Abstract
Species of Mycosphaerellaceae and Teratosphaeriaceae represent over 40% of the fungi identified on eucalypt leaves worldwide. These include some important pathogens that mainly cause leaf blight and spot, and result in increasingly negative impacts on global commercial eucalypt industries. Eucalyptus plantations are commonly cultivated in southern China for solid wood and pulp products. However, the species diversity and geographic distribution of Mycosphaerellaceae and Teratosphaeriaceae, associated with diseased plantation Eucalyptus leaves in China, have not been clarified. In this study, we conducted the first systematic surveys and sample collections of Mycosphaerellaceae- and Teratosphaeriaceae-like fungi from diseased plantation Eucalyptus leaves in southern China. In total, 558 isolates were obtained from 59 sampled sites in five provinces. One isolate was isolated from each tree. According to the disease symptoms, conidia morphological characteristics, and DNA sequence comparisons of ITS, tef1 and tub2 gene regions. The 558 isolates were identified as Teratosphaeria epicoccoides (312 isolates; 55.9%) and T. destructans (246 isolates, 44.1%). Both species were widely distributed in the sampled regions in southern China. The genotypes of T. epicoccoides and T. destructans were determined based on ITS, tef1, and tub2 sequences. The results showed that multiple genotypes of each species of T. epicoccoides and T. destructans exist in China. Additionally, isolates with multiple genotypes were obtained in all five sampled provinces. These results suggest that both T. epicoccoides and T. destructans are not clonal. This study proved that both T. epicoccoides and T. destructans are dominant species and widely distributed on diseased Eucalyptus leaves in southern China. The wide geographic distribution and potential high genetic diversity pose challenges for the disease management of Teratosphaeria leaf blight and leaf spot in China.
1. Introduction
In China, eucalypts plantations have expanded rapidly to meet the increasing demand for wood and pulp [1]. Occupying approximately 5.4 million hm2 in China in 2018, the plantations account for 6.8% of the national plantation area and provide more than one-third of the total national commercial timber production [2]. Eucalyptus plantations are mainly planted in the southern regions of China, especially in Guangxi, Guangdong, Yunnan, and Fujian Provinces. Mostly, selected genotypes of Eucalyptus urophylla × E. grandis hybrids are planted and grown [3].
In China, Eucalyptus plantations are primarily located in tropical and subtropical regions, where a diverse array of pathogenic fungi can easily proliferate [4]. Due to extensive contiguous planting and a monotonous genetic makeup, Eucalyptus plantations in China are highly susceptible to the rapid and widespread proliferation of diseases when affected by some pathogenic microorganisms [5,6]. Currently, important diseases affecting plantation Eucalyptus in China include bacterial wilt, caused by Ralstonia pseudosolanacearum [6,7,8]; stem canker/wilt, caused by species of Botryosphaeriaceae, Cryphonectriaceae, and Teratosphaeria zuluensis [4,9,10,11,12]; leaf blight/spot, caused by species of Teratosphaeriaceae and Mycosphaerellaceae [13]; Calonectria [14,15,16,17]; and Quambalaria [18,19]. With the exception of bacterial wilt, the other diseases are all caused by pathogenic fungi.
Globally, leaf diseases on Eucalyptus caused by the fungi of Mycosphaerellaceae and Teratosphaeriaceae are widely distributed [20,21,22,23]. In recent years, a number of novel species of Mycosphaerellaceae and Teratosphaeriaceae have been isolated from diseased Eucalyptus leaves and described [23,24,25], some of which are known to cause leaf diseases on Eucalyptus. According to the research findings of Crous et al. [23], species belonging to Mycosphaerellaceae and Teratosphaeriaceae demonstrate a high species diversity on a global scale. Species from these two families represent over 40% of the fungi identified on eucalypts leaves worldwide [23].
