhou | OR936315 | OR924345 | - |
| 59 | A. d. dafangensis | MT DF20230601001 | Dafang, Guizhou | OR936314 | OR924344 | - |
| 60 | A. d. wumengmontis | SWU 0009561 | Mt. Wumeng, Zhaotong, Yunnan, China | PX411479 | PX410826 | - |
| 61 | A. d. wumengmontis | SWU 0009562 | Mt. Wumeng, Zhaotong, Yunnan, China | PX411478 | PX410827 | - |
| 62 | A. minutus | IEBR 4342 | Muong La, Son La, Vietnam | - | - | MK941135 |
| 63 | A. minutus | TBU 06 | Muong La, Son La, Vietnam | - | - | MK941136 |
| 64 | A. chunganensis | SYS a004212 | Mt. Jinggang, Jinggang, Jiangxi | MK263263 | MG991914 | - |
| 65 | A. nyingchiensis | SYS a006679 | Lhunze, Xizang | MK573814 | MK568329 | - |
| 66 | A. cremnobatus | ROM 14528 | Khe Moi, Nghe An, Vietnam | DQ204477 | - | - |
| 67 | A. daiyunensis | SYS a001739 | Mt. Daiyun, Fujian, China | MK263243 | KX507328 | - |
| 68 | A. hainanensis | SYS a005281 | Mt. Wuzhi, Hainan, China | MK263281 | MG991916 | - |
| 69 | A. ricketti | WUSTW01 | Mt. Wugong, Jiangxi, China | KF956111 | KF956111 | KF956111 |
| 70 | A. marmoratus | KUHE 19089 | Chiang Mai, Thailand | AB211486 | - | AB259738 |
| 71 | A. spinapectoralis | ROM 37375 | Ngoc Linh vicinity, Kon Tum, Vietnam | MN953726 | MN961428 | - |
| 72 | A. cremnobatus | KIZ 011621 | Puhu National Reserve, Thanh Hoa, Vietnam | MN953672 | MN961368 | - |
| 73 | A. wangyufani | KIZ 014067 | Zayü, Tibet, China | MN953740 | MN961440 | - |
| 74 | A. cuongi | IEBR A.5140 | Tam Duong, Lai Chau, Vietnam | PX113529 | - | PX119668 |
| 75 | A. cuongi | IEBR A.5141 | Tam Duong, Lai Chau, Vietnam | PX113530 | - | PX119669 |
| 76 | Odorrana wuchuanensis | LBML 5230 | Libo, Guizhou, China | KU680791 | KU680791 | KU680791 |
| 77 | Odorrana margaretae | HNNU 1207003 | - | KJ815050 | KJ815050 | KJ815050 |
2.2. Molecular Analysis
We extracted genomic DNA from muscle or liver tissues using a DNA extraction kit (Tiangen Biotech (Beijing) Co., Ltd., Beijing, China). Segments of three mitochondrial genes, 16S ribosomal RNA (16S), cytochrome C oxidase subunit I (COI), and partial cytochrome b (Cytb) were amplified. The 16S primers were L3975 and H4551 [11], and the COI primers were Chmf4 and Chmr4 [12], and Cytb primers were Cytb-L and Cytb-H [13]. Amplification of the 16S, COI, and Cytb fragments was performed in a 25 μL volume reaction: initial denaturation at 94 °C for 5 min (95 °C for Cytb); 34 cycles (31 cycles for Cytb): denaturation for 30 s at 94 °C (95 °C for Cytb), annealing for 30 s at 55 °C (40 s for Cytb), extending for 30 s at 72 °C (1 min for Cytb); a final extension: 7 min at 72 °C (10 min for Cytb). Sequencing was conducted using a 3730xl DNA sequencer by Beijing Tsingke Biotech Co., Ltd. (Beijing, China).
In addition to the 27 sequences newly obtained in this study, 50 sequences were retrieved from GenBank, including two samples of “A. mantzorum (northern lineage)” from Lu et al. [9], currently recognized as “A. m. ssp. (northern lineage)” by Tang et al. [6]. The species Odorrana wuchuanensis and O. margaretae were selected as outgroups following Tang et al. [6]. Sequences of the species in the A. mantorum group were selected mostly following Lyu et al. [14] and supplemented with newly described species and subspecies. To reconstruct more stable trees, we also included one or two sequences from each species group of Amolops. In total, 77 sequences were used for phylogenetic analysis (Table 1).
The MAFFT algorithm [15] with its default parameters in MAFFT online service (https://mafft.cbrc.jp/alignment/server/index.html [accessed on 17 December 2025]) [16] was used to align the 16S (440 bp), COI (562 bp), and Cytb (756 bp) sequences, respectively, and AMAS [17] was used to concatenate the 16S, COI, and Cytb sequences of each specimen. Maximum-likelihood (ML) analysis was conducted using IQ-TREE [18,19]. The support values of the phylogenetic tree were assessed using 1000 bootstrap replicates (bootstrap support value, BSV). Bayesian inference (BI) was performed in MRBAYES 3.2.1 [20] based on the best partition scheme and substitution models: GTR + G for 16S, CO1 first codon, and Cytb third codon, HKY + I for COI second codon and Cytb first codon; GTR + I + G for CO1 third codon and Cytb second Codon, which were selected using ModelFinder [21] with Bayesian information criterion (BIC). Two independent runs were conducted during the BI analyses with 10,000,000 generations each, sampled every 1000 generations, with the first 25% of samples discarded as burn-in. We used a convergence diagnostic threshold of stopval = 0.01 to automatically terminate the MCMC run once convergence was achieved.
