Select Cover Crop Residue and Soil Microbiomes Contribute to Suppression of Fusarium Root and Crown Rot in Barley and Soybean
Abstract
1. Introduction
2. Materials and Methods
2.1. Field Trial Set Up
2.2. Metagenomic Analysis
2.3. Field Disease Assessment
2.4. Identification of Pathogenic Fusarium spp.
2.5. Greenhouse Trial
2.6. Statistical Analysis
3. Results
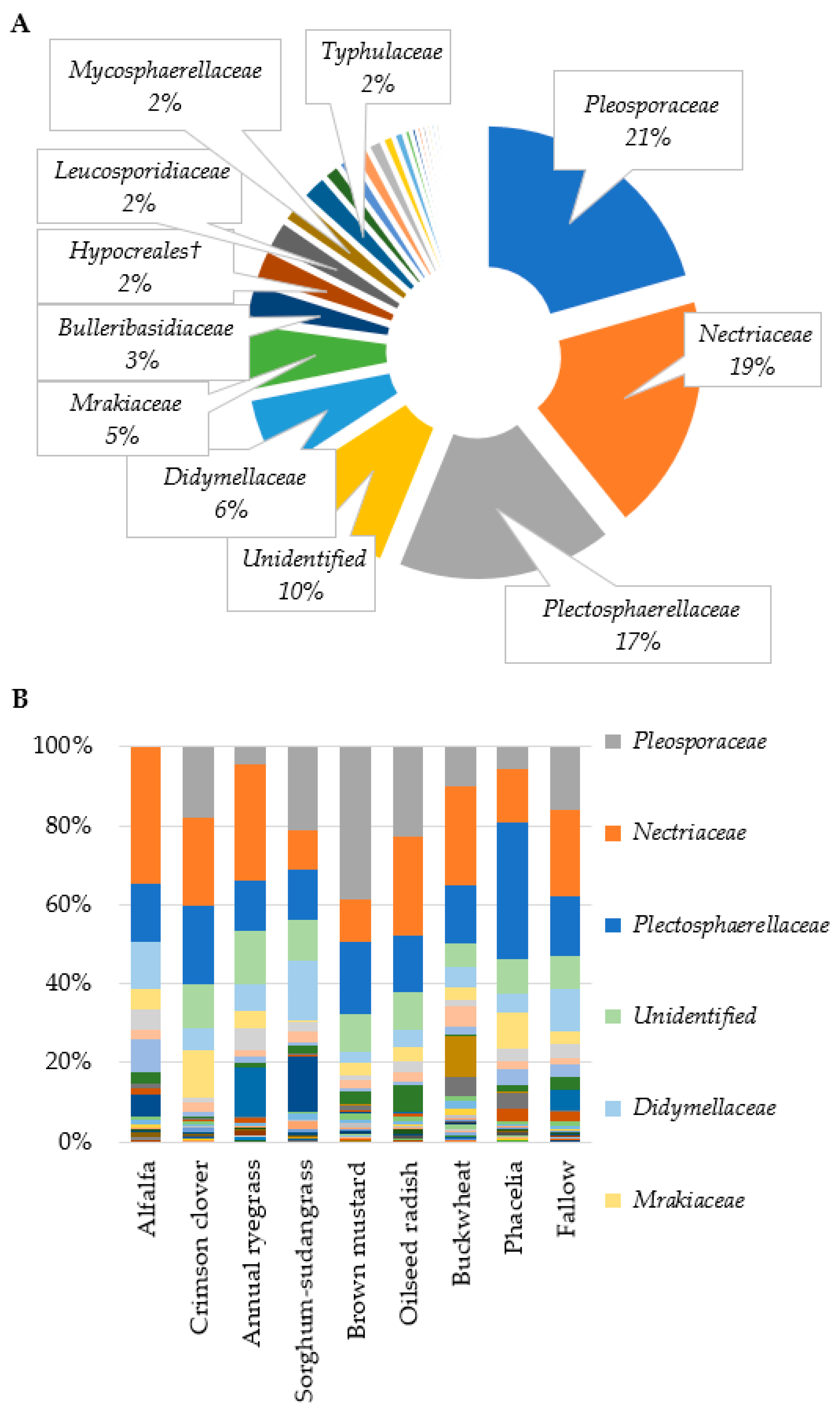
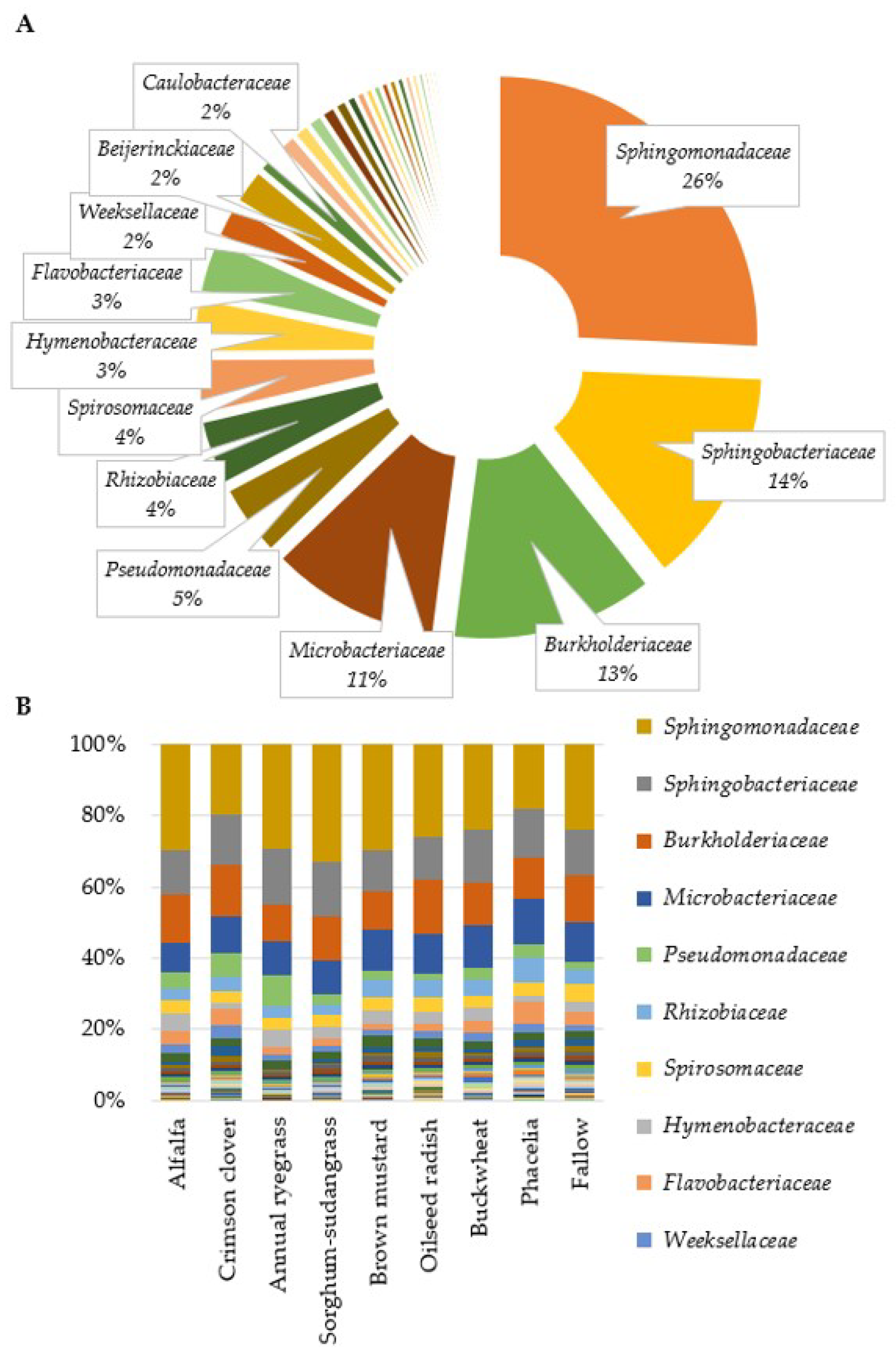
3.1. Sequencing and OTU Clustering
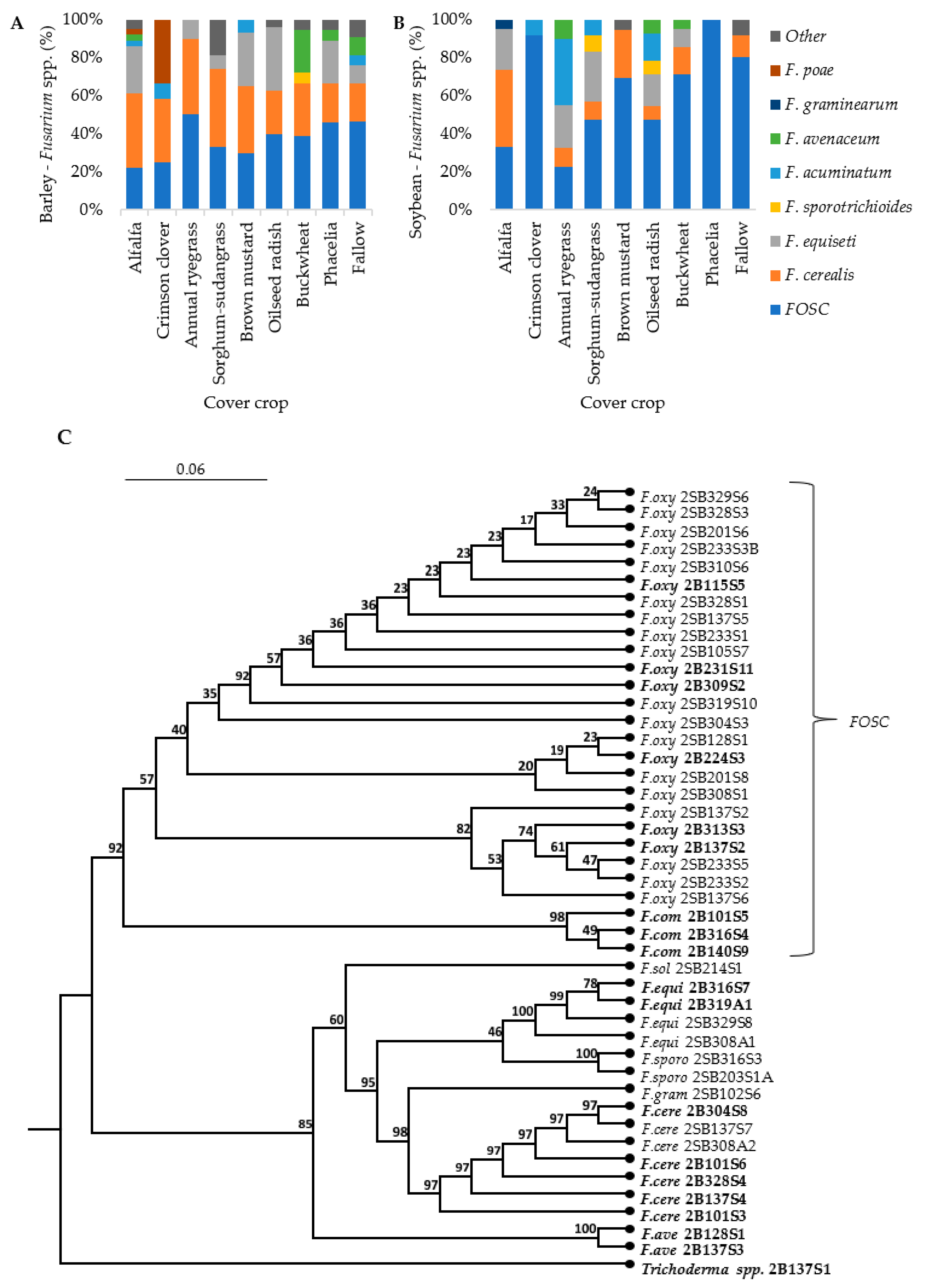
3.2. Fusarium Abundance in Soil and Residue
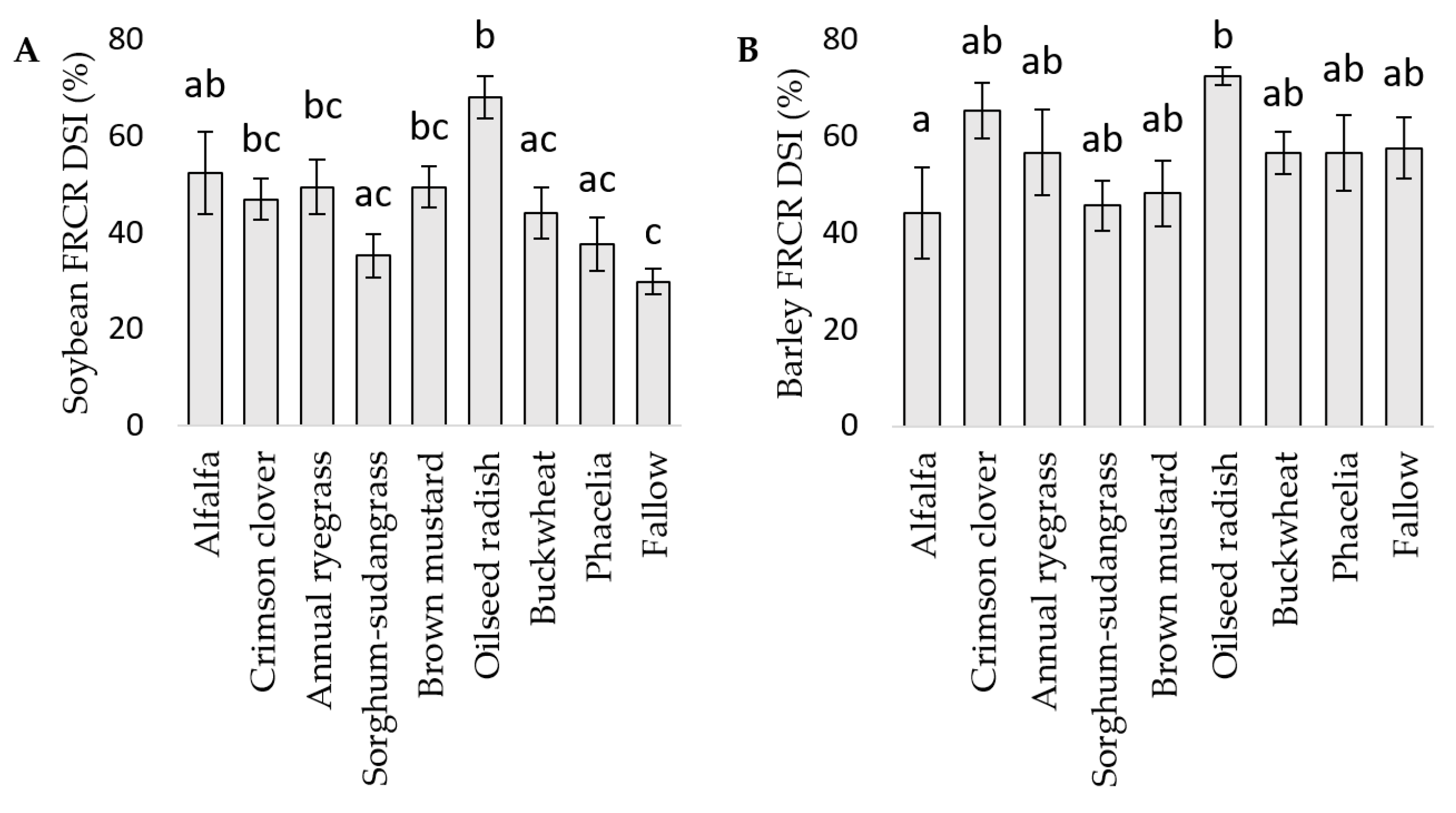
3.3. FRCR in Soybean and Barley in the Field
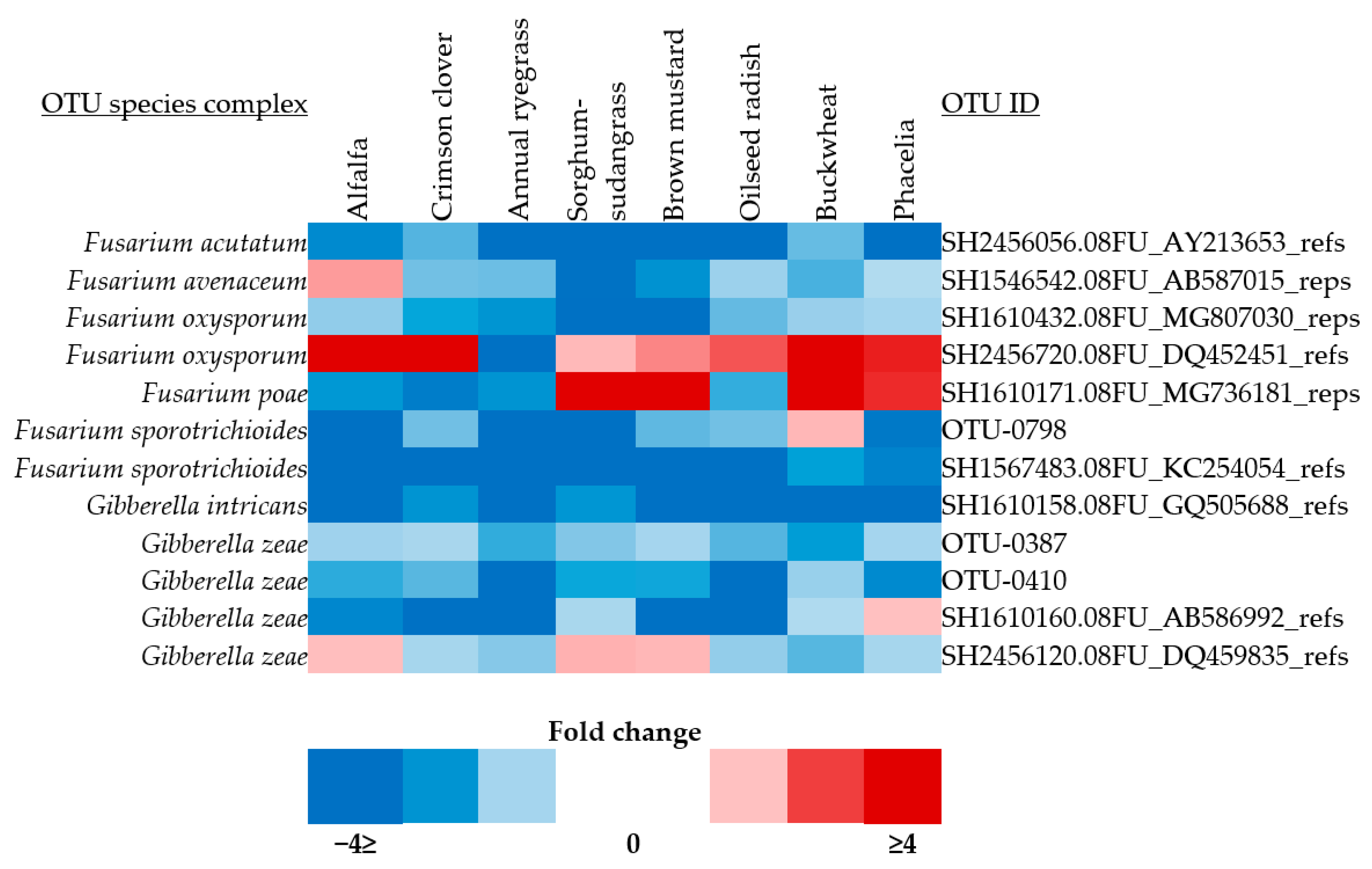
3.4. Pathogen Isolation and Phylogenetic Analysis
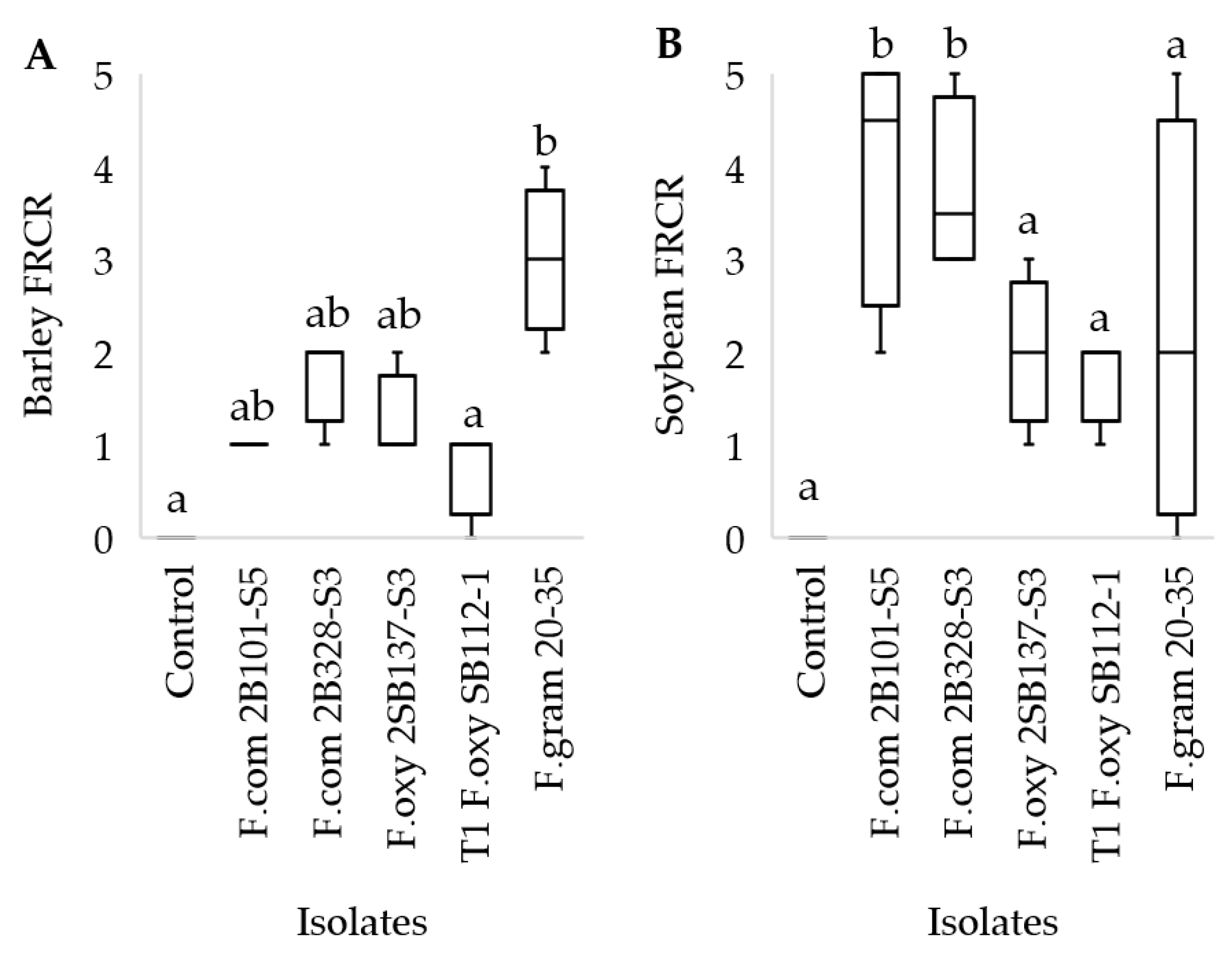
3.5. Fusarium Inoculum Virulence Assay
3.6. Disease-Suppressive Effects of Cover Crop Soils
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Leath, S.; Carroll, R.B. Use of ridge regression to predict yield reduction by Fusarium sp. in selected soybean cultivars. Can. J. Plant Pathol. 1985, 7, 58–66. [Google Scholar] [CrossRef]
