Abstract
This study aimed to isolate lactic acid bacteria (LAB) from wheat rhizosphere, to characterize their in vitro plant growth promoting activities and to differentiate plant-associated LAB from those associated with foods or human disease through comparative genomic analysis. Lactococcus lactis subsp. lactis and Enterococcus faecium were isolated using de Man-Rogosa-Sharpe (MRS) and Glucose Yeast Peptone (GYP) as enrichment culture media. Comparative genomic analyses showed that plant-associated LAB strains were enriched in genes coding for bacteriocin production when compared to strains from other ecosystems. Isolates of L. lactis and E. faecium did not produce physiologically relevant concentrations of the phyto-hormone indolacetic acid. All isolates solubilized high amount of phosphate and 12 of 16 strains solubilized potassium. E. faecium LB5, L. lactis LB6, LB7, and LB9 inhibited the plant pathogenic Fusarium graminearum to the same extent as two strains of Bacillus sp. However, the antifungal activity of the abovementioned LAB strains depended on the medium of cultivation and a low pH while antifungal activity of Bacillus spp. was independent of the growth medium and likely relates to antifungal lipopeptides. This study showed the potential of rhizospheric LAB for future application as biofertilizers in agriculture.
1. Introduction
Agriculture is an important economic sector in many countries and, according to FAO, 37% of the global land area is dedicated to agriculture [1]. Conventional agriculture produces high yields and seems to be the most appropriate solution to the estimated increase of the world population [2]. However, it relies on the use of chemical fertilizers, which are responsible for water and soil pollution, soil degradation and biodiversity loss [3]. Conventional agriculture also employs pesticides to control phytopathogens that cause the loss of an estimated 9–16% of important cereal crops such as wheat, rice, and maize [4].
Organic agriculture excludes use of synthetic chemical fertilizers, pesticides, and herbicides [5] and improves soil fertility through the incorporation of legumes and compost, determining an increase of specie biodiversity in the whole ecosystem [6]. Organic agriculture produces 13–34 % lower yields when compared to conventional cropping systems [7]. Eco-friendly and efficient solutions for pest control in organic agriculture may complement or substitute the use of pesticides in conventional agriculture.
The plant rhizosphere is rich in nutrients released by root exudates that create a suitable niche for proliferation of microorganisms. Microorganisms reach high cell densities and the rhizosphere is a complex ecosystem [8]. Organisms in the rhizosphere include bacteria, fungi, nematodes, algae, protozoa, arthropods, and archaea but bacteria are the most abundant (108–109 cells·g−1 of rhizospheric soil) [9]. Among bacteria, Gram-negative bacteria predominate [10]. Not all microorganisms populating the rhizosphere, termed “rhizobacteria”, have been characterized because it is not easy to culture them [11]. Many studies suggest that only 1–5% of total rhizobacteria are cultivable and abiotic stress could halve that number [8,9]. Some rhizobacteria improve plant growth and productivity through various mechanisms of action. Correspondingly, rhizobacteria are commercially used as plant growth promoting rhizobacteria (PGPR) or biofertilizers [12]. Depending on the basis of the mechanisms of action, PGPR are considered as biostimulants when they improve nitrogen fixation, phosphate and potassium solubilization, or produce phytohormones, or biocontrol agents when they produce antimicrobial compounds with activity against phytopathogens [13,14,15]. Introduction of PGPR in conventional agriculture is a strategy to reduce its environmental impact without decreasing plant productivity. The predominant genera used as PGPR are Bacillus and Pseudomonas [16]. Metagenomics analyses of plant microbiomes, including rhizobacteria, identified additional organisms including lactic acid bacteria (LAB); which are, however, hardly detectable because of their low abundance in the plant-soil ecosystem [17,18,19].
LAB are microaerophilic Gram-positive bacteria which play an important role in food fermentations but also include commensals of vertebrates and insects as well as pathogens [20]. Food fermenting LAB, particularly species grouped in the Lactobacillaceae, have a safe tradition of use in food [21], but this group also includes pathogens or opportunistic pathogens. For instance, Enterococcus faecium has evolved as vertebrate commensals and occur in food fermentations but specific lineages of E. faecium also are a leading cause of hospital-acquired infections with multi-drug resistant bacteria [22,23]. Strains of E. faecium which lack the virulence genes typical of hospital isolates have been approved as feed additives [24]. LAB also include plant-associated organisms, although their abundance and diversity in the rhizosphere is lower when compared to other bacteria [20]. Although they do not have a reputation of having as useful properties as bacilli and Pseudomonas species, some species of LAB were assessed with regards to their plant growth promoting activity and were reported to increase yield, improve seed germination [25], and the chlorophyll [26] and amino acid content [27] of plants. Some reports also suggested a role of LAB as biocontrol agents against soil-borne phytopathogens [25,28]. Nevertheless, the potential of LAB as PGPR is still under-explored. Therefore, the aims of this study were: (i) to isolate LAB from the rhizospheric soil collected from two cultivars of durum wheat during different growth stages; (ii) to determine whether any of the useful properties of the established PGPB are also found in LAB, by characterizing their plant growth promotion and protection activities in vitro; (iii) to evaluate whether the plant associated LAB differ from strains of the same species that are associated with foods and/or with humans.
2. Materials and Methods
2.1. Samples Collection
Two varieties of durum wheat (Triticum turgidum subsp. durum), Senatore Cappelli and MARAKAS, were grown with conventional farming systems at two sites in Southern Italy, Turi (40°91′ N, 17°04′ E) and in Gravina (40°49′ N, 16°18′ E). Turi soil (pH 7.95, electric conductivity (EC) 0.57 dS·m−1, soil organic carbon (SOC) 1.69%, soil organic matter (SOM) 2.91%) and Gravina in Puglia soil (pH 7.63, EC 0.61 dS·m−1, SOC 1.26%, SOM 2.17%) were characterized by a clay loam texture. Wheat rhizospheric soil samples were collected during four different growth stages (tillering, elongation, earing, and physiological maturity) between January and June, months during which the average monthly temperature ranges from 4 to 30 °C. For each field and growth stage, ten soil samples were randomly collected, for a total of 80 samples, using the following protocol: (i) eradicating manually to a depth of 0–20 cm and shaking plants to remove soil in excess; (ii) storage of plant’s roots individually in sterile bags and transport to laboratory at 4 °C; (iii) collection of soil attached to the roots using a sterile spatula; (iv) sieving (0.5 mm). Then, the soil samples were randomly pooled to form two sub-samples considered replicas, each one obtained from five plants.
2.2. Isolation of Lactic Acid Bacteria from Wheat Rhizospheric Soil
Lactic acid bacteria (LAB) were isolated from wheat rhizospheric soil upon enrichment, as described by Chen et al. [18]. One gram of rhizospheric wheat soil of each sub-sample was added to 5 mL of de Man, Rogosa, and Sharpe (MRS, pH 6.2) or Glucose Yeast Peptone (GYP, pH 7) broth, both supplemented with cycloheximide (0.1 g·L−1). MRS was purchased from Oxoid Ltd. (Basingstoke, UK), except when used for culturing bacteria before antifungal assay. GYP was prepared in laboratory and contained 20.0 g·L−1 glucose, 10.0 g·L−1 yeast extract, 10.0 g·L−1 peptone, 10.0 g·L−1 sodium acetate, 5 mL salt solution (40.0 g·L−1 MgSO4·7H2O, 1.6 g·L−1 MnSO4·4H2O, 2.0 g·L−1 FeSO4·7H2O, 2.0 g·L−1 NaCl) [18].
After 24 h of incubation at 30 °C, 1 mL of enrichment broth was diluted with Ringer solution 25% composed of 0.225 % NaCl w/v, 0.0105% KCl w/v, 0.0045% CaCl2 w/v, 0.005% NaHCO3 w/v, and 0.0034% citric acid w/v, and inoculated by pour-plate method on MRS and GYP agar plates. After incubation at 30 °C for 48 h, a number of colonies equal to the square root of the total number observed in plates coming from the highest dilution was randomly picked up. Gram-positive, catalase-negative, non-motile rod- and coccus-shaped LAB were cultivated in MRS or GYP at 30 °C for 24 h and re-streaked onto the same agar media. All isolates considered for further analyses were able to acidify the culture medium.
2.3. Molecular Biotyping and Identification of LAB Isolates
The isolates were typed by randomly amplified polymorphic DNA (RAPD)-PCR. Colony PCR was performed upon thermal shock of each colony and, subsequently, inserting the colony into the following reaction mix: 1× Reaction Buffer (EuroClone, Pero, MI, Italy), 2.5 mM MgCl2 (EuroClone), 0.2 mM each dNTP (EuroClone), 1 µM M13 primer (5′-GAGGGTGGCGGTTCT-3′) (Sigma-Aldrich, St. Louis, MO, USA) [29], 80 ng µL−1 bovine serum albumin (Sigma-Aldrich), 3 U of Taq DNA polymerase (EuroTaq, EuroClone). The amplification reactions were carried out using MyCycler™ thermal cycler (Bio-Rad Laboratories Inc., Hercules, CA, USA) and following the conditions described by Zapparoli et al. [30] with some modifications: 95 °C for 10 min, 35 cycles of 95 °C for 1 min, 45 °C for 30 s, 72 °C for 2 min, and a final extension of 72 °C for 5 min. The PCR products (3 μL) were separated by electrophoresis at 100 V for 60 min on a 1% (w/v) agarose gel, and the electrophoretic profile was detected by Bio-Rad Gel Doc 2000 System image detector and processed using the Image Lab software.
