Abstract
Reports on the use of 16S sequencing for the identification of bacteria in healthy animals are lacking. Bacterial contamination of bull semen can have a negative effect on the sperm quality. The aims of this study were threefold: to identify bacteria in the semen of healthy bulls using 16S sequencing; to investigate the differences in the bacterial community between individual bulls; and to establish if there was a relationship between the bacteria isolated and bull fertility. Semen from 18 bulls of known fertility was used for the DNA extraction and 16S sequencing; 107 bacterial genera were identified. The differences in the amplicon sequence variants (ASVs) and the numbers of genera between bulls were noted. Negative correlations (p < 0.05) between several bacterial genera with Curvibacter, Rikenellaceae RC9-gut-group and Dyella spp. were seen. Other negatively correlated bacteria were Cutibacterium, Ruminococcaceae UCG-005, Ruminococcaceae UCG-010 and Staphylococcus, all within the top 20 genera. Two genera, W5053 and Lawsonella, were enriched in bulls of low fertility; this is the first time that these bacteria have been reported in bull semen samples. The majority of the bacteria were environmental organisms or were species originating from the mucous membranes of animals and humans. The results of this study indicate that differences in the seminal microbiota of healthy bulls occur and might be correlated with fertility.
1. Introduction
One of the main reasons for introducing artificial insemination into animal breeding was to prevent the spread of infectious diseases between males and females [1]. Apart from the urogenital tract itself as a primary source of pathogens [2], there are critical points in semen processing where sperm samples can be contaminated with different types of microbiota [3,4,5]. Therefore, antibiotic use in semen for artificial insemination (AI) is considered to be essential. Different types of antibiotics, singly and in combination, are added to semen worldwide with the aim of decreasing or preventing microbial growth. Antibiotics used in bull semen cryopreservation are mainly based on the regulations given by government directives, e.g., the European Union [6]. However, despite the addition of antibiotics, bacteria can still be isolated from bull semen after thawing [7]. In addition, there is considerable concern about the emergence of antibiotic-resistant bacteria, particularly methicillin-resistant strains [8]. The identification of bacteria in semen samples is a primary step to the rational use of antibiotics and could be helpful in developing the next steps in the control of microbial contamination of bull semen [3].
The presence of bacteria in semen does not equate with infection and limited microbial activity does not necessarily affect sperm quality [9]. However, bacteria can have a direct negative affect on sperm quality, depending on the species and number. Negative correlations were reported between certain bacteria and sperm motility [10], viability, membrane integrity and acrosome reaction [11], sperm DNA fragmentation rates [12,13] and the total number of spermatozoa [14,15]. Givens and Marley [16] reported the presence of microorganisms that cause infertility and/or are transmitted via semen. This knowledge can facilitate the identification and exclusion of subclinically infected bulls from production. Bacteria present in the urogenital tract can influence fertility in cows [17]. Postpartum uterine disease caused by pathogenic bacteria not only reduces fertility but also decreases productivity [18]. Postpartum metritis and clinical endometritis caused subfertility in cows by increasing the time to first insemination, delayed conception and increasing the calving to conception interval [19,20]. Furthermore, postpartum clinical endometritis can cause infertility and consequently results in involuntary culling [21]. Marey et al. [22] reported that infections extending from the uterus to the oviduct induce an immune system disbalance that interferes with fertilization. Apart from an effect on the uterus itself, a uterine infection can lead to disruption in the secretion of gonadotropins [23], disruption in ovarian follicle growth and function [24] and a reduction in the oocyte quality [25] (cited from Sheldon and Owens [18]).
In contrast to these extensive studies in females, the presence and influence of bacteria on sperm quality and fertility in bulls have not been investigated to any great extent. However, the types of bacteria present in semen in various species such as stallions [2,26], boars [27] and rams [28] have been described as well as reports of the influence of bacteria on fertility and sperm quality parameters in humans [29,30]. Interest in the isolation and identification of the main bacteria in bull semen causing infertility began decades ago [31] using culture and identification by means of appearance and biochemical properties; other methods of identification were introduced as the science developed. In general, reports on the use of 16S sequencing for the identification of bacteriospermia in healthy animals are lacking, especially in bulls. The identification of individual opportunistic bacteria is of interest due to the possible transmission between bulls and from bulls to cows, causing economic losses to cattle production. However, there are few studies in which a full microbiological profiling of bull semen has been achieved.
The aims of the present study were to identify the bacteria in bull semen via 16S rRNA sequencing and to investigate the individual differences in bacterial type and number in bulls at a semen collection station. An additional aim was to examine the relationships between the bacterial community and the overall fertility of the bulls.
2. Materials and Methods
2.1. Animals and Semen Collection
Ejaculates were obtained from 18 Holstein bulls at an AI center (Animal Breeders’ Association of Estonia, Raplamaa, Estonia) where animals were kept according to national and international regulations. The age of the bulls ranged from 3 to 10 years. The bulls were kept on dry bedding (new sawdust added 4× /24 h) and, if necessary, cleaned by brushing before the semen collection. Semen collection with an artificial vagina is a routine agricultural practice and, therefore, does not require ethical approval according to Estonian law [32]; the bulls at the AI station were not considered to be experimental animals. Therefore, no special ethical permission was required.
The bulls were prepared for semen collection by allowing them to follow each other in a circular chute for 30–40 min; the bulls were able to make false mounts by jumping on the bull in front but the chute was not wide enough for the bulls to turn round. A sterilized artificial vagina lubricated with Bovivet Gel (Jørgen Kruuse A/S, Langeskov, Denmark) and a sterilized graduated collecting tube were used for the semen collection, which took place approximately 10 m from the laboratory separated by a glass wall. Within a minute after the collection, the sterile graduated tube containing the collected ejaculate was separated from the artificial vagina and passed to the laboratory personnel through a small hatch in the glass wall. All semen collection procedures were performed with sterile equipment and aseptic measures to avoid semen contamination. In the laboratory, an aliquot of 1 mL semen was transferred to an Eppendorf tube above an alcohol burner and stored in liquid nitrogen before transfer to −80 °C for storage until the bacterial DNA extraction was performed. The remainder of the semen samples were processed and used for the evaluation of the sperm motility by CASA. All samples with a total motility >90% and a progressive motility >80% were used for the routine inseminations. The CASA results are presented in Supplementary Table S1. The fertility performance of these bulls was estimated by non-return rates (NRRs) at 90 days post-AI, based on the outcome from the first insemination. In total, 48,469 females were inseminated, comprising 34,800 cows and 13,649 heifers. Miglior et al. [33] defined NRRs as the proportion of cows that are not seen in estrus again after insemination and are, therefore, considered to be potentially pregnant.
