Simple Summary
Creole animals are an important genetic resource due to their adaptation to adverse conditions, however, many of them have disappeared or have been reduced in number; therefore, their identification and preservation should be promoted. In the Mixteca zone of Oaxaca, México, there is a group of creole sheep with physical differences with respect to the other known breeds. The objective of the present study was to determine the degree of differentiation among the individuals of the population of creole sheep and the similarities with other breeds, as well as to measure their level of conservation and consanguinity. It was found that there is a group of sheep which is different from the presently known breeds and that they do not present high degree of consanguinity, and thus may be considered a new breed. Therefore, it is proposed that they are identified as the Chocholteca Creole breed, in honor of the ethnic group which inhabits this region. The present study is very important because it discovers a new genotype of sheep, which amplifies the genetic diversity. Therefore, further studies are needed, given that they are a potential alternative of meat and wool.
Abstract
Creole sheep in México have undergone crossbreeding, provoking the loss of genetic variability. The objective of the present study is to determine the intra-racial genetic diversity, the genetic relationship with other genotypes, and the populational substructure of the Oaxacan Creole sheep. Twenty-nine blood samples were obtained of Creole sheep of the Oaxaca Mixteca region in México. A genetic analysis was made with 41 microsatellites recommended for studies of genetic diversity in sheep. An analysis was made of genetic diversity, populational structure, and genetic distance with 27 other sheep populations. The study found 205 alleles with a range of 2 to 9 by locus and an effective number of 3.33. The intra-racial analysis showed a moderate genetic diversity with values of expected heterozygosity of 0.686 and observed of 0.756, a mean polymorphic information content of 0.609, and a mean coefficient of consanguinity of −0.002. In interracial genetic diversity for the coefficients of consanguinity, the values were FIS = 0.0774, FIT = 0.16993, and FST = 0.10028, showing an elevated genetic distance with other creole breeds, but close to Argentine Creole, to another Creole of México and the Spanish Merino. Its genetic structure showed that it does not have any populational subdivision nor mixes with the others analyzed. It is concluded that it is a distinct and isolated population and is proposed as the creole breed “Chocholteca” for its conservation.
1. Introduction
Sheep farming is one of the principal agricultural activities in the state of Oaxaca, Mexico. The Mixteca region has the highest number of production units [1,2], principally small scale as a system of family subsistence based on pasture grazing [3]. The flocks present in this region have been crossed with genetically improved commercial breeds to obtain better productive characteristics [4], provoking a crossbreeding and reduction of pure native creole breeds, placing them in a status of threatened species. In recent years, this process has caused the disappearance of 7% of the local breeds [5], as well as the loss of their genetic qualities, that give them resistance to the local environment, to diseases and allows them to feed with fodder lacking nutritional quality [6,7].
The High Mixteca zone, of the state of Oaxaca, inhabited by the Chocholteca ethnic group, still conserves a sheep population with no more than 100 animals that present morphological characteristics similar to those introduced over 500 years ago, as well as from other regions of México [8], which over the years became adapted to unfavorable environments and became the base of animal production. The study of this type of creole breeds has become a priority within the programs of sustainable management, in order to prevent their genetic deterioration or possible extinction.
At present, one of the tools most widely used is the characterization of production animals through the employment of molecular markers, such as microsatellites, in order to determine their purity or degree of crossbreeding with other breeds, the genetic diversity within the same population, the level of endogamy, etc. [9,10,11,12,13,14]. Therefore, the objective of the present study is to genetically characterize through microsatellites the Oaxacan Creole sheep, define whether it is a homogeneous breed and distanced from other native Spanish breeds, Creole breeds of América, as well as breeds common in México and Africa. This will make it possible to delineate strategies for recovering its population, as well as to reproduce it and conserve it as a genetic and economic resource for the Oaxacan producers.
2. Materials and Methods
2.1. Animals, Sampling, and Obtainment of DNA
Twenty-nine creole wool sheep (12 males, 17 females) of the High Mixteca, Oaxaca, México were included. The region is located in the coordinates 17°39′19” longitude west, 91°16′51” latitude north, at an altitude between 2100 and 2500 masl. The climate is humid temperate with rains in summer and an annual temperature of 14 to 20 °C (Figure 1). A sample was taken from each animal of 2 mL of blood from the jugular vein in tubes with EDTA anticoagulant and maintained in ice and transported to the Genetics and Animal Reproduction Laboratory of the FMVZ-UABJO. The samples were taken by a zootechnical veterinarian during the sanitary control of the flock; thus, the approval of an ethics committee was not required.
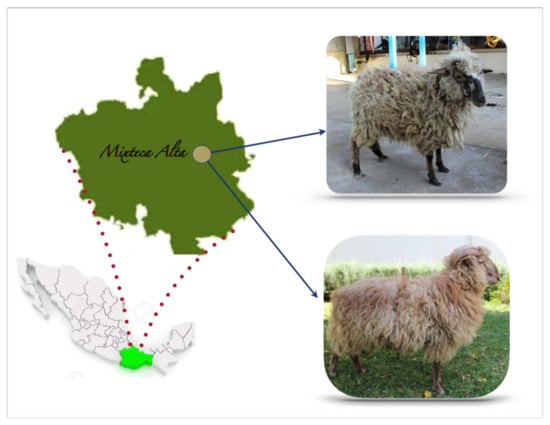
Figure 1.