Identification of LAB was carried out upon partial sequencing of 16S rRNA gene (V3-V4 regions), using the F357 forward and R1401 reverse primers [31]. The reaction mix contained 1× Wonder Taq Reaction Buffer (EuroClone), 50 pmol of each primer (Eurofins Genomics, Ebersberg, Germany), 80 ng·µL−1 bovine serum albumin, 3 U of Taq DNA polymerase. The PCR conditions were: 95 °C for 10 min, 35 cycles of 95 °C for 30 s, 63 °C for 30 s, 72 °C for 2 min and a final extension of 72 °C for 7 min. PCR amplicons of 1044 bp were purified using Wizard SV Gel and PCR Clean-Up System kit (Promega) following the manufacturer’s instructions. After quantification of amplicons through the spectrophotometer NanoDrop ND-1000, PCR products were sequenced by Eurofins Genomic.
2.4. In Vitro Characterization of Lactic Acid Bacteria
2.4.1. Quantification of 3-Indolacetic Acid
The strains were incubated in MRS and GYP broth, enriched by 0.2% L-Tryptophan, for 72 h at 28 °C. After incubation, the bacterial suspension was centrifuged at 5500× g for 15 min and 1 mL of cell-free supernatant was added to 4 mL Salkoswki’s reagent prepared as described by Gordon and Weber [32]: 36% H2SO4 solution containing 6.2 mM FeCl3. The mix was left in the dark at room temperature for 20 min, then the absorbance at 535 nm was read using a spectrophotometer (Ultrospec 4000 UV–visible Spectrophotometer, Pharmacia Biotech). The calibration curve was constructed using pure 3-indolacetic acid (Sigma-Aldrich) as standard at different concentration in the range of 2–30 mg·L−1.
2.4.2. Phosphate Solubilization and Phosphorus Quantification
A screening on NBRIP (National Botanical Research Institute’s Phosphate) agar (1% w/v glucose; 0.5% w/v Ca3(PO4)2; 0.5% w/v MgCl2·6H2O; 0.025% w/v MgSO4·7H2O; 0.02% w/v KCl; 0.01% w/v (NH4)2SO4; 1.5% w/v agar, pH 7) was performed to assay the ability of the strains to solubilize phosphate [33]. 10 µL of bacterial suspension, grown for 24 h in GYP or MRS, were spread in triplicates onto NBRIP agar plates and incubated for 15 days at 30 °C. The diameter of a halo zone around the colony was measured to calculate the Phosphate Solubilization Index (PSI).
After screening on NBRIP agar, quantification of solubilized phosphorous was carried out, following the colorimetric method of Watanabe and Olsen [34]. 100 µL of cell-free supernatant, harvested after 7 days of incubation in NBRIP broth at 28 °C, were added to 4 mL of a solution obtained dissolving 1.056 g of ascorbic acid in 200 mL 5N H2SO4 solution also containing 6.2 mg·mL−1 of ammonium molybdate and 0.14 mg·mL−1 of antimony potassium tartrate. Double distilled water was added until 100 mL of final volume and after 10 min of incubation at room temperature, the absorbance was read at 650 nm. The calibration curve was constructed using ascorbic acid at different concentration in the range 0–0.6 µg·mL−1.
2.4.3. Potassium Solubilization
Aleksandrov agar medium was used to assay the strains’ ability to solubilize potassium. The medium contained 3.5 g·L−1 glucose, 0.5 g·L−1 MgSO4·7 H2O, 0.1 g·L−1 CaCO3, 0.0005 g·L−1 FeCl3, 2 g·L−1 Ca3PO4, 12 g·L−1 agar and 1.0 g·L−1 mica powder, pH 7 [35]. Ten µL of bacterial suspension were spotted in triplicate on Aleksandrov agar and, after 4 days of incubation at 28 °C, the halo zone around the colony was measured to calculate solubilization index of potassium (KSI).
After screening on Aleksandrov agar, solubilized potassium was quantified, according to Yaghoubi Khanghahi et al. [36]. In detail, 100 µL of each bacterial suspension cultured for 24 h were inoculated, in triplicate, in 10 mL of Aleksandrov broth. After incubation (14 days at 28 °C), bacterial suspensions were centrifuged (4032× g, 20 min, 4 °C) and the supernatant was 0.2 μm filtered. Then, the supernatants were diluted 1:5 in double distilled water and 10 µL of a 100 mg·L−1 Gallium solution as internal standard were added. 10 μL of the abovementioned solution were spread onto a clean siliconized quartz disk and left to dry on a hot plate (SD300, Stuart) to form a circular thin film ready for analysis [37]. Quantitative analysis of potassium was carried out by total-reflection X-ray fluorescence spectrometry (TXRF), using an S2Picofox TXRF Spectrometer (Bruker Nano GmbH, Berlin, Germany) and spectra were analyzed by SPECTRA 7 software (Bruker Nano GmbH, Berlin, Germany). The intensity of the fluorescence emitted by the samples was measured, processed by referring to the internal standard, and converted in concentration of solubilized potassium [37].
2.4.4. Antifungal Activity
Aspergillus niger FUA5001, Penicillium roqueforti FUA5004, and Fusarium graminearum G-1, belonging to the Culture Collection of the University of Alberta (Edmonton, Canada), were used to test antifungal activity of LAB. Bacillus velezensis FUA2155 and Bacillus amyloliquefaciens Fad 82 [38], routinely cultured in Luria–Bertani (LB) broth, were used as positive controls because of the widespread ability of plant-associated bacilli to produce diverse antifungal lipopeptides [39]. Fungi were grown on Potato Dextrose Agar (PDA) at 25 °C in the dark for 7 days and then the spores were collected in sterile physiological solution. The suspension was filtered, and the number of the spores was counted using the hemocytometer (Fein-Optik, Jena, Germany). The suspension was first diluted until 1 × 104 spore/mL and then further diluted 1:30. Before the assay, bacterial strains were cultured in broths lacking sodium acetate: either GYP or laboratory-made MRS (mMRS: 10 g·L−1 peptone, 8 g·L−1 ‘Lab-Lemco’ powder, 4 g·L−1 yeast extract, 20 g·L−1 glucose, 1 mL·L−1 sorbitan mono-oleate, 2 g·L−1 dipotassium hydrogen phosphate, 2 g·L−1 triammonium citrate, 0.2 g·L−1 magnesium sulphate heptahydrate, 0.05 g·L−1 manganese sulphate tetrahydrate). The screening was performed in triplicate using 96-well plates. Each well contained 100 µL of culture medium routinely used for the bacteria tested, but lacking sodium acetate, and 15 µL of bacterial suspension cultured for 24 h. The plates were incubated at 30 °C for two days until bacterial growth; after that, 15 µL of a spore suspension were added. The 96-well plates were incubated at 25 °C, and day by day the mold growth was monitored against three controls consisting of only culture media (acidified to pH 4.5 using lactic acid) and spore suspension [40]. Antifungal activity was expressed as mean of the mold-free days and represented as a heatmap using R software.
2.4.5. Antibacterial Activity
Pantoea agglomerans NCPPB:2519, Pectobacterium rhapontici NCPPB:1578, Pseudomonas syringae NCPPB:281, Pectobacterium carotovorum subsp. carotovorum NCPPB:312, belonging to the National Collection of Plant Pathogenic Bacteria (Fera Science Ltd., London, UK), and Pseudomonas fulva, belonging to the Culture Collection of the University of Alberta, were used as indicators to test antibacterial activity of LAB. P. fulva was cultured in tryptic soy broth (TSB) at 37 °C for 24 h, whereas the other bacteria were cultured in nutrient broth (NB) at 25 °C for 24 h under shaking.
The screening was carried out in triplicate using the minimum inhibitory concentration (MIC) protocol [41]. In 96-well plates, 100 µL of culture medium of each phytopathogenic bacterial strain were added to each well. Then, 100 µL of cell-free supernatant of LAB collected from 24 h cultures were added in the wells of one column and diluted by serial two-fold dilutions. The plates were then inoculated with 50 µL of a cell suspension of the indicator strains that was diluted to 1–5 × 106 cfu/mL; one column remained uninoculated and served as negative control; one column was inoculated but contained no culture supernatant and served as positive control. Growth was assessed by measuring the optical density at 600 nm after 24 h of incubation (Biolog Inc., Hayward, CA, USA). A second test was performed as described above, with the only change of using the cell-free supernatant of LAB after pH adjustment to 7 with 5 M NaOH. pH values of suspension contained in each well were measured using a HI1083P electrode (Hanna Instruments Italia, Ronchi di Villafranca Padovana, Italy).