2.2. DNA Extraction
The DNA extraction was performed in the Clinical Sciences Research Laboratory at SLU using an AllPrep DNA/RNA/miRNA Universal Kit Cat No./ID 80224 (GIAGE, Germantown, Philadelphia, USA) following the manufacturer´s instructions for the protocol of the simultaneous purification of genomic DNA and total RNA, including miRNA from cells. In total, 10 µL of semen was used for the DNA extraction to reach the maximum amount of 1 × 107 cells, spermatozoa and bacteria according to the protocol. The sperm concentration was evaluated using a Nucleocounter SP100 (ChemoMetec, Allerød, Denmark) according to the manufacturer´s instructions. According to our calculations, 10 µL of semen contained on average 5 × 106 spermatozoa although the bacterial number was not calculated. All samples were centrifuged; the supernatant was removed and only pelleted cells were used. The purity and concentration of the DNA were tested using a NanoDrop 8000 Spectrophotometer (Thermo Scientific, Waltham (HQ), MA, USA). The DNA purity was considered adequate when the 260/280 ratio was between 1.7 and 1.9 and the concentrations were between 5.34 and 10.97 ng/µL. The DNA samples were stored at −80 °C until further preparation.
2.3. 16S rRNA Amplification and Sequencing
A two-step amplification protocol was used for the preparation of the V3–V4 16S gene region for Illumina sequencing. The details of the primers used and the cycling protocols are presented in Table 1. The reaction volume of the first step was 15 µL containing 4 ng of the DNA template, 0.25 µM Pro 341F and the same volume of Pro 805R primers with Nextera adaptor sequences (Illumina Inc., CA, USA) as well as 1 µg/µL BSA and 1 × Phusion Taq ready-to-use mix (New England Biolabs, MA, USA). For the second step, the reaction volume was 30 µL and contained 3 µL of the purified DNA template from the first PCR step, 0.20 µM tagged F and R primers with Nextera adaptor sequences and 1 µg/µL BSA and 1 × Phusion Taq ready-to-use mix. Both PCR steps were performed in duplicate and the reactions were pooled and purified between the steps using SeraMag Magnetic Carboxylate Modified particles in a ratio of 1:1. An Agilent Bioanalyzer was used for the quality check. The amplicons were eluted in 10 mM Tris with a pH of 8.5 and stored at −20 °C. The samples were sent for sequencing to SciLifeLab, SNP&SEQ Technology Platform, Uppsala University. Paired-end sequencing was performed on a MiSeq system (Illumina, San Diego, CA, USA) using kit V2. Approximately 40,000 to 60,000 paired-end reads of 250 bp length were obtained for all samples except the control sample (sterile water), which yielded only 13 reads after sequencing.

Table 1.
Primer combination and thermal cycling conditions used to quantify the 16S rRNA.
2.4. 16S Profiling
The analysis of the 16S rRNA sequencing data was performed using Nextflow pipeline ampliseq v.1.1.2 (https://github.com/nf-core/ampliseq, accessed on 24 November 2021). Briefly, raw sequencing reads were quality checked initially using FastQC [35] followed by the trimming of the primer sequences from the reads using cutadapt v.2.7 [36]. The raw sequencing data were cleaned from source contamination by running BLAT against the cow reference genome Bos taurus 8 available in the UCSC genome browser (https://genome.ucsc.edu/, accessed on 24 November 2021). The sequencing reads were denoised, dereplicated and filtered for chimeric sequences using DADA2 [37]. The denoised paired-end reads were truncated from position 229 (forward) and 215 (reverse) after a manual visualization of the sequencing error profile; all other reads shorter than the cutoff values were dropped. The truncated sequences were merged with at least a 20 bp overlap, resulting in exact amplicon sequence variants (ASVs). These ASVs were taxonomically classified from the phylum to species level clustered with 99% similarity using the SILVA v.132 database [38] by applying a Naive Bayes classifier implemented in QIIME 2 [39] trained on the preprocessed database. Following the taxonomic classification of the ASVs, the ASVs classified as a mitochondria or a chloroplast were removed. Only the ASVs with a minimum read frequency ≥5 in at least one sample were retained for a further analysis.
2.5. Statistical Analysis
The data analysis was performed using R v.3.3.1 software. Pearson correlation coefficients were calculated between the bacterial genera using the cor.test function in the R environment, with p < 0.05 being considered significant. The plotting was carried out in R using corrplot v.0.9. A linear discriminant analysis effect size (LEfSe) analysis of the microbial abundance between low and high fertility bulls was performed to detect the differences between the two groups and characterize the biomarkers. The groups were divided based on the NRR where the NRRs were <51% and >51% for low and high fertility bulls, respectively.
3. Results
The total amount of DNA in the pool was 1320 ng with a ratio of absorbance at A260/A280 at 1.89 and A260/A230 at 2.1 and a concentration of 24 ng/µL. In total, 107 bacterial genera were identified in 18 bull semen samples. The 20 most frequently seen bacterial genera are presented in Figure 1.
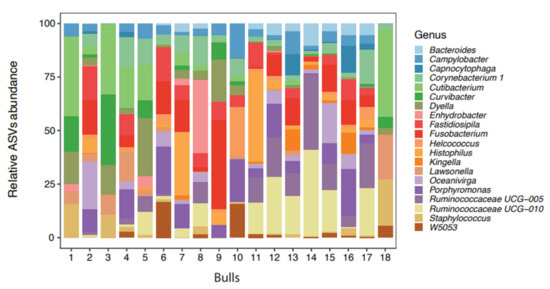
Figure 1.