- Smiley, R.W.; Gourlie, J.A.; Easley, S.A.; Patterson, L.M.; Whittaker, R.G. Crop damage estimates for crown rot of wheat and barley in the Pacific Northwest. Plant Dis. 2005, 89, 595–604. [Google Scholar] [CrossRef] [PubMed]
- Smith, S.N.; Snyder, W.C. Persistence of Fusarium oxysporum f. sp. vasinfectum. Phytopathology 1975, 65, 190–196. [Google Scholar] [CrossRef]
- Cotten, T.K.; Munkvold, G.P. Survival of Fusarium moniliforme, F. proliferatum, and F. subglutinans in maize stalk residue. Phytopathology 1998, 88, 550–555. [Google Scholar] [CrossRef]
- Todorović, I.; Moënne-Loccoz, Y.; Raičević, V.; Jovičić-Petrović, J.; Muller, D. Microbial diversity in soils suppressive to Fusarium diseases. Front. Plant Sci. 2023, 14, 1228749. [Google Scholar] [CrossRef] [PubMed]
- Martin, R.A.; Johnston, H.W. Effects and control of Fusarium diseases of cereal grains in the Atlantic Provinces. Can. J. Plant Pathol. 1982, 4, 210–216. [Google Scholar] [CrossRef]
- Wegulo, S.; Giesler, L.; Harveson, R.; Jackson-Ziems, T.A.; Liu, B.; Korus, K. Impacts of drought on disease development and management. In Crop Production Clinics Proceedings; University of Nebraska-Lincoln Extension, Institute of Agriculture and Natural Resources: Lincoln, NE, USA, 2013. [Google Scholar]