In China, several species of Mycosphaerellaceae and Teratosphaeriaceae have been implicated as a cause of Eucalyptus leaf spot/blight [26,27,28,29,30]. Of the foliar pathogens belonging to Mycosphaerellaceae and Teratosphaeriaceae isolated from Eucalyptus leaves in China, only T. destructans (causing Eucalyptus leaf blight) [26] and Pseudocercospora chiangmaiensis (responsible for Eucalyptus circular leaf spot) [28,31] were identified based on both DNA sequence comparisons and morphological characteristics. The identification of the remaining species causing leaf spot/blight relied solely on morphological features [32,33,34,35,36,37]. More than 600 Eucalyptus species are native to Australia, with only a few species endemic to Papua New Guinea, some parts of Indonesia and the Philippines [38,39]. Many fungi of Mycosphaerellaceae and Teratosphaeriaceae have been described from Eucalyptus foliage in Australia, and most were considered to be endemic [23,40,41]. Previous studies have shown that Mycosphaerellaceae and Teratosphaeriaceae exhibit high species diversity; species in these two families are the predominant fungi on diseased Eucalyptus leaves globally [23]. In China, research on the species diversity of Mycosphaerellaceae and Teratosphaeriaceae fungi on diseased Eucalyptus leaves is very limited, and the identification of some of these species is not accurate because of the lack of DNA sequence data [26,27,29,30]. Furthermore, there are limited systematic studies on their geographical distribution. Recently, we collected diseased Eucalyptus leaves with typical mature fruiting structures of Mycosphaerellaceae and Teratosphaeriaceae from Eucalyptus plantations in Yunnan, Guangxi, Hainan, Guangdong, and Fujian Provinces in southern China. We subsequently isolated these fungi. The aims of this study were to: (i) identify these fungi based on DNA sequence comparisons of multi-gene regions and morphological characteristics; and (ii) explore the geographical distribution characteristics of Mycosphaerellaceae and Teratosphaeriaceae species on diseased Eucalyptus leaves in southern China.
2. Materials and Methods
2.1. Disease Symptoms, Samples, and Fungal Isolations
The study was mainly conducted from July to October 2022, and from February to April 2023. The sampled regions had high temperatures (20 °C–35 °C) and high levels of humidity (60–90%). Diseases caused by Mycosphaerellaceae and Teratosphaeriaceae were surveyed on Eucalyptus plantations in the Guangxi, Guangdong, Yunnan, Hainan, and Fujian provinces in southern China. At most sampling sites, the trees were 1 to 2 years old. E. urophylla × E. grandis hybrids were dominant, although a few genotypes of E. urophylla, E. urophylla × E. tereticomis, and E. urophylla × E. pellita were also surveyed (Table 1). In the plantations, the leaves of trees infected by Mycosphaerellaceae and Teratosphaeriaceae fungi, resulting in leaf spot, vein delimitation, chlorosis, and intense defoliation were identified (Figure 1A,B). In some surveyed regions, all trees in the plantations were infected (Figure 1A,B). Based on the diseased symptoms, two groups of fungi causing different diseases were observed. The first group of fungi mainly infected mature and old leaves and produced abundant small spots on the infected leaves. The second group of fungi mainly infected the juvenile leaves, but affected some mature leaves as well, and resulted in water-soaked, chlorosis, and wrinkled symptoms on the infected leaves (Figure 1C–N). Disease symptoms of the first group of fungi were frequently observed in most surveyed Eucalyptus plantations. Disease symptoms with the second group of fungi were observed occasionally in some regions in southern China (Figure 1O–R). Diseased leaves with typical fruiting structures of Mycosphaerellaceae and Teratosphaeriaceae were collected from 3–59 trees, or approximately 30 trees, at the majority of sampling sites, depending on the area of the sampled plantation. Diseased leaf samples were transported to the laboratory for morphological examination, isolation, and further assessments.

Table 1.
Location details, collection information, and Eucalyptus tree plantation species of diseased leaf samples collected from 59 sites in five provinces.

Figure 1.