The MIC, representing the lowest dilution of supernatant able to totally inhibit phytopathogenic bacteria, was expressed as the ratio of the supernatant volume to the total volume (%, v/v).
2.5. Comparative Genomic Analysis
Genomes of two strains of Lactococcus lactis and nine strains of Enterococcus faecium were sequenced by paired end sequencing on a HiSeq2500 platform with 50-fold coverage by service of the Génome Québec Innovation Centre (Montreal, QC, Canada), followed by quality control with the FastQC tool, assembly using SPAdes and MeDuSa, and annotation on the rapid annotations using subsystems technology (RAST) server as described [42]. Genomic metadata for E. faecium and L. lactis was downloaded using NCBImeta [43]. From this data, 102 E. faecium strains were selected. Clinical strains (38) were rarified by selecting one strain from each depositing source. Plant strains were selected by selecting all NCBI strains from plant, soil or fresh water sources (35). The nine strains isolated in this study were included in the plant group (44). All available strains isolated from healthy non-domestic animals were included in the animal group (29). L. lactis strains (77) were split into dairy and plant groups. The 47 dairy strains were selected from isolates from backslopped dairy cultures, whereas the plant group consisted of all available strains isolated from plants (28) and two of the strains isolated in this study.
Comparative genomics was carried out after annotating the genomes with Prokka [44] and creating gene presence/absence table and core genome alignments with Roary [45]. Analysis of the enriched for genes in the different groups was carried out using Scoary [46]. The phylogenetic trees were generated from the core genome alignments using FastTree 2.1 [47] and the trees were visualized using ITOL [48]. Enterococcus faecalis V583 (GCF_000407305.1) and Lactococcus garvieae ATCC 49156 (GCF_000269925.1) were used as outgroups.
2.6. Statistical Analysis
Statistical analysis was performed using SigmaPlot 14.0. All assays were conducted in triplicate biological repeats. One-way analysis of variance (ANOVA) with LSD post hoc analysis were performed with SigmaPlot to determine significant differences at a 5% probability level.
3. Results
3.1. Isolation and Identification of Lactic Acid Bacteria from Wheat Rhizospheric Soil
One hundred and fifty-one bacteria were isolated from durum wheat rhizospheric soil during tillering (10 isolates), elongation (55), earing (71), and physiological maturity (15). Seventy-two bacteria were isolated using GYP medium and the other 79 through MRS medium, two standard media for isolation of lactic acid bacteria. Seven isolates were catalase-positive and discarded, all other isolates were Gram-positive, catalase-negative, acidifying cocci. Upon RAPD-typing, 18 different electrophoretic profiles were observed. One representative strains of each RAPD-profile was characterized by partial sequencing of 16S rRNA genes. Nine strains belonged to Lactococcus lactis subsp. lactis and the remaining nine to Enterococcus faecium (Table 1).

Table 1.
Lactic acid bacteria isolated from wheat rhizopsheric soil collected from two different experimental sites during three different growth stages.
3.2. Indolacetic Acid Production
L. lactis subsp. lactis LB9 was the higher producer of 3-indolacetic acid (IAA) (4.6 ± 0.4 mg·L−1) compared to others LAB (Figure 1). Lower values were found for L. lactis subsp. lactis LB6, LB7 and E. faecium LB16, LB23, LB24, and LB25. Values close to zero were obtained for the other strains.
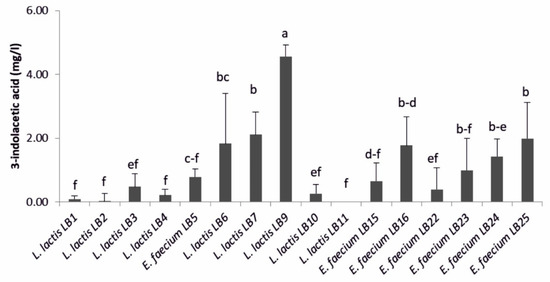
Figure 1.
3-indolacetic acid amount produced by lactic acid bacteria (n = 3). Means (±SD) followed by the similar letter(s) are not significantly different at the 5% probability level (LSD test).
3.3. Phosphate Solubilization and Phosphorus Quantification
All the LAB produced a phosphate solubilization halo, with PSI values ranging from 1.02 ± 0.03 to 1.87 ± 0.11 mm (Table 2). Most LAB were high phosphorous solubilizers, with values ranging from 200.0 ± 8.0 (L. lactis subsp. lactis LB7) to 388.2 ± 11.2 µg·mL−1 (E. faecium LB25). Only L. lactis subsp. lactis LB6 solubilized a lower amount of phosphorus (86.7 ± 16.0 µg·mL−1) (Figure 2).

Table 2.
Values of phosphate solubilization index measured for LAB strains isolated from wheat rhizospheric soil.
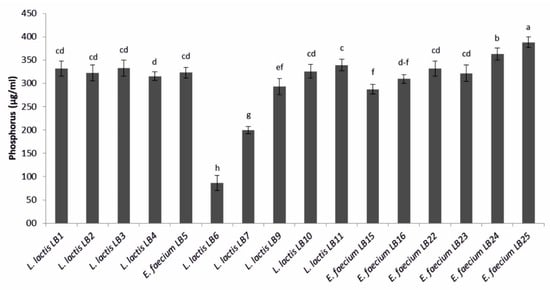
Figure 2.
Amount of phosphorus solubilized by lactic acid bacteria. Means (±SD) followed by the similar letter(s) are not significantly different at the 5% probability level (LSD test).
3.4. Potassium Solubilization
Twelve LAB solubilized potassium from mica powder. E. faecium LB15, LB16, LB23, LB24, and LB25 produced a halo zone whose diameter was greater than 2 mm (Table 3). For the other seven potassium solubilizing bacteria, potassium solubilization index (KSI) ranged from 1.7 ± 0.1 to 2.0 ± 0.3 mm.

Table 3.
Values of potassium solubilization index measured for LAB strains isolated from wheat rhizospheric soil and corresponding classification.
Among LAB showing a halo zone of potassium solubilization, strains of E. faecium were higher potassium solubilizers (4.6–5.7 mg·L−1) than L. lactis subsp. lactis (Figure 3). Among the L. lactis subsp. lactis strains tested, LB1 reached the highest value (3.2 ± 0.1 mg·L−1).
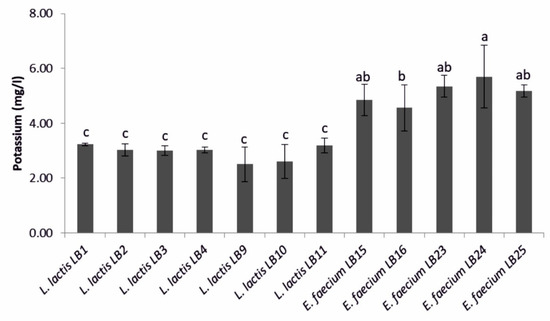
Figure 3.
Amount of potassium solubilized by the lactic acid bacteria strains that showed a halo zone of potassium solubilization. Means (±SD) followed by the similar letter(s) are not significantly different at the 5% probability level (LSD test).
3.5. Antifungal Activity
Bacillus velezensis FUA2155 and Bacillus amyloliquefaciens Fad 82 showed the strongest antifungal activity (Figure 4). E. faecium LB5 and L. lactis subsp. lactis LB6, LB7, LB9 exhibited inhibitory activity against Fusarium graminearum that was comparable the bacilli (Figure 4); however, activity of L. lactis culture supernatants against F. graminearum was observed only when cultured in mMRS but not when cultured in GYP. L. lactis subsp. lactis LB1, LB2, LB3, LB4, LB10, LB11 and E. faecium LB24, LB25 had a lower activity against F. graminearum when compared to Bacillus sp. strains. None of the LAB inhibited germination of A. niger spores, whereas Bacillus sp. strains were inhibitory to A. niger. All the LAB strains showed only weak inhibitory activity against P. roqueforti when compared to the two Bacillus sp. strains (Figure 4).
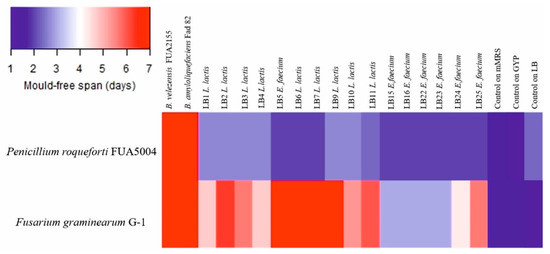
Figure 4.
Heatmap representing the average of the mold-free days (n = 3). Controls on mMRS, GYP, and LB broths refer to wells containing just broth acidified to pH 4.5 and fungal spore suspension. Bacillus velezensis FU2155 and Bacillus amyloliquefaciens Fad82 were cultured in LB broth. LAB were cultured keeping the same culture medium used for the isolation step: Lactococcus lactis and Enterococcus faecium LB5 were cultured in mMRS broth; the remaining E. faecium were cultured in GYP broth.
