Isolation of Bacillus amyloliquefaciens D39 and Identification of Its Antimicrobial Proteins Active Against Chestnut Blight
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Strains, Soil Samples, Chestnut Seedlings, and Culture Medium
2.2. Isolation and Identification of Antagonistic Bacteria
2.2.1. Isolation, Purification, and Preservation of Bacillus spp.
2.2.2. Screening of Antagonistic Bacteria
2.2.3. Identification of Antagonistic Bacteria
2.3. Analysis of the Antimicrobial Capability of Strain D39
2.3.1. Subculture Cultivation
2.3.2. Growth Curve and Antimicrobial Capability
2.3.3. Antimicrobial Spectrum
2.3.4. PCR detection of Biocontrol-Related Genes in Strain D39
2.3.5. Pot Experiments
2.4. Extraction and Purification of Antimicrobial Substances
2.4.1. Ammonium Sulfate Precipitation for Preliminary Extraction of Antimicrobial Substances
2.4.2. Separation and Purification of Antimicrobial Proteins and Determination of Protein
2.4.3. RPLC-MS and Data Analysis
2.4.4. Protein Structure Prediction
2.5. Statistical Analysis
3. Results
3.1. Isolation of Antagonistic Strain D39
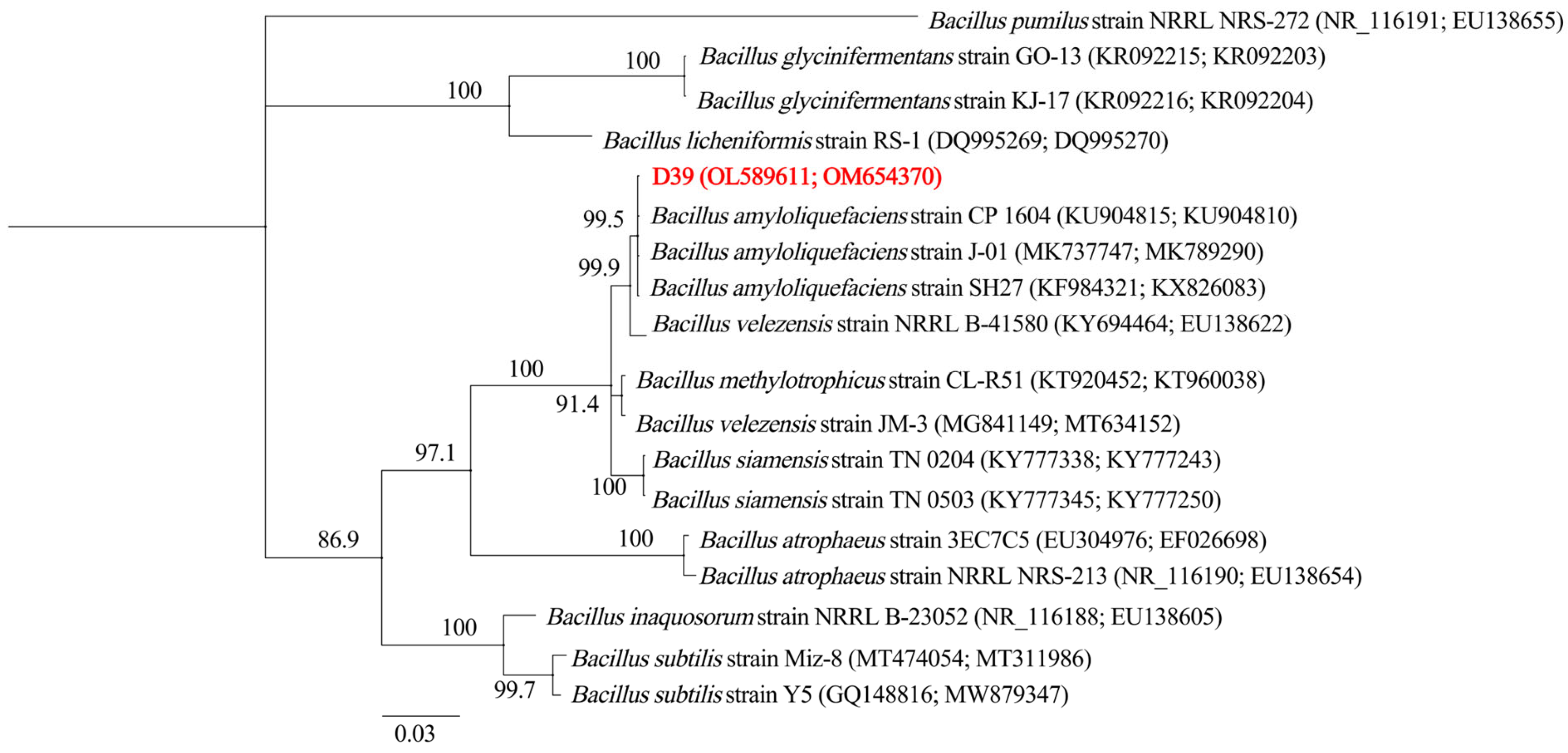
3.2. Identification of Strain D39 as B. amyloliquefaciens
3.3. Antimicrobial Activity of Strain D39
3.3.1. Subculturing
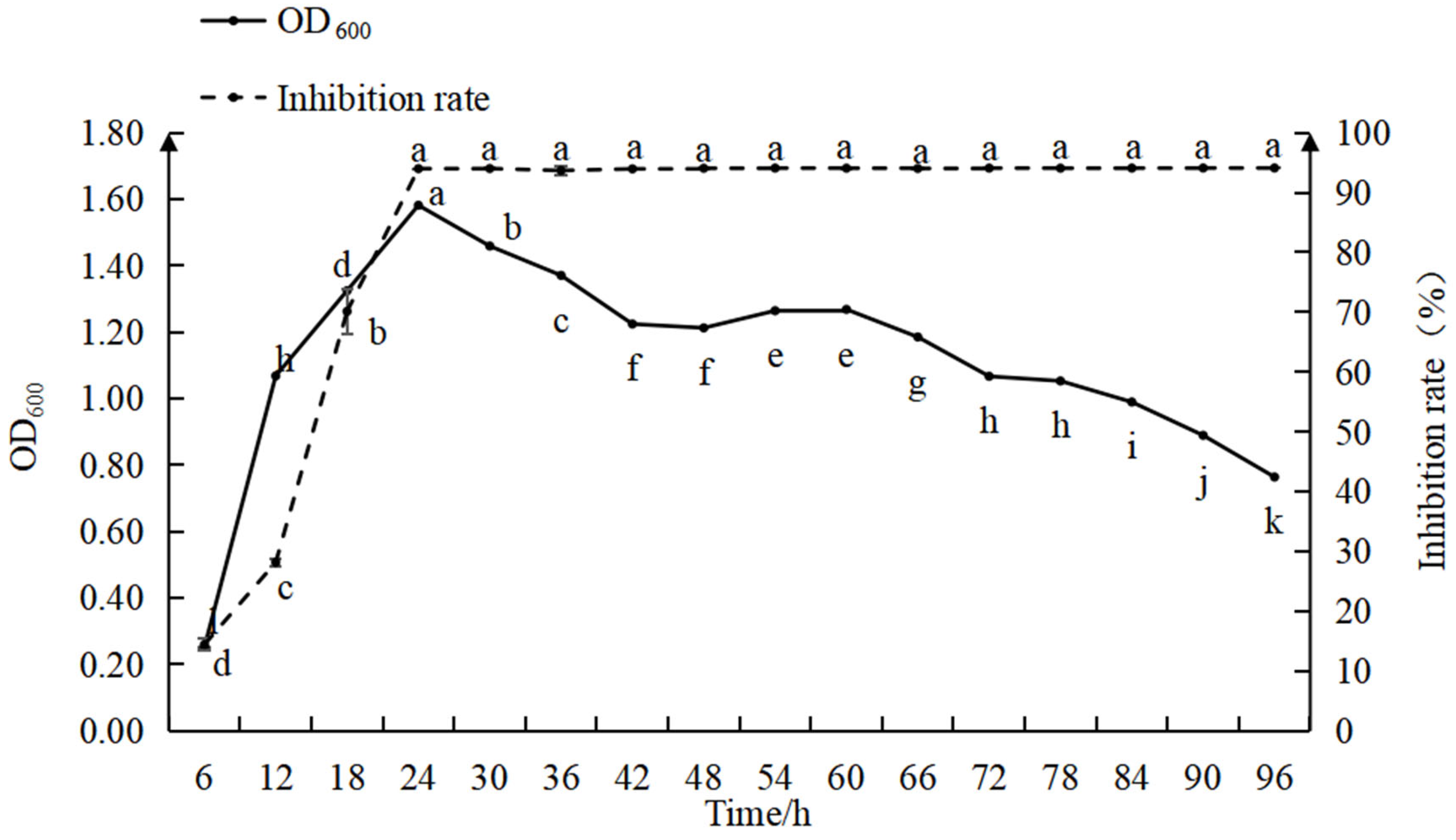
3.3.2. Growth Curve and Antimicrobial Activity
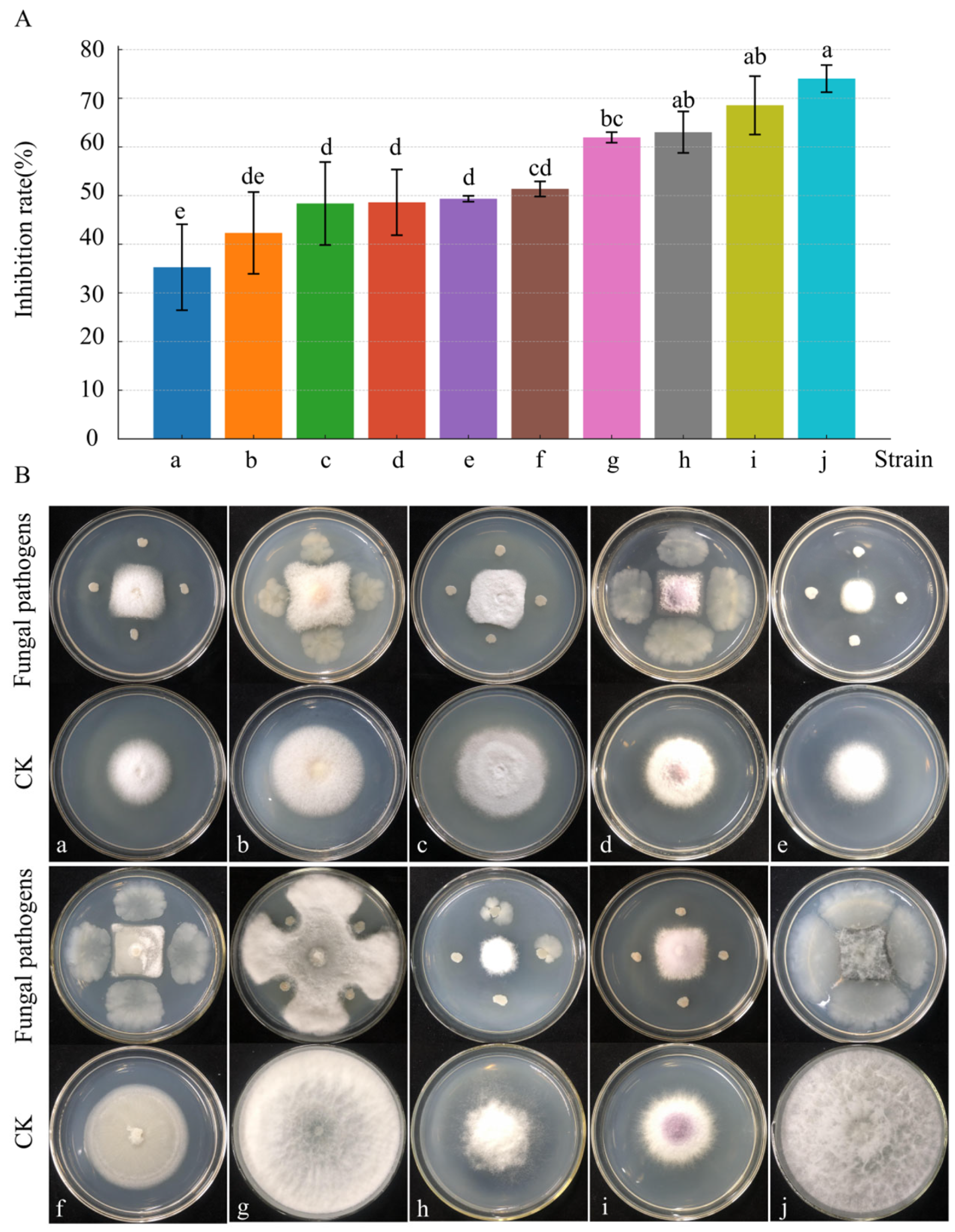
3.3.3. Broad-Spectrum Antifungal Activity of Strain D39
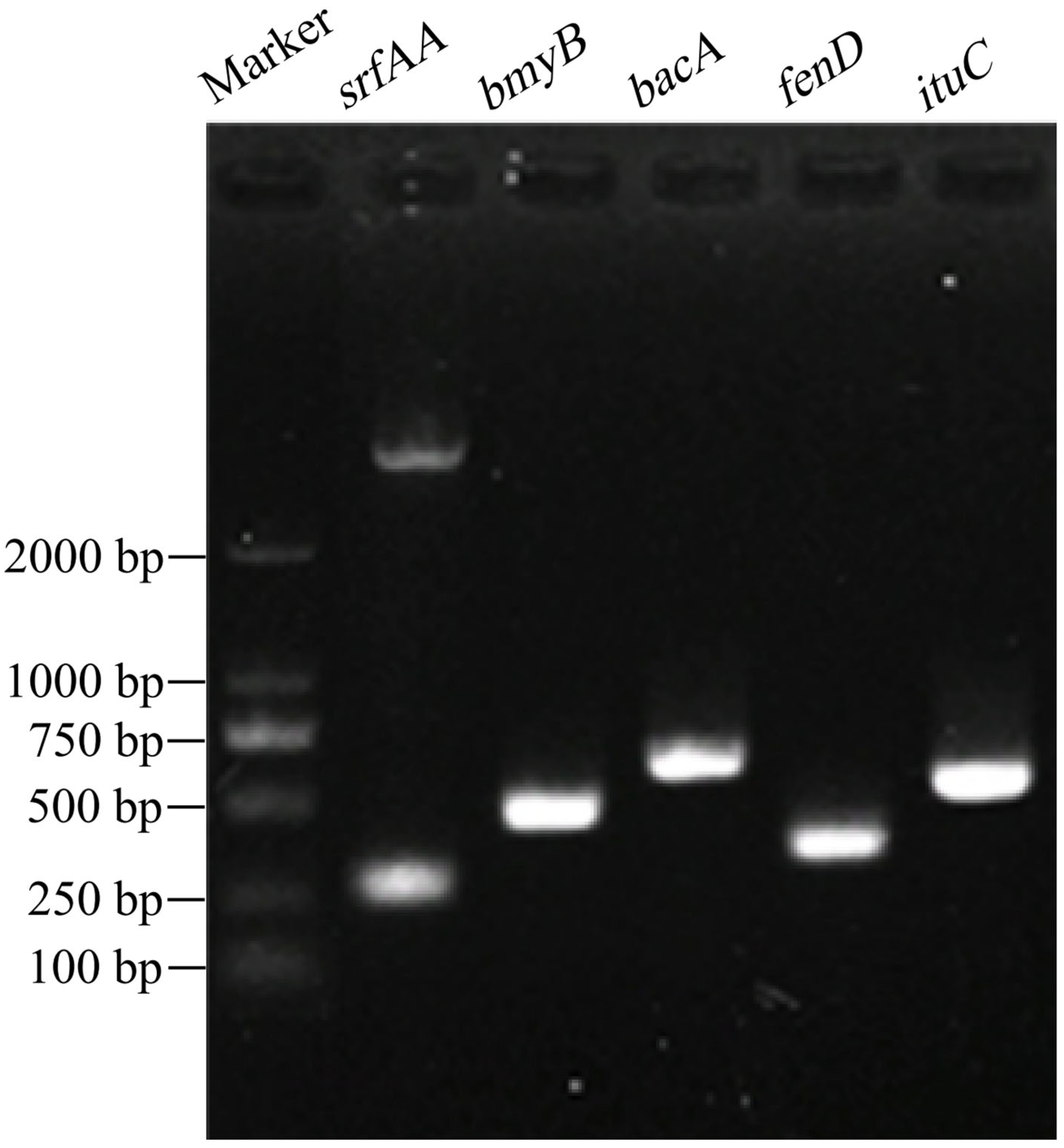
3.3.4. Detection of Functional Genes
3.4. Pot Experiment for Efficacy Evaluation
3.5. Characteristics of Antimicrobial Substances Produced by B. amyloliquefaciens D39
3.5.1. Effect of Different Ammonium Sulfate Saturation Levels on Protein Precipitation
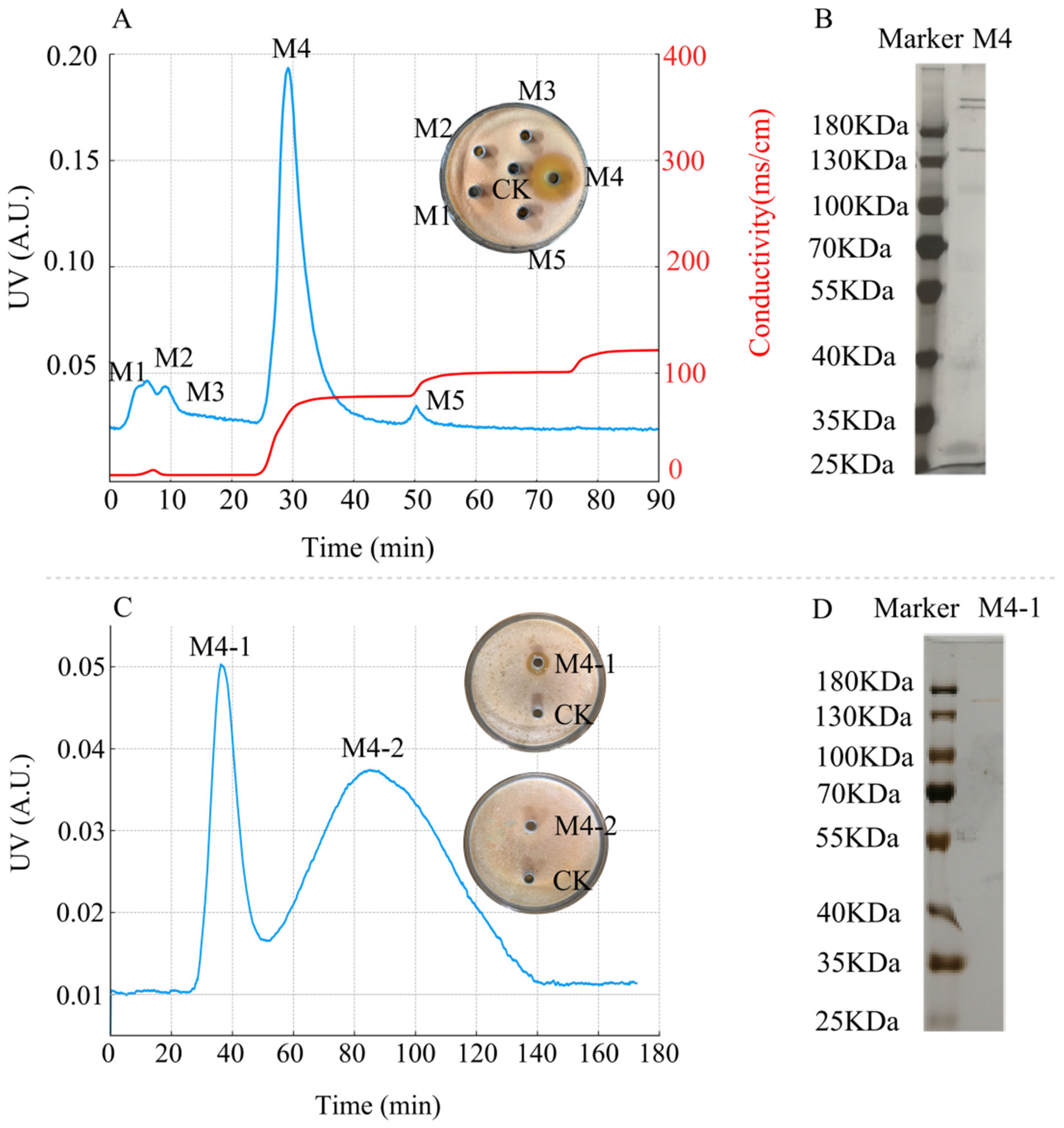
3.5.2. Results of DEAE Sepharose Fast Flow and Sephadex G-75 Chromatography
3.5.3. RPLC-MS Identification and Analysis Results
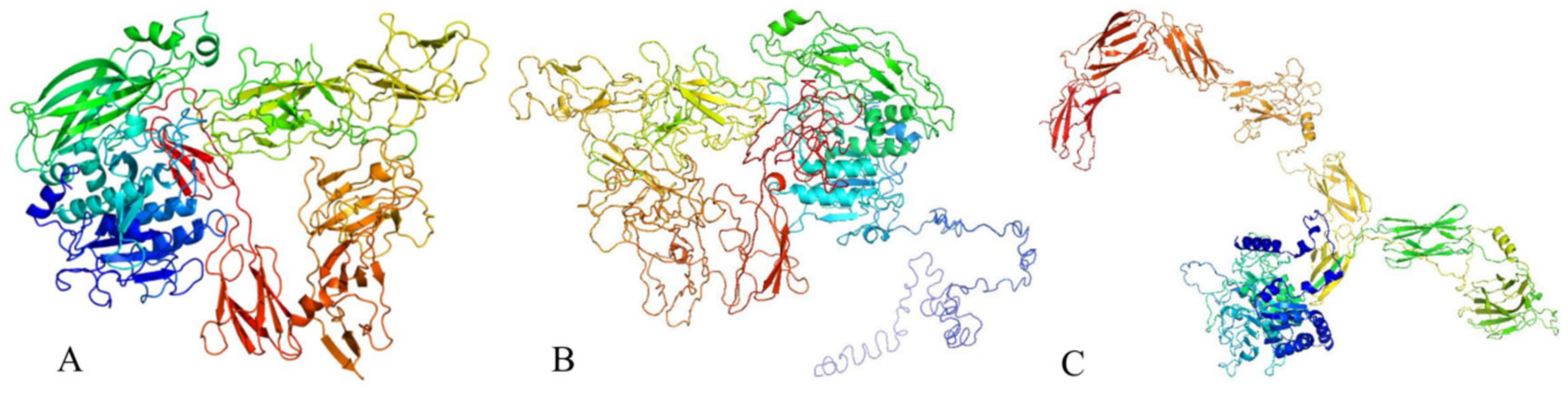
3.6. Protein Structure Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- He, L.; Min, Q.; Hong, C.; Zhang, Y. Features and socio-economic sustainability of traditional chestnut forestry landscape in china: A case of Kuancheng county, Hebei province. Land 2021, 10, 952. [Google Scholar] [CrossRef]
- Massantini, R.; Moscetti, R.; Frangipane, M.T. Evaluating progress of chestnut quality: A review of recent developments. Trends Food Sci. Technol. 2021, 113, 245–254. [Google Scholar] [CrossRef]
- Fernandes, P.; Colavolpe, M.B.; Serrazina, S.; Costa, R. European and American chestnuts: An overview of the main threats and control efforts. Front. Plant Sci. 2022, 13, 951844. [Google Scholar] [CrossRef] [PubMed]
- Anagnostakis, S.L. Chestnut blight: The classical problem of an introduced pathogen. Mycologia 1987, 79, 23–37. [Google Scholar] [CrossRef]
- Trapiello Vázquez, E.; Varela, G.; González, A. Chestnut blight control by agrochemicals in Castanea sativa under managed conditions. J. Plant Dis. Prot. 2015, 122, 120–124. [Google Scholar] [CrossRef]
- Cheradil, A.; Tarcali, G.; Csüllög, K.; Boukhili, M. Study of chemical control options against chestnut blight disease. Rev. Agric. Rural. Dev. 2022, 11, 20–25. [Google Scholar] [CrossRef]