- Howden, S.M.; Soussana, J.F.; Tubiello, F.N.; Chhetri, N.; Dunlop, M.; Meinke, H. Adapting agriculture to climate change. Proc. Natl. Acad. Sci. USA 2007, 104, 19691–19696. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Ye, W.; Wang, M.; Yan, X. Climate change and drought: A risk assessment of crop-yield impacts. Clim. Res. 2009, 39, 31–46. [Google Scholar] [CrossRef]
- Shah, K.K.; Modi, B.; Pandey, H.P.; Subedi, A.; Aryal, G.; Pandey, M.; Shrestha, J. Diversified crop rotation: An approach for sustainable agriculture production. Adv. Agric. 2021, 2021, 8924087. [Google Scholar] [CrossRef]
- Wagg, C.; van Erk, A.; Fava, E.; Comeau, L.P.; Mitterboeck, T.F.; Goyer, C.; Li, S.; McKenzie-Gopsill, A.; Mills, A. Full-season cover crops and their traits that promote agroecosystem services. Agriculture 2021, 11, 830. [Google Scholar] [CrossRef]
- Jatoe, J.B.D.; Yiridoe, E.K.; Weersink, A.; Clark, J.S. Economic and environmental impacts of introducing land use policies and rotations on Prince Edward Island potato farms. Land Use Policy 2008, 25, 309–319. [Google Scholar] [CrossRef]