Disease symptoms associated with Teratosphaeria epicoccoides and T. destructans on Eucalyptus plantations in southern China: (A,B) trees in 2-year-old Eucalyptus urophylla × E. grandis plantations associated with T. epicoccoides and T. destructans. Leaves of the whole trees were infected and resulted in intense defoliation in the tree growth season; (C,D) typical leaf spot, vein delimitation, and chlorosis symptoms on leaves of E. urophylla × E. grandis associated with T. epicoccoides. New leaves on the top of shoots emerged after infection (C); (E,F) all leaves of 0.5-year-old E. urophylla × E. grandis trees in one plantation infected by T. destructans; (G,H) typical disease symptoms on E. urophylla × E. grandis leaves caused by T. epicoccoides (G) and T. destructans (H); (I,J) leaf blighted after infection by T. destructans; (K,L) heavy sporulation of T. epicoccoides; (M,N) water-soaked and chlorosis symptoms caused by T. destructans on the adaxial (M) and abaxial (N) leaf surface; and (O–R) Four E. urophylla × E. grandis hybrid genotypes exhibiting chlorosis and leaf spot caused by T. destructans.
Fungal isolates in the conidiomata with the typical morphological characteristics of Mycosphaerellaceae and Teratosphaeriaceae [23] were isolated from diseased leaves. The conidia masses in the conidiomata were scattered onto 2% malt extract agar (MEA) (20 g malt extract powder and 20 g agar powder per liter of water; malt extract powder was obtained from the Beijing Shuangxuan Microbial Culture Medium Products factory, Beijing, China; the agar powder was obtained from Beijing Solarbio Science & Technology Co., Ltd., Beijing, China) with sterile needles under a stereoscopic microscope (AxioCam Stemi 2000C, Carl Zeiss, Jena, Germany). After incubation at 25 °C for 6–10 h, the germinated conidia were transferred individually onto fresh 2% MEA under the dissection microscope and incubated at 25 °C for four weeks to obtain single-conidium cultures. One single-conidium culture was obtained from leaves of each sampled tree. All the single-conidium cultures were deposited in the culture collection (CSF) of the Research Institute of Fast-growing Trees (RIFT), Chinese Academy of Forestry (CAF), in Zhanjiang, Guangdong Province, China.
2.2. DNA Extraction, PCR Amplification, and Sequencing
All isolates obtained were used for DNA extraction and sequence analyses. DNA was extracted from four-week-old cultures. Mycelia were scraped using a sterilized scalpel and transferred to 2.0 mL Eppendorf tubes. The total genomic DNA was extracted using the CTAB protocol, as described by van Burik et al. [42]. The extracted DNA was dissolved in 30 μL of TE buffer (1 M Tris-HCl and 0.5 M EDTA, pH = 8.0). To degrade the RNA, 2.5 μL RNase (10 mg/mL) was added at 37 °C for one hour. The DNA concentration was measured using a NanoDrop 2000 spectrometer (Thermo Fisher Scientific, Waltham, MA, USA).
Based on previous research results, DNA sequences of the internal transcribed spacer regions and intervening 5.8S nrRNA gene (ITS), partial translation elongation factor 1-alpha (tef1) gene, and partial β-tubulin (tub2) gene were used as reliable DNA barcodes to clearly distinguish species in Mycosphaerellaceae and Teratosphaeriaceae, especially species of Teratosphaeria [25,26,43,44]. The three partial gene regions of T. epicoccoides were amplified using the primer pairs V9G /ITS-4 [45], EF1-728F/EF2 [46,47] and BT2a/BT2b [48], respectively. The three partial gene regions of T. destructans were amplified using the primer pairs ITS-3 /ITS-4 [45], EF1-728F/EF1-986R [46] and BT2a/BT2b [48], respectively. The PCR procedure was conducted as described by Andjic and co-authors [26].
All the PCR products of all isolates obtained in this study were sequenced in both the forward and reverse directions with the same primers used in the PCR amplification. Sequence reactions were conducted at the Beijing Genomics Institute, Guangzhou, China. All sequences obtained were edited using MEGA v. 7.0 software [49], and were deposited in GenBank (https://www.ncbi.nlm.nih.gov, accessed on 6 December 2023). The ITS, tef1, and tub2 gene regions were sequenced for all isolates obtained in this study.
