Abstract
The control of transcription is poorly understood in dinoflagellates, a group of protists whose permanently condensed chromosomes are formed without histones. Furthermore, while transcriptomes contain a number of proteins annotated as transcription factors, the majority of these are cold shock domain proteins which are also known to bind RNA, meaning the number of true transcription factors is unknown. Here we have assessed the transcriptional response to light in the photosynthetic species Symbiodinium kawagutii. We find that three genes previously reported to respond to light using qPCR do not show differential expression using northern blots or RNA-Seq. Interestingly, global transcript profiling by RNA-Seq at LD 0 (dawn) and LD 12 (dusk) found only seven light-regulated genes (FDR = 0.1). qPCR using three randomly selected genes out of the seven was only able to validate differential expression of two. We conclude that there is likely to be less light regulation of gene expression in dinoflagellates than previously thought and suggest that transcriptional responses to other stimuli should also be more thoroughly evaluated in this class of organisms.
1. Introduction
Dinoflagellates are protists with an unusual chromatin structure [1]. Dinoflagellate chromosomes are permanently condensed, and can be observed with light microscopy using fluorescent DNA stains such as DAPI or propidinium iodide [2]. When observed using the electron microscope, individual chromosomes display a characteristic whorled banding pattern reminiscent of the bacterial nucleoid [3], and nucleosomes have never been observed [4]. The unusual chromatin structure has a number of molecular correlates. The histone proteins are at very low levels [5], and while one or two histones have been detected in several species [6,7], all four core histones have not yet been detected in any species. Instead of histones, dinoflagellates are thought to compact their DNA with a high level of divalent cations [8], histone-like proteins (HLP) [9], and a dinoflagellate/viral nucleoprotein (DVNP) [6].
The unusual dinoflagellate nuclear structure raises problems with respect to the mechanisms of both DNA replication and transcription. Little is known about replication, but many studies have examined changes in gene expression in response to light. Some of these studies use qPCR to examine specific genes. For example, rhodopsin in Prorocentrum was followed over a 14:10 L:D cycle and was observed to vary three-fold between LD 0 and LD 14 [10]. Similarly, transcripts encoding the oxygen evolving enzyme OEE1 in Symbiodinium were 2.5 fold more abundant at LD 12 than at LD 0 [11], while transcript levels encoding the large rubisco subunit rbcL were three fold higher at LD 12 than LD 0 [12], suggesting higher levels of transcription during the light. Levels of the thylakoid chlorophyll a-chlorophyll c2-peridinin-protein-complex (acpPC) were reported to be higher in dark phase than in light phase [13], suggesting that lack of light promotes expression of the light harvesting gene transcript.
Other experimental approaches have used high throughput expression measures such as microarrays or RNA-Seq. One of the earliest studies on differential transcription between day and night was carried out with Pyrocystis using microarrays programmed with about 3500 cDNAs [14]. About 80 differentially expressed genes (DEG) (~2%) were found to have a >2-fold difference between day and night in this species, with a maximum observed change of 2.5-fold. A similar microarray study comparing genes expressed during the day and night in Karenia brevis found 458 DEG among the 4629 genes examined (10%), with a significance threshold of p < 0.0001 and >1.7-fold change [15]. RNA-Seq studies in Symbiodinium microadriaticum found 67 DEG (0.1%) between day and night using DESeq with a false discovery rate (FDR) of 0.1 [16] and a maximum fold change of 160. A much more substantial number of DEG were noted in a study using Symbiodinium strain SSB01 24 h after a transfer from light to dark [17]. There were 1334 DEG (2.2%) when cells were grown phototrophically and 1739 DEG (2.9%) when cells were grown mixotrophically. These studies used duplicates (phototrophic growth) or triplicates (mixotrophic growth), but instead of an FDR = 0.1, the cutoff values for significance were p < 0.05 and a >1.5-fold change. Lastly, 131 DEG (0.17%) were found when samples of Lingulodinium polyedra taken every six hours were compared using an FDR of 0.1 [18], but northern blots analyses of a random selection of these showed no changes suggesting all were likely to be false positives.
The initial goal of our experiments was to identify a light regulated gene in S. kawagutii, so that potential regulatory elements in the promoter could be determined from the genome sequence [19], dissected, and the potential transcription factors involved identified. In one approach, we selected three genes whose transcripts had been previously been reported to be light regulated in Symbiodinium, and verified their expression levels using northern blots. In a second approach, we analysed global transcript levels at dawn and dusk by RNA-Seq. However, neither of these approaches successfully identified a light regulated gene, consistent with what has been observed with the dinoflagellate L. polyedra. This suggests that previous reports of light responsive genes may have overestimated their number, and further suggests that other reports of transcriptional responses may also benefit from additional verification.
2. Materials and Methods
2.1. Cell Cultures
Symbiodinium kawagutii (CCMP2468) was obtained from the National Center for Marine Algae and Microbiota (Boothbay Harbor, ME, USA) and cultured at 24 °C under a 12:12 light:dark cycle (40 µE m−2 s−1) in standard f/2 medium lacking silicate [20]. S. kawagutii has recently been renamed Fugacium kawagutii [21].
2.2. Microscopy
Cells were concentrated by centrifugation, then resuspended in a solution of 3% freshly made formaldehyde in seawater for 10 min then washed three times with fresh seawater. Cells were finally resuspended in phosphate buffered saline containing 0.05% Tween 20 and 1 µg/mL propidium iodide for 30 min. Images were taken using a Zeiss confocal microscope using a 63× objective in green (PI) and red (chlorophyll) channels. 3D reconstructions were made using Fiji [22].
2.3. RNA Extraction and Northern Blots
For the high light condition, S. kawagutii cells in fresh normal culture medium were transferred to 350 μmol of photons m−2 s−1 high light (HL) for 24 h. S. kawagutii cells were harvested from LD0 (beginning of light), LD12 (beginning of darkness), and HL (24 h in constant light) cultures. Total RNA was extracted with Trizol as described [23], the quantity and quality assessed by spectrophotometry and then stored at −80 °C. S. kawagutii RBCL, AcpPC, OEE1, and Actin sequences were acquired from the genome sequence (http:web.malab.cn/symka_new). Primers were used to amplify the sequence from a first strand cDNA reaction product using S. kawagutii total RNA using the ProtoScript First Strand cDNA synthesis kit (Invitrogen, Burlington, ON, Canada). The identity of all PCR products was confirmed by sequencing.