2.3. Morphological Analysis
Morphological characters and measurements followed Fei et al. [22], Jiang et al. [23], and Lyu et al. [14]. In total, 13 measurements were taken using slide calipers to the nearest 0.1 mm: (1) SVL, snout-vent length, from tip of snout to posterior margin of vent; (2) HL, head length, from tip of snout to the articulation of the jaw; (3) HW, head width, at the commissure of the jaws; (4) SL, snout length, from tip of snout to the anterior corner of the eye; (5) INS, internasal space, the distance between the nostrils; (6) IOS, interorbital space, the shortest distance between the upper eyelids; (7) UEW, upper eyelid width, maximum width of upper eyelid; (8) ED, eye diameter, horizontal distance from the anterior corner to the posterior corner of the eye; (9) LAHL, length of forearm and hand, from the tip of digit III to elbow joint; (10) HAL, hand length, from the proximal border of the outer palmar tubercle to the tip of digit III; (11) TL, tibia length, from the outer surface of the flexed knee to the heel; (12) TFL, length of tarsus and foot, from proximal end of tarsus to tip of toe IV; (13) FL, foot length, from proximal end of inner metatarsal tubercle to tip of toe IV.
Morphometric analyses were conducted for the putative new subspecies, A. m. xinduqiao and A. m. mantzorum. We measured specimens for numerical taxonomic analyses, including 10 adult males of the putative new subspecies, 10 adult males of A. m. mantzorum, and 15 adult males of A. m. xinduqiao. To avoid errors due to different measurement personnel, all 11 measurements were conducted by Y.-J.G. To reduce the impact of allometry, the logarithm (base 10) of the ratio of each measured value to SVL was used in the analyses. Mann–Whitney U test in SPSS (statistics 25.0) (In the “Two Independent Samples” option of the “Nonparametric Tests” option of the software, we ticked the “Mann–Whitney U” option box) was used to test for significant differences between the three subspecies of A. mantzorum, with significance level set at p < 0.05 (significant) and p < 0.01 (highly significant). Principal component analysis (PCA) in Origin (Pro) (Version 2024) (the “Correlation Matrix” option box in the “Analyze” option was ticked) was used to assess whether the subspecies were separated in morphometric space.
For morphological comparisons, we directly examined specimens of six species within the Amolops mantzorum group (detailed in Appendix A). For the remaining 12 species for which specimens were not examined, morphological data were obtained from the literature.
3. Results
3.1. Molecular Phylogeny
The reconstructed phylogenies are presented in Figure 2. Both the ML and BI trees show relatively weak support values for several major clades of previously identified species groups (e.g., A. lifanensis of A. mantrorum group clustered out of its clade), indicating that the complex evolutionary history of the A. mantzorum group remains unresolved. A total of 22 newly collected samples (A. mantzorum ssp., the new lineage) from along the Yalong River clustered together as one lineage with well support (BSV 99, BPP 1.00). Samples of A. m. xinduqiao, A. m. mantzorum, and “A. m. ssp. (northern lineage)” each formed distinct monophyletic lineages: the A. m. xinduqiao lineage with strong support in both the ML and BI tree (BSV 99, BPP 1.00), the A. m. mantzorum lineage with strong support in both the ML and BI trees (BSV 99, BPP 0.99), and the “A. m. ssp. (northern lineage)” with strong support in both the BI and ML trees (BSV 100, BPP 1.00) (Figure 2). These four monophyletic lineages formed a clade with strong support in both the ML and BI trees (BSV 100), BPP 1.00) (Figure 2).
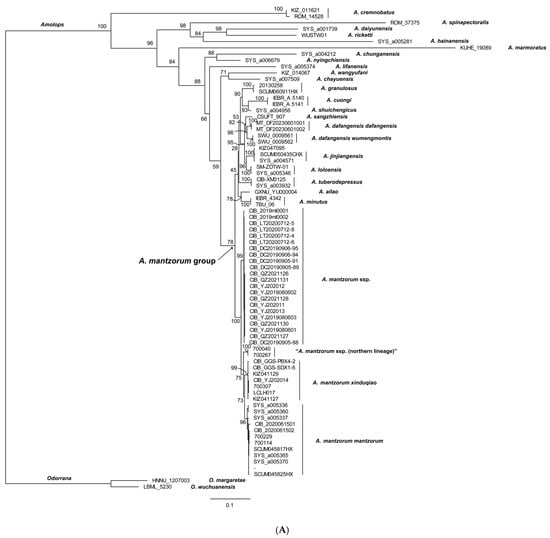
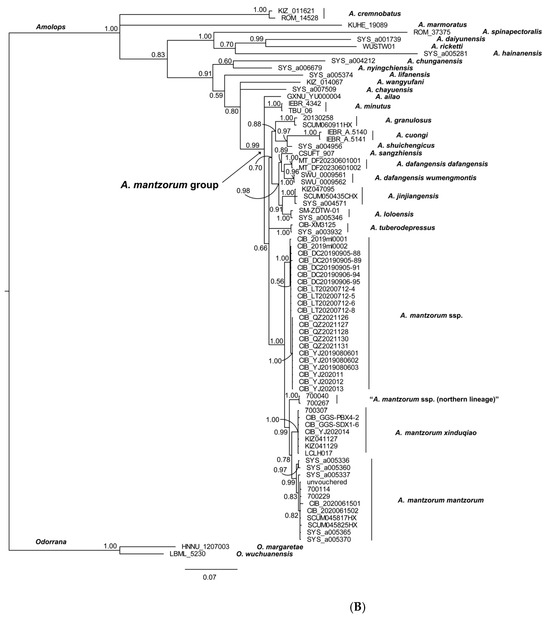
Figure 2.