Oaxacan Creole sheep and their geographic location. A female is shown on the right (above) and a male (below) of the Oaxacan Creole sheep. The Chocholteca Creole Sheep has a great adaptive capacity to cold climate and pastures with low fodder. In an extensive grazing system, adult males and females reach 33.75 kg and 26.5 kg weight, respectively; if the feeding system improves, it is possible to find males of 60 kg and females of 38 kg. Males have 64 cm height at withers, 9 cm more than females. They can reproduce throughout the year, but it is possible to detect seasonality in the rainy season. They are medium-sized with a straight facial profile. They have long, open wool that invades the forehead. There are two types differentiated by the layer color: white and cardinal. White sheep with brown coffee spots on limbs and face are predominant in the herd. A small number of specimens show a cardinal layer (black and white hairs) with black spots on the face and limbs. Males have two horn types: athropy and spiral-shaped. Females lack horns.
One-hundred microliters were taken from each sample and placed in filter paper with the individual dates. These were left to dry at room temperature and thus conserved until their analysis in the Applied Molecular Genetics Laboratory of the University of Córdoba, Spain. The DNA of each sample was obtained employing the Walsh technique [15], using the chelate Chelex 100 (Bio-Rad, Hercules, CA, USA) and maintained at −20 °C until its analysis.
2.2. Genotyping through Microsatellites
In the analysis, 41 microsatellites were used (BM1818, BM1824, BM6506, BM6526, BM8125, CD5, CMSS66, CRSD247, D5S2, ETH010, ETH225, HSC, ILSTS005, ILSTS008, ILSTS011, ILSTS87, INRA005, INRA006, INRA023, INRA035, INRA049, INRA063, INRA132, INRA172, MAF065, MAF214, McM042, McM527, OarAE129, OarCP20, OarCP34, OarCP49, OarFCB11, OarFCB20, OarFCB304, RM006, SPS113, SPS115, TGLA053, TGLA122, TGLA126), which have been utilized for the identification of other breeds of sheep and are recommended by the Food and Agriculture Organization and the International Society of Animal Genetics (FAO/ISAG). Each microsatellite was amplified by PCR and the obtained fragments were identified by means of 4-capillary electrophoresis in an automatic sequencer ABI 3130XL (Applied Biosystem, Foster City, CA, USA), using the marker “Genescan® 400 HD ROX size Standard” (Thermofisher, Waltham, MA, USA) to corroborate the sizes of the amplified microsatellites. The analysis of fragments and allelic typification were made using the programs Genescan Analysis® 3.1.2 (Fischer Scientific, Waltham, MA, USA) and Genotyper® 2.5.2 (Fischer Scientific), respectively.
2.3. Intra-Racial Genetic Diversity
The average number of alleles was determined per locus (MNA), the allelic frequency (Ae), the expected and observed heterozygosis (He), (Ho), along with polymorphic information contained (PIC) using the programs MICROSATELLITE TOOLKIT software of excel [16] and CERVUS 3.0.3 [17]. The effective number of alleles was calculated using the program PopGene [18]; the coefficient of consanguinity, with a confidence interval of 95%, in the program GENETIX v 4.05 [19]; and a Hardy–Weinberg equilibrium test was performed using the program GENEPOP v 3.1c [20], which applies an exact Fisher test by Markov Chain Monte Carlo (MCMC) method [21] and the correction of Bonferroni.
2.4. Interracial Genetic Diversity
For the analysis of genetic differentiation of the Oaxacan Creole sheep, it was compared with 27 breeds (Table 1), including some native Spanish, American Creole, African, and breeds common in México. For this purpose, we used the database of the Laboratory of Applied Molecular Genetics of Animal Breeding Consulting S.L. and of the Biovis Consortium (https://biovis.jimdo.com/breeds/ accessed on 21 October 2019). The intellectual property of the data of these breeds belongs to investigators of the network CONBIAND (https://biovis.jimdo.com/investigadores/ accessed on 21 October 2019). The F statistics of Wright [22] were calculated for the coefficient of consanguinity (FIT), as well as the coefficient of genetic differentiation (FST) and the endogamy coefficient (FIS), and a Factorial Analysis of Correspondence was also made using the program GENETIX v 4.05 [19]. The Genetic Distance was calculated (DA) [23] in the program POPULATIONS [24]. With the values of distance obtained, a phylogenic network of union of neighbors using the program SPLITSTREE [25] was made to represent the distance among the breeds.

Table 1.
Sheep breeds used in this study. The table shows the number of individuals used, their provenance, and the acronym for each one.