Northern blotting analysis was performed as described [18], 10 µg total RNA was electrophoresed on a denaturing agarose gel. The RNAs were transferred onto a nylon membrane (HybondTM-H+; Amersham Pharmacia Biotechnology, Piscataway, NJ, USA) and cross-linked by UV. PCR generated probes were labeled with [α-32P] ATP (BLU512H, Perkin Elmer, Woodbridge, OA, Canada) for hybridization. Membranes were hybridized at 65 °C for 16 h and were then washed twice at 65 °C for 15 min. The radiolabeled membranes were exposed to a phosphoscreen for 24 h and revealed by Typhoon Imager.
2.4. RNA Sequencing
Quality control, library construction, and Illumina sequencing were performed on RNA samples prepared in triplicate from S. kawagutii at LD 0 and LD 12 at McGill University and Genome Quebec Innovation Centre (Montreal, QC, Canada). Between 36 and 57 million paired end reads were recovered for each of the six samples. Raw sequence reads are available from NCBI using the accession number PRJNA517819.
The unigene list used for read mapping was downloaded from the S. kawagutii genome resources (http://web.malab.cn/symka_new/). This unigene list, containing 70,987 sequences, as well as the six sets of paired-end Illumina sequence reads, were uploaded to the Galaxy web platform at usegalaxy.org. The reads were trimmed using TrimGalore and read counts for all sequences in the unigene list were determined using Salmon [24]. Statistical significance was estimated using DESeq2 running in R [25].
2.5. Quantitative PCR
cDNAs were prepared from S. kawagutii RNAs extracted from the cells collected at four-hour intervals over an LD cycle plus high light cultures using ProtoScript First Strand cDNA Synthesis Kit and an oligo(dT) primer (Invitrogen, Burlington, ON, Canada). Specific primers were designed for SymkaALLUN13501, SymkaALLUN19088, SymkaALLUN64909, and Actin. qPCR analysis was performed in a ViiA7 Real-Time PCR System (Applied Biosystem, Burlington, ON, Canada) using SYBR green qPCR Master Mix (Thermo Fisher, St. Laurent, QC, Canada). Gene specific primers (250 nM) and cDNA (150 ng) were used in a total volume of 10 µl. Triplicate samples from each of three biological replicates amplified using 10 min at 95 °C, followed by 35 cycles of 15 s at 95 °C, 1 min at 60 °C, and 35 s at 68 °C, followed by a melt curve stage from 60 °C to 95 °C to verify the absence of non-specific amplification.
For gene expression analysis, cycle threshold (Ct) values were obtained from the ViiA7 Real-Time PCR software (Thermo Fisher, St. Laurent, QC, Canada). Student’s t-test was used to verify the statistical significance of the data.
3. Results
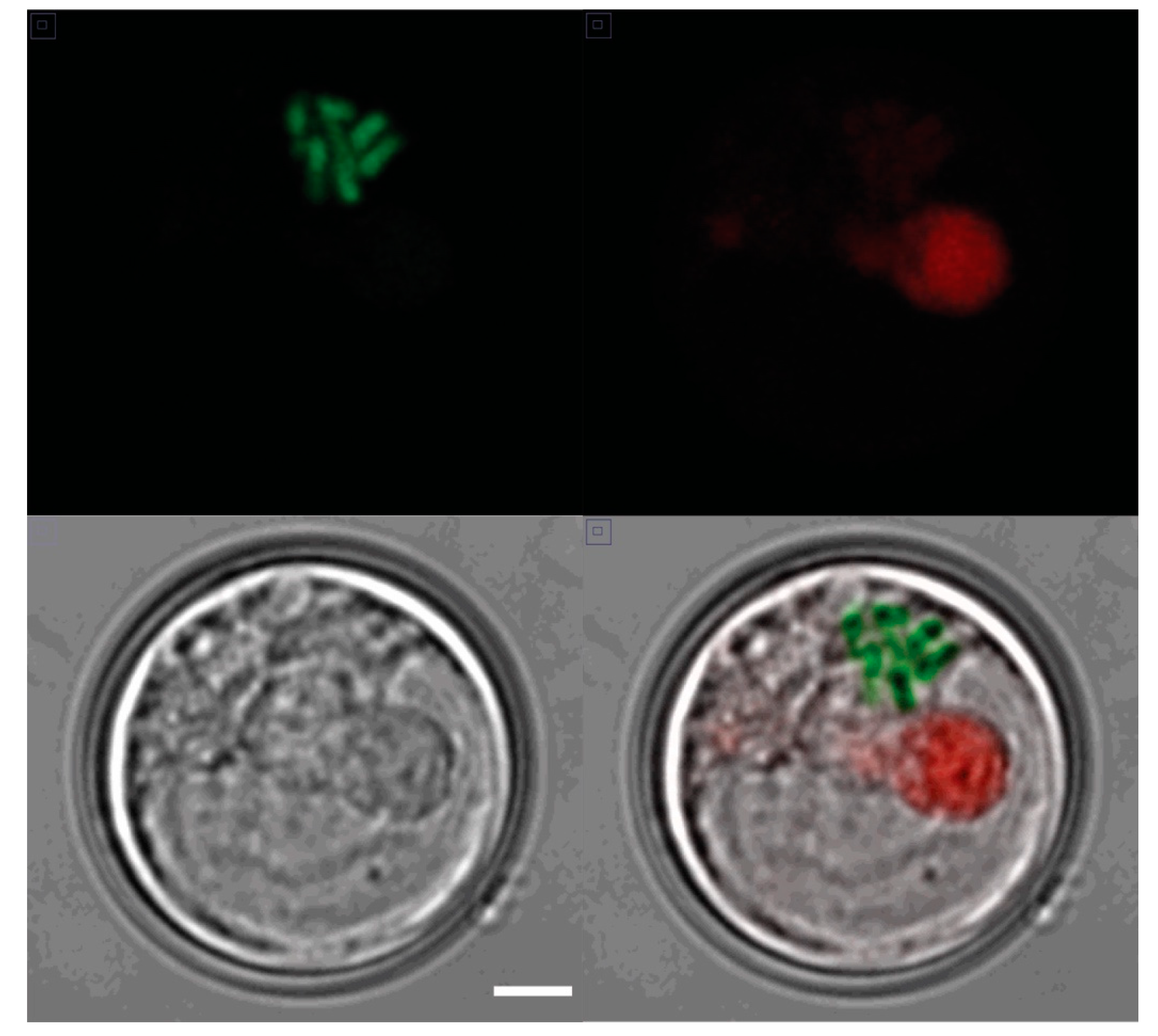
S. kawagutii has a typical dinoflagellate chromosome structure. Cells at all times have visibly condensed chromosomes (Figure 1) that appear superficially similar to mitotic chromosomes in other cells. This compact structure suggests that transcription is likely to be challenging, since more typical eukaryotic cells transcription rates decrease during mitosis when the chromatin is more condensed [26].