The molecular phylogenetic tree inferred from 1758 bp of 16S-COI-Cytb genes (440 bp of 16S gene, 562 bp of COI gene, and 756 bp of Cytb gene). (A). Maximum-likelihood tree (numbers near branches are BSV), and (B). Bayesian inference tree (numbers near branches are BPP), respectively. The vouchers to the right of branches correspond to the voucher numbers in Table 1.
3.2. Morphological Comparisons Within Subspecies of A. mantzorum
Comparisons among the new lineage from along the Yalong River, A. m. xinduqiao, and A. m. mantzorum are presented herein. Morphological data on “A. m. ssp. (northern lineage)” are missing.
The new lineage can be distinguished from A. m. xinduqiao by the absence of a tympanum (vs. small but distinct) (Figure 3), distinct and strong vomerine teeth (vs. small, two tiny rows) (Figure 4), and larger body size in adult females (SVL 56.7–63.7 mm [n = 10] vs. 48.5–56.6 mm [n = 15]) (Table 2); and from A. m. mantzorum by the absence of a tympanum (vs. small but distinct) (Figure 3), and smaller body size in adult males (SVL 42.3–51.2 mm [n = 10] vs. SVL 51.4–57.8 mm [n = 10]) (Table 2).
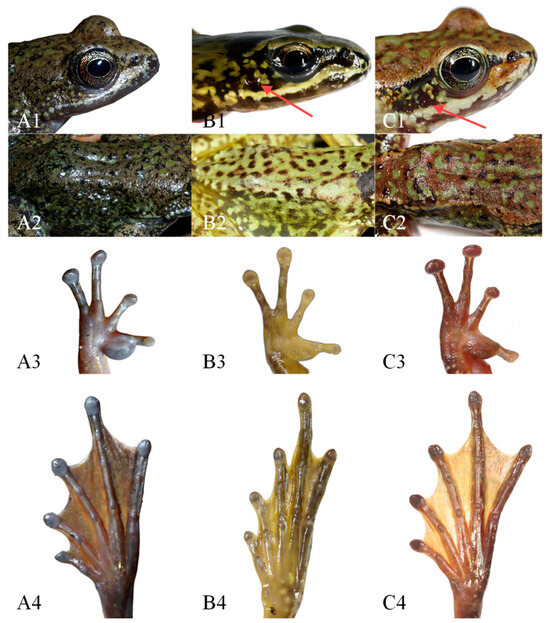
Figure 3.
Comparisons of morphological characters of the new lineage (Amolops mantzorum ssp.), A. m. xinduqiao, and A. m. mantzorum in life. (A): A. m. ssp. (CIB QZ2021131); (B): A. m. xinduqiao (CIB GGS-PBX4-2); (C): A. m. mantzorum (CIB GGS-DWX2-6). (1): Head in dorsolateral view (red arrow points to the tympanum); (2): dorsolateral fold; (3): ventral view of hand; (4): ventral view of foot.
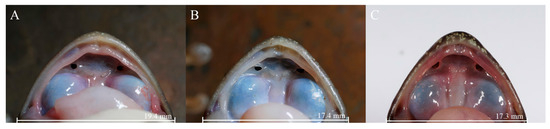
Figure 4.
Comparisons of vomerine teeth of the new lineage (Amolops mantzorum ssp.) and A. m. xinduqiao. (A): A. m. ssp. (CIB QZ2021126, female); (B): A. m. ssp. (CIB QZ2021130, male); (C): A. m. xinduqiao (CIB GGS-PBX4-1, female).

Table 2.
The main morphological characters of species within the Amolops mantzorum group. “-” indicates no data; “*” indicates the data resource is this study.
Amolops m. xinduqiao is distinguished from A. m. mantzorum by the smaller body size in adult males (SVL 40.3–46.7 mm [n = 15] vs. 51.4–57.8 mm [n = 10]) and adult females (SVL 48.5–56.6 mm [n = 15] vs. 59.0–72.0 mm [n = 10]) (Table 2).
3.3. Morphometric Analysis
Based on the measurements of 13 characters of the three lineages (the new lineage, A. m. xinduqiao, and A. m. mantzorum) (Table 3), the results of comparisons among them (Table 4) showed that: (1) two characters were significantly different between the new lineage and A. m. xinduqiao: SVL and SL/SVL; four characters were highly significantly different between them: UEW/SVL, ED/SVL, HAL/SVL, and TFL/SVL; (2) three characters were significantly different between the new lineage and A. m. mantzorum: HW/SVL, INS/SVL, and IOS/SVL, and four characters were highly significantly different between them: SVL, HL/SVL, SL/SVL, and TL/SVL; (3) one character was significantly different between A. m. xinduqiao and A. m. mantzorum: TL/SVL, and ten characters were highly significantly different between them: SVL, HL/SVL, HW/SVL, SL/SVL, INS/SVL, IOS/SVL, UEW/SVL, ED/SVL, HAL/SVL, and TFL/SVL.

Table 3.
Measurements (in mm; mean ± SD (range)) of the male type series of the new lineage (Amolops mantzorum ssp.), A. m. xinduqiao, and A. m. mantzorum.

Table 4.
Morphometric comparisons among males of the new lineage (Amolops mantzorum ssp.), A. m. xinduqiao, and A. m. mantzorum. p-values are results from Mann–Whitney U tests on each character between subspecies. (* p-value < 0.05; ** p-value < 0.01).
In the PCA of males, the eigenvalues of the first three principal components (PC1, PC2, and PC3) were more than 1.0, and their total variation accounted for 69.5% (Table 5); therefore, the first three PCs could be used for elucidating their taxonomic significance. In the score plot of PCA (Figure 5), the plot of PC1 plus PC2 showed that the new lineage was almost separated from A. m. xinduqiao and A. m. mantzorum, though with some overlap, while the two lineages A. m. xinduqiao and A. m. mantzorum were well-separated. These differences were largely contributed by PC1 (Figure 5), to which SL and ED had higher contributions (Table 5). The plot of PC1 versus PC3 showed that the three lineages were distinctly separated from each other. These differences were contributed by combinations of PC1 and PC3 (Figure 5), of which HAL and FL had higher contributions to PC3 (Table 5). These results demonstrate that the three lineages exhibit distinct morphological differentiation.