2.5. Genetic Structure
For the analysis of the genetic substructure among individuals of the Oaxacan Creole sheep, a Bayesian algorithm was made with the program STRUCTURE v 2.1 [26] that uses a model based on the chain method of Monte Carlo Markov (MCMC) for estimating the distribution of coefficient of mixture of each individual (q). A total of 100,000 iterations of Burn-in were made with 300,000 iterations of a chain of MCMC.
3. Results
3.1. Intra-Species Genetic Diversity
The population analyzed of the Oaxacan Creole sheep presented a polymorphic character with respect to the allelic frequencies corresponding to the 41 microsatellites analyzed. A total of 205 alleles were found, with a minimum of two in the markers ETH010 and ILSTS008, and a maximum of nine in the marker OarCP49, showing a moderate allelic diversity in the population (Table 2).

Table 2.
Allelic frequencies of the 41 microsatellites in the population of Oaxacan Creole sheep.
In the analysis of genetic variability (Table 3), the Oaxacan Creole sheep presented a mean number of alleles of 5, an effective number of 3.327, expected values of heterozygosity (He) minimum 0.25 for ET010 and maximum of 0.963 for TGLA122, with a mean of 0.685; an observed heterozygosity (Ho) minimum of 0.0223 in ETH010 and maximum of 0.841 in OarCP49, with a mean of 0.671 by a direct recount.

Table 3.
Genetic results of the microsatellites in the Oaxacan Creole sheep. The table shows the values obtained for the number of alleles detected and expected, the expected and observed heterozygosity, the values of the coefficient of endogamy, and the deviation from the Hardy–Weinberg equilibrium.
The values of Polymorphic Information Content (PIC) in the population presented 8 markers with a value less than 0.5 and 33 with a value above 0.5 considered very informative for the characterization of genetic variability. The marker ETH010 is the only uninformative marker showing a PIC of < 0.25.
The value for the frequency of the coefficient of consanguinity (FIS) had a range of −0.301 (BM6526) to 0.332 (SPS115) with a mean of −0.022 (−0.087–0.002), utilizing 10,000 re-samplings and with a confidence interval of 95%, manifesting that this population does not have a significant deviation of Hardy–Weinberg equilibrium. When the Bonferroni correction was made, it was found that the markers BM6506, BM6526, CD5, ETH010, INRA063, TGLA053, and TGLA122 have a significant defect (p < 0.05) of homozygotes with negative values of FIS, presenting a deviation of the Hardy–Weinberg equilibrium.
3.2. Interracial Genetic Diversity
Parameters of genetic diversity of the 28 breeds are compared in Supplement Table S1. The Oaxacan Creole sheep, when compared with native Spanish breeds, American Creoles, African breeds, and breeds more common in México, through a Factorial analysis of Correspondence, showed an elevated genetic differentiation, presenting statistical values of FIS = 0.0774 (0.05353–0.10389), FIT = 0.16993 (0.14598–0.19770), and FST = 0.10028 (0.09159–0.10917). In the Factorial Correspondence Analysis (Figure 2), the Oaxacan Creole sheep present a higher genetic distancing with the sheep of African origin (axis 2), followed by the hair sheep and being closer to the creoles of American and Spanish breeds (axis 1).
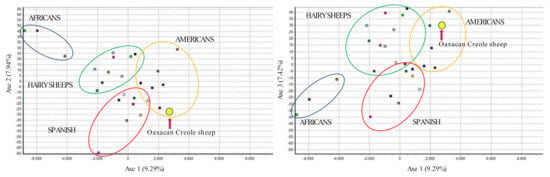
Figure 2.
Factor of correspondence in the 28 sheep studied. The Oaxacan Creole sheep is shown in the yellow circle and indicated with a red arrow.
Analyzing the genetic distance (DA) and the coefficients of genetic differentiation with respect to other populations (FST) ( Supplement Table S2), the Oaxacan Creole presents the lowest values with some creole sheep of America (DA = 0.122–0.198; FST = 0.114–0.174), Africa (DA = 0.194–0.163; FST = 0.140–0.175), and the highest values with the creole of Brazil (DA = 0.247; FST = 0.218) and Palmera of Spain (DA = 0.248; FST = 0.220).
The representation of DA genetic distances through a neighbor-net graphic shows that the Oaxacan Creole sheep are grouped in the same branch as the Argentine Creole and share the same trunk of origin with the Creole of Mexico, Spanish Merino, and Chilean Merino (Figure 3), even though their genetic distances are high. Furthermore, interestingly, it is observed that it has a high genetic distance with the creole sheep of Chiapas, Mexico, which is the most popular creole sheep in Mexico.