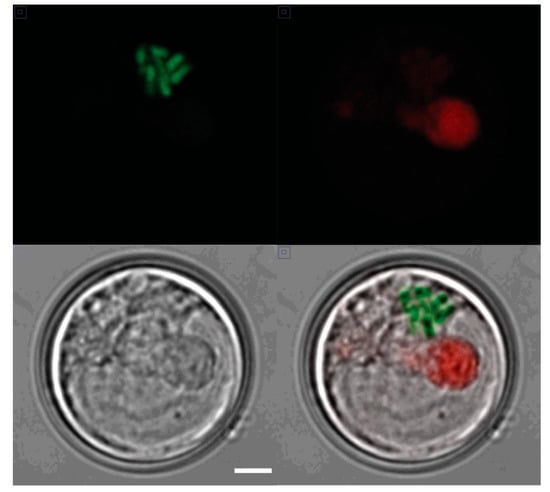
Figure 1.
Condensed chromosomes in an interphase S. kawagutii cell. A confocal image of a single cell taken in (upper left) the green channel (PI staining of chromosomes), (upper right) the red channel (natural chlorophyll fluorescence), and (lower left) a DIC (Nomarski) image (scale bar 1 µm for all panels). A merged image is shown in the lower right.
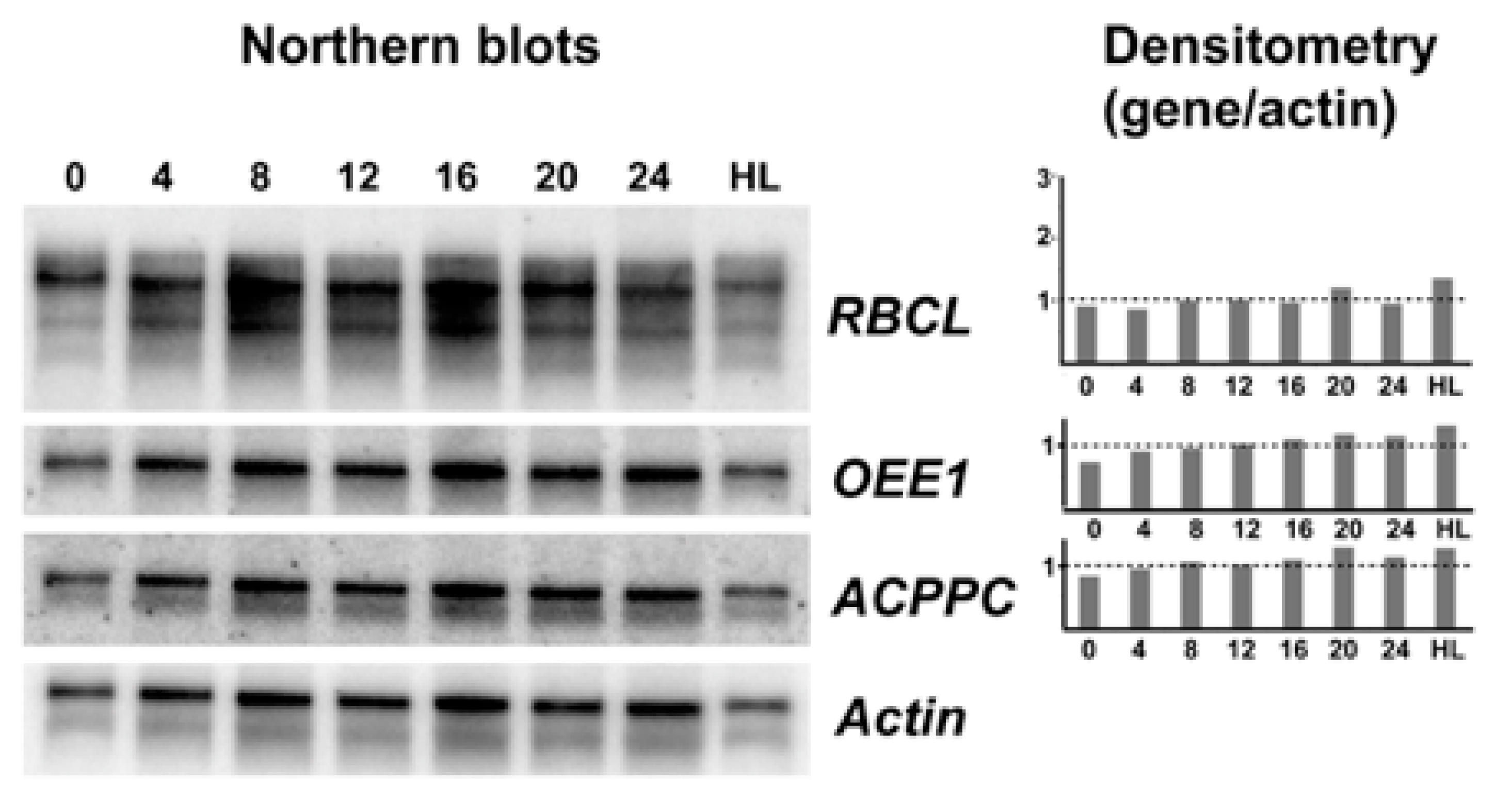
In a first attempt to identify light responsive genes in S. kawagutii, examples were selected from the literature. We selected oxygen evolving enzyme (OEE1) where transcript levels changed in abundance by 2.5-fold between LD 0 and LD 12 [11], the large rubisco subunit rbcL where transcript levels were three-fold higher at LD 12 than LD 0 [12], and the thylakoid chlorophyll a-chlorophyll c2-peridinin-protein-complex (acpPC) where transcript levels were roughly three-fold higher in dark phase than in light phase [13]. Actin was chosen as a reference because it is not regulated by light in Lingulodinium [18] or as shown here by RNA-Seq in S. kawagutii. We amplified probes for these sequences from S. kawagutii, and used the probes to asses transcript levels at four-hour intervals over an LD cycle, as well as a culture left under high light conditions. In no case were different transcript levels observed (Figure 2). We conclude there is no support for the hypothesis that transcription of these three genes responds to light.
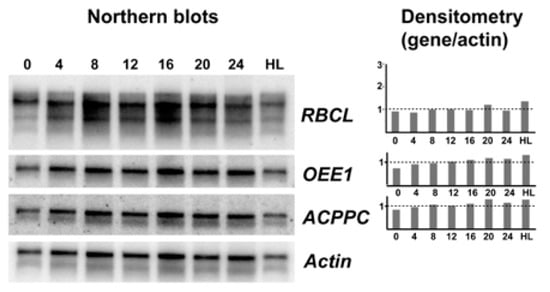
Figure 2.