- Naz, I.; Khan, R.A.A.; Masood, T.; Baig, A.; Siddique, I.; Haq, S. Biological control of root knot nematode, Meloidogyne incognita, in vitro, greenhouse and field in cucumber. Biol. Control 2021, 152, 104429. [Google Scholar] [CrossRef]
- Ayilara, M.; Adeleke, B.; Akinola, S.; Fayose, C.A.; Adeyemi, U.; Gbadegesin, L.; Omole, R.; Johnson, R.; Uthman, Q.; Babalola, O. Biopesticides as a promising alternative to synthetic pesticides: A case for microbial pesticides, phytopesticides, and nanobiopesticides. Front. Microbiol. 2023, 14, 1040901. [Google Scholar] [CrossRef]
- Hashem, A.H.; Abdelaziz, A.M.; Attia, M.S.; Abd-Elsalam, K.A. Biocontrol potential of endophytic fungi against postharvest grape pathogens. In Fungal Endophytes Volume II: Applications in Agroecosystems and Plant Protection; Abd-Elsalam, K.A., Hashem, A.H., Eds.; Springer Nature Singapore: Singapore, 2025; pp. 509–530. [Google Scholar]
- Abdelaziz, A.M.; Hashem, A.H.; El-Sayyad, G.S.; El-Wakil, D.A.; Selim, S.; Alkhalifah, D.H.M.; Attia, M.S. Biocontrol of soil borne diseases by plant growth promoting rhizobacteria. Trop. Plant Pathol. 2023, 48, 105–127. [Google Scholar] [CrossRef]
- Ahmad, F.; Tomada, S.; Poonsiri, T.; Baric, S. Molecular genetic variability of Cryphonectria hypovirus 1 associated with Cryphonectria parasitica in South Tyrol (Northern Italy). Front. Microbiol. 2024, 15, 1291542. [Google Scholar] [CrossRef]
- Coelho, V.; Nunes, L.; Gouveia, E. Short and long term efficacy and prevalence of Cryphonectria parasitica hypovirulent strains released as biocontrol agents of chestnut blight. Eur. J. Plant Pathol. 2021, 159, 769–781. [Google Scholar] [CrossRef]
- Robin, C.; Lanz, S.; Soutrenon, A.; Rigling, D. Dominance of natural over released biological control agents of the chestnut blight fungus Cryphonectria parasitica in south-eastern France is associated with fitness-related traits. Biol. Control 2010, 53, 55–61. [Google Scholar] [CrossRef]
- Diamandis, S. Management of chestnut blight in Greece using hypovirulence and silvicultural interventions. Forests 2018, 9, 492. [Google Scholar] [CrossRef]
- Double, M.; Jarosz, A.; Fulbright, D.; Baines, A.; Macdonald, W. Evaluation of two decades of Cryphonectria parasitica hypovirus introduction in an American chestnut stand in Wisconsin. Phytopathology 2018, 108, 702–710. [Google Scholar] [CrossRef]
- Double, M.; Nuss, D.; Rittenour, W.; Holaskova, I.; Short, D.; Kasson, M.; Macdonald, W. Long-term field study of transgenic hypovirulent strains of Cryphonectria parasitica in a forest setting. For. Pathol. 2017, 47, e12367. [Google Scholar] [CrossRef]
- Lawson, S.; Ebrahimi, A.; Mckenna, J. Differing responses to Cryphonectria parasitica at two Indiana locations. Forests 2021, 12, 794. [Google Scholar] [CrossRef]
- Rodríguez-Molina, M.D.C.; García-García, M.; Osuna, M.; Gouveia, E.; Serrano-Pérez, P. Various population structures of Cryphonectria parasitica in Cáceres (Spain) determine the feasibility of the biological control of chestnut blight with hypovirulent strains. Agronomy 2023, 13, 1208. [Google Scholar] [CrossRef]
- Peever, T.L.; Liu, Y.C.; Wang, K.; Hillman, B.I.; Foglia, R.; Milgroom, M.G. Incidence and diversity of double-stranded RNAs occurring in the chestnut blight fungus, Cryphonectria parasitica, in China and Japan. Phytopathology 1998, 88, 811–817. [Google Scholar] [CrossRef]