Table 5.
Factor loadings of the first three principal components among Amolops mantzorum ssp., A. m. xinduqiao, and A. m. mantzorum.
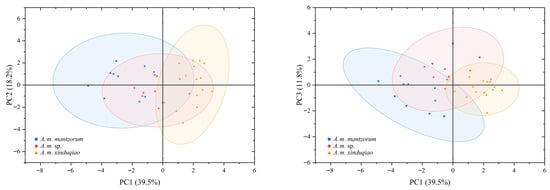
Figure 5.
The score plot of PCA for morphological characteristics of Amolops mantzorum ssp., A. m. xinduqiao, and A. m. mantzorum.
3.4. Comparisons Between the New Lineage and Other Species Within the A. mantzorum Group
The morphological differences within the A. mantzorum group are summarized in Table 2. The new lineage can be clearly distinguished from A. tuberodepressus, A. granulosus, A. ailao, A. minutus, and A. dafangensis by the absence of a tympanum; differs from A. tuberodepressus by head width slightly larger than head length (vs. head slightly longer than broad); differs from A. granulosus by head width slightly larger than the head length (vs. head length slightly larger than head width), and the absence of vocal sacs (vs. present); differs from A. ailao by larger body size, SVL 42.3–51.2 mm in adult males and 56.7–63.7 mm in adult females (vs. 33.0–35.1 mm in males and 41.3 mm in female), head width slightly larger than head length (vs. head slightly longer than wide), and distinct and strong vomerine teeth (vs. absent); differs from A. minutus by larger body size, SVL 42.3–51.2 mm in adult males and 56.7–63.7 mm in adult females (vs. 29.70–36.42 mm in males and 38.47–50.22 mm in females), head width slightly larger than head length (vs. head length larger than head width), and the absence of dorsolateral folds (vs. very poorly developed); differs from A. shuichengicus by head width slightly larger than head length (vs. head length slightly larger than head width), and the absence of dorsolateral folds (vs. present); differs from A. jinjiangensis by head width slightly larger than head length (vs. head length slightly larger than head width), and the absence of dorsolateral folds (vs. present); differs from A. loloensis by smaller body size, SVL 42.3–51.2 mm in adult males and 56.7–63.7 mm in adult females (vs. 54.5–62.0 mm in males and 69.5–77.5 mm in females), head width slightly larger than head length (vs. head length almost equal to head width), distinct and strong vomerine teeth (vs. absent), and distinct and blunt canthus rostralis (vs. indistinct); differs from A. sangzhiensis by head width slightly larger than head length (vs. head length about equal to or larger than head width); differs from A. lifanensis by smaller body size, SVL 42.3–51.2 mm in adult males and 56.7–63.7 mm in adult females (vs. 52.0–56.0 mm in males and 61.0–79.0 mm in females), head width slightly larger than head length (vs. head length almost equal to width), and distinct and blunt canthus rostralis (vs. indistinct); differs from A. dafangensis by head width slightly larger than head length (vs. head length larger than head width slightly).
3.5. Taxonomic Account

Figure 6.
Amolops mantzorum feiye ssp. nov. (holotype, adult male, CIB QZ2021131). (A): Dorsolateral view; (B): head dorsolateral view; (C): ventral view; (D): dorsal foot; (E): ventral hand. Scale bars equal 10 mm.

Figure 7.
Comparisons of morphological characters of different adult individuals of Amolops mantzorum feiye ssp. nov. (the new lineage). (A): Dorsolateral view; (B): head in dorsolateral view; (C): ventral view of hand. (1): Female adult CIB QZ2021126 (SVL: 59.8 mm); (2): Female adult CIB QZ2021127 (SVL: 60.1 mm); (3): Female adult CIB QZ2021128 (SVL: 61.4 mm); (4): Male adult CIB QZ2021130 (SVL: 48.7 mm). Scale bar in (A) equals 10 mm. B and C not to scale.
Holotype. CIB QZ2021131, adult male, collected in Bajiaolou Village (30.0729° N, 101.1398° E; alt. 2841 m), Bajiaolou Town, Yajiang County, Sichuan Province, China by Shengchao Shi, Peng Yan, and Shun Ma on 20 August 2021.
Paratypes. Nineteen adult specimens (Appendix A (1)). One male (CIB QZ2021130) and three females (CIB QZ2021126, CIB QZ2021127, and CIB QZ2021128) were collected at the same time and locality as the holotype; two males (CIB YJ2019080601, CIB YJ2019080602) and one female (CIB YJ2019080603) were collected at the same locality as the holotype on 6 August 2019; two males (CIB YJ202012 and CIB YJ202013) and one female (CIB YJ202011) were collected in Nimazong Village, Zhusang Town, Yajiang County, Sichuan Province, China on 8 July 2020; four females (CIB LT20200712-4, CIB LT20200712-5, CIB LT20200712-6, CIB LT20200712-8) were collected in Dewu Town, Litang County, Sichuan Province, China on 12 July 2020; four males (CIB DC20190905-89, CIB DC20190905-91, CIB DC20190906-94, CIB DC20190906-95) and one female (CIB DC20190905-88) were collected in Daocheng County, Sichuan Province, China on 5–6 September 2019.