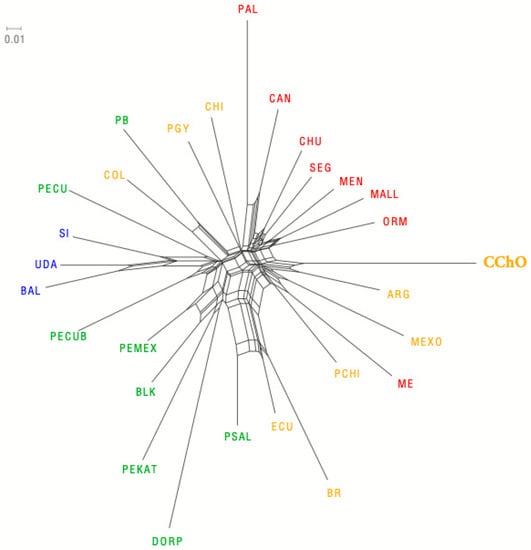
Figure 3.
Representation of the DA genetic distances in a neighbor-net between the Oaxacan Creole and 27 other sheep populations. In yellow, Creole breeds; in red, Spanish breeds; in blue, African breeds; in green, Hairy sheep. ARG: Argentine Creole, BAL: Balami, BLK: Blackbelly, BR: Brazil Creole, CAN: Canarian, CChO: Creole Chocholteca Oaxaca, CHI: Chiapas Creole, CHU: Churra, COL: Colombian Creole, DORP: Dorper, ECU: Ecuador Creole, MALL: Mallorquina, ME: Merino Español, MEN: Menorquina, MEXO: México, ORM: Roja Mallorquina, PAL: Palmera, PB: Pelibuey, PCHI: Merino Chileno, PECU: Pelibuey Ecuador, PECUB: Pelibuey Cuban, PGY: Creole Paraguay, PEKAT: Katahdin, PEMEX: Pelibuey Mexicano, PSAL: Pelibuey El Salvador, UDA: UDA.
3.3. Genetic Structure of the Oaxacan Creole Sheep
To determine the genetic substructure in the population of Oaxacan Creole sheep, efficiently assign individuals within the flock and that do not present any degree of crossbreeding with other populations, an analysis was made in the program STRUCTURE v 2.1, using a Bayesian algorithm of the program that calculates the distribution posteriori of each coefficient of a mixture of each individual (q) (Figure 4). This analysis shows each individual as a vertical bar and each color represents the proportion, in percentage, of the genome of the populations analyzed, making it possible to identify from which ancestral population the Oaxacan Creole sheep is from.
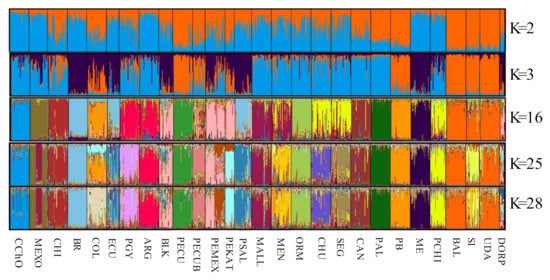
Figure 4.
Genetic structure of the 28 sheep populations analyzed. The figure shows the graphic representation of the populational grouping when it is compared with a different number of populations (K): K = 2, K = 3, K = 16, K = 25 (optimum) and K = 28. In the X-axis the populations occupied are numbered (Table 1), where the 1 is the Oaxacan Creole.
When two populations are analyzed (K2) (African vs. Merino), the Oaxacan Creole is grouped with those that descend from the Merino. The analysis of K3 shows the difference from the Spanish Merino and those that carry a greater genetic load of their ancestors (dark blue). When utilizing 10 populations (K10), more than 85% of the individuals of the Oaxacan Creole were assigned to the same group (data not shown). When compared with 16 populations, it is separated as an independent group, even from the other Mexican creoles. In the analysis of 25 (K25) (statistically optimum number) and 28 (K28) populations, 86.8% of the individuals are assigned to the same group, where no subdivision or substructure of the population is observed. Therefore, it can be considered a homogeneous flock without a populational substructure or genetic mixing with the rest of the breeds used.
4. Discussion
Sheep breeding in Mexico, in order to satisfy the productive, economic, and social needs of the producers and the population, has opted for improving flocks through the introduction of improved breeds and has carried out crossbreeding with no established control [27]. However, these practices have caused an erosion in the native genetic resources, increasing diversity and provoking their extinction [4,5,6,7]. Presently, programs have been established for the identification and conservation of native zoogenic resources for productive species [28,29]. One of the fundamental tools for this purpose is the molecular markers, principally the microsatellites, which facilitate identification, estimation of genetic diversity, detection of genes with qualitative features, and assisted selection [30].
In the present study, 29 sheep of the Oaxacan Creoles population, corresponding to one-third of the entire population, were genetically characterized, utilizing 41 microsatellites recommended by the FAO/ISAG for sheep [31]. These organizations recommend the minimum use of 25 markers for establishing genetic intra and interracial relationships of domestic animals. According to Nei et al. [23], the standard error of genetic diversity is lower when more loci are used. Furthermore, it is considered that the optimum sample size is 30, thus this study has a high degree of reliability due to the number of individuals and markers employed.