- Xiong, Q.; Zhang, L.; Waletich, J.; Zhang, L.; Zhang, C.; Xinyue, Z.; Qian, Y.; Zhang, Z.; Wang, Y.; Cheng, Q. Characterization of the papain-like protease p29 of the hypovirus CHV1-CN280 in its natural host fungus Cryphonectria parasitica and nonhost fungus Magnaporthe oryzae. Phytopathology 2018, 109, 736–747. [Google Scholar] [CrossRef]
- Liu, Y.C.; Milgroom, M.G. High diversity of vegetative compatibility types in Cryphonectria parasitica in Japan and China. Mycologia 2007, 99, 279–284. [Google Scholar] [CrossRef]
- Robin, C.; Anziani, C.; Cortesi, P. Relationship between biological control, incidence of hypovirulence, and diversity of vegetative compatibility types of Cryphonectria parasitica in France. Phytopathology 2000, 90, 730–737. [Google Scholar] [CrossRef] [PubMed]
- Yan, B.; Li, Z.; Huang, H.; Qin, L. Genetic diversity and population differentiation of chestnut blight fungus, Cryphonectria parasitica, in China as revealed by RAPD. Biochem. Genet. 2007, 45, 487–506. [Google Scholar] [CrossRef] [PubMed]
- Romon Ochoa, P.; Forster, J.; Chitty, R.; Gorton, C.; Lewis, A.; Eacock, A.; Kupper, Q.; Rigling, D.; Pérez-Sierra, A. Canker development and biocontrol potential of CHV-1 infected English isolates of Cryphonectria parasitica is dependent on the virus concentration and the compatibility of the fungal inoculums. Viruses 2022, 14, 2678. [Google Scholar] [CrossRef] [PubMed]
- Ke, X.; Wu, Z.; Liu, Y.; Liang, Y.; Du, M.; Li, Y. Isolation, antimicrobial effect and metabolite analysis of Bacillus amyloliquefaciens ZJLMBA1908 against citrus canker caused by Xanthomonas citri subsp. citri. Microorganisms 2023, 11, 2928. [Google Scholar] [CrossRef]
- Miljaković, D.; Marinković, J.; Balešević-Tubić, S. The significance of Bacillus spp. in disease suppression and growth promotion of field and vegetable crops. Microorganisms 2020, 8, 1037. [Google Scholar] [CrossRef]
- Karačić, V.; Miljaković, D.; Marinković, J.; Ignjatov, M.; Milošević, D.; Tamindzic, G.; Ivanović, M. Bacillus species: Excellent biocontrol agents against tomato diseases. Microorganisms 2024, 12, 457. [Google Scholar] [CrossRef]
- Harun-Or-Rashid, M.; Kim, H.; Yeom, S.; Yu, H.; Manir, M.M.; Moon, S.; Kang, Y.J.; Chung, Y.R. Bacillus velezensis YC7010 enhances plant defenses against brown planthopper through transcriptomic and metabolic changes in rice. Front. Plant Sci. 2018, 9, 1904. [Google Scholar] [CrossRef]
- Lahlali, R.; Ezrari, S.; Radouane, N.; Kenfaoui, J.; Esmaeel, Q.; El Hamss, H.; Belabess, Z.; Barka, E.A. Biological control of plant pathogens: A global perspective. Microorganisms 2022, 10, 596. [Google Scholar] [CrossRef]
- Luo, Y.; Cheng, Y.; Yi, J.; Zhang, Z.; Luo, Q.; Zhang, D.; Li, Y. Complete genome sequence of industrial biocontrol strain Paenibacillus polymyxa HY96-2 and further analysis of its biocontrol mechanism. Front. Microbiol. 2018, 9, 1520. [Google Scholar] [CrossRef]
- Dutilloy, E.; Arguelles Arias, A.; Richet, N.; Guise, J.; Duban, M.; Leclère, V.; Selim, S.; Jacques, P.; Jacquard, C.; Clément, C.; et al. Bacillus velezensis BE2 controls wheat and barley diseases by direct antagonism and induced systemic resistance. Appl. Microbiol. Biotechnol. 2024, 108, 64. [Google Scholar] [CrossRef]