2.3. Multi-Gene Phylogenetic Analyses and Species Identification
All isolates obtained in this study were genotyped by their ITS, tef1, and tub2 sequences. Based on the genotypes generated by the ITS, tef1, and tub2 sequences, sequences of two isolates for each ITS–tef1–tub2 genotype were selected for phylogenetic analyses. Isolates with the same genotype were considered to be the same species.
The preliminary identities of the isolates obtained in this study were determined by conducting a standard nucleotide BLAST search using all the generated sequences of ITS, tef1, and tub2. The BLAST results indicated that the isolates obtained in this study were grouped in the genus Teratosphaeria. The sequences of the type strains closely related to the Teratosphaeria isolates sequenced in the current study were downloaded from the NCBI database (http://www.ncbi.nlm.nih.gov/ accessed on 15 October 2023) and used for phylogenetic analyses (Table 3).

Table 3.
Isolates from other studies used in phylogenetic analyses in this study.

Table 2.
Isolates sequenced and used for phylogenetic analyses in this study.
Table 2.
Isolates sequenced and used for phylogenetic analyses in this study.
| Sampling Site No. a | Province | Isolate No. b,c | Sample and Isolate Information d | Species | GenBank Accession No. e | Genotype g | ||
|---|---|---|---|---|---|---|---|---|
| ITS | tef1 | tub2 | ||||||
| 1 | Yunnan | CSF25654 | 20230315-2-(2) | T. destructans | OR961532 | OR980975 | OR973016 | AAA |
| 1 | Yunnan | CSF25655 | 20230315-2-(3) | T. destructans | OR961533 | OR980976 | OR973017 | AAA |
| 1 | Yunnan | CSF25656 c | 20230315-2-(4) | T. destructans | OR961534 | OR980977 | OR973018 | BAA |
| 1 | Yunnan | CSF25657 | 20230315-2-(6) | T. destructans | OR961535 | OR980978 | OR973019 | AAA |
| 1 | Yunnan | CSF25658 | 20230315-2-(7) | T. destructans | OR961536 | OR980979 | OR973020 | AAA |
| 1 | Yunnan | CSF25659 c | 20230315-2-(8) | T. epicoccoides | OR961537 | OR980980 | OR973021 | ABA |
| 1 | Yunnan | CSF25660 | 20230315-2-(9) | T. destructans | OR961538 | OR980981 | OR973022 | AAA |
| 1 | Yunnan | CSF25661 | 20230315-2-(10) | T. destructans | OR961539 | OR980982 | OR973023 | AAA |
| 1 | Yunnan | CSF25686 | 20230315-2-(12) | T. epicoccoides | OR961540 | OR980983 | OR973024 | AAA |
| 1 | Yunnan | CSF25687 | 20230315-2-(13) | T. epicoccoides | OR961541 | OR980984 | OR973025 | ECA |
| 1 | Yunnan | CSF25688 | 20230315-2-(15) | T. epicoccoides | OR961542 | OR980985 | OR973026 | ECA |
| 1 | Yunnan | CSF25689 | 20230315-2-(16) | T. epicoccoides | OR961543 | OR980986 | OR973027 | ACA |
| 1 | Yunnan | CSF25690 | 20230315-2-(17) | T. epicoccoides | OR961544 | OR980987 | OR973028 | AAA |
| 1 | Yunnan | CSF25691 | 20230315-2-(19) | T. epicoccoides | OR961545 | OR980988 | OR973029 | AAA |