Distribution of the 20 most abundant bacteria genera in the semen from 18 bulls (amplicon sequence variants) identified by 16 rRNA sequencing.
The bacteria listed among the top 20 for each bull varied considerably; there was also a considerable variation between bulls in both the number of bacterial genera present (ranging from 12 to 89) and in the bacterial count (ranging from 27,827 to 247,273) (Table 2). The highest number of genera identified was in the sample from bull 12, which contained 89 bacterial genera of which 19 came from the 20 most frequently seen. In contrast, Bull 3 was the least colonized, with only 12 bacterial genera present.

Table 2.
Number of top 20 and total genera, counts, total counts and non-return rate (NRR, %) per bull.
The 20 most frequently seen bacterial genera, starting with the most frequent, were: Porphyromonas, Fusobacterium, Ruminococcaceae UCG-010, Fastidiosipila, Ruminococcaceae UCG-005, Cutibacterium, Histophilus, Oceanivirga, Corynebacterium 1, Campylobacter, W5053, Dyella, Staphylococcus, Lawsonella, Helcococcus, Bacteroides, Capnocytophaga, Curvibacter, Kingella and Enhydrobacter, which are presented in Figure 1 as the relative ASV abundance.
There were negative correlations between several bacterial genera (Figure 2). The majority of the bacteria that showed negative correlations were from the top 20 genera (Figure 1) and included Curvibacter, Cutibacterium, Dyella, Ruminococcaceae UCG-005, Ruminococcaceae UCG-010 and Staphylococcus spp. In addition, Curvibacter, Rikenellaceae RC9-gut-group and Dyella spp. (brown spots) were negatively correlated in most cases.
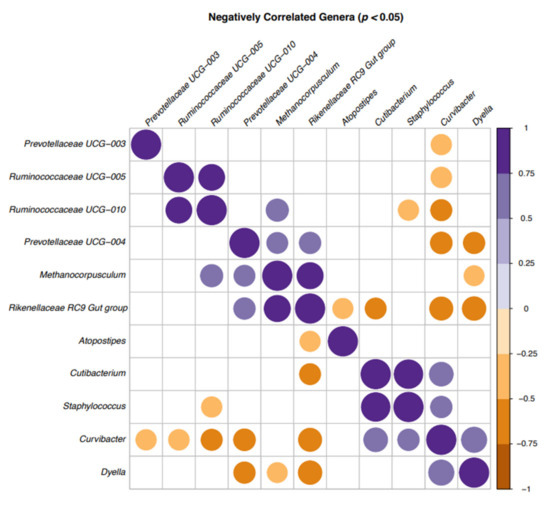
Figure 2.
Correlation plot of the bacteria in bull semen identified by 16S sequencing in a correlation matrix showing the significant correlations (p < 0.05) between genera: positive (purple) and negative (brown). Blank cells indicate non-significant correlations.
In the bar plot (Figure 3), the taxa (including the genus and family) with significant differences between the groups were detected by a LEfSe analysis with a log-10 transformed LDA (linear discriminant analysis) threshold score of 2.0 and a significant p-value <0.05. The ASV with a higher LDA score indicated that the ASV was more important according to the LEfSe in discriminating between the low and high individuals. Two genera, W5053 and Lawsonella, were enriched in the low fertility group.
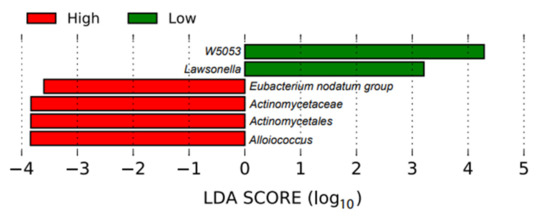
Figure 3.
Linear discriminant analysis effect size plot. This plot shows the enriched taxa that were significantly different between the high fertility (red) and low fertility (green) bulls.
4. Discussion
In this study, 16S sequencing of bull semen microbiomes was performed with the aim of identifying the most common bacteria in the semen of healthy bulls. The determination of the most common bacteria genera can enable the development of appropriate control methods.
The isolation and identification of bacteria by commercial microbiological methods are more difficult compared with metagenomics analyses. First, it may not be possible to isolate all the bacteria in a sample. The isolation and identification of bacteria by culture-dependent methods require different culture conditions for different type of bacteria such as the media, temperature, presence or absence of oxygen and time of incubation. The growth of a few bacteria may be inhibited by competition between bacterial species or bacterial overgrowth. Traditional culture-dependent morphological identification methods are time-consuming and do not enable all bacteria to be identified [40]. Although the development of MALDI-TOF (matrix-assisted laser desorption ionization-time of flight) mass spectrometry can help in the process of bacterial identification after culturing, the possibility of identifying isolates depends on the information in the database for the instrument [2]. In contrast, 16S sequencing does not require a bacterial culture and enables the identification of large numbers of bacteria present in a sample [26].
To our knowledge, this is the first comprehensive metagenomic study of the seminal microbiome of healthy bulls. During the introduction of artificial insemination as an animal husbandry technique, there was a need to inhibit bacterial growth in bull semen [41]. Later, culture-dependent studies were conducted to isolate the bacteria responsible for bull infertility [42] and this technique was also used in more recent studies of the microbiological evaluation of frozen semen samples [43]. During the past few years, the identification of individual bacteria from preputial mucosa [44,45] and bull semen [46] was performed with metagenomic methods. Interestingly, there were no significant correlations between the preputial microbial community of bulls of different ages, breed, diet or co-housing [47]. In other species, several studies on culture-independent methods of identification of the normal microbiota in semen have been reported. Stallion semen microbiota was studied by conventional methods of identification as well as MALDI-TOF [2] and recently by 16S sequencing [26]. Al-Kass et al. [26] reported that large numbers of bacterial genera could be identified in stallion semen using metagenomic analyses by 16S sequencing. Based on those studies, variations in bacteria genera and their number between individual stallions and between countries were identified. However, the researchers agreed that the identified bacteria in both stallions and bulls [26,47] were mostly environmental in origin.