4.1. Intra-Racial Genetic Diversity
It was found that the population has a mean of alleles (NAM) of 5 and an effective number (Ae) of 3.327, parameters which in conjunction with heterozygosity, which indicate the genetic variability within a population. The value of NAM is low compared with that of the sheep breeds of Colombia (14.27) [11], of China (13.59) [32], and of the creole of Chiapas (14.19) [9], but it is similar to that of the creole breeds of Argentina (Corrientes: 7.53; Santiago del Estero: 6.16; Salta: 6.7; Buenos Aires: 5.73) [33], to the creole of Paraguay (7.25) [34], to Italian breeds (Cornigliese: 6,64; Bergamasca: 5.56), and to the Spanish Merino (7.41) [35]. The values of Ae are similar to those found by Peña et al. [33] in 4 populations of Argentine creole sheep (Corrientes: 4.078; Santiago del Estero: 4.036; Salta: 3.856; Buenos Aires: 3.193) and to those reported in 13 breeds of sheep (3.64–4.43) of Colombia by Ocampo et al. [11].
The heterozygosity in the Oaxacan Creole is considered from medium to high according to what was reported by Martínez [33], given that it showed a mean of He = 0.686 and Ho = 0672, values similar to those presented by the creole of Chiapas, Mexico (He = 0.624–0712; Ho−0.606–0.666) [9], to the creoles of Argentina (He = 0.676; Ho = 0.685 [33] and the Spanish Merino (He = 0.661; Ho = 0.620 [36]. On the other hand, it is lower than the creole breed of Paraguay (He = 0.73) [34]. In addition, the Oaxacan Creole in the estimation of endogamy level [22] had a mean value of PIC = 0.609 and a mean consanguinity of FIS = 0.022, showing polymorphic values of moderate to high and a moderate genetic diversity [14], lower than those present in Colombian sheep (PIC = 0.74; FIS = 0.09) [11], but similar for the creole sheep of Chiapas, Mexico (FIS = 0.026–0.076) [9], the Argentinian creoles (FIS = 0.030 [33], and the Spanish Merino (FIS = 0.074). Therefore, it can be concluded that the Oaxacan Creole is a population with a moderate intra-racial diversity without a significant deviation from the Hardy–Weinberg equilibrium, similar to other creole breeds of America and the Spanish Merino. Furthermore, the presence of markers with values of FIS very far from zero (reduction of heterozygosity) in the Oaxacan Creole sheep may be due to the low number of individuals in the population, which favors endogamy, added to the lack of planning of crosses between unrelated individuals.
4.2. Inter-Racial Genetic Diversity
Genetic diversity among populations can be analyzed using two variables, the statistic of the coefficient of genetic differentiation (FST) and the measurement of the genetic distance among them (DA) [37]. Estimating these values makes it possible to distinguish how genetically isolated a population is, which infer its phylogeny and generate strategies for its conservation.
The Oaxacan Creole sheep, in the analysis of genetic differentiation with the 27 populations utilized, showed a mean value of FST−0.1003. This is similar to what was reported by Quiroz et al. [9] for the creole of Chiapas (FST = 0.1) and to that reported between the Argentinian creoles Santiago de Estero and Buenos Aires (0.104) [33]. According to the Wright classification [38], this value is contemplated as a moderate degree of genetic differentiation; but according to what was reported for domestic sheep breeds [9], it is considered that the Oaxacan Creole has a high degree of genetic differentiation in comparison to what was found by Rendón et al. [39] for Spanish breeds (FST = 0.07), to that of Merino breeds of different countries (0.03) reported by Diez-Tascón et al. [36], and to that estimated among the Colombian creoles (0.01–0.047) [13].
The analysis of the genetic relationships among the 28 populations in a neighbor-net built with DA genetic distances showed the separation of 4 groups: grouping the Oaxacan Creole with another creole of Mexico, the Chilean Merino, the Spanish Merino, and with the lowest DA, with the Argentinian creole. This analysis makes it possible to suggest that the Oaxacan Creole conserves certain ancestry to the breeds introduced during the colonization [40], and by its DA with the Spanish Merino and the Chilean Merino. On the other hand, it is grouped in the same phylogenic trunk as the Argentinian Creole and to the other creole of Mexico, suggesting the same origin, although with elevated genetic differences, and an origin different from that of the creole of Chiapas. This diversity or genetic differentiation of the Oaxacan Creole could be due to its geographic localization and to the management established by the producer, resulting in reproductive isolation, thus allowing the conservation of its genetic characteristics.
4.3. Genetic Structure
This analysis provides information for estimating the proportion that each individual or population carries the genome of its parents or ancestors, making it possible to distinguish if there is a mixing of breeds or not in order to assign individuals or homogeneous populations. This analysis helped to confirm what was observed in the dendrogram of the union of neighbors, revealing that the Oaxacan Creole sheep is a homogeneous population, with the ancestral influence of the Merino; without a populational substructure or mixing of its genome with any of the 27 populations analyzed, even with the other populations of the country, thus separating them as an independent breed.
Finally, the data obtained show that the sheep flock subjected to study in the region of the High Mixteca of Oaxaca is a creole breed different from those analyzed, maintaining its genetic merit. Therefore, its recognition is proposed as Chocholteca Creole breed for the zone in which it has been conserved and in honor of the ethnic group which inhabits this region. Furthermore, this recognition will help to promote its conservation and to implement populational recovery programs in conjunction with the producer and his cultural system.