- Aiyer, H.; Fofana, B.; Fraser, T.; Caldwell, C.; McKenzie-Gopsill, A.; Mills, A.; Foster, A. Choice of cover crop influences soil fungal and bacterial communities in Prince Edward Island, Canada. Can. J. Microbiol. 2022, 68, 465–482. [Google Scholar] [CrossRef]
- Bainard, L.D.; Navarro-Borrell, A.; Hamel, C.; Braun, K.; Hanson, K.; Gan, Y. Increasing the frequency of pulses in crop rotations reduces soil fungal diversity and increases the proportion of fungal pathotrophs in a semiarid agroecosystem. Agric. Ecosyst. Environ. 2017, 240, 206–214. [Google Scholar] [CrossRef]
- Alori, E.T.; Glick, B.R.; Babalola, O.O. Microbial phosphorus solubilization and its potential for use in sustainable agriculture. Front. Microbiol. 2017, 8, 971. [Google Scholar] [CrossRef] [PubMed]
- Abbasi, P.A.; Renderos, W.; Fillmore, S. Soil incorporation of buckwheat as a pre-plant amendment provides control of Rhizoctonia damping-off and root rot of radish and Pythium damping-off and root rot of cucumber. Can. J. Plant Pathol. 2019, 41, 24–34. [Google Scholar] [CrossRef]
- Noronha, C. Crop rotation as a management tool for wireworms in potatoes. In Proceedings of the IOPC/WPRS Working Group “Insect Pathogens and Entomopathogenic Nematodes”, Innsbruck, Austria, 19–23 June 2011; Volume 66, pp. 467–471. [Google Scholar]