3.6. Antibacterial Activity
Neutralized culture supernatant of any of the strains of L. lactis and E. faecium showed no inhibitory activity against the tested phytopathogenic bacteria. The cell-free supernatant of seven strains of L. lactis was weakly inhibitory against the Pantoea, Pseudomonas, and Pectobacterium species if the pH remained unadjusted, with MIC values ranging from 2 to 17% (Table S1). Supernatant of E. faecium was not inhibitory against phytopathogenic bacteria even if the pH of the culture supernatant remained unadjusted.
3.7. Comparative Genomic Analysis between Plant Associated Lactococci and Enterococci and Strains Dairy or Clinical Isolates of the Same Species
To determine whether plant associated lactococci differ from those strains that are used in back-slopped dairy fermentations, comparative genomic analysis was performed on a total of 77 strains of L. lactis, isolated from two different ecosystems: two genomes of strains isolated in this study; 28 genomes, available on NCBI, of strains isolated from plants; and 47 genomes, available on NCBI, of strains isolated from either back-slopped dairy starter cultures or cheese produced with back-slopped cultures. The two lactococcal strains (LB6 and LB7) were randomly selected from the nine strains isolated in this study for genome sequencing. Likewise, comparative genomic analysis was performed on a total of 111 strains of E. faecium, isolated from different ecosystems: 9 genomes belonged to as many strains isolated in this study; the other 102 genomes were available on NCBI and, among these, 35 belonged to as many strains isolated from plant, soil or fresh water, 38 belonged to clinical isolates and 29 genomes to strains from healthy and non-domesticated animals. AntiSMASH identified genes coding for production of multiple antifungal lipopeptides in the genome of B. velezensis FUA 2155 and B. amyloliquefaciens Fad 82 (Table S2) but not in any of the genome of L. lactis and E. faecium. Core genome phylogenetic trees of L. lactis and E. faecium are shown in Figure 5.
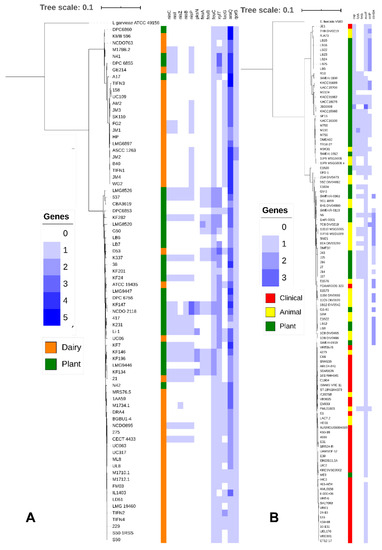
Figure 5.
(A) Core genome tree of 77 Lactococcus lactis genomes. Genomes from L. lactis strains isolated from Dairy (Orange) and Plants (Green) are indicated, Lactococcus garvieae ATCC 49156 was included as an outgroup. A heatmap was generated indicating copy number of genes involved in nisin production (nisC, nisI, nisZ, nisB, and nisP), surfactin production (PksN), iron uptake (feuA, feuB, and feuC), xylose utilization (xylT), lactose utilization (lacG), arabinose utilization (araQ) and phenolic metabolism (qorB). (B) Core genome tree of 111 Enterococcus faecium genomes. Genomes from E. faecium strains isolated from clinical sources (Red), animals (Yellow), and plants (Green) are indicated, Enterococcus faecalis V583 is included as an outgroup. A heatmap was generated indicating the copy number of genes involved in cold shock response (csp), iron uptake (fetA, fetB), microcin (mccF), enterocin (entP), and carnobacteriocin (cbnBA).
Dairy isolates of L. lactis strains were dispersed in the tree but clustered in areas, representing L. lactis subsp. cremoris (upper cluster) and L. lactis subsp. lactis (lower cluster) (Figure 5A). Plant isolates of L. lactis were identified in both subspecies but predominantly belonged to L. lactis subsp. lactis. Over 2000 genes were differentially distributed in dairy and plant isolates of L. lactis (Benjamini-Hochberg adjusted p < 0.05) (Table S3). A majority of these genes were hypothetical proteins without known function, followed by genes coding for carbohydrate metabolism. Genes enriched in dairy isolates of L. lactis included a plasmid-encoded lactose PTS system, genes of the tagatose pathway for galactose utilization, genes coding for components of the oligopeptide transporter Opp, and genes coding for the manganese transporter MtnH and cystathionine-γ-lyase (Table S3 and Figure 5A). Genes that were enriched in plant associated lactococci include genes encoding for nisin biosynthesis (nisC, nisI, nisZ, nisB, and nisP), disaccharide and pentose metabolism (e.g., xylT and araQ), cystathionine-β-lyase, agmatine deiminase, iron uptake systems (feuA, feuB, and feuC), genes related to synthesis of the sideropore enterobactin, oleate hydratase, the quaternary ammonia compound resistance gene qacC, the lactose permease lacF, and the quinone oxidoreductase qorB (Table S3 and Figure 5A).
Clinical isolates of E. faecium were concentrated to a single phylogenetic clade (Figure 5B) although few clinical isolates also clustered in other clades. Plant isolates and isolates from wild animals that have little or no exposure to humans or antibiotics were represented on the tree without clear separation (Figure 5B). Strains of enterococci isolated from wheat rhizosphere predominantly, but not exclusively, clustered together (Figure 5B), likely reflecting that most but not all of these isolates were obtained from the same field (Table 1). E. faecium LB8 and LB12, isolated in this study, fell in the same phylogenetic clade as the clinical isolates. When comparing clinical and plant isolates of E. faecium, the gene content of 681 genes was differentially distributed (Benjamini-Hochberg adjusted p < 0.05) (Table S4); 606 genes were differentially distributed between clinical and animal isolates but only 400 genes were differentially distributed between plant and animal isolates (Table S4). Genes that were differentially distributed between clinical and plant isolates predominantly related to carbohydrate transport and metabolism but also included over 50 IS-transposases (Table S4 and Figure 5B). Genes that were enriched in plant associated enterococci also included genes coding for a cold shock protein (csp), the iron transport system FetB, and genes related to the synthesis of the bacteriocins microcin (mccF), enterocin P (entB), and carnobacteriocin A (cbnBA) (Table S4 and Figure 5B).
4. Discussion
Only few studies elucidated the role of lactic acid bacteria (LAB) in the rhizosphere and their plant growth promoting properties [18,19]. In our study, LAB were isolated from rhizospheric soil collected from two different cultivars of durum wheat during different growth stages. L. lactis subsp. lactis and E. faecium were the only two species of LAB found. Studies on microbial communities associated with roots showed that the growth stages [49,50,51], genotype [49,50], and soil type [51] influence their diversity. Among these factors, soil type is the main force driving changes in the community, followed by growth stage [51]. LAB represent only a small part of the wheat rhizosphere microbial community [52,53], therefore, enrichment methods are necessary to isolate them from soil [18]. In addition, LAB are fastidious microorganisms and are more easily detected in soils cultivated with fruit trees or, more in general, soils rich in carbon sources [54]. However, carbon sources from root exudates may also support growth of LAB in rhizospheric soil [55].
Research on L. lactis and E. faecium focused mainly on their role as dairy starter cultures and as nosocomial pathogens, respectively. L. lactis subsp. lactis and L. lactis subsp. cremoris are major components of mesophilic dairy starter cultures, which are maintained by back-slopping [56] but the organism is thought to originate from plant-associated habitats. The adaptation of L. lactis to the dairy environment relates to the acquisition of lactose metabolism, proteolysis and peptide transport; these genes are often encoded on plasmids [57,58]. Our comparative genomic analyses confirm these prior observations; the availability of a large number of genomes of lactococci of plant origin additionally allowed to identify genes that are enriched in plant-associated strains. Dairy isolates clustered in two distinct phylogenetic clades presenting L. lactis subsp. cremoris and L. lactis subsp. lactis; the close relationship of dairy isolates may reflect the fact that dairy cultures are descendants of only few strains that were isolated in the U.S. and Denmark [59]. Plant-associated strains were enriched in genes coding for disaccharide and pentose metabolism and were more likely to encode genes for synthesis of nisin variants. Plant strains also encoded for a polyketide synthase (PKS). The closest homologue to the PKS in L. lactis is the surfactin synthase in B. subtilis (GenBank accession no. Q04747). Because six out of 30 genomes of plant-associated strains also included a surfactin thioesterase, these strains are likely capable of producing the peptide antibiotic surfactin [60]. Plant-associated lactococci also encoded for agmatine deiminase (AgDI). Agmatine is the product of decarboxylation of arginine by Enterobacteriaceae; among Lactobacilli, AgDI is found only in the plant-associated genera Levilactobacillus, Secundilactobacillus, and Lentilactobacillus [61]. Plant tissue injury activates polyphenol oxidases and lipoxygenases which catalyse quinones and lipoperoxides as part of the plant stress response [62,63]. Plant-associated lactococci strains encoded a different ortholog of the quinone oxidoreductase QorB than dairy isolates. Moreover, seven plant strains encoded for oleate hydratase, which synthesizes antifungal oxylipins from linoleic acid [64].