5. Conclusions
The Chocholteca Creole sheep has been maintained as an isolated breed, conserving genetic characteristics since it was introduced to America, and does not present features of crossbreeding with the new lines of improved sheep or other creoles of America, or breeds of Spain and Africa.
This creole sheep has in its population a moderate–high genetic diversity, with a homogeneous structure, and does not present a subdivision or genetic substructure.
It can be considered as a creole breed different from the others of the American continent and Spain, establishing itself as an important new zoogenic resource. Therefore, it is necessary to establish reproductive management for its conservation.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/ani11041172/s1, Table S1. Parameters of genetic diversity of the 28 breeds. The table shows values corresponding to Ae, Ho He, Fis and Fis IC from each sheep used in the analisys; Table S2: Genetic Distances (DA) are shown in red and the coefficient of genetic differentiation (FST) in blue between pairs of the populations used. The values for Oaxacan Creole are highlighted in black.
Author Contributions
Conceptualization, H.M.R.-M., J.H.-B., and T.S.-R.; investigation, T.S.-R. and H.M.R.-M.; methodology, A.M.-M. (Amparo Martínez-Martínez); original writing of the project, H.M.R.-M., A.M.-M. (Araceli Mariscal-Méndez), and T.S.-R.; revision and editing of the text, J.H.-B., A.M.-M. (Araceli Mariscal-Méndez), and M.A.-C.; administration of the project, H.M.R.-M., J.H.-B., and T.S.-R.; intellectual property of genetic database, Consorcio Biovis. All authors have read and agreed to the published version of the manuscript.
Funding
This investigation did not receive any external funds for its execution.
Institutional Review Board Statement
Ethical approval was waived due blood sample was taken by a zootechnical veterinarian during the sanitary control of the flock.
Data Availability Statement
The data in this study was microsatellite analysis by using known specific primers. No new data were created on this study. Data sharing is not applicable to this article.
Acknowledgments
The authors are grateful for the funding provided by the director of the School of Veterinary Medicine and Animal Science and by the rectory of the Universidad Autónoma Benito Juárez of Oaxaca. We give special thanks to the producer Trinidad García of the High Mixteca, Oaxaca, for conserving the breed and allowing us to utilize it in the present work. Consorcio Biovis: Edgar Lenin Aguirre Riofrío (Centro Biotecnología Reproductiva Animal, Universidad Nacional de Loja, Loja, Ecuador; edgar.aguirre@unl.edu.ec); Luz Angela Alvarez Franco. (Universidad Nacional de Colombia, Sede Palmira, CR 32 no. 12-00 Chapinero, Palmira, Valle del Cauca, Colombia. laalvarezf@unal.edu.co); Juan Capote. (ICIA. Tenerife. Spain. jcapote1@gmail.com); María Antonia Revidatti. (Departamento de Producción Animal, Facultad de Ciencias Veterinarias, Universidad Nacional del Nordeste, Corrientes, Argentina. toninacereza@hotmail.com); Beatriz López de los Santos. (EA group S. Coop. Villanueva de la Serena, Badajoz. Spain. beatrizlopez@eagroup.coop); Raúl Perezgrovas Garza. (Universidad Autónoma de Chiapas. Chiapas. México. rgrovas@unach.mx); O. Roberto Martínez López. (Universidad Nacional de Asunción. Paraguay. agrotulo@yahoo.com); Agueda Pons. (Serveis de Millora Agrària i Pesquera. Islas Baleares. Spain. apons@semilla-caib.es); Jorge Quiroz Valiente. (Instituto Nacional de Investigaciones Forestales, Agrícolas y Pecuarias, México, jorgequirozvaliente@gmail.com); Julio César Vargas Burgos. (Departamento de la Unidad de Producción y Comercialización, Centro de Investigación Posgrado y Conservación Amazónica, Universidad Estatal Amazónica, 54830 Pastaza, Ecuador; jvargasburgos@yahoo.com).
Conflicts of Interest
The authors declare that they have no conflicts of interest.
References
- Secretaría de Agricultura, Ganadería, Desarrollo Rural, Pesca y Alimentación (SAGARPA). 2016. Padrón Ganadero Nacional. Available online: https://inehrm.gob.mx/recursos/Libros/SAGARPA.pdf (accessed on 1 August 2020).
- Servicio de Información Agroalimentaria y Pesquera (SIAP). Acciones Y Programas Para La Producción Pecuaria. 2018. Available online: https://www.gob.mx/siap/acciones-y-programas/produccion-pecuaria (accessed on 1 August 2020).