- Peters, R.D.; Sturz, A.V.; Carter, M.R.; Sanderson, J.B. Crop rotation can confer resistance to potatoes from Phytophthora erythroseptica attack. Can. J. Plant Sci. 2005, 85, 523–528. [Google Scholar] [CrossRef]
- McKenzie-Gopsill, A.; Mills, A.; MacDonald, A.N.; Wyand, S. The importance of species selection in cover crop mixture design. Weed Sci. 2022, 70, 436–447. [Google Scholar] [CrossRef]
- Atlantic Grains Council (AGC). Crop Statistics. 2021. Available online: https://atlanticgrainscouncil.ca/crop-statistics/ (accessed on 28 December 2023).
- Sturz, A.V.; Johnston, H.W. Characterization of Fusarium colonization of spring barley and wheat produced on stubble or fallow soil. Can. J. Plant Pathol. 1985, 7, 270–276. [Google Scholar] [CrossRef]
- Backhouse, D.; Abubakar, A.A.; Burgess, L.W.; Dennisc, J.I.; Hollaway, G.J.; Wildermuth, G.B.; Wallwork, H.; Henry, F.J. Survey of Fusarium species associated with crown rot of wheat and barley in eastern Australia. Australas. Plant Pathol. 2004, 33, 255–261. [Google Scholar] [CrossRef]
- Ivic, D. Pathogenicity and potential toxigenicity of seed-borne Fusarium species on soybean and pea. J. Plant Pathol. 2014, 96, 541–551. [Google Scholar] [CrossRef]