- Kandil, E.K.; Nofel, M.M.; Abdelaziz, A.M.; Mansour, M.M.; Attia, M.S. Effectual role of plant growth-promoting fungi and fosthiazate in controlling tomato root-knot nematode infection: In vivo and in vitro studies. Physiol. Mol. Plant Pathol. 2024, 134, 102463. [Google Scholar] [CrossRef]
- Daigham, G.E.; Mahfouz, A.Y.; Abdelaziz, A.M.; Nofel, M.M.; Attia, M.S. Protective role of plant growth-promoting fungi Aspergillus chevalieri OP593083 and Aspergillus egyptiacus OP593080 as biocontrol approach against Alternaria leaf spot disease of Vicia faba plant. Biomass Convers. Biorefin. 2024, 14, 23073–23089. [Google Scholar] [CrossRef]
- Tianhui, Z.; Fanglian, L. Identification and control of Chinese chestnut blight in Sichuan province. J. Sichuan For. Sci. Technol. 2008, 1, 50–53. [Google Scholar] [CrossRef]
- Ma, W.; Zhu, T.; Han, S. Molecular detection of Cryphonectria parasitica based on internal transcribed spacer (its) sequences. Plant Prot. 2015, 41, 125–130. [Google Scholar] [CrossRef]
- Liu, C.; Yin, X.; Wang, Q.; Peng, Y.; Ma, Y.; Liu, P.; Shi, J. Antagonistic activities of volatiles produced by two Bacillus strains against Monilinia fructicola in peach fruit. J. Sci. Food Agric. 2018, 98, 5756–5763. [Google Scholar] [CrossRef]
- Yilmaz, M.; Soran, H.; Beyatli, Y. Antimicrobial activities of some Bacillus spp. Strains isolated from the soil. Microbiol. Res. 2006, 161, 127–131. [Google Scholar] [CrossRef]
- Fan, H.; Li, S.; Zeng, L.; He, P.; Xu, S.; Bai, T.; Huang, Y.; Guo, Z.; Zheng, S. Biological control of Fusarium oxysporum f. sp. cubense tropical race 4 using natively isolated Bacillus spp. YN0904 and YN1419. J. Fungi 2021, 7, 795. [Google Scholar] [CrossRef]
- Liu, H.; An, M.; Si, H.; Shan, Y.; Xu, C.; Hu, G.; Xie, Y.; Liu, D.; Li, S.; Qiu, R.; et al. Identification of cyclic dipeptides and a new compound (6-(5-hydroxy-6-methylheptyl)-5,6-dihydro-2h-pyran-2-one) produced by Streptomyces fungicidicus against Alternaria solani. Molecules 2022, 27, 5649. [Google Scholar] [CrossRef]
- Xiuzhu, D.; Miaoying, C. Common bacterial system identification manual; Science Press: Beijng, China, 2001; pp. 364–398. [Google Scholar]
- Sun, P.; Cui, J.; Jia, X.; Wang, W. Isolation and characterization of Bacillus amyloliquefaciens L-1 for biocontrol of pear ring rot. Hortic. Plant J. 2017, 3, 183–189. [Google Scholar] [CrossRef]
- Zhang, D.; Gao, F.; Jakovlić, I.; Zou, H.; Zhang, J.; Li, W.; Wang, G. Phylosuite: An integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Mol. Ecol. Resour. 2020, 20, 348–355. [Google Scholar] [CrossRef]
- Ronquist, F.; Teslenko, M.; van der Mark, P.; Ayres, D.L.; Darling, A.; Höhna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. Mrbayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef] [PubMed]
- Mora, I.; Cabrefiga, J.; Montesinos, E. Antimicrobial peptide genes in Bacillus strains from plant environments. Int. Microbiol. 2011, 14, 213–223. [Google Scholar] [CrossRef] [PubMed]
- Ma, Q.; Cong, Y.; Feng, L.; Liu, C.; Yang, W.; Xin, Y.; Chen, K. Effects of mixed culture fermentation of Bacillus amyloliquefaciens and Trichoderma longibrachiatum on its constituent strains and the biocontrol of tomato Fusarium wilt. J. Appl. Microbiol. 2022, 132, 532–546. [Google Scholar] [CrossRef] [PubMed]