Diagnosis. Amolops mantzorum feiye ssp. nov. is distinguished from all other congeners by the following combination of characters: (1) moderate body size, SVL 42.3–51.2 mm in males (n = 10), and 56.7–63.7 mm in females (n = 10); (2) tympanum absent; (3) head width slightly larger than head length; (4) vomerine teeth distinct and strong; (5) male forearm relatively strong, inner side of first finger with a developed nuptial pad; (6) supernumerary tubercles below the base of fingers II, III, and IV distinct; (7) circummarginal grooves present on tips of outer three fingers, absent on finger I; (8) dorsolateral fold absent but dorsolateral fold-like glands thick and flat; (9) vocal sac absent in males.
Description of Holotype. Medium body size, SVL 47.4 mm; flat head, head width slightly larger than head length (HL/HW ratio 0.98); snout rounded, canthus rostralis distinct and blunt, loreal region concave and oblique; internasal space greater than interorbital space (INS/IOS ratio 1.28), interorbital space greater than the maximum width of upper eyelid (IOS/UEW ratio 1.52); eyes convex in dorsal view, moderate size (ED/HL ratio 0.34); tympanum absent; vomerine teeth distinct and strong; tongue deeply notched posteriorly; vocal sac absent.
Forelimbs: forearm robust, the length of forearm and hand approximately half of body length (LAHL/SVL ratio 0.47); fingers slender, all four fingers’ tips expanded into disks, and the order of finger length is I < II < IV < III; circummarginal grooves present on tips of outer three fingers, absent on finger I; subarticular tubercles distinct; supernumerary tubercles below the base of fingers II, III, and IV distinct; inner metacarpal tubercle oval, two outer metacarpal tubercles present; fringe on fingers absent; a developed nuptial pad on the inner side of the finger I.
Hindlimbs: long and thin, heels overlapping when hind limbs flexed and held perpendicular to body; all five toe tips expanded into disks; the order of toe length I < II < III < V < IV, the subarticular tubercles well-developed; entirely webbed on all toes except toe IV; inner metatarsal tubercle, outer metatarsal tubercle absent.
Skin: dorsal surface relatively smooth; supratympanic fold absent; dorsolateral fold absent but dorsolateral fold-like glands thick and flat; ventral surface smooth except for slightly flattened tubercles on basal ventral surface of thigh.
Color in life. Entire dorsal surface grayish brown with dense green stripes and black spots in the middle of the dorsal surface; upper lip grayish black, lower lip grayish white; a large area of green stripes on the lateral body, black stripes and brownish green stripes on the dorsal surface of the forelimbs, and reddish brown patches on the dorsal surface of hindlimbs; ventral surface of body opaque milky white; ventral surface of limbs red with a small amount of pale yellow spots; ventral surface of hands reddish gray; toe webs brown.
Color in preservative. Entire dorsal surface dark gray with black patches scattered; sides light grayish brown; ventral surface milky white without mottling; crotch slightly dark; dorsal surface of limbs grayish brown, with black stripes scattered, and ventral surface light gray.
Secondary sexual characters. Males obviously smaller than females; the forearm of males is relatively strong, and males have a gray nuptial pad on the inner side of finger I (Figure 7).
Variation. The dorsal surfaces of two individuals (CIB QZ2021126 and CIB YJ2019080602) from Yajiang County are light brown rather than grayish brown as in the holotype; in two individuals (CIB QZ2021127 and CIB YJ2019080603), ventral surfaces of hands are milky white rather than reddish gray as in the holotype; in three individuals (CIB QZ2021126, CIB QZ2021128 and CIB YJ2019080603), the supernumerary tubercle below the base of finger III and the lower subarticular tubercle on finger III are separate, rather than linked together as in the holotype.
In terms of body size (mean SVL), among males, the individuals from Yajiang County are larger than individuals from Daocheng County (SVL 47.8 mm [n = 6] vs. 45.2 mm [n = 4]). Among females, individuals from Yajiang County are larger than individuals from Litang County and Daocheng County (SVL 61.0 mm [n = 5] vs. 60.5 mm [n = 1] vs. 56.7 mm [n = 4]). All measurements of type series specimens are summarized in Table 6.

Table 6.
Measurements (in mm; mean ± SD [range]) of adult specimens (type series) of Amolops mantzorum feiye ssp. nov.
Etymology. The specific epithet “feiye” is named after Prof. Liang Fei and his wife Prof. Changyuan Ye, combining their first names “Fei” and “Ye”. Prof. Fei and Prof. Ye have made significant contributions to Chinese herpetology. We suggest the English common name “Feiye’s torrent frog” and the Chinese common name “费叶湍蛙 (in Chinese Pinyin: fèi yè tuān wā)”.
Distribution and ecology. At present, A. m. feiye ssp. nov. is known from Yajiang County, Daocheng County, Muli County, and Litang County in Sichuan Province (Figure 1). Specimens were found on rocks exposed to water or along the river (2800–2900 m a.s.l.), surrounded by shrubs or tussocks. The rivers crossed villages and roads where we collected these specimens.
4. Discussion
Amolops m. xinduqiao was previously recognized as “A. kangtingensis” in Cai & Zhao [1] (Kangding population), Lu et al. [9], and Zhang et al. [10]. However, “A. kangtingensis” was described based on specimens from multiple localities. Fei et al. [2] re-examined the holotype of “A. kangtingensis” and classified it as A. mantzorum (currently A. m. mantzorum), rendering “A. kangtintensis” an invalid name that should be synonymized with A. mantzorum (currently A. m. mantzorum). They proposed a new name, A. xinduqiao (currently A. m. xinduqiao) for the population studied by the above authors as distinct from A. m. mantzorum.