In order to understand the composition of the bull semen bacterial community, it is important to discuss its potential sources. Three of the most common genera identified in our study were Porphyromonas, Fusobacterium and Ruminococcaceae UCG-010 respectively. This finding is in agreement with the study of Wickware et al. [47] where Porphyromonas and Fusobacterium were the most abundant genera in the preputial microbiota of Hereford bulls. These two genera, together with Bacterioides, were among the most abundant genera from the upper respiratory and oral mucosal membranes of healthy calves in the first month of life [48,49]. In contrast, the anaerobic bacteria Porphyromonas, Fusobacterium and Fastidiosipila were highly prevalent in most cases of lameness caused by foot lesions [50]. Their presence in semen is probably due to their common occurrence in the environment and subsequent colonization of mucosal membranes.
In the study of Klein-Jöbstl et al. [49] on the microbiota of newborn calves and their mothers, Ruminococcaceae was the most abundant type in cow fecal and vaginal samples. In the same study, Enhydrobacter was the most dominant in the colostrum on the first day postpartum; this bacterium appeared in our top 20 isolated bacteria, albeit in low numbers. Ruminococcaceae UCG-005 and Ruminococcaceae UCG-010, ranked in the top 10 isolated genera in our study, were also present in the samples of healthy skin in the studies of Bay et al. [50]. Coryneform bacteria Corynebacterium and Cutibacterium are distributed in the environment in soil and water [51]. These bacteria, as well as Staphylococci, are commensals and colonizers of the skin and mucous membranes in animals [52]. Dyella, Helicococcus, Capnocytophaga and Kingella are commensal bacteria isolated from the human respiratory tract [53] as well as the oral cavity of humans and animals [54] and are part of the skin flora [55]. Although they are considered to be commensals, all of them were isolated from a patient with severe clinical symptoms; this is the first report of their occurrence in bull semen. Their influence on sperm quality parameters is unknown.
Histophilosis is a common disease in North American cattle in the form of septicemia with a high risk of infection and sudden death in calves. A pathogenic form of Histophila somni was isolated from the prepuce of a healthy bull and from the vagina of cows with a clinical manifestation of granular vulvovaginitis and abortion [56]. Bovine genital campylobacteriosis caused by Campylobacter fetus veneralis or Campylobacter fetus fetus is a venereal disease of cattle characterized by infertility, mucopurulent endometritis, early embryonic death and occasionally abortion in systemically healthy cows. As well as venereal transmission, Campylobacter fetus fetus can be transmitted by AI in contaminated semen as well as by contaminated instruments. Infections in young bulls can be transient in contrast to older animals with established chronic infections, which may be due to differences in the preputial and penile epithelial surfaces of the lumen and within the crypts in the older animals and the microaerophilic environment that deeper crypts may provide. Campylobacter is one of the most demanding bacteria to culture due to its requirement for microaerophilic or anaerobic conditions as well as the need to be cultured immediately after sampling [57]. Therefore, it may be missed when culturing under conventional microbiological conditions. Cagnoli et al. [11] described a significant negative influence of both Campylobacter fetus veneralis and Campylobacter fetus fetus bacteria species on bull semen quality parameters.
Metagenomic analyses, especially 16S rDNA sequencing, allows the identification of bacteria with unusual phenotypic profiles, rare bacteria, slow growing bacteria and bacteria that cannot be cultured. Furthermore, 16S sequencing can facilitate the definitions of the etiologies of infectious diseases as well as aiding clinicians to choose the most effective antibiotics and determining the duration of the treatment. However, the interpretation of the results can be challenging even for clinical microbiologists [40].
Farahani et al. [58] conducted a systematic review study and meta-analysis of bacteria identified from fertile and infertile men and their influence on sperm quality and fertility parameters. Major differences in the bacterial presence of fertile and infertile men were identified with different sperm quality parameters as well as the negative and positive effects of the individual bacteria on these parameters. Apparent positive effects of Lactobacillus spp. on the sperm morphology in addition to a protective effect against Pseudomonas and opportunistic pathogens were highlighted. Boud et al. [9] reported that Pasteurella spp. abundance was increased in sperm samples with poor motility. Although the latter study showed that the bacterial content might not have an influence on the fertility of men, specific bacterial genera had an impact on sperm morphology and motility. There are few studies on the influence of the bacterial community in semen on sperm quality and fertility outcomes in veterinary medicine. There are a few studies of bacterial influence on sperm quality parameters but usually fertility data are lacking [59]. Cryopreservation does not necessarily reduce the bacterial count. In the study of Reda et al. [60], the bacterial content of cryopreserved semen was evaluated. Their study showed a negative effect of an increased bacterial content on sperm motility, viability and morphology with differences between ejaculates although no differences between the bulls were detected. When boar semen was stored for five days in the presence of an antibiotic, there was an increase in the number of certain bacteria, which was associated with a decrease in sperm motility [61].
In this study, we were interested in the potential associations between the bacteria in the male reproductive tract and the overall fertility of the semen from these bulls. We were not studying the effects of these bacteria on the sperm quality during subsequent storage, which is a different topic. Our results showed that two genera, W5053 and Lawsonella, were enriched in the semen samples from the low fertility bull group. These genera were also present among the 20 most abundant bacteria. Bell at al. [62] described Lawsonella spp. as Gram-positive, partially acid-fast, non-spore-forming, anaerobic, catalase-positive and pleomorphic bacteria. Three strains were isolated from human abscesses, which were determined to represent a novel genus (Lawsonella clevelandensis gen. nov., sp. nov.). Genus W5053 is also a novel bacteria; more comprehensive information is lacking. However, a higher abundance of both genera were seen in HIV-infected patients without chronic respiratory diseases [63]. The presence of these two bacterial genera in the semen samples of the bulls with a lower fertility potential is of interest for future research with regard to the origin and potential microbial influence on the sperm quality.
The diversity, number and interaction between the bacteria found in this study put 16S rDNA sequencing high on the list of the methods of choice for the diagnostics of bacterial contamination based on its objectivity and reliability. As most of the bacteria present in bull semen samples originate from the environment or from the mucosa of animals and humans, there is a need for a more effective management of the critical control points during semen collection. However, as 16S sequencing provides information about the presence of bacterial DNA in samples and not specifically about bacterial viability, it can only indicate the likely presence of the bacteria rather than the actual cause of a fertility issue. A possible interaction between the bacteria found in bull semen with a low fertility potential and host bacteria complex interactions would be of interest in future research.