- Hernández-Bautista, J.; Salinas-Rios, T.; Rodríguez-Magadán, H.M.; Aquino-Cleto, M.; Mariscal-Méndez, A.; Ortiz-Muñoz, I.Y. Características que determinan el sistema de producción ovina en el estado de Oaxaca, México. Rev. Mex. De Agroecosistemas 2017, 4, 38–47. [Google Scholar]
- Martínez-Peña, M.; Villagómez-Cortés, J.A.; Mora-Brito, A.H. Rentabilidad del sistema de producción ovina en el bajo Mixe, Oaxaca, México. Agrociencia 2018, 52, 107–122. [Google Scholar]
- Food and Agriculture Organization of the United Nations. The Stage of the World´s Biodiversity for Food and Agriculture; Bélanger, J., Pilling, D., Eds.; FAO Commission on genetics resources for Food and Agriculture Assessment: Rome, Italy, 2019; p. 572. [Google Scholar]
- Martínez, R.D. Prejuicios que afectan a bovinos y ovinos criollos en Argentina. Revista AICA 2015, 5, 26–35. [Google Scholar]
- Martínez, S.R.; Vásquez, R.R. Evaluación de la conservación y comportamiento productivo del banco de germoplasma de la especie ovina en Colombia. Anim. Genet. Resour. Inf. 2005, 36, 33–45. [Google Scholar]
- Alonso, A.R.; Ulloa-Arvisu, R.; Gayosso-Vázquez, A. Mitochondrial DNA sequence analisys of the Mexican creole sheep (Ovis aries) reveals a narrow Iberian maternal origin. Mitochondrial DNA Part A 2017, 28, 793–800. [Google Scholar] [CrossRef] [PubMed]
- Quiroz, J.; Martinez, M.A.; Zaragoza, L.; Perezgrovas, R.; Vega-Pla, J.L.; Delgado, J.V. Genetic characterization of the autochthonous sheep populations from Chiapas, México. Livest. Sci. 2008, 116, 156–161. [Google Scholar] [CrossRef]
- Delgado, J.V.; Martínez, M.A.; Acosta, A.; Álvarez, L.A.; Armstrong, E.; Camacho, E.; Cañón, J.; Cortés, O.; Dunner, S.; Landi, V.; et al. Genetic characterization of Latin-America creole cattle using microsatellite markers. Anim. Genet. 2011, 43, 2–10. [Google Scholar] [CrossRef]
- Ocampo, R.; Cardona, H.; Martínez, R. Genetic diversity of Colombian sheep by microsatellite markers. Chileanjar 2015, 76, 40–47. [Google Scholar] [CrossRef][Green Version]
- Aguirre-Riofrio, L.; Maza-Tandazo, T.; Quezada-Padilla, M.; Albito-Balcazar, O.; Flores-González, A.; Camacho-Enriquez, O.; Martínez-Martínez, A.; BioGoat Consortium; Delgado-Bermejo, J.V. Genetic characterization of the “Chusca Lojana” a creole goat reared in Ecuador, and its relationship with other Goat Breeds. Animals 2020, 10, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Vivas, N.A.; Landi, V.; Muñoz, F.J.; Bustamante, M.Y.; Álvarez, L.F. Genetic diversity of Colombian Creole sheep. J. MVZ Cordoba 2020, 25, e2185. [Google Scholar]
- Yatkin, S.; Özdil, F.; Özkan, E.Ü.; Genc, S.; Kaplan, S.; Kemal, E.G.; Arat, S.; Ihsan, M.S. Genetic Characterization of native donkey (Equus asinus) populations of Turkey using microsatellite markers. Animals 2020, 10, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Walsh, P.S.; Metzger, D.A.; Higuchi, R. Chelex 100 as a medium for simple extraction of DNA for PCR-based typing from forensic material. BioTechniques 1991, 10, 506–513. [Google Scholar] [CrossRef] [PubMed]
- Park, S.D.E. Trypanotolerance in West African Cattle and the Population Genetic Effects of Selection; University of Dublin: Dublin, Ireland, 2001. [Google Scholar]
- Kalinowski, S.T.; Taper, M.L.; Marshall, T.C. Revising how the computer program CERVUS accommodates genotyping error increases success in paternity assignment. Mol. Ecol. 2007, 16, 1099–1106. [Google Scholar] [CrossRef] [PubMed]
- Yeh, F.C.; Boyle, T.J.B. Population genetic analysis of co-dominant and dominant markers and quantitative traits. Belg. J. Bot. 1997, 130, 129–157. [Google Scholar]
- Belkhir, K.; Borsa, P.; Chikhi, L.; Raufaste, N.; Bonhomme, F. Genetix: 4.05 Logiciel Sous WindowsTM Pour la Genetique Des Populations; Université de Montpellier: Montpellier, France, 2003. [Google Scholar]
- Raymond, M.; Rousset, F. GENEPOP (Version 1.2): Population genetics software for exact test and ecumenicism. J. Hered. 1995, 86, 248–249. [Google Scholar] [CrossRef]
- Guo, S.W.; Thompson, E.A. Performing the exact test of Hardy-Weinberg proportions for multiple alleles. Biometrics 1992, 48, 361–372. [Google Scholar] [CrossRef]
- Wright, S. Theory of gene frequencies. In Evolution and genetics of populations; University of Chicago: Chicago, IL, USA, 1969; pp. 291–293. [Google Scholar]
- Nei, M.; Tajima, F.; Tateno, Y. Accuracy of Estimated Phylogenetic Trees from Molecular Data. II. Gene Frequency Data. J. Mol. Evol. 1983, 19, 153–170. [Google Scholar] [CrossRef]
- Langella, O. Populations 1.2.28 CNRS UPR9034 1999. Available online: http//www.cnrs-gif.fr/pge/bioinfo/populations/index.php (accessed on 21 October 2019).