| 2 | Yunnan | CSF25640 | 20230315-1-(1) | T. destructans | OR961546 | OR980989 | OR973030 | AAA |
| 2 | Yunnan | CSF25641 | 20230315-1-(2) | T. destructans | OR961547 | OR980990 | OR973031 | AAA |
| 2 | Yunnan | CSF25642 | 20230315-1-(3) | T. destructans | OR961548 | OR980991 | OR973032 | AAA |
| 2 | Yunnan | CSF25643 | 20230315-1-(4) | T. destructans | OR961549 | OR980992 | OR973033 | AAA |
| 2 | Yunnan | CSF25644 | 20230315-1-(5) | T. destructans | OR961550 | OR980993 | OR973034 | AAA |
| 2 | Yunnan | CSF25645 | 20230315-1-(6) | T. destructans | OR961551 | OR980994 | OR973035 | AAA |
| 2 | Yunnan | CSF25646 | 20230315-1-(7) | T. destructans | OR961552 | OR980995 | OR973036 | AAA |
| 2 | Yunnan | CSF25647 | 20230315-1-(8) | T. destructans | OR961553 | OR980996 | OR973037 | AAA |
| 2 | Yunnan | CSF25648 | 20230315-1-(9) | T. destructans | OR961554 | OR980997 | OR973038 | AAA |
| 2 | Yunnan | CSF25649 | 20230315-1-(10) | T. destructans | OR961555 | OR980998 | OR973039 | AAA |
| 2 | Yunnan | CSF25650 | 20230315-1-(11) | T. destructans | OR961556 | OR980999 | OR973040 | AAA |
| 2 | Yunnan | CSF25651 | 20230315-1-(12) | T. destructans | OR961557 | OR981000 | OR973041 | AAA |
| 2 | Yunnan | CSF25652 | 20230315-1-(13) | T. destructans | OR961558 | OR981001 | OR973042 | AAA |
| 2 | Yunnan | CSF25653 | 20230315-1-(14) | T. destructans | OR961559 | OR981002 | OR973043 | AAA |
| 2 | Yunnan | CSF25683 c | 20230315-1-(24) | T. epicoccoides | OR961560 | OR981003 | OR973044 | AAA |
| 2 | Yunnan | CSF25685 | 20230315-1-(29) | T. epicoccoides | OR961561 | OR981004 | OR973045 | AAA |
| 3 | Yunnan | CSF25671 | 20230314-2-(12) | T. epicoccoides | OR961562 | OR981005 | OR973046 | AAA |
| 3 | Yunnan | CSF25672 | 20230314-2-(13) | T. epicoccoides | OR961563 | OR981006 | OR973047 | ACA |
| 3 | Yunnan | CSF25673 | 20230314-2-(14) | T. epicoccoides | OR961564 | OR981007 | OR973048 | AAA |
| 3 | Yunnan | CSF25674 | 20230314-2-(15) | T. epicoccoides | OR961565 | OR981008 | OR973049 | EAA |
| 3 | Yunnan | CSF25675 | 20230314-2-(16) | T. epicoccoides | OR961566 | OR981009 | OR973050 | AAA |
| 3 | Yunnan | CSF25676 | 20230314-2-(17) | T. epicoccoides | OR961567 | OR981010 | OR973051 | AAA |
| 4 | Yunnan | CSF25637 | 20230314-1-(4) | T. destructans | OR961568 | OR981011 | OR973052 | AAA |
| 4 | Yunnan | CSF25638 | 20230314-1-(10) | T. destructans | OR961569 | OR981012 | OR973053 | AAA |
| 4 | Yunnan | CSF25639 | 20230314-1-(13) | T. destructans | OR961570 | OR981013 | OR973054 | AAA |
| 5 | Yunnan | CSF25662 | 20230315-3-(1) | T. destructans | OR961571 | OR981014 | OR973055 | AAA |
| 5 | Yunnan | CSF25663 | 20230315-3-(2) | T. destructans | OR961572 | OR981015 | OR973056 | AAA |
| 5 | Yunnan | CSF25664 | 20230315-3-(3) | T. destructans | OR961573 | OR981016 | OR973057 | AAA |
| 5 | Yunnan | CSF25665 | 20230315-3-(4) | T. destructans | OR961574 | OR981017 | OR973058 | AAA |