Amolops m. mantzorum represents the population along the Dadu River Basin according to Fei et al. [2], which also corresponds to the “A. mantzorum central population” of Lu et al. [9]. Samples from Hongya and Dayi represent one of the four distinct genetic populations of A. mantzorum identified by Cai & Zhao [1]. In our study, molecular phylogeny including samples from these two localities indicates that this population should be recognized as the nominate A. m. mantzorum. According to Zhang et al. [10], the distribution of A. m. mantzorum also includes Kangding, Luding, Yingjing, Hanyuan, Baoxing, Tianquan, and Chongzhou.
Geographically, A. mantzorum has a continuous distribution west of the Sichuan Basin, but four lineages can be separated based on their distribution patterns The northern lineage (“A. m. ssp. [northern lineage]”) is restricted to the east of the Min River. Amolops m. mantzorum is distributed from the Min River to the Dadu River. The new subspecies has nearly overlapping distribution with A. m. xinduqiao near site 23 (Figure 1) at the Yalong River, but samples were collected from different villages, and samples of A. m. xinduqiao were collected above 3000 m elevation, whereas samples of the new subspecies were collected at approximately 2800–2900 m. Geographical barriers formed by large rivers and different elevational preferences may have contributed to this separation of genetic lineages.
The description of the new subspecies has clarified species delimitation within Amolops mantzorum. The four monophyletic lineages recognized in this study are consistent with the four genetic populations revealed by Cai & Zhao [1]. Except for “A. m. ssp. (northern lineage)”, three of them have been described and exhibit morphological divergence. However, the delimitation of the four lineages relies solely on mitochondrial markers ([1]; this sudy). Separation in mitochondrial phylogeny is sometimes not supported by nuclear markers (e.g., Lu et al. [9]), but it can be confirmed after deeper genomic investigations [3,4]. Morphologicaly, the new subspecies can be clearly separated from the other two recognized subspecies by its distinct and strong vomerine teeth (vs. small in both A. m. mantzorum and A. m. xinduqiao), and the invisible tympanum (vs. distinct in both). The differences between A. m. mantrozum and A. m. xinduqiao are also recognized in body size.
Currently, controversy persists regarding whether these subspecies represent independent species (i.e., [28,31,32]). Given the limitations of mitochondrial genes in DNA barcoding and phylogenetic estimation [33,34,35], further studies—including a detailed description of “A. m. ssp. (northern lineage)”, and a more robust phylogeny based on SNP data—would benefit efforts to resolve this controversy.
5. Conclusions
Based on mitochondrial phylogeny and morphology, we clearly identified and described the third subspecies of Amolops mantzorum. The new subspecies, together with A. m. mantzorum, A. m. xinduqiao, and “A. m. ssp. (northern lineage)” forms four monophyletic lineages within A. mantzorum. Given that taxonomists do not fully agree on the placement of subspecies in these lineages, which remains controversial, future studies based on genome data and further surveys—especially on the “A. m. ssp. (northern lineage)”—should be prioritized.
Supplementary Materials
The following supporting information can be downloaded at https://www.mdpi.com/article/10.3390/ani16010055/s1, Figure S1: Dorsolateral view of Amolops mantzorum feiye ssp. nov. (the new lineage); Figure S2: Head dorsolateral view of Amolops mantzorum feiye ssp. nov. (the new lineage).
Author Contributions
Conceptualization, T.Q. and Y.-J.G.; methodology, Y.-J.G. and S.-C.S.; software, Y.-J.G.; validation, Y.-J.G., S.-C.S. and J.-P.J.; formal analysis, Y.-J.G.; investigation, T.Q., Y.-J.G. and S.-C.S.; resources, Y.-J.G., K.T., S.-C.S., D.Z., S.M., Y.-M.H. and F.X.; data curation, Y.-J.G. and S.-C.S.; writing—original draft preparation, T.Q., Y.-J.G.; writing—review and editing, S.-C.S., K.T., S.M., D.Z.,Y.-M.H., F.X. and J.-P.J.; visualization, Y.-J.G. and S.-C.S.; supervision, J.-P.J.; project administration, J.-P.J.; funding acquisition, J.-P.J. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the National Key Research and Development Programme of China (2022YFF1301401), and China Biodiversity Observation Networks (Sino BON—Amphibian & Reptile).
Institutional Review Board Statement
All animal protocols in this study were reviewed and approved on 6 March 2017 by the Animal Ethical and Welfare Committee of Chengdu Institute of Biology, Chinese Academy of Sciences (permit number: 2017-AR-JJP-03).
Informed Consent Statement
Not applicable.
Data Availability Statement
All data are provided in the main text or Supplementary Materials.
Acknowledgments
We thank Peng Yan, Gang Wang, Xiuqin Lin, Liang Xu for help in filed work. We also thank the Administration of Gongga Mountain National Nature Reserve for help in field work.
Conflicts of Interest
The authors declare no conflicts of interest.
Appendix A. Specimens Examined
(1) Amolops mantzorum feiye ssp. nov. (n = 20): Sichuan Province, China:
Yajiang County: Bajiaolou Town: Bajiaolou Village (type locality): CIB QZ2021131 (holotype), CIB QZ2021130, CIB QZ2021126– 28, and CIB YJ2019080601–03;
Yajiang County: Zhusang Town: Nimazong Village: CIB YJ202011–13;
Daocheng County: Shengmu Town: Rihuo Village: CIB DC20190905-88–89, and CIB DC20190905-91;
Daocheng County: Mula Town: Maize Village: CIB DC20190906-94–95;
Litang County: Dewu Town: CIB LT20200712-4–6 and CIB LT20200712-8.
(2) A. m. xinduqiao (n = 18): Sichuan Province, China:
Kangding City: Jiagenba Town, Geridi Village: CIB GGS-PBX4-1–2;
Kangding City: Xinduqiao Town (type locality): CIB 80I0692 (holotype), CIB 80I0704, CIB 80I0725, CIB 80I0729–30, CIB 80I0737–44, CIB 80I0747, and CIB 80I0775;
Yajiang County: Shuiluo Town: Wangxia Village: CIB YJ202014.