Northern blot analysis of three potentially light regulated transcripts. A representative sample of northern blots (n = 4) using either an rbcL, an oee1, an acppc, or an Actin cDNA as a probe. RNA was prepared from samples taken every four hours from cells grown under a normal 12:12 LD cycle as well as from cells grown under high light (note that LD 0 and LD 24 should be identical). At right, densitometric scans for the top three probes are shown relative to the Actin signal.
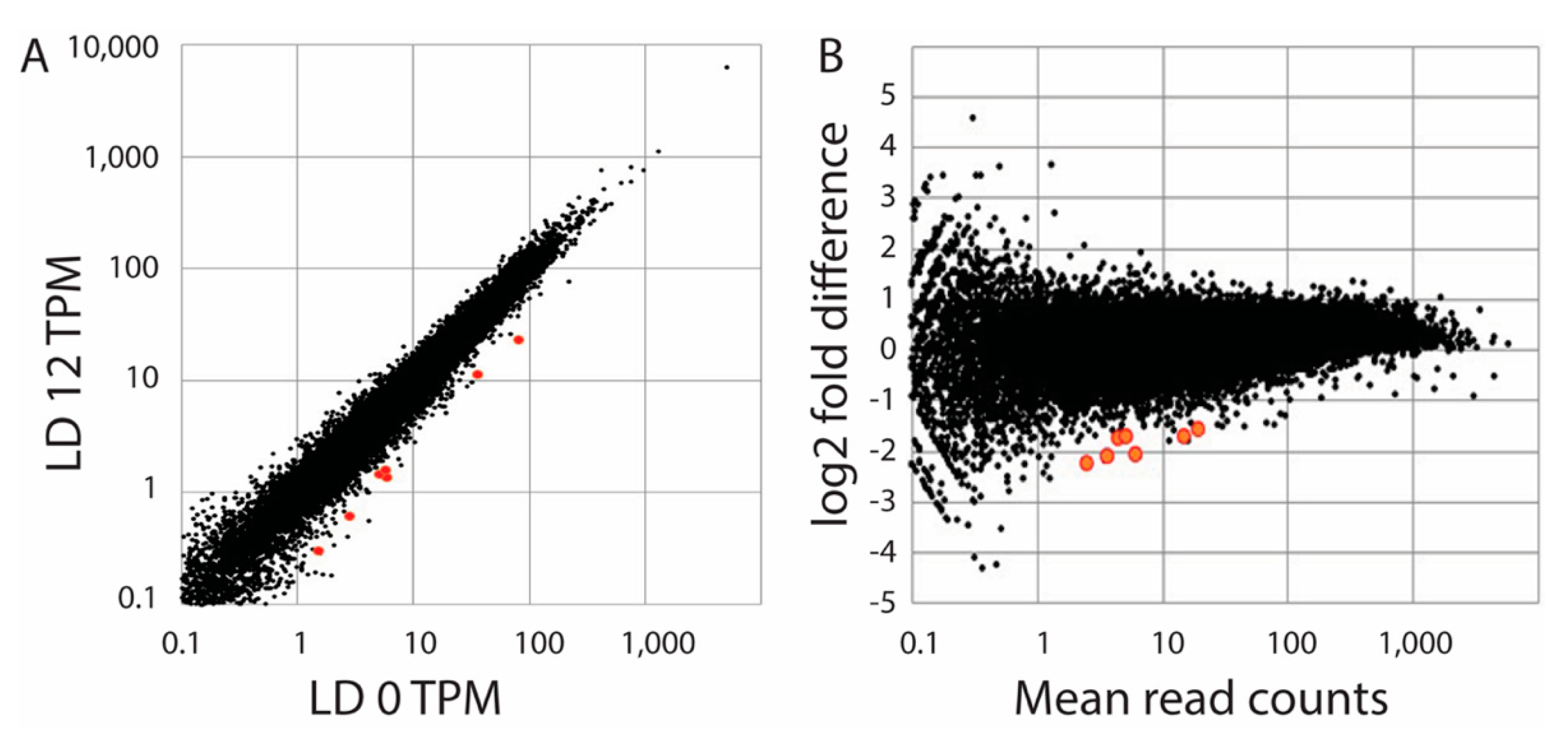
As a second attempt to identify light responsive genes, we prepared RNA samples in triplicate from S. kawagutii at LD 0 (dawn) and LD 12 (dusk). We reasoned that any light responsive genes would accumulate during the light period, and these would thus have higher levels at the end of the light phase. We compared read counts using the DESeq with a Benjamini–Hochberg correction (FDR = 0.1) to determine significant changes. A total of 7 changes (0.01%) were observed, all with higher levels at LD 0 than at LD 12 (Figure 3). Since all seven were higher at LD 0, this suggested that if these were truly light-regulated genes they would be induced by darkness or inhibited by light. These seven sequences were identified by BLAST searches (Table 1), and none correspond to the three sequences tested by northern blots. When the stringency of statistical significance was increased by setting the FDR to 0.05, only one of these was observed to display a statistically significant change. When a Bonferroni correction was applied instead of the Benjamini–Hochberg correction, four genes showed significant changes with p < 0.05, and one with p < 0.01. We conclude the number of significant changes in transcript levels is very low.
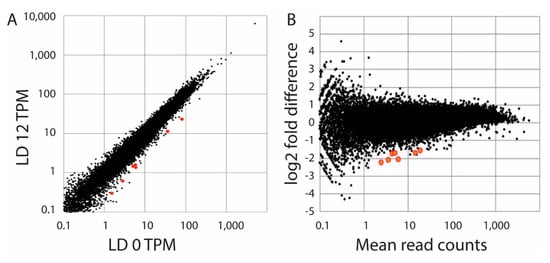
Figure 3.
Comparison of transcript levels at LD 0 and LD 12. (A) A plot of read counts (as TPM, or transcripts per million) as the average of three samples at LD 12 are compared with the average of samples at LD 0. (B) An MA plot (fold-difference as a function of mean read count) is shown for triplicate samples at each of the two times as determined by DESeq2. The 7 sequences determined to be significantly different (p-adjust < 0.05; FDR = 0.1) are shown in red in both plots and are higher at LD 0 than at LD 12.

Table 1.
Best BLAST hit for the seven potentially light regulated genes identified (False Discovery Rate FDR = 0.1).