- Bradford, M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Matsumoto, H.; Haniu, H.; Komori, N. Determination of protein molecular weights on SDS-PAGE: Methods and protocols. In Electrophoretic Separation of Proteins; Kurien, B.T., Scofield, R.H., Eds.; Methods in Molecular Biology Series; Springer: Berlin/Heidelberg, Germany, 2019; pp. 101–105. [Google Scholar] [CrossRef]
- Gasteiger, E.; Hoogland, C.; Gattiker, A.; Duvaud, S.; Wilkins, M.R.; Appel, R.D.; Bairoch, A. Protein identification and analysis tools on the ExPASy server. In The Proteomics Protocols Handbook; Walker, J.M., Ed.; Springer: Berlin/Heidelberg, Germany, 2005; pp. 571–607. [Google Scholar] [CrossRef]
- Buchan, D.W.A.; Jones, D.T. The PSIPRED protein analysis workbench: 20 years on. Nucleic Acids Res. 2019, 47, W402–W407. [Google Scholar] [CrossRef]
- Kelley, L.A.; Mezulis, S.; Yates, C.M.; Wass, M.N.; Sternberg, M.J.E. The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 2015, 10, 845–858. [Google Scholar] [CrossRef]
- Du, Z.; Su, H.; Wang, W.; Ye, L.; Wei, H.; Peng, Z.; Anishchenko, I.; Baker, D.; Yang, J. The trRosetta server for fast and accurate protein structure prediction. Nat. Protoc. 2021, 16, 5634–5651. [Google Scholar] [CrossRef]
- Yang, J.; Yan, R.; Roy, A.; Xu, D.; Poisson, J.; Zhang, Y. The I-TASSER suite: Protein structure and function prediction. Nat. Methods 2015, 12, 7–8. [Google Scholar] [CrossRef]
- Xu, J.; Zhang, Y. How significant is a protein structure similarity with TM-score = 0.5? Bioinformatics 2010, 26, 889–895. [Google Scholar] [CrossRef]
- Chen, K.; Tian, Z.; Luo, Y.; Cheng, Y.; Long, C.A. Antagonistic activity and the mechanism of Bacillus amyloliquefaciens DH-4 against citrus green mold. Phytopathology 2018, 108, 1253–1262. [Google Scholar] [CrossRef]
- Li, H.; Li, C.; Song, X.; Li, J.; Zhang, P.; Sun, F.; Geng, Z.; Liu, X. Isolation and identification of antagonistic Bacillus amyloliquefaciens HSE-12 and its effects on peanut growth and rhizosphere microbial community. Front. Microbiol. 2023, 14, 1274346. [Google Scholar] [CrossRef]
- Zhou, Q.; Fu, M.; Xu, M.; Chen, X.; Qiu, J.; Wang, F.; Yan, R.; Wang, J.; Zhao, S.; Xin, X.; et al. Application of antagonist Bacillus amyloliquefaciens NCPSJ7 against botrytis cinerea in postharvest Red Globe grapes. Food Sci. Nutr. 2020, 8, 1499–1508. [Google Scholar] [CrossRef]
- Jiang, C.H.; Wu, F.; Yu, Z.Y.; Xie, P.; Ke, H.J.; Li, H.W.; Yu, Y.Y.; Guo, J.H. Study on screening and antagonistic mechanisms of Bacillus amyloliquefaciens 54 against bacterial fruit blotch (BFB) caused by Acidovorax avenae subsp. Citrulli. Microbiol. Res. 2015, 170, 95–104. [Google Scholar] [CrossRef]
- Zhang, Y.; Fan, Y.; Dai, Y.; Jia, Q.; Guo, Y.; Wang, P.; Shen, T.; Wang, Y.; Liu, F.; Guo, W.; et al. Crude lipopeptides produced by Bacillus amyloliquefaciens could control the growth of Alternaria alternata and production of Alternaria toxins in processing tomato. Toxins 2024, 16, 65. [Google Scholar] [CrossRef]
- Duan, Y.; Chen, R.; Zhang, R.; Jiang, W.; Chen, X.; Yin, C.; Mao, Z. Isolation, identification, and antibacterial mechanisms of Bacillus amyloliquefaciens QSB-6 and its effect on plant roots. Front. Microbiol. 2021, 12, 746799. [Google Scholar] [CrossRef]