Braga et al. [64] reported that the bacteria of different genera have an influence on microbial community modulation. This microbial coexistence occurs via chemical mediators among bacteria and also between microbes and hosts [65]. Such an interaction may cause alterations in the host physiology [64].
Deines et al. [66] also reported that a host-bacteria interaction, i.e., a host-environment and a bacteria–bacteria interaction, influences the coexistence of microbial species. This study showed that the competitive effect of Curvibacter depends on direct contact and indicated that rare microbial community members might be relevant for achieving a native community composition and carrying capacity. Although the genus Curvibacter was first mentioned by Ding and Yakota [67], who described three species isolated from well water as the source of origin, there is no previous documentation of its presence in bull semen or other sources. That Curvibacter was negatively correlated with other bacterial genera in most cases in this study indicated that this bacterium could be the focus for further research on the influence of the semen microbiota on the fertility of healthy bulls.
The second most negatively correlated bacteria in this study was Rikenellaceae RC9-gut-group. It belongs to the Rikenellacea family that are recently identified bacteria described as being challenging to culture [68]. This bacterial genus was previously identified in the digestive tract and fecal samples of different animals but not in other types of samples including bull semen. An increased abundance of this bacteria was found in an inflamed human digestive tract but there was no direct indication of their association with disease [69]. The study of Bálingt A et al. [70] showed that Rikenellaceae RC9-gut-group with other bacteria increased the sensitivity of the gut to inflammation. Based on the fact that these organisms can be challenging to culture, our current knowledge about these bacteria is based on the information gained from large scale sequencing studies.
The results of the present study indicated that differences in the bacterial microbiota of healthy bulls occur and might be associated with the fertility potential of the bull. Most of the identified bacteria were environmental in origin, indicating that a focus on how bulls are housed and how the semen is processed could help to reduce the bacterial abundance in commercial semen doses. The processing of bull semen should always be performed with a high level of hygiene and microbiological control.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/microorganisms9122431/s1: Table S1: CASA results for the sperm samples from 18 bulls.
Author Contributions
Conceptualization, A.C. and J.M.M.; methodology, A.C., A.N., Y.G., T.H., P.P. and J.M.M.; investigation, A.C., A.N. and Y.G.; data curation, A.C. and A.N.; writing—original draft preparation, A.C.; writing—review and editing, A.C., A.N., Y.G., T.H., P.P. and J.M.M.; resources, J.M.M. and P.P.; supervision, J.M.M.; project administration, J.M.M.; funding acquisition, J.M.M. and A.C. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by FORMAS, Stockholm (project number 2017-00957, awarded to JMM), KSLA (grant number GFS2020-0058).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The sequencing data generated in this study were deposited in the European Nucleotide Archive (ENA) under accession number PRJEB47651.
Acknowledgments
We thank the personnel at the Kehta cattle breeding station, Animal Breeders’ Association of Estonia, for providing the bull semen samples. Sequencing was performed by the SNP&SEQ Technology Platform in Uppsala. The facility is part of the National Genomics Infrastructure (NGI) Sweden and Science for Life Laboratory. The SNP&SEQ Platform is also supported by the Swedish Research Council and the Knut and Alice Wallenberg Foundation. We thank SLU Bioinformatics Infrastructure (SLUBI) for the management and processing of the sequencing data. Support from NBIS (National Bioinformatics Infrastructure Sweden) is gratefully acknowledged. This project was supported by the UMBLA platform at SLU.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses or interpretation of data; in the writing of the manuscript or in the decision to publish the results.
References
- Foote, H.R.; Siddhartha, L.S.; Parks, E.J. Artificial insemination. In Reference Module in Food Science; Cornell University: Ithaca, NY, USA; Elsevier: Amsterdam, The Netherlands, 2020. [Google Scholar]
- Al-Kass, Z.; Eriksson, E.; Bagge, E.; Wallgren, M.; Morrell, J.M. Microbiota of semen from stallions in Sweden identified by MALDI-TOF. Vet. Anim. Sci. 2020, 10, 100143. [Google Scholar] [CrossRef]
- Thibier, M.; Guerin, B. Hygienic aspects of storage and use of semen for artificial insemination. Anim. Reprod. Sci. 2000, 62, 233–251. [Google Scholar] [CrossRef]
- Bielanski, A. Disinfection procedures for controlling microorganisms in the semen and embryos of humans and farm animals. Theriogenology 2007, 68, 1–22. [Google Scholar] [CrossRef] [PubMed]
- Sannat, C.; Nair, A.; Sahu, S.B.; Sahasrabudhe, S.A.; Kumar, A.; Gupta, A.K.; Shende, R.K. Critical sources of bacterial contamination and adoption of standard sanitary protocol during semen collection and processing in Semen Station. Vet. World 2015, 8, 631–635. [Google Scholar] [CrossRef] [PubMed]
- Council Directive 88/407/EEC of 18 June 1988 Laying down the Animal Health Requirements Applicable to Intra-Community Trade in and Imports of Deep-Frozen Semen of Domestic Animals of the Bovine Species Annex C2. Available online: https://www.eumonitor.eu/9353000/1/j9vvik7m1c3gyxp/vhckkm8vsrz6 (accessed on 23 November 2021).