| 5 | Yunnan | CSF25666 | 20230315-3-(5) | T. destructans | OR961575 | OR981018 | OR973059 | AAA |
| 5 | Yunnan | CSF25667 | 20230315-3-(6) | T. destructans | OR961576 | OR981019 | OR973060 | AAA |
| 5 | Yunnan | CSF25668 | 20230315-3-(7) | T. destructans | OR961577 | OR981020 | OR973061 | AAA |
| 5 | Yunnan | CSF25669 | 20230315-3-(8) | T. destructans | OR961578 | OR981021 | OR973062 | AAA |
| 5 | Yunnan | CSF25670 | 20230315-3-(9) | T. destructans | OR961579 | OR981022 | OR973063 | AAA |
| 5 | Yunnan | CSF25698 | 20230315-3-(16) | T. epicoccoides | OR961580 | OR981023 | OR973064 | AAA |
| 5 | Yunnan | CSF25699 | 20230315-3-(17) | T. epicoccoides | OR961581 | OR981024 | OR973065 | ACA |
| 5 | Yunnan | CSF25700 | 20230315-3-(18) | T. epicoccoides | OR961582 | OR981025 | OR973066 | EAA |
| 5 | Yunnan | CSF25701 | 20230315-3-(19) | T. epicoccoides | OR961583 | OR981026 | OR973067 | EAA |
| 5 | Yunnan | CSF25702 | 20230315-3-(21) | T. epicoccoides | OR961584 | OR981027 | OR973068 | ACA |
| 6 | Yunnan | CSF25703 | 20230316-2-(1) | T. epicoccoides | OR961585 | OR981028 | OR973069 | DIA |
| 6 | Yunnan | CSF25704 c | 20230316-2-(2) | T. epicoccoides | OR961586 | OR981029 | OR973070 | DIA |
| 6 | Yunnan | CSF25705 | 20230316-2-(4) | T. epicoccoides | OR961587 | OR981030 | OR973071 | DIA |
| 6 | Yunnan | CSF25706 | 20230316-2-(6) | T. epicoccoides | OR961588 | OR981031 | OR973072 | DIA |
| 6 | Yunnan | CSF25707 | 20230316-2-(7) | T. epicoccoides | OR961589 | OR981032 | OR973073 | DIA |
| 6 | Yunnan | CSF25708 | 20230316-2-(8) | T. epicoccoides | OR961590 | OR981033 | OR973074 | DIA |
| 7 | Yunnan | CSF25713 | 20230317-1-(1) | T. epicoccoides | OR961591 | OR981034 | OR973075 | AAA |
| 7 | Yunnan | CSF25714 | 20230317-1-(2) | T. epicoccoides | OR961592 | OR981035 | OR973076 | AAA |
| 7 | Yunnan | CSF25715 | 20230317-1-(4) | T. epicoccoides | OR961593 | OR981036 | OR973077 | EAA |
| 7 | Yunnan | CSF25716 | 20230317-1-(6) | T. epicoccoides | OR961594 | OR981037 | OR973078 | EAA |
| 7 | Yunnan | CSF25717 | 20230317-1-(7) | T. epicoccoides | OR961595 | OR981038 | OR973079 | ECA |
| 7 | Yunnan | CSF25718 | 20230317-1-(8) | T. epicoccoides | OR961596 | OR981039 | OR973080 | AAA |
| 8 | Yunnan | CSF25725 | 20230318-1-(1) | T. epicoccoides | OR961597 | OR981040 | OR973081 | ECA |
| 8 | Yunnan | CSF25726 | 20230318-1-(2) | T. epicoccoides | OR961598 | OR981041 | OR973082 | EAA |
| 8 | Yunnan | CSF25727 c | 20230318-1-(3) | T. epicoccoides | OR961599 | OR981042 | OR973083 | FCA |
| 8 | Yunnan | CSF25728 c | 20230318-1-(4) | T. epicoccoides | OR961600 | OR981043 | OR973084 | CAA |
| 8 | Yunnan | CSF25729 | 20230318-1-(5) | T. epicoccoides | OR961601 | OR981044 | OR973085 | EAA |
| 8 | Yunnan | CSF25730 | 20230318-1-(6) | T. epicoccoides | OR961602 | OR981045 | OR973086 | EAA |