- Wang, X.; Liang, L.; Shao, H.; Ye, X.; Yang, X.; Chen, X.; Shi, Y.; Zhang, L.; Xu, L.; Wang, J. Isolation of the novel strain Bacillus amyloliquefaciens F9 and identification of lipopeptide extract components responsible for activity against Xanthomonas citri subsp. citri. Plants 2022, 11, 457. [Google Scholar] [CrossRef]
- Ye, J.; Wu, H.; Feng, L.; Huang, Q.; Li, Q.; Liao, W.; Wu, J.C. Characterization of Bacillus amyloliquefaciens PM415 as a potential bio-preserving probiotic. Arch. Microbiol. 2024, 206, 222. [Google Scholar] [CrossRef]
- Chen, L.; Chang, S.; Zhao, L.; Li, B.; Zhang, S.; Yun, C.; Wu, X.; Meng, J.; Li, G.; Guo, S.; et al. Biosynthesis of a water solubility-enhanced succinyl glucoside derivative of luteolin and its neuroprotective effect. Microb. Biotechnol. 2022, 15, 2401–2410. [Google Scholar] [CrossRef]
- Lee, A.; Cheng, K.C.; Liu, J.R. Isolation and characterization of a Bacillus amyloliquefaciens strain with zearalenone removal ability and its probiotic potential. PLoS ONE 2017, 12, e182220. [Google Scholar] [CrossRef]
- Joshi, R.; Mcspadden Gardener, B.B. Identification and characterization of novel genetic markers associated with biological control activities in Bacillus subtilis. Phytopathology 2006, 96, 145–154. [Google Scholar] [CrossRef]
- Li, S.; Xu, J.; Fu, L.; Xu, G.; Lin, X.; Qiao, J.; Xia, Y. Biocontrol of wheat crown rot using Bacillus halotolerans QTH8. Pathogens 2022, 11, 595. [Google Scholar] [CrossRef] [PubMed]
- Ji, C.; Zhang, M.; Kong, Z.; Chen, X.; Wang, X.; Ding, W.; Lai, H.; Guo, Q. Genomic analysis reveals potential mechanisms underlying promotion of tomato plant growth and antagonism of soilborne pathogens by Bacillus amyloliquefaciens BA13. Microbiol. Spectr. 2021, 9, e161521. [Google Scholar] [CrossRef] [PubMed]
- Niazi, A.; Manzoor, S.; Asari, S.; Bejai, S.; Meijer, J.; Bongcam-Rudloff, E. Genome analysis of Bacillus amyloliquefaciens subsp. plantarum UCMB5113: A rhizobacterium that improves plant growth and stress management. PLoS ONE 2014, 9, e104651. [Google Scholar] [CrossRef]
- Luo, L.; Zhao, C.; Wang, E.; Raza, A.; Yin, C. Bacillus amyloliquefaciens as an excellent agent for biofertilizer and biocontrol in agriculture: An overview for its mechanisms. Microbiol. Res. 2022, 259, 127016. [Google Scholar] [CrossRef]
- Chen, X.; Zhang, Y.; Fu, X.; Li, Y.; Wang, Q. Isolation and characterization of Bacillus amyloliquefaciens PG12 for the biological control of apple ring rot. Postharvest Biol. Technol. 2016, 115, 113–121. [Google Scholar] [CrossRef]
- Huang, J.; Wei, Z.; Tan, S.; Mei, X.; Shen, Q.; Xu, Y. Suppression of bacterial wilt of tomato by bioorganic fertilizer made from the antibacterial compound producing strain Bacillus amyloliquefaciens HR62. J. Agric. Food. Chem. 2014, 62, 10708–10716. [Google Scholar] [CrossRef]
- Wingfield, P.T. Protein precipitation using ammonium sulfate. Curr. Protoc. Protein Sci. 2016, 84, 1F–3F. [Google Scholar] [CrossRef]
- Lv, B.; Zhao, X.; Guo, Y.; Li, S.; Sun, M. Serine protease CrKP43 interacts with MAPK and regulates fungal development and mycoparasitism in Clonostachys chloroleuca. Microbiol. Spectr. 2023, 11, e02448-23. [Google Scholar] [CrossRef]