- Zampieri, D.; Santos, V.G.; Braga, P.A.C.; Ferreira, R.C.; Ballottin, D.; Tasic, L.; Basso, A.C.; Sanches, B.V.; Pontes, J.H.F.; Pereira da Silva, B.; et al. Microorganisms in cryopreserved semen and culture media used in the in vitro production (IVP) of bovine embryos identified by matrix-assistedlaser desorption ionization mass spectrometry (MALDI-MS). Theriogenology 2013, 80, 337–345. [Google Scholar] [CrossRef]
- Catry, B.; Van Duijkeren, E.; Pomba, M.C.; Greko, C.; Moreno, M.A.; Pyörälä, S.; Ruzauskas, M.; Sanders, P.; Threlfall, E.J.; Ungemach, F.; et al. Reflection paper on MRSA in food-producing and companion animals: Epidemiology and control options for human and animal health. Epidemiol. Infect. 2010, 138, 626–644. [Google Scholar] [CrossRef] [PubMed]
- Baud, D.; Pattaroni, C.; Vulliemoz, N.; Castella, V.; Marsland, J.B.; Stojanov, M. Sperm microbiota and its impact on semen parameters. Front. Microbial. 2019, 10, 234. [Google Scholar] [CrossRef]
- Del Porto, G.B.; Derrick, F.C.; Bannister, E.R. Bacterial effect on sperm motility. Urology 1975, 5, 638–639. [Google Scholar] [CrossRef]
- Cagnoli, C.I.; Chiapparrone, M.L.; Cacciato, C.S.; Rodríguez, M.G.; Aller, J.F.; Catena, M.D.C. Effects of Campylobacter fetus on bull sperm quality. Microb. Pathog. 2020, 149, 104486. [Google Scholar] [CrossRef] [PubMed]
- González-Marín, C.; Roy, R.; López-Fernández, C.; Diez, B.M.; Carabaño, M.J.; Fernández, J.L.; Kjelland, M.E.; Moreno, J.F.; Gosálvez, J. Bacteria in bovine semen can increase sperm DNA fragmentation rates: A kinetic experimental approach. Anim. Reprod. Sci. 2011, 123, 139–148. [Google Scholar] [CrossRef] [PubMed]
- Marchiani, S.; Baccani, I.; Tamburrino, L.; Mattiuz, G.; Nicolò, S.; Bonaiuto, C.; Panico, C.; Vignozzi, L.; Antonelli, A.; Rossolini, G.; et al. Effects of common gram-negative pathogens causing male genitourinary-tract infections on human sperm functions. Sci. Rep. 2021, 11, 19177. [Google Scholar] [CrossRef] [PubMed]
- Oghbaei, H.; Rastgar Rezaei, Y.; Nikanfar, S.; Zarezadeh, R.; Sadegi, M.; Latifi, Z.; Nouri, M.; Fattahi, A.; Ahmadi, Y.; Bleisinger, N. Effects of bacteria on male fertility: Spermatogenesis and sperm function. Life Sci. 2020, 256, 117891. [Google Scholar] [CrossRef] [PubMed]
- Eini, F.; Kutenaei, M.A.; Zareei, F.; Dastjerdi, Z.S.; Shirzeyli, M.H.; Salehi, E. Effect of bacterial infection on sperm quality and DNA fragmentation in subfertile men with Leukocytospermia. BMC Mol. Cell Biol. 2021, 22, 42. [Google Scholar] [CrossRef]
- Givens, M.D.; Marley, M.S.D. Pathogens that cause infertility of bulls or transmission via semen. Theriogenology 2008, 70, 504–507. [Google Scholar] [CrossRef] [PubMed]
- Appiah, M.O.; Wang, J.; Lu, W. Microflora in the reproductive tract of cattle: A Review. Agriculture 2020, 10, 232. [Google Scholar] [CrossRef]
- Sheldon, I.M.; Owens, S.E. Postpartum uterine infection and endometritis in dairy cattle. Anim. Reprod. 2018, 14, 622–629. [Google Scholar] [CrossRef]
- Fourichon, C.; Seegers, H.; Malher, X. Effect of disease on reproduction in the dairy cow: A metaanalysis. Theriogenology 2000, 53, 1729–1759. [Google Scholar] [CrossRef]
- Borsberry, S.; Dobson, H. Periparturient diseases and their effect on reproductive performance in five dairy herds. Vet. Rec. 1989, 124, 217–219. [Google Scholar] [CrossRef]
- LeBlanc, S.J.; Duffield, T.F.; Leslie, K.E.; Bateman, K.G.; Keefe, G.P.; Walton, J.S.; Johnson, W.H. Defining and diagnosing postpartum clinical endometritis and its impact on reproductive performance in dairy cows. J. Dairy Sci. 2002, 85, 2223–2236. [Google Scholar] [CrossRef]
- Marey, M.A.; Yousef, M.S.; Kowsar, R.; Hambruch, N.; Shimizu, T.; Pfarrer, C.; Miyamoto, A. Local immune system in oviduct physiology and pathophysiology: Attack or tolerance? Domest. Anim. Endocrinol. 2016, 56, 204–211. [Google Scholar] [CrossRef]
- Karsch, F.J.; Battaglia, D.F.; Breen, K.M.; Debus, N.; Harris, T.G. Mechanisms for ovarian cycle disruption by immune/inflammatory stress. Stress 2002, 5, 101–112. [Google Scholar] [CrossRef]
- Sheldon, I.M.; Noakes, D.E.; Rycroft, A.N.; Pfeiffer, D.U.; Dobson, H. Influence of uterine bacterial contamination after parturition on ovarian dominant follicle selection and follicle growth and function in cattle. Reproduction 2002, 123, 837–845. [Google Scholar] [CrossRef] [PubMed]
- Bromfield, J.J.; Sheldon, I.M. Lipopolysaccharide initiates inflammation in bovine granulosa cells via the TLR4 pathway and perturbs oocyte meiotic progression in vitro. Endocrinology 2011, 152, 5029–5040. [Google Scholar] [CrossRef]
- Al-Kass, Z.; Guo, Y.; Vinnere Pettersson, O.; Niazi, A.; Morrell, J.M. Metagenomic analysis of bacteria in stallion semen. Anim. Reprod. Sci. 2020, 221, 106568. [Google Scholar] [CrossRef] [PubMed]
- Althouse, G.C.; Lu, K.G. Bacteriospermia in extended porcine semen. Theriogenology 2005, 63, 573–584. [Google Scholar] [CrossRef] [PubMed]
- Yániz, J.L.; Marco-Aguado, M.A.; Mateos, J.A.; Santolaria, P. Bacterial contamination of ram semen, antibiotic sensitivities, and effects on sperm quality during storage at 15°C. Anim. Reprod. Sci. 2010, 122, 142–1499. [Google Scholar] [CrossRef] [PubMed]
- Sanocka-Maciejewska, D.; Ciupińska, M.; Kurpisz, M. Bacterial infection and semen quality. J. Reprod. Immunol. 2005, 67, 51–56. [Google Scholar] [CrossRef]
- Enwuru, C.A.; Iwalokun, B.; Enwuru, V.N.; Ezechi, O.; Oluwadun, A. The effect of presence of facultative bacteria species on semen and sperm quality of men seeking fertility care. Afr. J. Urol. 2016, 22, 213–222. [Google Scholar] [CrossRef][Green Version]
- Prince, P.W.; Almquist, J.O.; Reid, J.J. Bacteriological studies of bovine semen. II. The incidence of specific types of bacteria and the relation to fertility. J. Dairy Sci. 1949, 32, 849–855. [Google Scholar] [CrossRef]
- Riigi Teataja. Loomakaitseseadus—Riigi Teataja. Available online: https://www.riigiteataja.ee/akt/LoKS (accessed on 24 September 2021).