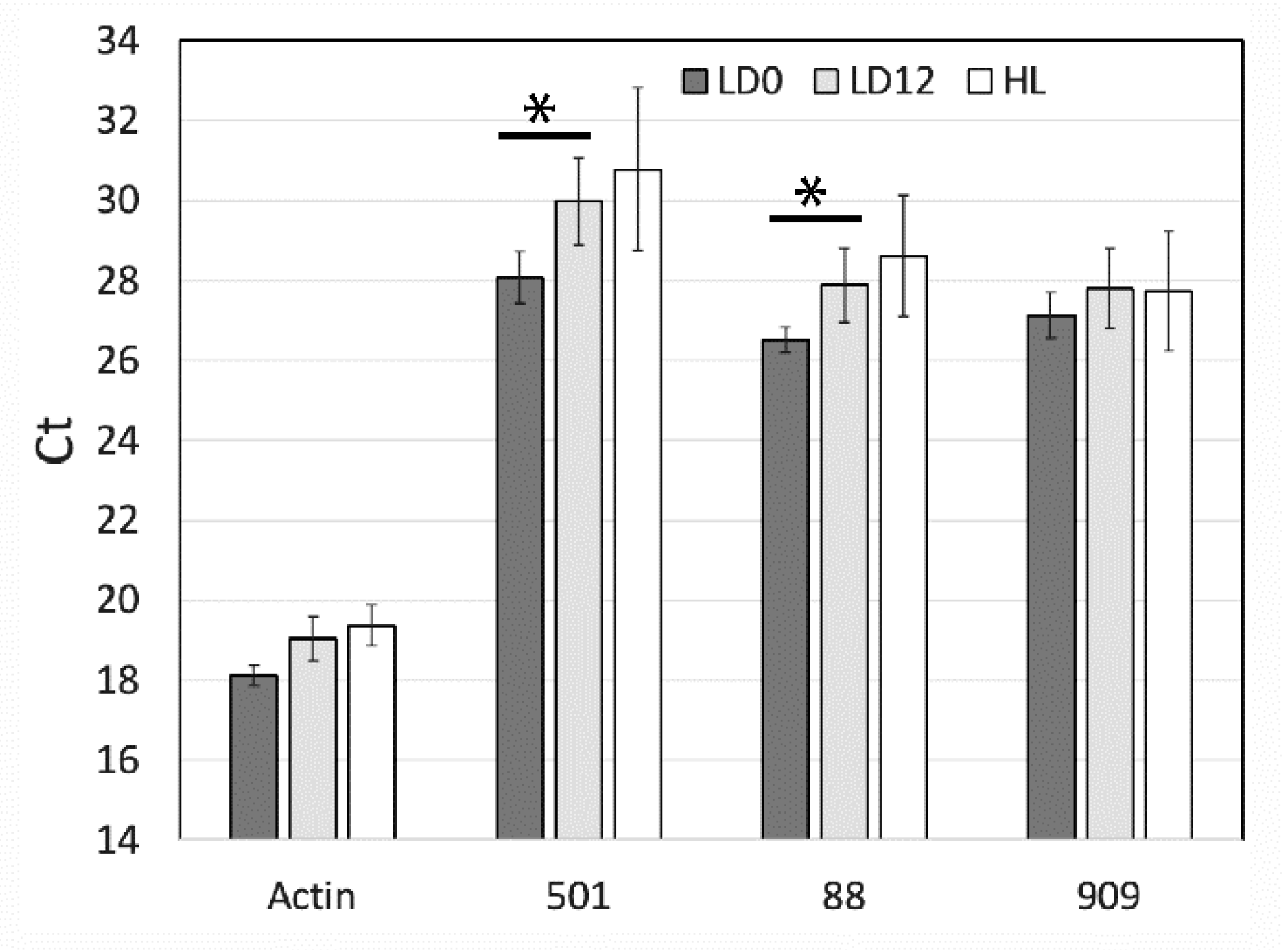
To validate the differential expression of the seven genes detected by RNA-Seq, we performed qPCR to assess the relative levels of three randomly selected genes (SymkaALLUN13501, SymkaALLUN19088, and SymkaALLUN64909). Assays were performed in triplicate for each of three biological replicates, and only two of these (ALLUN13501 and ALLUN19088) showed a significant difference between the two times (Figure 4). Since lower Ct values reflect higher transcript levels (i.e., transcript levels for these two genes are slightly higher at LD 0, as also found by RNA-Seq), we conclude that at least some of the seven genes with different levels as measured by RNA-Seq may reflect real differences in transcript levels. We note, however, that the fold difference appears smaller than that predicted by RNA-Seq.
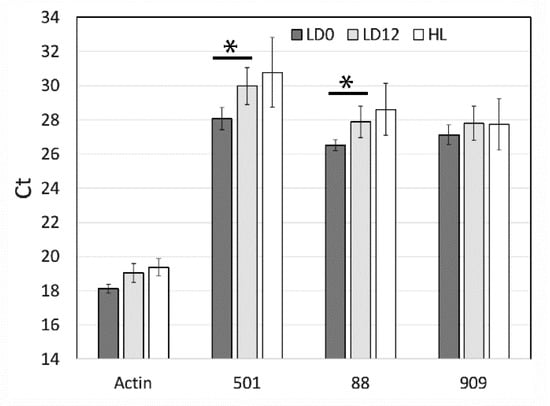
Figure 4.
qPCR analysis of 3 selected light-regulated genes from RNA sequencing analysis. Ct values were obtained for three RNA-Seq predicted regulated genes (501, SymkaALLUN13501; 88, SymkaALLUN19088; 909, SymkaALLUN64909) as well as Actin as a control for the amount of cDNA. Triplicate samples from each of three biological replicates were averaged for LD 0 (samples were in the dark for 12 h), LD 12 (samples were in the light for 12 h), and for samples kept under constant high light for 24 h. Comparisons marked with * are significant at p < 0.01 using student’s t-test, respectively.