- Knight, N.L.; Sutherland, M.W. Assessment of Fusarium pseudograminearum and F. culmorum biomass in seedlings of potential host cereal species. Plant Dis. 2017, 101, 2116–2122. [Google Scholar] [CrossRef] [PubMed]
- Abdelmagid, A.; Hafez, M.; Lawley, Y.; Adam, L.R.; Daayf, F. First Report of Fusarium cerealis Causing Root Rot on Soybean. Plant Dis. 2018, 102, 2638–2639. [Google Scholar] [CrossRef]
- Abdelmagid, A.; Hafez, M.; Soliman, A.; Adam, L.R.; Daayf, F. First report of Fusarium sporotrichioides causing root rot of soybean in Canada and detection of the pathogen in host tissues by PCR. Can. J. Plant Pathol. 2021, 43, 527–536. [Google Scholar] [CrossRef]
- Knight, N.L.; Sutherland, M.W. Histopathological assessment of Fusarium pseudograminearum colonization of cereal culms during crown rot infections. Plant Dis. 2016, 100, 252–259. [Google Scholar] [CrossRef] [PubMed]
- Aiyer, H. Effects of Cover Crops on the Soil Microbiome and Carry over Impact on FRCR in Barley and Soybean. Master’s Thesis, Dalhousie University, Halifax, NS, Canada, 10 January 2022. Available online: http://hdl.handle.net/10222/81187 (accessed on 28 December 2023).