- Huson, D.H.; Bryant, D. Application of phylogenetic networks in evolutionary studies. Mol. Biol. Evol. 2006, 23, 254–267. [Google Scholar] [CrossRef]
- Pritchard, J.K.; Stephens, M.; Donnelly, P. Inference of population structure using multilocus genotype data. Genetics 2000, 155, 945–949. [Google Scholar]
- Arriaga-Jordán, C.M.; Pedraza-Fuentes, A.M.; Nava-Bernal, E.G.; Chávez-Mejía, M.C.; Castelán-Ortega, O.A. Livestock agrodiversity of Mazahua smallholder Campesino system in the highlands of central México. Hum. Ecol. 2005, 33, 821–845. [Google Scholar] [CrossRef]
- Food and Agriculture Organization (FAO). The State of the World´s Animal Genetic Resources for Food and Agriculture; Dafydd Pilling, D., Rischkowsky, B., Eds.; FAO: Rome, Italy, 2007; p. 39. [Google Scholar]
- Delgado, J.V.; Camacho, M.E. A Latinoamerican experience in the conservation of zoogenetic resources and traditional management system. Ital. J. Anim. Sci. 2007, 6, 120–121. [Google Scholar] [CrossRef]
- Sabir, J.; Mutwakil, M.; EL-Hanafy, A.; Al-Hejin, A.; Sadek, M.A.; Abou-Alsoud, M.; Qureshi, M.; Saini, K.; Ahmed, M. Appling molecular tools for improving livestock performance: From DNA markers to next generation sequencing technologies. J. Food Agric. Environ. 2014, 12, 351–363. [Google Scholar]
- Food and Agriculture Organization of the Unated Nations/International Society of Animal Genetics (FAO/ISAG). Secondary Guidelines for Development of National Farm Animal Genetic Resources Management Plans; FAO: Rome, Italy, 2004; p. 24. [Google Scholar]
- Zhong, T.; Han, J.; Guo, J. Genetic diversity of Chinese indigenous sheep breeds inferred from microsatellite markers. Small Rumin. Res. 2010, 90, 88–94. [Google Scholar] [CrossRef]
- Peña, S.; Martínez, M.A.; Villegas, C.E.; Aulicino, M.; Género, E.R.; Giovambattista, G.; Martínez, R.D. Caracterización Genética de cuatro poblaciones de ovinos criollos de Argentina. J. Bag 2017, 28, 43–55. [Google Scholar]
- Ochipinti, G.; Núñez, L.; Cazal, C.; Samudio, A.; Castro, L.; Ramírez, L.; León, D.; Martínez, M.A.; Oka-Obara, A.; Landi, V.; et al. Diversidad genética en ovejas de los humedades de la región oriental de Paraguay. Actas Iberoam. Conserv. Anim. 2012, 2, 227–230. [Google Scholar]
- Ceccobelli, S.; Karsli, T.; Di Lorenzo, P.; Marozzi, G.; Landi, V.; Sarti, F.M.; Sabbioni, A.; Lasagna, E. Genetic diversity of Cornigliese sheep breed using STR markers. Small Rumin. Res. 2015, 123, 62–69. [Google Scholar] [CrossRef]
- Diez-Tascón, C.; Littlejohn, R.P.; Almeida, P.A.R.; Crawford, A.M. Genetic variation within the Merino sheep breed: Analysis of closely related populations using microsatellites. Anim. Genet. 2000, 31, 243–251. [Google Scholar] [CrossRef] [PubMed]
- Baumung, R.; Simianer, H.; Hoffmann, I. Genetic diversity studies in farm animals—A survey. J. Anim. Breed Genet. 2004, 121, 361–373. [Google Scholar] [CrossRef]
- Wright, S. Evolutions, and the Genetics of Populations. Vol 4. Variability within and among Natural Populations; University of Chicago Press: Chicago, IL, USA, 1978. [Google Scholar]
- Rendo, F.; Iriondo, M.; Jugo, B.M.; Mazón, L.I.; Aguirre, A.; Vicario, A.; Estonba, A. Tracking diversity and differentiation in six sheep breeds from the North Iberian Peninsula through DNA variation. Small Rumin. Res. 2004, 52, 195–202. [Google Scholar] [CrossRef]
- Rodero, A.; Delgado, J.V.; Rodero, E. Primitive Andalusian livestock and their implications in the discovery of America. Arch. Zootec. 1992, 41, 383–400. [Google Scholar]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).