- Ling, L.; Cheng, W.; Jiang, K.; Jiao, Z.; Luo, H.; Yang, C.; Pang, M.; Lu, L. The antifungal activity of a serine protease and the enzyme production of characteristics of Bacillus licheniformis TG116. Arch. Microbiol. 2022, 204, 601. [Google Scholar] [CrossRef]
- Koehler Leman, J.; Szczerbiak, P.; Renfrew, P.D.; Gligorijevic, V.; Berenberg, D.; Vatanen, T.; Taylor, B.C.; Chandler, C.; Janssen, S.; Pataki, A.; et al. Sequence-structure-function relationships in the microbial protein universe. Nat. Commun. 2023, 14, 2351. [Google Scholar] [CrossRef]
- Wang, Q.; Wang, B.; Xu, Z.; Wu, J.; Zhao, P.; Li, Z.; Wang, S.; Huang, J.; Cui, S. PSSM-distil: Protein secondary structure prediction (PSSP) on low-quality PSSM by knowledge distillation with contrastive learning. In Proceedings of the Thirty-Fifth AAAI Conference on Artificial Intelligence (AAAI-21), Vancouver, Canada, 2–9 February 2021; pp. 617–625. [Google Scholar] [CrossRef]
- Wüthrich, K. Protein structure determination in solution by nuclear magnetic resonance spectroscopy. Science 1989, 243, 45–50. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y. A glimpse of structural biology through X-ray crystallography. Cell 2014, 159, 995–1014. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Shuai, Y.; Yang, Q.; Zhang, F.; Zeng, M.; Li, M. A comprehensive computational benchmark for evaluating deep learning-based protein function prediction approaches. Brief. Bioinform. 2024, 25, bbae050. [Google Scholar] [CrossRef] [PubMed]
- Kuhlman, B.; Bradley, P. Advances in protein structure prediction and design. Nat. Rev. Mol. Cell Biol. 2019, 20, 681–697. [Google Scholar] [CrossRef]




| Strain Number | Primary Screening | Secondary Screening | |
|---|---|---|---|
| Inhibition Rate (%) | Inhibition Zone Width (cm) | Inhibition Zone Diameter (cm) | |
| D18 | 75.41 ± 1.56 ef | 0.42 ± 0.14 f | − |
| D22 | 81.32 ± 0.62 bcd | 1.08 ± 0.07 bc | 2.28 ± 0.18 ab |
| D30 | 65.76 ± 0.62 g | 0.24 ± 0.06 fg | − |
| D36 | 64.93 ± 2.89 g | 0.13 ± 0.01 g | − |
| D37 | 73.23 ± 4.36 f | 0.34 ± 0.15 f | − |
| D38 | 83.19 ± 0.62 abc | 1.08 ± 0.07 bc | − |
| D39 | 85.06 ± 0.62 ab | 0.7 ± 0.13 e | 2.39 ± 0.07 a |
| D40 | 84.13 ± 0.93 abc | 0.83 ± 0.07 de | 2.05 ± 0.04 b |
| D41 | 82.15 ± 2.94 bcd | 1.06 ± 0.15 bc | − |
| D42 | 80.39 ± 1.25 cd | 1 ± 0.09 cd | 2.1 ± 0.14 b |
| D43 | 81.14 ± 1.97 bcd | 1.24 ± 0.1 b | − |
| D45 | 78.00 ± 1.90 de | 1.24 ± 0.08 b | 2.13 ± 0.32 ab |
| D52 | 78.69 ± 3.12 de | 1.01 ± 0.15 cd | − |
| D55 | 87.45 ± 2.07 a | 1.72 ± 0.02 a | − |
| Test | Reaction |
|---|---|
| Starch hydrolysis | + |
| Aerobic test | + |
| Catalase reaction | + |
| Voges–Proskauer | + |
| Nitrate reduction reaction | + |
| Methyl red | − |
| Citrate utilization | − |
| Gelatin liquefaction | + |
| Phenylalanine deaminase | − |
| Gram-stain | + |
| Gene | Size (bp) | GenBank Accession Number | Best Matches in GenBank | Scientific Name | Query Cover | Percent Identity |
|---|---|---|---|---|---|---|
| srfAA | 177 | OM830959 | CP053376 | B. amyloliquefaciens | 89% | 95.57% |
| bmyB | 340 | OM830955 | KP453869 | B. amyloliquefaciens | 97% | 98.5% |
| bacA | 461 | OM830956 | MG800648 | B. subtilis | 95% | 100% |
| fenD | 242 | OM830957 | KP453873 | B. amyloliquefaciens | 93% | 99.12% |