- Legge, W.G.; Tucker, J.R.; Fetch, T.G., Jr.; Haber, S.; Menzies, J.G.; Tekauz, A.; Turkington, T.K.; Savard, M.E. AAC Synergy barley. Can. J. Plant Sci. 2014, 94, 797–803. [Google Scholar] [CrossRef]
- Nilsson, R.H.; Larsson, K.-H.; Taylor, A.F.S.; Bengtsson-Palme, J.; Jeppesen, T.S.; Schigel, D.; Kennedy, P.; Picard, K.; Glöckner, F.O.; Tedersoo, L.; et al. The UNITE database for molecular identification of fungi: Handling dark taxa and parallel taxonomic classifications. Nucleic Acids Res. 2019, 47, D259–D264. [Google Scholar] [CrossRef]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2012, 41, D590–D596. [Google Scholar] [CrossRef]
- Nguyen, N.H.; Song, Z.; Bates, S.T.; Branco, S.; Tedersoo, L.; Menke, J.; Schilling, J.S.; Kennedy, P.G. FUNGuild: An open annotation tool for parsing fungal community datasets by ecological guild. Fungal Ecol. 2016, 20, 241–248. [Google Scholar] [CrossRef]
- Louca, S.; Parfrey, L.W.; Doebeli, M. Decoupling function and taxonomy in the global ocean microbiome. Science 2016, 353, 1272–1277. [Google Scholar] [CrossRef]
- Chekali, S.; Gargouri, S.; Ben Hammouda, M.; M’hamed, H.C.; Nasraoui, B. Incidence of Fusarium foot and root rot of cereals under conservation agriculture in north west Tunisia. Phytopathol. Mediterr. 2019, 58, 95–102. [Google Scholar]
- Ellis, M.L.; Broders, K.D.; Paul, P.A.; Dorrance, A.E. Infection of soybean seed by Fusarium graminearum and effect of seed treatments on disease under controlled conditions. Plant Dis. 2011, 95, 401–407. [Google Scholar] [CrossRef]
- Chiang, K.; Liu, H.; Bock, C.H. A discussion on disease severity index values: Warning on inherent errors and suggestions to maximize accuracy. Ann. Appl. Biol. 2017, 171, 139–154. [Google Scholar] [CrossRef]
- Leslie, J.F.; Summerell, B.A. The Fusarium Laboratory Manual; John Wiley & Sons: Hoboken, NJ, USA, 2008. [Google Scholar]
- White, T.J.; Bruns, T.; Lee, S.; Taylor, J.W. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In PCR Protocols: A Guide to Methods and Applications; Innis, M.A., Gelfand, D.H., Sninsky, J.J., White, T.J., Eds.; Academic Press, Inc.: New York, NY, USA, 1990; pp. 315–322. [Google Scholar]
- O’Donnell, K.; Kistler, H.C.; Cigelnik, E.; Ploetz, R.C. Multiple evolutionary origins of the fungus causing Panama disease of banana: Concordant evidence from nuclear and mitochondrial gene genealogies. Proc. Natl. Acad. Sci. USA 1998, 95, 2044–2049. [Google Scholar] [CrossRef]
- Altschul, S.F.; Madden, T.L.; Schäffer, A.A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef]
- Koch, R. Untersuchungen uber bakterien V. Die aetiologie der milzbrand-krankheit, begrunder auf die entwicklungegeschichte Bacillus anthracis. Beitr. Biol. Pflanz. 1877, 2, 277–310. [Google Scholar] [CrossRef]
- Zhou, Q.; Li, N.; Chang, K.F.; Hwang, S.F.; Strelkov, S.E.; Conner, R.L.; McLaren, D.L.; Fu, H.; Harding, M.W.; Turnbull, G.D. Genetic diversity and aggressiveness of Fusarium species isolated from soybean in Alberta, Canada. Crop Prot. 2018, 105, 49–58. [Google Scholar] [CrossRef]
- Johnstone, E.; Matters, R.; Foster, A. Causal species of Fusarium head blight of spring wheat and winter wheat in Prince Edward Island in 2020. Canadian Plant Disease Survey 2021 Volume 101: Disease Highlights 2020. Can. J. Plant Pathol. 2021, 43, 92–94. [Google Scholar] [CrossRef]
- Ellis, M.L.; Jimenez, D.R.C.; Leandro, L.F.; Munkvold, G.P. Genotypic and phenotypic characterization of fungi in the Fusarium oxysporum species complex from soybean roots. Phytopathology 2014, 104, 1329–1339. [Google Scholar] [CrossRef] [PubMed]
- Gentosh, D.T.; Kyryk, M.M.; Gentosh, I.D.; Pikovskyi, M.Y.; Polozhenets, V.M.; Stankevych, S.V.; Nemerytska, L.V.; Zhuravska, I.A.; Zabrodina, I.V.; Zhukova, L.V. Species compositions of root rot agents of spring barley. Ukr. J. Ecol. 2020, 10, 106–109. [Google Scholar]
- Chen, D.; Nahar, K. First Report of Fusarium commune Causing Root Rot of Field Peas in Canada. Plant Dis. 2023, 107, 2259. [Google Scholar] [CrossRef] [PubMed]
- Smolinska, U.; Morra, M.J.; Knudsen, G.R.; James, R.L. Isothiocyanates produced by Brassicaceae species as inhibitors of Fusarium oxysporum. Plant Dis. 2003, 87, 407–412. [Google Scholar] [CrossRef] [PubMed]
- Nallanchakravarthula, S.; Marupakula, S.; Alström, S.; Finlay, R.D.; Mahmood, S. Changes in the root fungal microbiome of strawberry following application of residues of the biofumigant oilseed radish. Appl. Soil Ecol. 2021, 168, 104116. [Google Scholar] [CrossRef]
- Alabouvette, C. Fusarium wilt suppressive soils: An example of disease-suppressive soils. Australas. Plant Pathol. 1999, 28, 57–64. [Google Scholar] [CrossRef]