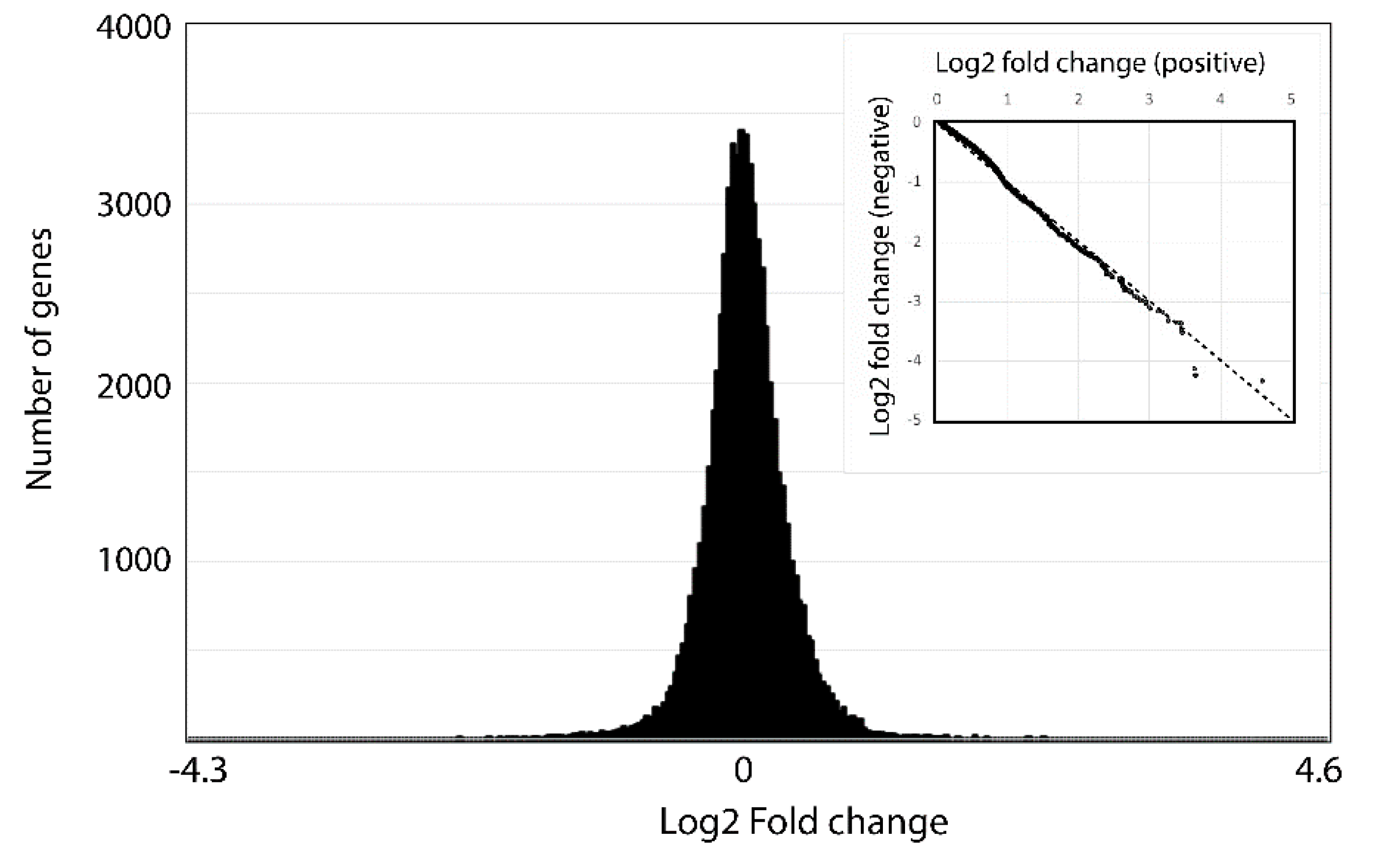
Finally, to gain a global picture of the different fold changes detected, significant or not, we plotted the number of times different fold changes were observed as a function of the fold change (Figure 5). This analysis reveals a normal distribution of fold changes within the data set. To test the symmetry of the bell curve, positive fold changes were plotted as a function of negative fold changes (Figure 5 inset). The resulting curve is essentially a straight line with a slope of −1. The few exceptions to the linear relationship do not correspond to the genes classified as significant by DESeq. We conclude there is no overall bias for either positive or negative changes in transcript abundance between the two times examined.
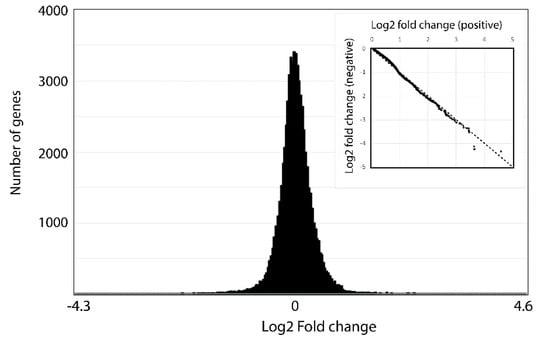
Figure 5.
Fold changes are equally distributed. A histogram showing of the number of times a given log2 fold change is found in the data shows a normal distribution. The inset, showing a plot of the positive vs. negative log2 fold changes, is essentially an unbiased straight line indicating few genes are differentially expressed at one of the two times.
4. Discussion
In many of the studies reporting differential gene expression as a result of light, only a single method was used to measure transcript abundance. For example, qPCR, northern blots, microarrays, or RNA-Seq have been used in individual studies but were not, with few exceptions, combined in the same study. One notable exception in Lingulodinium first used RNA-Seq to identify DEG and then verified a random selection of these using northern blots. Since northern blots failed to confirm the RNA-Seq-derived DEG, it was concluded all were likely to be false positives. This underscores the importance of validating high-throughput approaches, and suggests that it would be beneficial when several methods are combined to test for DEG.
The RNA-Seq experiments reported here used DESeq2 to identify DEG, with the threshold for significance determined by a false discovery rate (FDR) of 0.1. The FDR method, developed by Benjamini and Hochberg, uses a statistical method to restrain the number of false positives to a fixed percentage of the total positives, and thus provides increased confidence that significant changes are in large datasets are likely true positives [27]. The FDR can be thought of as a method for using lower p-values to determine significance when datasets become larger. For example, using a dataset with 100 values, of which 5 are really significant, a p-value of 0.05 would mean there are 5 false positives detected among 95 non-significant values, thus corresponding to a false discovery rate of 50% among the ten positives. The false discovery rate climbs when either the number of really significant values decreases or the number of non-significant values increases, the latter being a direct consequence of using large datasets such as those produced by RNA-Seq. In our study, when the FDR was fixed at 0.1, seven genes with significant difference were found. However, the number of significant differences decreases to 1 using a more stringent FDR of 0.05. It has been shown that the number of false positives recovered is considerably higher than the number expected [28]. This would agree with our observation that only two thirds of the DEG tested by qPCR were also found to show significant differences. Thus, in the light of the small number of significant changes found in our RNA-Seq experiment, we suggest that there are likely no real significant changes in transcript levels brought about by the changes in light intensity in our experiment. This would then agree with the lack of significant changes in transcript abundance over the course of the daily LD cycle using the dinoflagellate L. polyedra [18].