E. faecium is usually associated with gastrointestinal tract of humans and animals [65] but is also relevant as a nosocomial pathogen causing life-threatening systemic infections. Most isolates from infected patients can be assigned to a single phylogenetic clade [23,66]. These observations were confirmed in our analyses, which also confirmed that mobile genetic elements, particularly transposases, strongly contribute to the diversification of plant strains, animal commensals, and human clinical isolates. Comparable to lactococci, genes coding for bacteriocin production were more frequent in plant-associated enterococci, indicating that antimicrobial activity against closely related strains [67] contributes to ecological fitness in plant ecosystems. Plant growth promotion properties were previously documented for a soil isolate of E. faecium, which secreted gibberellins and 3-indolacetic acid (IAA), thus improving the length and biomass of rice shoots [68].
That E. faecium LB8 and LB12 were closely related to clinical isolates of the same species and were thus not considered for further characterization. Investigating in vitro the properties of the LAB isolated from wheat rhizospheric soil was the first step to understand their potential plant growth promoting activity. We tested the capacity by LAB to produce IAA, because it is one of the main PGP traits of bacteria inhabiting rhizosphere [13]. No isolate produced IAA in physiologically relevant concentrations, in agreement with previous studies which also included also LAB [69,70]. On the other hand, Mussa et al. [71] reported that one strain of Enterococcus casseliflavus, isolated from grass pea rhizospheric soil, synthesized IAA at a concentration of ca. 56 µg mL−1, corresponding to about one order of magnitude higher than the best IAA producer among the LAB subjected to this research. Many rhizobacteria, including Pseudomonas sp. [72] Bacillus sp. [73] and, especially Enterobacter sp. [74,75,76] synthesize this phyto-hormone at concentrations of more than 100 µg·mL−1.
Phosphorus (P) is an important macronutrient for plant growth, but most of the soil phosphorus is not available for plants because it is either immobilized in soil in organic forms such as inositol phosphates or phosphonates or precipitated in inorganic form including apatites or Ca, Fe, and Al phosphates [77]. Even phosphorus applied as external input is quite often transformed into one of the unavailable forms [78]. Soil microorganisms play an important role in the mineralization and solubilization of organic and inorganic P respectively [79]. Under our experimental conditions, all the 16 LAB strains solubilized high amount of phosphate, comparable to strains of Enterococcus sp. isolated from grass pea rhizospheric soil [71]. The release of organic acids from LAB and the consequent acidification of broth (4.5 < pH < 4.9) could explain the observed solubilization of P. Indeed, previous researches showed inverse correlation between phosphate solubilization and pH of culture broth, varying from weak–moderate [71] to strong [80]. On the other hand, de Lacerda et al. [81] reported no significant correlation between pH and calcium phosphate solubilization. The same authors suggested that the ability of L. lactis to solubilize different sources of organic and inorganic P could be related to gene sequences possibly coding for two types of alkaline phosphatase, enzymes that catalyze the mineralization of phosphates [81].
Potassium (K) is another essential element for plants and its deficiency has a negative impact on roots, seeds, growth, and yields [82]. Although its concentration in soil solution is higher than P (generally 2–5 mg·L−1), most of K is in fixed forms [83]. Two potassium-solubilizing rhizobacteria (KSR), isolated from K-bearing minerals, and belonging to Bacillus sp., were proposed as an alternative to chemical K-containing fertilizers [82]. As far as we know, first our work showed LAB ability to solubilize K from mica powder. Strains of E. faecium proved to be better K-solubilizers than L. lactis subsp. lactis. The K-solubilizing activity of LAB could be due to formation of lactic acid and the corresponding acidification.
Defending plants from soil-borne pathogens is another feature of many rhizobacteria, being Pseudomonas and Bacillus currently the genera most used as biocontrol agents [84]. The antibacterial and antifungal activity of LAB is exploited in food production [67,85] but their use as biocontrol agents in crops is limited to in vitro studies [86] and to very few in situ applications. Cell-free supernatant of Levilactobacillus brevis JJ2P inhibited Zymoseptoria tritici in wheat seedling test, reducing the development of wheat leaf blotch [87]. Consortia of lactobacilli were inoculated in corn kernels, together with Fusarium verticilloides, a mycotoxigenic fungus causing ear rot. Compared to corn plants inoculated just with the phytopathogenic fungus, lactobacilli decreased disease severity and increased yield performance and the seed weight [88]. In a greenhouse experiment, a strain of L. lactis subsp. lactis was applied to foliar branches of holy basil inoculated with conidia of Alternaria sp. After 60 days post inoculation, the percentage of leaf blight in plants inoculated with the lactococcal strain was lower than the plants inoculated just with Alternaria sp. In field conditions, where basil plants had not been deliberately inoculated with Alternaria sp., plants inoculated with Lc. lactis subsp. lactis also showed a lower disease incidence than untreated control plants [89]. In the current study, three strains of lactococci and one E. faecium strain inhibited in vitro growth of the phytopathogenic fungus F. graminearum to an extent that was comparable to the two Bacillus sp. strains that were used as positive controls. However, the antifungal activity depended on the culture medium used for LAB. We may hypothesize that the four LAB strains synthesized cyclic dipeptides at concentrations higher than the MIC during growth on MRS [85], but not during growth on GYP. The two Bacillus strains displayed potent antifungal activity also against P. roqueforti and A. niger, and both strains harboured genes encoding for synthesis of several antifungal lipopeptides [39]. Remarkably, genes related to biosynthesis of surfactin in Bacillus subtilis were identified in six genomes of plant-associated lactococci. Surfactin does not display strong antifungal activity but was reported to act synergistically with other antifungal compounds [90] and thus may enhance the antifungal activity of bacilli.
Food preservation with LAB relies mainly on the acidification; bacteriocins specifically enhance the inhibitory activity of LAB towards Gram-positive bacteria, but bacteriocins of LAB are generally inactive against Gram-negative bacteria [67,91]. Comparative genomic analyses presented in this study identified bacteriocin production as a part of the plant-associated lifestyle of lactococci and enterococci but also documented only weak and pH-dependent antibacterial activity with Gram-negative phytopathogens as indicator strains. Lactic acid production, however, permeabilizes the outer membrane of Gram-negative bacteria and increases their sensitivity to hydrophobic inhibitors [92]. Lactic acid production in association with the production of other antibacterial compounds may account for the protective effect of a LAB consortium against papaya infection with Erwinia mallotivora [93].
5. Conclusions
This study showed that, besides the well-recognized PGPB, LAB isolated from wheat rhizosphere may have potential plant growth promotion activities, too. No strain possessed all the plant growth promoting activity at the highest level and therefore ongoing research is focusing on setting-up a LAB consortium for future application as biofertilizer in conventional and organic agriculture. LAB showed only weak inhibitory activity against plant pathogenic fungi and Gram-negative bacteria, but may synergistically enhance the activity of compounds produced by other members of the rhizosphere. Plant growth promotion and efficacy of biocontrol should obviously be confirmed under outdoor conditions, where cultivar and soil type, among others, could affect plant response [94]. Comparative genomics also allowed to differentiate LAB with potential plant growth promotion and biocontrol activity from food-related strains of L. lactis and from strains of E. faecium isolated from humans.
Supplementary Materials
The following are available online at https://www.mdpi.com/2076-2607/9/1/78/s1, Table S1: MIC (% vol/vol) values of the cell-free supernatant of L. lactis subsp. lactis strains against phytopathogenic bacteria; Table S2: Secondary metabolites produced by Bacillus spp; Table S3: genes’ distribution between dairy and plant isolates of L. lactis; Table S4: genes’ distribution between clinical and animal isolates of E. faecium.
Author Contributions
M.D.A., F.M., and C.C. designed the project; S.S. performed the experiments and wrote the original draft; D.J.S. compared the genomes of microorganisms and wrote the part of the manuscript dedicated to them; M.Y.K. analyzed the data; F.M., M.D.A., M.G., and C.C. reviewed and edited the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
Sabrina Strafella acknowledges the Italian Ministry of University and Research (MUR) and the University of Bari Aldo Moro (D.R. 993, March 29th, 2018) for the Ph.D. grant no. 5 “CUP: H92H18000370006”, co-funded by European Union–FSE-FESR, PON Ricerca e Innovazione 2014–2020, Ph.D. Program “Soil and Food Sciences”, DOT1302942, XXXIII Cycle. The Natural Science and Engineering Research Council of Canada (NSERC) provided funding under the Discovery program. Michael Gänzle acknowledges funding from the Canada Research Chairs Program.
Data Availability Statement
Data is contained within the article or supplementary material.
Acknowledgments
The authors thank Nuanyi Liang for her mentorship about antifungal and antibacterial assays.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Available online: http://www.fao.org/faostat/en/#country (accessed on 20 June 2020).