- Weston, L.A.; Harmon, R.; Mueller, S. Allelopathic potential of sorghum-sudangrass hybrid (sudex). J. Chem. Ecol. 1989, 15, 1855–1865. [Google Scholar] [CrossRef]
- Fernandez, M.R.; Huber, D.; Basnyat, P.; Zentner, R.P. Impact of agronomic practices on populations of Fusarium and other fungi in cereal and noncereal crop residues on the Canadian Prairies. Soil Tillage Res. 2008, 100, 60–71. [Google Scholar] [CrossRef]
- Yang, P.; Chen, Y.; Wu, H.; Fang, W.; Liang, Q.; Zheng, Y.; Olsson, S.; Zhang, D.; Zhou, J.; Wang, Z.; et al. The 5-oxoprolinase is required for conidiation, sexual reproduction, virulence and deoxynivalenol production of Fusarium graminearum. Curr. Genet. 2018, 64, 285–301. [Google Scholar] [CrossRef]
- Goswami, R.S.; Kistler, H.C. Heading for disaster: Fusarium graminearum on cereal crops. Mol. Plant Pathol. 2004, 5, 515–525. [Google Scholar] [CrossRef]

| Primer | Sequence (5′-3′) | Locus | Conditions |
|---|---|---|---|
| ITS1F * | CTT GGT CAT TTA GAG GAA GTA A | ITS1 | 5 min—95 °C ----40 cycles---- Denaturation: 30 s—94 °C Annealing: 30 s—52 °C Elongation: 1 min—72 °C 8 min—72 °C |
| ITS4 * | TCCTCCGCTTATTGATATGC | ||
| EF1 ** | ATG GGT AAG GA(A/G) GAC AAG AC | TEF-1α | 8 min—95 °C -----35 cycles----- Denaturation: 30 s—95 °C Annealing: 60 s—53 °C Elongation: 1 min—72 °C 5 min—72 °C |
| EF2 ** | GGA (G/A)GT ACC AGT (G/C)AT CAT GTT |
| Bacteria | Fungi | ||
|---|---|---|---|
| Sequencing Platform | PacBio Sequel | Illumina MiSeq | |
| Target region | Full 16S rRNA | ITS1 | |
| Number of samples | 54 | 54 | |
| Targeted reads per sample | 5000 | 90,000 | |
| Raw data | Total nucleotides in data sets | 286,168,882 | 2,945,618,500 |
| Total reads in data sets | 193,619 | 11,782,472 | |
| Mean read length | 1478 bp | 250 bp | |
| Mean reads per sample | 3586 | 218,193 | |
| Quality control | Total reads after trimming | 189,923 (98.1%) | 5,891,123 (99.9%) * |
| Post trimming mean reads | 3517 | 218,192 | |
| OTU Clustering | Reads in OTUs | 174,085 (89.9%) | 4,924,314 (83.6%) |
| Total predicted OTUs | 17,170 | 8588 | |
| Mean OTU length | 1454 bp | 282 bp | |
| Filtered OTUs | Reads in OTU after filtering | 130,412 (67.4%) | 4,217,277 (71.6%) |
| Total OTUs after filtering | 1067 | 898 |
| Alfalfa | Crimson Clover | Annual Ryegrass | Sorghum-Sudangrass | Brown Mustard | Oilseed Radish | Buckwheat | Phacelia | Fallow | |
|---|---|---|---|---|---|---|---|---|---|
| Alfalfa | - | NS | NS | NS | NS | NS | NS | NS | NS |
| Crimson clover | NS | - | 0.04 | 0.03 | 0.05 | NS | 0.03 | NS | NS |
| Annual ryegrass | NS | NS | - | NS | 0.03 | 0.01 | 0.02 | 0.01 | 0.02 |
| Sorghum-sudangrass | NS | NS | 0.02 | - | NS | 0.05 | NS | 0.01 | 0.03 |
| Brown mustard | NS | NS | 0.02 | NS | - | NS | NS | 0.03 | NS |
| Oilseed radish | NS | NS | 0.01 | NS | NS | - | 0.05 | NS | NS |
| Buckwheat | NS | NS | 0.02 | NS | NS | NS | - | 0.04 | 0.03 |
| Phacelia | 0.05 | NS | 0.01 | 0.01 | NS | NS | 0.05 | - | NS |
| Fallow | NS | NS | NS | NS | NS | NS | NS | NS | - |
| Functional Group | Family | Barley FRCR | Soybean FRCR |
|---|---|---|---|
| plant pathogen | Enterobacteriaceae | NS | NS |
| Pseudomonadaceae | |||
| ureolysis | Isosphaeraceae | NS | 0.5418 |
| Acetobacteraceae | |||
| Beijerinckiaceae | |||
| Rhizobiaceae | |||
| Burkholderiaceae | |||
| Methylophilaceae | |||
| Pseudomonadaceae | |||
| Xanthomonadaceae | |||
| animal parasites or symbionts | Acetobacteraceae | 0.3263 | NS |
| Enterobacteriaceae | |||
| Pseudomonadaceae | |||
| Xanthomonadaceae | |||
| human associated | Acetobacteraceae | 0.3079 | NS |
| Enterobacteriaceae | |||
| Xanthomonadaceae |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Aiyer, H.S.; McKenzie-Gopsill, A.; Mills, A.; Foster, A.J. Select Cover Crop Residue and Soil Microbiomes Contribute to Suppression of Fusarium Root and Crown Rot in Barley and Soybean. Microorganisms 2024, 12, 404. https://doi.org/10.3390/microorganisms12020404
Aiyer HS, McKenzie-Gopsill A, Mills A, Foster AJ. Select Cover Crop Residue and Soil Microbiomes Contribute to Suppression of Fusarium Root and Crown Rot in Barley and Soybean. Microorganisms. 2024; 12(2):404. https://doi.org/10.3390/microorganisms12020404
Chicago/Turabian StyleAiyer, Harini S., Andrew McKenzie-Gopsill, Aaron Mills, and Adam John Foster. 2024. "Select Cover Crop Residue and Soil Microbiomes Contribute to Suppression of Fusarium Root and Crown Rot in Barley and Soybean" Microorganisms 12, no. 2: 404. https://doi.org/10.3390/microorganisms12020404
APA StyleAiyer, H. S., McKenzie-Gopsill, A., Mills, A., & Foster, A. J. (2024). Select Cover Crop Residue and Soil Microbiomes Contribute to Suppression of Fusarium Root and Crown Rot in Barley and Soybean. Microorganisms, 12(2), 404. https://doi.org/10.3390/microorganisms12020404