Our RNA-Seq experiment indicating there are no light induced transcripts has methodological differences with other reports in the literature suggesting the opposite. For example, an RNA-Seq study with Symbiodinium microadriaticum that showed 67 DEG when day and night were compared using DESeq with a false discovery rate (FDR) of 0.1 [16] used single samples rather than triplicate samples (Table 2). When we perform DESeq with an FDR = 0.1 using only one of three samples for each of the two time points, DESeq recovers 55 DEG instead of the seven DEG found when triplicate samples are used. Thus, in the S. microadriaticum study, insufficient replication may have exaggerated the number of light responsive transcripts. Another RNA-Seq study using Symbiodinium strain SSB01 looked at the number of DEG 24 h after a transfer from light to dark [17]. Here, 1334 DEG were found using cells grown phototrophically and 1739 DEG when cells were grown mixotrophically. These studies used duplicates (phototrophic growth) or triplicates (mixotrophic growth), but, instead of an FDR = 0.1, the cut-off values for significance were p < 0.05 and a fold change >1.5-fold. In our experiment, using triplicate samples with a similar cut-off value would result in 789 DEG instead of seven. Thus, the Symbiodinium SSB01 study had an exaggerated number of DEG because the cut-off criteria were not as stringent as using an FDR of 0.1. Both replicated samples and appropriate statistical analysis of significance are required for correct interpretation of RNA-Seq data.

Table 2.
Differentially expressed genes (DEG) identified in different dinoflagellates after different treatments.
It is important to emphasize that we do not propose dinoflagellates are incapable of transcriptional responses. However, in view of the experiments reported here, we believe it may be worthwhile re-examining the transcriptional response of dinoflagellates to stimuli other than light. A logical prediction from the permanently condensed chromatin that characterises dinoflagellate chromosomes is that transcriptional regulation is likely to be more difficult than in other cells. We thus suggest it may be important to verify transcriptional responses observed by a single method by using a complementary technique. Certainly, the finding of a true transcriptional response will be an important part in dissecting the molecular machinery that underpins this process in the dinoflagellates.
Author Contributions
Conceptualization, B.Z. and D.M.; methodology, B.Z. and S.D.-B.; software, B.S.; validation, B.Z.; writing—original draft preparation, B.Z. and D.M.; writing—review and editing, B.Z., B.S., S.D.-B. and D.M.; supervision, D.M.; project administration, D.M.; funding acquisition, D.M.
Funding
The financial support of the Canadian National Science and Engineering Research Council is gratefully acknowledged (grant number 171382-03 to D.M.).
Conflicts of Interest
The authors declare no conflict of interest.
References
- Spector, D. Dinoflagellate nuclei; Academic Press: New York, NY, USA, 1984; pp. 107–147. [Google Scholar]
- Roy, S.; Morse, D. Transcription and maturation of mrna in dinoflagellates. Microorganisms 2013, 1, 71–99. [Google Scholar] [CrossRef] [PubMed]
- Soyer, M.O.; Haapala, O.K. Electron microscopy of rna in dinoflagellate chromosomes. Histochemistry 1974, 42, 239–246. [Google Scholar] [CrossRef] [PubMed]
- Bodansky, S.; Mintz, L.B.; Holmes, D.S. The mesokaryote gyrodinium cohnii lacks nucleosomes. Biochem. Biophys. Res. Commun. 1979, 88, 1329–1336. [Google Scholar] [CrossRef]
- Roy, S.; Morse, D. A full suite of histone and histone modifying genes are transcribed in the dinoflagellate lingulodinium. PLoS ONE 2012, 7, e34340. [Google Scholar] [CrossRef] [PubMed]
- Gornik, S.G.; Ford, K.L.; Mulhern, T.D.; Bacic, A.; McFadden, G.I.; Waller, R.F. Loss of nucleosomal DNA condensation coincides with appearance of a novel nuclear protein in dinoflagellates. Curr. Biol. 2012, 22, 2303–2312. [Google Scholar] [CrossRef] [PubMed]
- Beauchemin, M.; Morse, D. A proteomic portrait of dinoflagellate chromatin reveals abundant RNA-binding proteins. Chromosoma 2017, 127, 1–15. [Google Scholar] [CrossRef]
- Levi-Setti, R.; Gavrilov, K.L.; Rizzo, P.J. Divalent cation distribution in dinoflagellate chromosomes imaged by high-resolution ion probe mass spectrometry. Eur. J. Cell Biol. 2008, 87, 963–976. [Google Scholar] [CrossRef]
- Wong, J.T.; New, D.C.; Wong, J.C.; Hung, V.K. Histone-like proteins of the dinoflagellate crypthecodinium cohnii have homologies to bacterial DNA-binding proteins. Eukaryot Cell 2003, 2, 646–650. [Google Scholar] [CrossRef]
- Shi, X.; Li, L.; Guo, C.; Lin, X.; Li, M.; Lin, S. Rhodopsin gene expression regulated by the light dark cycle, light spectrum and light intensity in the dinoflagellate prorocentrum. Front. Microbiol. 2015, 6, 555. [Google Scholar] [CrossRef]
- Sorek, M.; Yacobi, Y.Z.; Roopin, M.; Berman-Frank, I.; Levy, O. Photosynthetic circadian rhythmicity patterns of symbiodinium, [corrected} the coral endosymbiotic algae. Proc. R Soc. B 2013, 280, 20122942. [Google Scholar] [CrossRef]
- Mayfield, A.B.; Hsiao, Y.Y.; Chen, H.K.; Chen, C.S. Rubisco expression in the dinoflagellate symbiodinium sp. Is influenced by both photoperiod and endosymbiotic lifestyle. Mar. Biotechnol. 2014, 16, 371–384. [Google Scholar] [CrossRef]
- Boldt, L.; Yellowlees, D.; Leggat, W. Measuring symbiodinium sp. Gene expression patterns with quantitative real-time pcr. In Proceedings of the 11th International Coral Reef Symposium (ICRS), Ft. Lauderdale, FL, USA, 7–11 July 2008; pp. 118–122. [Google Scholar]