| ituC | 396 | OM830958 | KT781920 | B. subtilis | 95% | 98.95% |
| Time | Treatment | Bacterial Fermentation Broth, | CK | Significance Analysis |
|---|---|---|---|---|
| Before prevention and treatment | Incidence rate (%) | 26.67 ± 3.33 | 27.78 ± 1.92 | NS |
| Day 10 | Incidence rate (%) | 40.00 ± 3.33 | 88.89 ± 1.92 | Treatment: * Time: * |
| Disease index | 21.11 ± 0.96 | 62.22 ± 2.54 | ||
| Control effect (%) | 66.07 ± 1.55 | − | − | |
| Day 20 | Incidence rate (%) | 58.89 ± 10.71 | 98.89 ± 1.92 | Treatment: * Time: * |
| Disease index | 25.56 ± 3.85 | 87.78 ± 0.00 | ||
| Control effect (%) | 70.89 ± 4.38 | − | − |
| No. | Accession | −10lgP | Cov. (%) | Unique Peptides | Avg. Mass | Description |
|---|---|---|---|---|---|---|
| 1 | A0A5C8IVR9 | 245.12 | 29 | 10 | 38,666 | Aminopeptidase YhfE |
| 2 | A0A6M9ZGL4 | 197.99 | 10 | 5 | 86,631 | Peptidase G2 |
| 3 | A0A6M9ZD65 | 170.03 | 13 | 5 | 64,115 | Gamma-glutamyltransferase |
| 4 | A0A1Y0XB26 | 168.71 | 19 | 4 | 39,375 | Cellulase |
| 5 | A0A5C8IRJ3 | 162.51 | 21 | 7 | 45,709 | Peptidase T |
| 6 | A0A6M9ZCG3 | 158.94 | 6 | 4 | 154,192 | S8 family serine peptidase |
| 7 | A0A5C8IRC6 | 158.65 | 21 | 5 | 20,619 | Spore coat protein GerQ |
| 8 | A0A5C8II39 | 134.65 | 35 | 6 | 27,110 | Uncharacterized protein |
| 9 | A0A5C8IP87 | 120.05 | 11 | 4 | 50,053 | Dihydrolipoyl dehydrogenase |
| 10 | A0A6M9ZDC1 | 115.73 | 19 | 6 | 48,236 | S8 family peptidase |
| 11 | A0A5C8IRK7 | 111.76 | 5 | 3 | 77,011 | Catalase |
| 12 | A0A6M9ZHR5 | 101.54 | 3 | 4 | 139,053 | Nitrate reductase (quinone) |
| 13 | A0A6M9ZII5 | 81.53 | 14 | 2 | 17,975 | Stress protein |
| 14 | Q9F9Q4 | 81.53 | 6 | 1 | 19,564 | Stress protein |
| 15 | A0A5C8IUA2 | 72.75 | 21 | 4 | 17,600 | Spore coat protein |
| 16 | A0A6M9ZCB4 | 72.24 | 3 | 1 | 49,735 | Dihydrolipoyl dehydrogenase |
| 17 | A0A6M9ZGS6 | 66.97 | 10 | 2 | 42,518 | N-acetylglucosamine-6-phosphate deacetylase |
| 18 | A0A5C8IVB4 | 64.26 | 11 | 5 | 54,544 | Catalase |
| 19 | A0A1Y0X703 | 63.15 | 8 | 2 | 30,200 | D-aminopeptidase |
| 20 | A0A5C8IK53 | 62.68 | 11 | 3 | 39,282 | Glutamyl aminopeptidase |
| 21 | A0A6M9ZKZ3 | 60.73 | 5 | 1 | 19,298 | Type 1 glutamine amidotransferase |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Deng, T.; Wang, L.; Zhu, T. Isolation of Bacillus amyloliquefaciens D39 and Identification of Its Antimicrobial Proteins Active Against Chestnut Blight. Microorganisms 2025, 13, 1302. https://doi.org/10.3390/microorganisms13061302
Deng T, Wang L, Zhu T. Isolation of Bacillus amyloliquefaciens D39 and Identification of Its Antimicrobial Proteins Active Against Chestnut Blight. Microorganisms. 2025; 13(6):1302. https://doi.org/10.3390/microorganisms13061302
Chicago/Turabian StyleDeng, Tingting, Linmin Wang, and Tianhui Zhu. 2025. "Isolation of Bacillus amyloliquefaciens D39 and Identification of Its Antimicrobial Proteins Active Against Chestnut Blight" Microorganisms 13, no. 6: 1302. https://doi.org/10.3390/microorganisms13061302
APA StyleDeng, T., Wang, L., & Zhu, T. (2025). Isolation of Bacillus amyloliquefaciens D39 and Identification of Its Antimicrobial Proteins Active Against Chestnut Blight. Microorganisms, 13(6), 1302. https://doi.org/10.3390/microorganisms13061302