- Zhang, X.; Davidson, E.A.; Mauzerall, D.L.; Searchinger, T.D.; Dumas, P.; Shen, Y. Managing nitrogen for sustainable development. Nature 2015, 528, 51–59. [Google Scholar] [CrossRef]
- Reganold, J.; Wachter, J. Organic agriculture in the twenty-first century. Nat. Plants. 2016, 2, 15221. [Google Scholar] [CrossRef] [PubMed]
- Oerke, E.C. Crop losses to pests. J. Agric. Sci. 2006, 144, 31–43. [Google Scholar] [CrossRef]
- Trewavas, A. Urban myths of organic farming. Nature 2001, 410, 409–410. [Google Scholar] [CrossRef]
- Niggli, U. Sustainability of organic food production: Challenges and innovations. Proc. Nutr. Soc. 2015, 74, 83–88. [Google Scholar] [CrossRef]
- Seufert, V.; Ramankutty, N.; Foley, J.A. Comparing the yields of organic and conventional agriculture. Nature 2012, 485, 229–232. [Google Scholar] [CrossRef]
- Mendes, R.; Garbeva, P.; Raaijmakers, J.M. The rhizosphere microbiome: Significance of plant beneficial, plant pathogenic, and human pathogenic microorganisms. FEMS Microbiol. Rev. 2013, 37, 634–663. [Google Scholar] [CrossRef]
- Glick, B.R. Plant Growth-Promoting Bacteria: Mechanisms and Applications. Scientifica 2012, 2012, 963401. [Google Scholar] [CrossRef]
- Kennedy, A.C.; de Luna, L.Z. Rhizosphere. In Encyclopedia of Soils in the Environment; Hillel, D., Ed.; Elsevier Ltd.: Pullman, WA, USA, 2005; pp. 399–406. [Google Scholar] [CrossRef]
- Backer, R.; Rokem, J.S.; Ilangumaran, G.; Lamont, J.; Praslickova, D.; Ricci, E.; Subramanian, S.; Smith, D.L. Plant Growth-Promoting Rhizobacteria: Context, Mechanisms of Action, and Roadmap to Commercialization of Biostimulants for Sustainable Agriculture. Front. Plant Sci. 2018, 9, 1473. [Google Scholar] [CrossRef]
- Lucy, M.; Reed, E.; Glick, B.R. Applications of free living plant growth-promoting rhizobacteria. Antonie Leeuwenhoek 2004, 86, 1–25. [Google Scholar] [CrossRef]
- Pii, Y.; Mimmo, T.; Tomasi, N.; Terzano, R.; Cesco, S.; Crecchio, C. Microbial interactions in the rhizosphere: Beneficial influences of plant growth-promoting rhizobacteria on nutrient acquisition process. A review. Biol. Fertil. Soils 2015, 51, 403–415. [Google Scholar] [CrossRef]
- Figueiredo, M.d.V.; Bonifacio, A.; Rodrigues, A.C.; de Araujo, F.F. Plant Growth-Promoting Rhizobacteria: Key Mechanisms of Action. In Microbial-Mediated Induced Systemic Resistance in Plants; Choudhary, D., Varma, A., Eds.; Springer: Singapore, 2016; pp. 23–37. [Google Scholar] [CrossRef]
- Lamont, J.R.; Wilkins, O.; Bywater-Ekegard, M.; Smith, D.L. From yogurt to yield: Potential applications of lactic acid bacteria in plant production. Soil Biol. Biochem. 2017, 111, 1–9. [Google Scholar] [CrossRef]
- Beneduzi, A.; Ambrosini, A.; Passaglia, L.M. Plant growth-promoting rhizobacteria (PGPR): Their potential as antagonists and biocontrol agents. Genet. Mol. Biol. 2012, 35, 1044–1051. [Google Scholar] [CrossRef] [PubMed]
- Lagos, M.L.; Maruyama, F.; Nannipieri, P.; Mora, M.L.; Ogram, A.; Jorquera, M.A. Current overview on the study of bacteria in the rhizosphere by modern molecular techniques: A mini‒review. J. Soil Sci. Plant Nutr. 2015, 15, 504–523. [Google Scholar] [CrossRef]
- Chen, Y.S.; Yanagida, F.; Shinohara, T. Isolation and identification of lactic acid bacteria from soil using an enrichment procedure. Lett. Appl. Microbiol. 2005, 40, 195–200. [Google Scholar] [CrossRef]
- Fhoula, I.; Najjari, A.; Turki, Y.; Jaballah, S.; Boudabous, A.; Ouzari, H. Diversity and Antimicrobial Properties of Lactic Acid Bacteria Isolated from Rhizosphere of Olive Trees and Desert Truffles of Tunisia. Biomed. Res. Int. 2013, 2013, 405708. [Google Scholar] [CrossRef]
- Duar, R.M.; Lin, X.B.; Zheng, J.; Martino, M.E.; Grenier, T.; Pérez-Muñoz, M.E.; Leulier, F.; Gänzle, M.; Walter, J. Lifestyles in transition: Evolution and natural history of the genus Lactobacillus. FEMS Microbiol. Rev. 2017, 41, S27–S48. [Google Scholar] [CrossRef]
- EFSA BIOHAZ Panel (EFSA Panel on Biological Hazards); Koutsoumanis, K.; Allende, A.; Alvarez-Ordonez, A.; Bolton, D.; Bover-Cid, S.; Chemaly, M.; Davies, R.; de Cesare, A.; Hilbert, F.; et al. Statement on the update of the list of QPS-recommended biological agents intentionally added to food or feed as notified to EFSA 11: Suitability of taxonomic units notified to EFSA until September. EFSA J. 2020, 18, e05965. [Google Scholar] [CrossRef]
- Franz, C.M.A.P.; Stiles, M.E.; Schleifer, K.H.; Holzapfel, W.H. Enterococci in foods—a conundrum for food safety. Int. J. Food Microbiol. 2003, 88, 105–122. [Google Scholar] [CrossRef]
- van Tyne, D.; Gilmore, M.S. Friend turned foe: Evolution of enterococcal virulence and antibiotic resistance. Annu. Rev. Microbiol. 2014, 68, 337–356. [Google Scholar] [CrossRef]
- EFSA FEEDAP Panel (EFSA Panel on Additives and Products or Substances used in Animal Feed). Scientific Opinion on the safety and efficacy of Enterococcus faecium (NCIMB 10415, DSM 22502, ATCC 53519 and ATCC 55593) as silage additives for all animal species. EFSA J. 2013, 11, 3363. Available online: https://efsa.onlinelibrary.wiley.com/doi/pdf/10.2903/j.efsa.2013.3363 (accessed on 1 July 2020).
- Konappa, N.M.; Maria, M.; Uzma, F.; Krishnamurthy, S.; Nayaka, S.C.; Niranjana, S.R.; Chowdapp, S. Lactic acid bacteria mediated induction of defense enzymes to enhance the resistance in tomato against Ralstonia solanacearum causing bacterial wilt. Sci. Hortic. 2016, 207, 183–192. [Google Scholar] [CrossRef]
- Shrestha, A.; Kim, B.S.; Park, D.H. Biological control of bacterial spot disease and plant growth-promoting effects of lactic acid bacteria on pepper. Biocontrol. Sci. Technol. 2014, 24, 763–779. [Google Scholar] [CrossRef]
- Kang, S.M.; Radhakrishnan, R.; You, Y.H.; Khan, A.L.; Park, J.M.; Lee, S.M.; Lee, I.J. Cucumber performance is improved by inoculation with plant growth promoting microorganisms. Acta Agric. Scand. Sect. B Soil Plant Sci. 2015, 65, 36–44. [Google Scholar] [CrossRef]
- Daranas, N.; Roselló, G.; Cabrefiga, J.; Donati, I.; Francés, J.; Badosa, E.; Spinelli, F.; Montesinos, E.; Bonaterra, A. Biological control of bacterial plant diseases with Lactobacillus plantarum strains selected for their broad-spectrum activity. Ann. Appl. Biol. 2019, 174, 92–105. [Google Scholar] [CrossRef] [PubMed]
- Stendid, J.; Karlsson, J.O.; Hogberg, N. Intra-specific genetic variation in Heterobasidion annosum revealed by amplification of minisatellite DNA. Mycol. Res. 1994, 98, 57–63. [Google Scholar] [CrossRef]
- Zapparoli, G.; Torriani, S.; Dell’aglio, F. Differentiation of Lactobacillus sanfranciscensis strains by randomly amplified polymorphic DNA and pulsed-field gel electrophoresis. FEMS Microbiol. Lett. 1998, 166, 324–332. [Google Scholar] [CrossRef][Green Version]
- Yu, Z.; Morrison, M. Comparisons of different hypervariable regions of rrs genes for use in fingerprinting of microbial communities by PCR-denaturing gradient gel electrophoresis. Appl. Environ. Microbiol. 2004, 70, 4800–4806. [Google Scholar] [CrossRef]
- Gordon, S.A.; Weber, R.P. Colorimetric estimation of indoleacetic acid. Plant Physiol. 1951, 26, 192–195. [Google Scholar] [CrossRef]