(3) A. m. mantzorum (n = 14): Sichuan Province, China:
Baoxing County (type locality): Wulong Town: Dongsheng Village: CIB 2020061501–02, 04–05;
Shimian County: Caoke Town: CIB GGS-CKX1-2–3;
Shimian County: Caoke Town: Heping Village: CIB GGS-CKX5-1–3;
Luding County: Tianba Town: CIB GGS-TBX4-1 and CIB GGS-TBX4-2;
Luding County: Dewei Town: Zawei Village: CIB GGS-DWX2-6;
Kangding City: CIB GGS-GZ2-01–02.
(4) A. loloensis (n = 7): Sichuan Province, China:
Baoxing County: Wulong Town: Dongsheng Village: CIB 2020061506–07;
Shimian County: Caoke Town: Heping Village: CIB GGS-CKX5-5;
Luding County: Tianba Town: CIB GGS-TBX4-2 and CIB GGS-TBX4-7–9.
(5) A. jinjiangensis (n = 10): Yunnan Province, China:
Deqin County (type locality): CIB 5334220036, CIB 5334220039–40, CIB 5334220042, CIB 5334220093–97, and CIB 5334220146.
(6) A. tuberodepressus (n = 7): Yunnan Province, China:
Jingdong County (type locality): CIB JDA01–02, CIB JD20230725001–02, CIB JDH01–03.
References
- Cai, H.X.; Zhao, E.M. The validity of four Amolops species in Hengduan Mountains China. Sichuan J. Zool. 2008, 27, 483–488, (In Chinese with English Abstract). [Google Scholar]
- Fei, L.; Ye, C.Y.; Wang, Y.F.; Jiang, K. A new species of the genus Amolops (Anura: Ranidae) from high-altitude Sichuan, southwestern China, with a discussion on the taxonomic status of Amolops kangtingensis. Zool. Res. 2017, 38, 138–145. [Google Scholar]
- Wu, Y.H.; Yan, F.; Stuart, B.L.; Prendini, E.; Suwannapoom, C.; Dahn, H.A.; Zhang, B.L.; Cai, H.X.; Xu, Y.B.; Jiang, K.; et al. A combined approach of mitochondrial DNA and anchored nuclear phylogenomics sheds light on unrecognized diversity, phylogeny, and historical biogeography of the torrent frogs, genus Amolops (Anura: Ranidae). Mol. Phylogenet. Evol. 2020, 148, 106789. [Google Scholar] [CrossRef]
- Zeng, Z.C.; Liang, D.; Li, J.X.; Lyu, Z.T.; Wang, Y.Y.; Zhang, P. Phylogenetic relationships of the Chinese torrent frogs (Ranidae: Amolops) revealed by phylogenomic analyses of AFLP-Capture data. Mol. Phylogenet. Evol. 2020, 146, 106753. [Google Scholar] [CrossRef] [PubMed]
- Dufresnes, C.; Litvinchuk, S.N. Diversity, distribution and molecular species delimitation in frogs and toads from the Eastern Palaearctic. Zool. J. Linn. Soc. 2022, 195, 695–760. [Google Scholar] [CrossRef]
- Tang, S.J.; Sun, T.; Liu, S.; Luo, S.D.; Yu, G.H.; Du, L. A new species of cascade frog (Anura: Ranidae: Amolops) from central Yunnan, China. Zool. Lett. 2023, 9, 15. [Google Scholar] [CrossRef]
- Frost, D.R. Amphibian Species of the World, Version 6.2, an Online Reference; American Museum of Natural History: New York, NY, USA. Available online: https://amphibiansoftheworld.amnh.org (accessed on 13 February 2025).
- Li, P.Y.; Li, J.Y.; Zhang, W.; Yi, X.B.; Yuan, Z.Y.; Huang, J.K.; Liu, X.L. Integrated evidence reveals a new subspecies of Amolops Cope, 1865 (Anura, Ranidae) from northeastern Yunnan, China. ZooKeys 2025, 1257, 39–54. [Google Scholar] [CrossRef] [PubMed]
- Lu, B.; Bi, K.; Fu, J.Z. A phylogeographic evaluation of the Amolops mantzorum species group: Cryptic species and plateau uplift. Mol. Phylogenet. Evol. 2014, 73, 40–52. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.H.; Yuan, S.Q.; Xia, Y.; Zeng, X.M. Species delimitation of Amolops kangtingensis. Sichuan J. Zool. 2015, 34, 801–809, (In Chinese with English Abstract). [Google Scholar]
- Simon, C.; Frati, F.; Beckenbach, A.; Crespi, B.; Liu, H.; Flook, P. Evolution, weighting and phylogenetic utility of mitochondrial gene sequences and a compilation of conserved polymerase chain reaction primers. Ann. Entomol. Soc. Am. 1994, 87, 651–701. [Google Scholar] [CrossRef]
- Che, J.; Chen, H.M.; Yang, J.X.; Jin, J.Q.; Jiang, K.; Yuan, Z.Y.; Murphy, R.W.; Zhang, Y.P. Universal COI primers for DNA barcoding amphibians. Mol. Ecol. Resour. 2012, 12, 247–258. [Google Scholar] [CrossRef]
- Wu, X.Y. Genetic Structure and Phylogeography of Amolops wuyiensis Based on Mitochondrial DNA Cytb Gene. Master’s Thesis, Anhui Normal University, Wuhu, China, 2015. [Google Scholar]
- Lyu, Z.T.; Zeng, Z.C.; Wan, H.; Yang, J.H.; Li, Y.L.; Pang, H.; Wang, Y.Y. A new species of Amolops (Anura: Ranidae) from China, with taxonomic comments on A. liangshanensis and Chinese populations of A. marmoratus. Zootaxa 2019, 4609, 247–268. [Google Scholar] [CrossRef]
- Kuraku, S.; Zmasek, C.M.; Nishimura, O.; Katoh, K. aLeaves facilitates on-demand exploration of metazoan gene family trees on MAFFT sequence alignment server with enhanced interactivity. Nucleic Acids Res. 2013, 41, W22–W28. [Google Scholar] [CrossRef]
- Katoh, K.; Rozewicki, J.; Yamada, K.D. MAFFT online service: Multiple sequence alignment, interactive sequence choice and visualization. Brief. Bioinfom. 2019, 20, 1160–1166. [Google Scholar] [CrossRef]
- Borowiec, M.L. AMAS: A fast tool for alignment manipulation and computing of summary statistics. PeerJ 2016, 4, e1160. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, L.; Schmidt, H.A.; von Haeseler, A.; Minh, B.Q. IQ-TREE: A Fast and Effective Stochastic Algorithm for Estimating Maximum-Likelihood Phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef]
- Wong, T.K.F.; Ly-Trong, N.; Ren, H.; Banos, H.; Roger, A.J.; Susko, E.; Bielow, C.; De Maio, N.; Goldman, N.; Hahn, M.W.; et al. IQ-TREE 3: Phylogenomic Inference Software Using Complex Evolutionary Models. Ecoevorxiv. 2025. Available online: https://ecoevorxiv.org/repository/view/8916/ (accessed on 18 December 2025).