- Okamoto, O.K.; Hastings, J.W. Novel dinoflagellate circadian-clock genes identified through microarray analysis of a phase shifted clock. J. Phycol. 2003, 39, 1–9. [Google Scholar] [CrossRef]
- van Dolah, F.M.; Lidie, K.B.; Morey, J.S.; Brunelle, S.A.; Ryan, J.C.; Monroe, E.A.; Haynes, B.L. Microarray analysis of diurnal and circadian regulated genes in the florida red-tide dinoflagellate karenia brevis (dinophyceae). J. Phycol. 2007, 43, 741–752. [Google Scholar] [CrossRef]
- Baumgarten, S.; Bayer, T.; Aranda, M.; Liew, Y.J.; Carr, A.; Micklem, G.; Voolstra, C.R. Integrating microrna and mrna expression profiling in symbiodinium microadriaticum, a dinoflagellate symbiont of reef-building corals. BMC Genom. 2013, 14, 704. [Google Scholar] [CrossRef]
- Xiang, T.; Nelson, W.; Rodriguez, J.; Tolleter, D.; Grossman, A.R. Symbiodinium transcriptome and global responses of cells to immediate changes in light intensity when grown under autotrophic or mixotrophic conditions. Plant. J. 2015, 82, 67–80. [Google Scholar] [CrossRef] [PubMed]
- Roy, S.; Beauchemin, M.; Dagenais-Bellefeuille, S.; Letourneau, L.; Cappadocia, M.; Morse, D. The lingulodinium circadian system lacks rhythmic changes in transcript abundance. BMC Biol. 2014, 12, 107. [Google Scholar] [CrossRef]
- Lin, S.; Cheng, S.; Song, B.; Zhong, X.; Lin, X.; Li, W.; Li, L.; Zhang, Y.; Zhang, H.; Ji, Z.; et al. The symbiodinium kawagutii genome illuminates dinoflagellate gene expression and coral symbiosis. Science 2015, 350, 691–694. [Google Scholar] [CrossRef] [PubMed]
- Guillard, R.R.L.; Ryther, J.H. Studies on marine planktonic diatoms: Cyclotella nana hufstedt and denotula confervacea (cleve) gran. Can. J. Microbiol. 1962, 8, 229–239. [Google Scholar] [CrossRef] [PubMed]
- LaJeunesse, T.C.; Parkinson, J.E.; Gabrielson, P.W.; Jeong, H.J.; Reimer, J.D.; Voolstra, C.R.; Santos, S.R. Systematic revision of symbiodiniaceae highlights the antiquity and diversity of coral endosymbionts. Curr. Biol. 2018, 28, 2570–2580. [Google Scholar] [CrossRef]
- Schindelin, J.; Arganda-Carreras, I.; Frise, E.; Kaynig, V.; Longair, M.; Pietzsch, T.; Preibisch, S.; Rueden, C.; Saalfeld, S.; Schmid, B.; et al. Fiji: An open-source platform for biological-image analysis. Nat. Methods 2012, 9, 676–682. [Google Scholar] [CrossRef]
- Beauchemin, M.; Roy, S.; Daoust, P.; Dagenais-Bellefeuille, S.; Bertomeu, T.; Letourneau, L.; Lang, B.F.; Morse, D. Dinoflagellate tandem array gene transcripts are highly conserved and not polycistronic. Proc. Natl. Acad. Sci. USA 2012, 109, 15793–15798. [Google Scholar] [CrossRef] [PubMed]
- Patro, R.; Duggal, G.; Love, M.I.; Irizarry, R.A.; Kingsford, C. Salmon provides fast and bias-aware quantification of transcript expression. Nat. Methods 2017, 14, 417–419. [Google Scholar] [CrossRef] [PubMed]
- Anders, S.; Huber, W. Differential expression analysis for sequence count data. Genome Biol. 2010, 11, R106. [Google Scholar] [CrossRef] [PubMed]
- Palozola, K.C.; Donahue, G.; Liu, H.; Grant, G.R.; Becker, J.S.; Cote, A.; Yu, H.; Raj, A.; Zaret, K.S. Mitotic transcription and waves of gene reactivation during mitotic exit. Science 2017, 358, 119–122. [Google Scholar] [CrossRef] [PubMed]
- Benjamini, Y.; Hochberg, Y. Controlling the fasle discovery rate: A practical and powerful approach to multiple testing. J. R. Statist. Soc. B 1995, 57, 289–300. [Google Scholar]
- Rocke, D.M.; Ruan, L.; Zhang, Y.; Gossett, J.J.; Durbin-Johnson, B.; Aviran, S. Excess false positive rates in methods for differential gene expression analysis using rna-seq data. Biorxiv 2015. [Google Scholar] [CrossRef]
- Barshis, D.J.; Ladner, J.T.; Oliver, T.A.; Palumbi, S.R. Lineage-specific transcriptional profiles of Symbiodinium spp. Unaltered by heat stress in a coral host. Mol. Biol. Evol. 2014, 31, 1343–1352. [Google Scholar] [CrossRef]
- Gierz, S.L.; Foret, S.; Leggat, W. Transcriptomic analysis of thermally stressed Symbiodinium reveals differential expression of stress and metabolism genes. Front. Plant sci. 2017, 8, 271. [Google Scholar] [CrossRef]
- Roy, S.; Letourneau, L.; Morse, D. Cold-induced cysts of the photosynthetic dinoflagellate Lingulodinium polyedrum have an arrested circadian bioluminescence rhythm and lower levels of protein phosphorylation. Plant Physiol. 2014, 164, 966–977. [Google Scholar] [CrossRef]
- Cooper, J.T.; Sinclair, G.A.; Wawrik, B. Transcriptome analysis of Scrippsiella trochoidea CCMP 3099 reveals physiological changes related to nitrate depletion. Front Microbiol. 2016, 7, 639. [Google Scholar] [CrossRef]
- Moustafa, A.; Evans, A.N.; Kulis, D.M.; Hackett, J.D.; Erdner, D.L.; Anderson, D.M.; Bhattacharya, D. Transcriptome profiling of a toxic dinoflagellate reveals a gene-rich protist and a potential impact on gene expression due to bacterial presence. PLoS ONE 2010, 5, e9688. [Google Scholar] [CrossRef] [PubMed]
- Lowe, C.D.; Mello, L.V.; Samatar, N.; Martin, L.E.; Montagnes, D.J.; Watts, P.C. The transcriptome of the novel dinoflagellate Oxyrrhis marina (alveolata: Dinophyceae): Response to salinity examined by 454 sequencing. BMC Genom. 2011, 12, 519. [Google Scholar] [CrossRef] [PubMed]
- Morey, J.S.; Monroe, E.A.; Kinney, A.L.; Beal, M.; Johnson, J.G.; Hitchcock, G.L.; van Dolah, F.M. Transcriptomic response of the red tide dinoflagellate, Karenia brevis, to nitrogen and phosphorus depletion and addition. BMC Genom. 2011, 12, 346. [Google Scholar] [CrossRef] [PubMed]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).