- Nautiyal, C.S. An Efficient Microbiological Growth Medium for Screening Phosphate Solubilizing Microorganisms. FEMS Microbiol. Lett. 1999, 170, 265–270. [Google Scholar] [CrossRef]
- Watanabe, F.S.; Olsen, S.R. Test of an ascorbic acid method for determining phosphorus in water and NaHCO3 extracts from soils. Soil Sci. Soc. Am. J. 1965, 29, 677–678. [Google Scholar] [CrossRef]
- Aleksandrov, V.G.; Blagodyr, R.N.; Live, I.P. Phosphorus acid isolation from apatite produced by silicate bacteria. Microchem. J. 1967, 29, 111–114. [Google Scholar]
- Yaghoubi Khanghahi, M.; Ricciuti, P.; Allegretta, I.; Terzano, R.; Crecchio, C. Solubilization of insoluble zinc compounds by zinc solubilizing bacteria (ZSB) and optimization of their growth conditions. Environ. Sci. Pollut. Res. 2018, 25, 25862–25868. [Google Scholar] [CrossRef]
- Towett, E.K.; Shepherd, K.D.; Cadisch, G. Quantification of total element concentrations in soils using total X-ray fluorescence spectroscopy (TXRF). Sci. Total Environ. 2013, 463, 74–388. [Google Scholar] [CrossRef]
- Li, Z.; Schottroff, F.; Simpson, D.J.; Gänzle, M.G. The copy number of the spoVA2mob operon determines pressure resistance of Bacillus endospores. Appl. Environ. Microbiol. 2019, 85, e01596-19. [Google Scholar] [CrossRef] [PubMed]
- Penha, R.O.; Vandenberghe, L.P.S.; Faulds, C.; Soccol, V.T.; Soccol, C.R. Bacillus lipopeptides as powerful pest control agents for a more sustainable and healthy agriculture: Recent studies and innovations. Planta 2020, 251, 70. [Google Scholar] [CrossRef]
- Liang, N.; Dacko, A.; Tan, A.K.; Xiang, S.; Curtis, J.M.; Gänzle, M.G. Structure-function relationships of antifungal monohydroxy unsaturated fatty acids (HUFA) of plant and bacterial origin. Food Res. Int. 2020, 134, 109237. [Google Scholar] [CrossRef]
- Magnusson, J.; Schnürer, J. Lactobacillus coryniformis subsp. coryniformis strain Si3 produces a broad-spectrum proteinaceous antifungal compound. Appl. Environ. Microbiol. 2001, 67, 1–5. [Google Scholar] [CrossRef]
- Gaur, G.; Oh, J.-H.; Filannino, P.; Gobbetti, M.; van Pijkeren, J.-P.; Gänzle, M.G. Genetic determinants of hydroxycinnamic acid metabolism in heterofermentative lactobacilli. Appl. Environ. Microbiol. 2020, 86, e02461-19. [Google Scholar] [CrossRef]
- Eaton, K. NCBImeta: Efficient and comprehensive metadata retrieval from NCBI databases. J. Open Source Softw. 2020, 5, 1990. [Google Scholar] [CrossRef]
- Seemann, T. Prokka: Rapid prokaryotic genome annotation. Bioinformatics 2014, 30, 2068–2069. [Google Scholar] [CrossRef] [PubMed]
- Page, A.J.; Cummins, C.A.; Hunt, M.; Wong, V.K.; Reuter, S.; Holden, M.T.G.; Fookes, M.; Falush, D.; Keane, J.A.; Parkhill, J. Roary: Rapid large-scale prokaryote pan genome analysis. Bioinformatics 2015, 31, 3691–3693. [Google Scholar] [CrossRef] [PubMed]
- Brynildsrud, O.; Bohlin, J.; Scheffer, L.; Eldholm, V. Rapid scoring of genes in microbial pan-genome-wide association studies with Scoary. Genome Biol. 2016, 17, 238. [Google Scholar] [CrossRef] [PubMed]
- Price, M.N.; Dehal, P.S.; Arkin, A.P. FastTree 2—Approximately maximum-likelihood trees for large alignments. PLoS ONE 2010, 5, e9490. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree of Life (iTOL) v4: Recent updates and new developments. Nucleic Acids Res. 2019, 47, W256–W259. [Google Scholar] [CrossRef]
- Micallef, S.A.; Channer, S.; Shiaris, M.P.; Colón-Carmona, A. Plant age and genotype impact the progression of bacterial community succession in the Arabidopsis rhizosphere. Plant Signal. Behav. 2009, 4, 777–780. [Google Scholar] [CrossRef]
- Inceoglu, O.; Salles, J.F.; van Overbeek, L.; van Elsas, J.D. Effects of plant genotype and growth stage on the betaproteobacterial communities associated with different potato cultivars in two fields. Appl. Environ. Microbiol. 2010, 76, 3675–3684. [Google Scholar] [CrossRef]
- Schlemper, T.R.; Leite, M.F.A.; Lucheta, A.R.; Shimels, M.; Bouwmeester, H.J.; van Veen, J.A.; Kuramae, E.E. Rhizobacterial community structure differences among sorghum cultivars in different growth stages and soils. FEMS Microbiol. Ecol. 2017, 93. [Google Scholar] [CrossRef]
- Yaghoubi Khanghahi, M.; Murgese, P.; Strafella, S.; Crecchio, C. Soil Biological Fertility and Bacterial Community Response to Land Use Intensity: A Case Study in the Mediterranean Area. Diversity 2019, 11, 211. [Google Scholar] [CrossRef]
- Kavamura, V.N.; Robinson, R.J.; Hughes, D.; Clark, I.; Rossmann, M.; de Melo, I.S.; Hirsch, P.R.; Mendes, R.; Mauchline, T.H. Wheat dwarfing influences selection of the rhizosphere microbiome. Sci. Rep. 2020, 10, 1452. [Google Scholar] [CrossRef]
- George, F.; Daniel, C.; Thomas, M.; Singer, E.; Guilbaud, A.; Tessier, F.J.; Revol-Junelles, A.M.; Borges, F.; Foligné, B. Occurrence and Dynamism of Lactic Acid Bacteria in Distinct Ecological Niches: A Multifaceted Functional Health Perspective. Front. Microbiol. 2018, 9, 2899. [Google Scholar] [CrossRef]
- Kiely, P.D.; Haynes, J.M.; Higgins, C.H.; Franks, A.; Mark, G.L.; Morrissey, J.P.; O’Gara, F. Exploiting new systems-based strategies to elucidate plant-bacterial interactions in the rhizosphere. Microb. Ecol. 2006, 51, 257–266. [Google Scholar] [CrossRef] [PubMed]
- Hutkins, R.W. Microbiology and Technology of Fermented Foods, 2nd. ed.; Hutkins, R.W., Ed.; IFT Press: Chigaco, IL, USA, 2019. [Google Scholar]
- Kelleher, P.; Bottacini, F.; Mahony, J.; Kilcawley, K.N.; van Sinderen, D. Comparative and functional genomics of the Lactococcus lactis taxon; insights into evolution and niche adaptation. BMC Genom. 2017, 18, 267. [Google Scholar] [CrossRef] [PubMed]
- Wels, M.; Siezen, R.; van Hijum, S.; Kelly, W.J.; Bachmann, H. Comparative genome analysis of Lactococcus lactis indicates niche adaptation and resolves genotype/phenotype disparity. Front. Microbiol. 2019, 10, 4. [Google Scholar] [CrossRef] [PubMed]
- Salama, M.; Sandine, W.; Giovannoni, S. Development and application of oligonucleotide probes for identification of Lactococcus lactis subsp Cremoris. Appl. Environ. Microbiol. 1991, 57, 1313–1318. [Google Scholar] [CrossRef] [PubMed]
- Carrillo, C.; Teruel, J.A.; Aranda, F.J.; Ortiz, A. Molecular mechanism of membrane permeabilization by the peptide antibiotic surfactin. Biochim. Biophys. Acta Biomembr. 2003, 1611, 91–97. [Google Scholar] [CrossRef]
- Zheng, J.; Wittouck, S.; Salvetti, E.; Franz, C.M.A.P.; Harris, H.M.B.; Mattarelli, P.; O’toole, P.W.; Pot, B.; Vandamme, P.; Walter, J.; et al. A taxonomic note on the genus Lactobacillus: Description of 23 novel genera, emended description of the genus Lactobacillus beijerinck 1901, and union of Lactobacillaceae and Leuconostocaceae. Int. J. Syst. Evol. Micr. 2020, 70, 2782–2858. [Google Scholar] [CrossRef]
- Burton, S.G. Biocatalysis with polyphenol oxidase: A review. Catal. Today 1994, 22, 459–487. [Google Scholar] [CrossRef]
- Prasad, A.; Sedlářová, M.; Kale, R.S.; Pospíšil, P. Lipoxygenase in singlet oxygen generation as a response to wounding: In vivo imaging in Arabidopsis thaliana. Sci. Rep. 2017, 7, 9831. [Google Scholar] [CrossRef]