- Ronquist, F.; Teslenko, M.; Mark, P.; Ayres, D.L.; Darling, A.; Höhna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef]
- Kalyaanamoorthy, S.; Minh, B.Q.; Wong, T.K.F.; von Haeseler, A.; Jermiin, L.S. ModelFinder: Fast model selection for accurate phylogenetic estimates. Nat. Method. 2017, 14, 587–589. [Google Scholar] [CrossRef] [PubMed]
- Fei, L.; Hu, S.Q.; Ye, C.Y.; Huang, Y.Z. Fauna Sinica Amphibia, Vol. 2: Anura; Science Press: Beijing, China, 2009; pp. 1–958. (In Chinese) [Google Scholar]
- Jiang, K.; Wang, K.; Yan, F.; Xie, J.; Zhou, D.H.; Liu, W.L.; Jiang, J.P.; Li, C.; Che, J. A new species of the genus Amolops (Amphibia: Ranidae) from southeastern Tibet, China. Zool. Res. 2016, 37, 31–40. [Google Scholar]
- Fei, L.; Hu, S.Q.; Ye, C.Y.; Huang, Y.Z. Fauna Sinica Amphibia, Vol. 3: Anura Ranidae; Science Press: Beijing, China, 2009; pp. 959–1847. (In Chinese) [Google Scholar]
- Liu, W.Z.; Yang, D.T. A new species of Amolops (Anura: Ranidae) from Yunnan, China, with a discussion of karyological diversity in Amolops. Herpetologica 2000, 56, 231–238. [Google Scholar]
- Orlov, N.L.; Ho, C.T. Two new species of cascade ranids of Amolops genus (Amphibia: Anura: Ranidae) from Lai Chau Province (northwest Vietnam). Russian J. Herpetol. 2007, 14, 211–228. [Google Scholar]
- Su, C.Y.; Yang, D.T.; Li, S.M. A new species of Amolops from the Hengduan Shan Mountains. Acta Herpetol. Sin. 1986, 5, 204–206. [Google Scholar]
- Qian, T.Y.; Xiang, J.J.; Jiang, J.P.; Yang, D.D.; Gui, J. A new species of the Amolops mantzorum group (Anura: Ranidae: Amolops) from northwestern Hunan Province, China. Asian Herpetol. Res. 2023, 14, 54–64. [Google Scholar]
- Fei, L. Atlas of Amphibians in China (Field Edition); Henan Science and Technology Press: Zhengzhou, China, 2020; pp. 549–562. (In Chinese) [Google Scholar]
- Li, S.Z.; Liu, J.; Ke, X.C.; Cheng, G.; Wang, B. A new species of Amolops (Amphibia, Anura, Ranidae) from Guizhou Province, China. ZooKeys 2024, 1189, 33–54. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Zhang, D.R.; Hou, S.B.; Wu, Y.H. Annual Review: Taxonomic Changes of Herpetofauna from China in 2022. AmphibiaChina 2022, 1–16. Available online: https://www.amphibiachina.org/news/scientifictrends/321–2023 (accessed on 3 November 2025).
- AmphibiaChina. The Database of Chinese Amphibians. Kunming Institute of Zoology (CAS), Kunming, Yunnan, China. Available online: http://www.amphibiachina.org/ (accessed on 26 February 2025).
- Chan, K.O.; Hertwig, S.T.; Neokleous, D.N.; Flury, J.M.; Brown, R.M. Widely used, short 16S rRNA mitochondrial gene fragments yield poor and erratic results in phylogenetic estimation and species delimitation of amphibians. BMC Ecol. Evol. 2022, 22, 37. [Google Scholar] [CrossRef]
- Hupało, K.; Copilaș-Ciocianu, D.; Leese, F.; Weiss, M. Morphology, nuclear SNPs and mate selection reveal that COI barcoding overestimates species diversity in a Mediterranean freshwater amphipod by an order of magnitude. Cladistics 2022, 39, 129–143. [Google Scholar] [CrossRef]
- Vences, M.; Miralles, A.; Dufresnes, C. Next-generation species delimitation and taxonomy: Implications for biogeography. J. Biogeogr. 2024, 51, 1709–1722. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license.