- Miglior, F.; Pizzi, F.; Guaita, N. Effect of environmental factors on non return rate in Italian Holstein-Friesians. Interbull. Bull. 1997, 27, 106–108. [Google Scholar]
- Takahashi, S.; Tomita, J.; Nishioka, K.; Hisada, T. Development of a prokaryotic universal primer for simultaneous analysis of bacteria and archaea using next-generation sequencing. PLoS ONE 2014, 9, e105592. [Google Scholar] [CrossRef] [PubMed]
- Andrews, S. A quality control tool for high throughput sequence data. FASTQC 2010, 532, 1. [Google Scholar]
- Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 2011, 17, 10–12. [Google Scholar] [CrossRef]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef] [PubMed]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2013, 41, D590–D596. [Google Scholar] [CrossRef]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef] [PubMed]
- Woo, P.C.; Lau, S.K.; Teng, J.L.; Tse, H.; Yuen, K.Y. Then and now: Use of 16S rDNA gene sequencing for bacterial identification and discovery of novel bacteria in clinical microbiology laboratories. Clin. Microbiol. Infect. 2008, 14, 908–934. [Google Scholar] [CrossRef]
- Gunsalus, I.C.; Salisbury, G.W.; Willett, E.L. The bacteriology of bull semen. J. Dairy Sci. 1941, 24, 911–919. [Google Scholar] [CrossRef]
- Bush, L.J.; Ludwick, T.M.; Ferguson, L.C.; Ely, F. The effect of bacteria on the fertility of bovine semen. J. Dairy Sci. 1950, 33, 633–638. [Google Scholar] [CrossRef]
- Mitra, J.; Chowdhury, S.; Panda, S.; Chakraborty, M.; Singha, A. Microbiological evaluation of bovine frozen semen samples in West Bengal, India. Explor. Anim. Med. Res. 2016, 6, 185–191. [Google Scholar]
- Balqis, U.; Hambal, M.; Admi, M.; Safika, S.; Meutia, N.; Sugit, S.; Dasrul; Abdullah, M.A.N.; Ferasyi, T.R.; Lubis, T.M.; et al. Escherichia fergusonii identified in preputial swabs from healthy Aceh cattle by phylogenetic 16S rRNA analysis. Malays. J. Microbiol. 2018, 14, 229–235. [Google Scholar] [CrossRef]
- Hambal, M.; Admi, M.; Safika, S.; Sari, W.E.; Ferasyi, T.R.; Dasrul, D.; Balqis, U.; Darmawi, D. Identification of Staphylococcus species isolated from preputium of Aceh cattle based on 16S rRNA gene sequences analysis. Vet. World 2019, 12, 1540–1545. [Google Scholar] [CrossRef] [PubMed]
- Haapala, V.; Pohjanvirta, T.; Vähänikkilä, N.; Halkilahti, J.; Simonen, H.; Pelkonen, S.; Soveri, T.; Simojoki, H.; Autio, T. Semen as a source of Mycoplasma bovis mastitis in dairy herds. Vet. Microbiol. 2018, 216, 60–66. [Google Scholar] [CrossRef]
- Wickware, C.L.; Johnson, T.A.; Koziol, J.H. Composition and diversity of the preputial microbiota in healthy bulls. Theriogenology 2020, 145, 231–237. [Google Scholar] [CrossRef]
- Lima, S.F.; Teixeira, A.G.V.; Higgins, C.H.; Lima, F.S.; Bicalho, R.C. The upper respiratory tract microbiome and its potential role in bovine respiratory disease and otitis media. Sci. Rep. 2016, 6, 29050. [Google Scholar] [CrossRef] [PubMed]
- Klein-Jöbstl, D.; Quijada, N.M.; Dzieciol, M.; Feldbacher, B.; Wagner, M.; Drillich, M.; Schmitz-Esser, S.; Mann, E. Microbiota of newborn calves and their mothers reveals possible transfer routes for newborn calves’ gastrointestinal microbiota. PLoS ONE 2019, 14, e0220554. [Google Scholar] [CrossRef] [PubMed]
- Bay, V.; Griffiths, B.; Carter, S.; Evans, N.J.; Lenzi, L.; Bicalho, R.C.; Oikonomou, G. 16S rRNA amplicon sequencing reveals a polymicrobial nature of complicated claw horn disruption lesions and interdigital phlegmon in dairy cattle. Sci. Rep. 2018, 8, 15529. [Google Scholar] [CrossRef]
- Kim, R.; Reboli, C.A. Mandell, Douglas, and Bennett’s Principles and Practice of Infectious Diseases, 8th ed.; ESEVIRA Saunders: Philadelphia, PA, USA, 2015; Volume 2, pp. 2373–2382. [Google Scholar]
- Foster, A.P. Staphylococcal skin disease in livestock. Vet. Dermatol. 2012, 23, 342–351. [Google Scholar] [CrossRef]
- Duus, L.M.; Høiby, N.; Wang, M.; Schiøtz, O.; Nørskov-Lauritsen, N. Bacteria of the genus Dyella can chronically colonise the airways of patients with cystic fibrosis and elicit a pronounced antibody response. Int. J. Med. Microbiol. 2013, 303, 267–269. [Google Scholar] [CrossRef]
- Piau, C.; Arvieux, C.; Bonnaure-Mallet, M.; Jolivet-Gougeon, A. Capnocytophaga spp. involvement in bone infections: A review. Int. J. Antimicrob. Agents 2013, 41, 509–515. [Google Scholar] [CrossRef]