- Chen, Y.Y.; Liang, N.Y.; Curtis, J.M.; Gänzle, M.G. Characterization of linoleate 10-hydratase of Lactobacillus plantarum and novel antifungal metabolites. Front. Microbiol. 2016, 7. [Google Scholar] [CrossRef] [PubMed]
- Fiore, E.; van Tyne, D.; Gilmore, M.S. Pathogenicity of Enterococci. Microbiol. Spectr. 2019, 7, GPP3-0053-2018. [Google Scholar] [CrossRef] [PubMed]
- Lebreton, F.; Manson, A.L.; Saavedra, J.T.; Straub, T.J.; Earl, A.M.; Gilmore, M.S. Tracing the Enterococci from Paleozoic Origins to the Hospital. Cell 2017, 169, 849–861.e13. [Google Scholar] [CrossRef] [PubMed]
- Ross, P.; Morgan, S.; Hill, C. Preservation and fermentation: Past, present and future. Int. J. Food Microbiol. 2002, 79, 3–16. [Google Scholar] [CrossRef]
- Lee, K.E.; Radhakrishnan, R.; Kang, S.M.; You, Y.H.; Joo, G.J.; Lee, I.J.; Ko, J.H.; Kim, J.H. Enterococcus faecium LKE12 Cell-Free Extract Accelerates Host Plant Growth via Gibberellin and Indole-3-Acetic Acid Secretion. J. Microbiol. Biotechnol. 2015, 25, 1467–1475. [Google Scholar] [CrossRef] [PubMed]
- Giassi, V.; Kiritani, C.; Kupper, K.C. Bacteria as growth-promoting agents for citrus rootstocks. Microbiol. Res. 2016, 190, 46–54. [Google Scholar] [CrossRef] [PubMed]
- Mohite, B. Isolation and characterization of indole acetic acid (IAA) producing bacteria from rhizospheric soil and its effect on plant growth. J. Soil Sci. Plant Nutr. 2013, 13, 638–649. [Google Scholar] [CrossRef]
- Mussa, A.; Million, T.; Assefa, F. Rhizospheric bacterial isolates of grass pea (Lathyrus sativus L.) endowed with multiple plant growth promoting traits. J. Appl. Microbiol. 2018, 125, 1786–1801. [Google Scholar] [CrossRef]
- Malik, D.K.; Sindhu, S.S. Production of indole acetic acid by Pseudomonas sp.: Effect of coinoculation with Mesorhizobium sp. Cicer on nodulation and plant growth of chickpea (Cicer arietinum). Physiol. Mol. Biol. Plants 2011, 17, 25–32. [Google Scholar] [CrossRef]
- Wagi, S.; Ahmed, A. Bacillus spp.: Potent microfactories of bacterial IAA. PeerJ 2019, 7, e7258. [Google Scholar] [CrossRef]
- Park, J.; Radhakrishnan, R.; Kang, S.M.; Lee, I.J. IAA Producing Enterobacter sp. I-3 as a Potent Bio-herbicide Candidate for Weed Control: A Special Reference with Lettuce Growth Inhibition. Indian J. Microbiol. 2015, 55, 207–212. [Google Scholar] [CrossRef]
- Bose, A.; Kher, M.M.; Nataraj, M.; Keharia, H. Phytostimulatory effect of indole-3-acetic acid by Enterobacter cloacae SN19 isolated from Teramnus labialis (L. f.) Spreng rhizosphere. Biocatal. Agric. Biotechnol. 2016, 6, 128–137. [Google Scholar] [CrossRef]
- Scagliola, M.; Pii, Y.; Mimmo, T.; Cesco, S.; Ricciuti, P.; Crecchio, C. Characterization of plant growth promoting traits of bacterial isolates from the rhizosphere of barley (Hordeum vulgare L.) and tomato (Solanum lycopersicon L.) grown under Fe sufficiency and deficiency. Plant Physiol. Biochem. 2016, 107, 187–196. [Google Scholar] [CrossRef] [PubMed]
- Shen, J.; Yuan, L.; Zhang, J.; Li, H.; Bai, Z.; Chen, X.; Zhang, W.; Zhang, F. Phosphorus Dynamics: From Soil to Plant. Plant Physiol. 2011, 156, 997–1005. [Google Scholar] [CrossRef] [PubMed]
- Schachtman, D.P.; Reid, R.J.; Ayling, S.M. Phosphorus Uptake by Plants: From Soil to Cell. Plant Physiol. 1998, 116, 447–453. [Google Scholar] [CrossRef]
- Sharma, S.B.; Sayyed, R.Z.; Trivedi, M.H.; Gobi, T.A. Phosphate solubilizing microbes: Sustainable approach for managing phosphorus deficiency in agricultural soils. Springerplus 2013, 2, 587. [Google Scholar] [CrossRef] [PubMed]
- Muleta, D.; Assefa, F.; Borjesson, E.; Granhall, U. Phosphate-solubilising rhizobacteria associated with Coffee arabica L. in natural forests of South Western Ethiopia. J. Saudi Soc. Agric. Sci. 2013, 12, 73–84. [Google Scholar] [CrossRef]
- de Lacerda, J.R.M.; Da Silva, T.F.; Vollú, R.E.; Marques, J.M.; Seldin, L. Generally recognized as safe (GRAS) Lactococcus lactis strains associated with Lippia sidoides Cham. are able to solubilize/mineralize phosphate. Springerplus 2016, 5, 828. [Google Scholar] [CrossRef]
- Meena, V.S.; Maurya, B.R.; Verma, J.P.; Aeron, A.; Kumar, A.; Kim, K.; Bajpai Vivek, K. Potassium solubilizing rhizobacteria (KSR): Isolation, identification, and K-release dynamics from waste mica. Ecol. Eng. 2015, 81, 340–347. [Google Scholar] [CrossRef]
- Etesami, H.; Emami, S.; Alikhani, H.A. Potassium solubilizing bacteria (KSB): Mechanisms, promotion of plant growth, and future prospects—A review. J. Soil Sci. Plant Nutr. 2017, 17, 897–911. [Google Scholar] [CrossRef]
- Santoyo, G.; del Carmen Orozco-Mosqueda, M.; Govindappa, M. Mechanisms of biocontrol and plant growth-promoting activity in soil bacterial species of Bacillus and Pseudomonas: A review. Biocontrol Sci. Technol. 2012, 22, 855–872. [Google Scholar] [CrossRef]
- Axel, C.; Zannini, E.; Arendt, E.K. Mold spoilage of bread and its biopreservation: A review of current strategies for bread shelf life extension. Crit. Rev. Food Sci. 2017, 57, 3528–3542. [Google Scholar] [CrossRef]
- Gajbhiye, M.H.; Kapadnis, B.P. Antifungal-activity-producing lactic acid bacteria as biocontrol agents in plants. Biocontrol Sci. Technol. 2016, 26, 1451–1470. [Google Scholar] [CrossRef]
- Lynch, K.M.; Zannini, E.; Guo, J.; Axel, C.; Arendt, E.K.; Kildea, S.; Coffey, A. Control of Zymoseptoria tritici cause of Septoria tritici blotch of wheat using antifungal Lactobacillus strains. J. Appl. Microbiol. 2016, 121, 485–494. [Google Scholar] [CrossRef] [PubMed]
- Kharazian, Z.A.; Jouzani, G.S.; Aghdasi, M.; Khorvash, M.; Zamani, M.; Mohammadzadeh, H. Biocontrol potential of Lactobacillus strains isolated from corn silages against some plant pathogenic fungi. Biol. Control. 2017, 110, 33–43. [Google Scholar] [CrossRef]
- Barman, S.; Ghosh, R.; Dalal, D.; Mandal, N.C. Suppression of Leaf Blight of Ocimum sanctum L. Using Lactic Acid Bacteria as Novel Bio-control Agent. Proc. Natl. Acad. Sci. India Sect. B Biol. Sci. 2018, 88, 1389–1397. [Google Scholar] [CrossRef]
- Liu, X.; Ren, B.; Gao, H.; Liu, M.; Dai, H.; Song, F.; Yu, Z.; Wang, S.; Hu, J.; Kokare, C.R.; et al. Optimization for the production of surfactin with a new synergistic antifungal activity. PLoS ONE 2012, 7, e34430. [Google Scholar] [CrossRef]
- McAuliffe, O.; Ross, R.P.; Hill, C. Lantibiotics: Structure, biosynthesis and mode of action. FEMS Microbiol. Rev. 2001, 25, 285–308. [Google Scholar] [CrossRef] [PubMed]
- Alakomi, H.L.; Skyttä, E.; Saarela, M.; Mattila-Sandholm, T.; Latva-Kala, K.; Helander, I.M. Lactic acid permeabilizes gram-negative bacteria by disrupting the outer membrane. Appl. Environ. Microbiol. 2000, 66, 2001–2005. [Google Scholar] [CrossRef]
- Taha, M.D.M.; Jaini, M.F.M.; Saidi, N.B.; Rahim, R.A.; Shah, U.K.M.; Hashim, A.M. Biological control of Erwinia mallotivora, the causal agent of papaya dieback disease by indigenous seed-borne endophytic lactic acid bacteria consortium. PLoS ONE 2019, 14, e0224431. [Google Scholar] [CrossRef]
- Kildea, S.; Ransbotyn, V.; Khan, M.R.; Fagan, B.; Leonard, G.; Mullins, E.; Doohan, F.M. Bacillus megaterium shows potential for the biocontrol of Septoria tritici blotch of wheat. Biol. Control 2008, 47, 37–45. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).