- Vergne, A.; Guérin, F.; Lienhard, R.; Le Coustumier, A.; Daurel, C.; Isnard, C.; Marty, N.; Poyart, C.; Cattoir, V. Identification and Clinical Significance of Helcococcus kunzii in Human Samples. J. Clin. Microbiol. 2015, 53, 2703–2705. [Google Scholar] [CrossRef] [PubMed]
- Headley, S.A.; Voltarelli, D.; de Oliveira, V.H.; Bronkhorst, D.E.; Alfieri, A.F.; Filho, L.C.; Okano, W.; Alfieri, A.A. Association of Histophilus somni with spontaneous abortions in dairy cattle herds from Brazil. Trop. Anim. Health Prod. 2015, 47, 403–413. [Google Scholar] [CrossRef] [PubMed]
- Michi, A.N.; Favetto, P.H.; Kastelic, J.; Cobo, E.R. A review of sexually transmitted bovine trichomoniasis and campylobacteriosis affecting cattle reproductive health. Theriogenology 2016, 85, 781–791. [Google Scholar] [CrossRef] [PubMed]
- Farahani, L.; Tharakan, T.; Yap, T.; Ramsay, W.J.; Jayasena, N.C.; Minhas, S. The semen microbiome and its impact on sperm function and male fertility: A systematic review and meta-analysis. Andrology 2021, 9, 115–144. [Google Scholar] [CrossRef]
- Varela, E.; Rey, J.; Plaza, E.; Muñoz de Propios, P.; Ortiz-Rodríguez, J.M.; Álvarez, M.; Anel-López, L.; Anel, L.; De Paz, P.; Gil, M.C.; et al. How does the microbial load affect the quality of equine cool-stored semen? Theriogenology 2018, 114, 212–220. [Google Scholar] [CrossRef]
- Reda, A.A.; Almaw, G.; Abreha, S.; Tadeg, W.; Tadesse, B. Bacteriospermia and sperm quality of cryopreserved bull semen used in artificial insemination of cows in South Wollo zone, Ethiopia. Vet. Med. Int. 2020, 2020, 2098315. [Google Scholar] [CrossRef]
- Gączarzewicz, D.; Udała, J.; Piasecka, M.; Błaszczyk, B.; Stankiewicz, T. Bacterial Contamination of boar semen and its relationship to sperm quality preserved in commercial extender containing gentamicin sulfate. Pol. J. Vet. Sci. 2016, 19, 451–459. [Google Scholar] [CrossRef]
- Bell, M.E.; Bernard, K.A.; Harrington, S.M.; Patel, N.B.; Tucker, T.A.; Metcalfe, M.G.; McQuiston, J.R. Lawsonella clevelandensis gen. nov., sp. nov., a new member of the suborder Corynebacterineae isolated from human abscesses. Int. J. Sys.t Evol. Microbiol. 2016, 66, 2929–2935. [Google Scholar] [CrossRef]
- Flygel, T.T.; Sovershaeva, E.; Claassen-Weitz, S.; Hjerde, E.; Mwaikono, K.S.; Odland, J.Ø.; Ferrand, R.A.; Mchugh, G.; Gutteberg, T.J.; Nicol, M.P.; et al. Composition of gut microbiota of children and adolescents with perinatal human immunodeficiency virus infection taking antiretroviral therapy in Zimbabwe. J. Infect. Dis. 2020, 221, 483–492. [Google Scholar]
- Braga, R.M.; Dourado, M.N.; Araujo, W.L. Microbial interactions: Ecology in a molecular perspective. Braz. J. Microbiol. 2016, 47, 86–98. [Google Scholar] [CrossRef]
- Niehaus, L.; Boland, I.; Liuz, M.; Chen, K.; Fu, D.; Henckel, C.; Chaung, K.; Miranda, S.E.; Dyckman, S.; Crum, M.; et al. Microbial coexistence through chemical-mediated interactions. Nat. Commun. 2019, 10, 2052. [Google Scholar] [CrossRef] [PubMed]
- Deines, P.; Hammerschmidt, K.; Bosch, T. Microbial species coexistence depends on the host environment. mBio 2020, 11, e00807-20. [Google Scholar] [CrossRef] [PubMed]
- Ding, L.; Yokota, A. Proposals of Curvibacter gracilis gen. nov., sp. nov. and Herbaspirillum putei sp. nov. for bacterial strains isolated from well water and reclassification of [Pseudomonas] huttiensis, [Pseudomonas] lanceolata, [Aquaspirillum] delicatum and [Aquaspirillum] autotrophicum as Herbaspirillum huttiense comb. nov., Curvibacter lanceolatus comb. nov., Curvibacter delicatus comb. nov. and Herbaspirillum autotrophicum comb. nov. Int. J. Syst. Evol. Microbiol. 2004, 54, 2223–2230. [Google Scholar] [PubMed]
- Graf, J. The Family Rikenellaceae. In The Prokaryotes: Other Major Lineages of Bacteria and The Archaea; Rosenberg, E., DeLong, E.F., Lory, S., Stackebrandt, E., Thompson, F., Eds.; Springer: Berlin/Heidelberg, Germany, 2014; Volume 68, pp. 857–859. [Google Scholar]
- Cai, B.; Pan, J.; Chen, H.; Chen, X.; Ye, Z.; Yuan, H.; Sun, H.; Wan, P. Oyster polysaccharides ameliorate intestinal mucositis and improve metabolism in 5-fluorouracil-treated S180 tumour-bearing mice. Carbohydr. Polym. 2021, 256, 117545. [Google Scholar] [CrossRef] [PubMed]
- Bálint, A.; Farkas, K.; Méhi, O.; Kintses, B.; Vásárhelyi, B.M.; Ari, E.; Pál, C.; Madácsy, T.; Maléth, J.; Szántó, K.J.; et al. Functional anatomical changes in ulcerative colitis patients determine their gut microbiota composition and consequently the possible treatment outcome. Pharmaceuticals 2020, 13, 346. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).