| 9 | Yunnan | CSF25737 | 20230318-2-(1) | T. epicoccoides | OR961603 | OR981046 | OR973087 | AAA |
| 9 | Yunnan | CSF25738 | 20230318-2-(2) | T. epicoccoides | OR961604 | OR981047 | OR973088 | EAA |
| 9 | Yunnan | CSF25739 | 20230318-2-(3) | T. epicoccoides | OR961605 | OR981048 | OR973089 | EAA |
| 9 | Yunnan | CSF25740 | 20230318-2-(4) | T. epicoccoides | OR961606 | OR981049 | OR973090 | AAA |
| 9 | Yunnan | CSF25741 | 20230318-2-(5) | T. epicoccoides | OR961607 | OR981050 | OR973091 | EAA |
| 9 | Yunnan | CSF25742 | 20230318-2-(6) | T. epicoccoides | OR961608 | OR981051 | OR973092 | EAA |
| 10 | Yunnan | CSF25750 | 20230318-3-(1) | T. epicoccoides | OR961609 | OR981052 | OR973093 | EAA |
| 10 | Yunnan | CSF25751 | 20230318-3-(2) | T. epicoccoides | OR961610 | OR981053 | OR973094 | EAA |
| 10 | Yunnan | CSF25752 | 20230318-3-(3) | T. epicoccoides | OR961611 | OR981054 | OR973095 | EAA |
| 10 | Yunnan | CSF25753 | 20230318-3-(4) | T. epicoccoides | OR961612 | OR981055 | OR973096 | CAA |
| 10 | Yunnan | CSF25754 | 20230318-3-(7) | T. epicoccoides | OR961613 | OR981056 | OR973097 | AAA |
| 10 | Yunnan | CSF25755 | 20230318-3-(8) | T. epicoccoides | OR961614 | OR981057 | OR973098 | AAA |
| 11 | Yunnan | CSF25763 | 20230319-1-(2) | T. epicoccoides | OR961615 | OR981058 | OR973099 | EAA |
| 11 | Yunnan | CSF25764 | 20230319-1-(3) | T. epicoccoides | OR961616 | OR981059 | OR973100 | ACA |
| 11 | Yunnan | CSF25765 | 20230319-1-(5) | T. epicoccoides | OR961617 | OR981060 | OR973101 | AAA |
| 11 | Yunnan | CSF25766 | 20230319-1-(6) | T. epicoccoides | OR961618 | OR981061 | OR973102 | CAA |
| 11 | Yunnan | CSF25767 | 20230319-1-(7) | T. epicoccoides | OR961619 | OR981062 | OR973103 | AAA |
| 12 | Guangxi | CSF25066 c | 20220703-1-(1) | T. epicoccoides | OR961620 | OR981063 | OR973104 | ABA |
| 12 | Guangxi | CSF25067 | 20220703-1-(3) | T. destructans | OR961621 | OR981064 | OR973105 | CAA |
| 12 | Guangxi | CSF25068 | 20220703-1-(4) | T. destructans | OR961622 | OR981065 | OR973106 | CAA |
| 12 | Guangxi | CSF25069 | 20220703-1-(5) | T. destructans | OR961623 | OR981066 | OR973107 | AAA |
| 12 | Guangxi | CSF25070 | 20220703-1-(6) | T. destructans | OR961624 | OR981067 | OR973108 | CAA |
| 12 | Guangxi | CSF25071 | 20220703-1-(8) | T. destructans | OR961625 | OR981068 | OR973109 | AAA |
| 12 | Guangxi | CSF25072 c | 20220703-1-(10) | T. epicoccoides | OR961626 | OR981069 | OR973110 | XDA |
| 12 | Guangxi | CSF25218 c | 20220703-1-(12) | T. epicoccoides | OR961627 | OR981070 | OR973111 | AAA |
| 12 | Guangxi | CSF25219 | 20220703-1-(14) | T. epicoccoides | OR961628 | OR981071 | OR973112 | CAA |
| 12 | Guangxi | CSF25220 | 20220703-1-(15) | T. epicoccoides | OR961629 | OR981072 | OR973113 | ECA |
| 12 | Guangxi | CSF25222 | 20220703-1-(18) | T. epicoccoides | OR961630 | OR981073 | OR973114 | CAA |
| 12 | Guangxi | CSF25223 | 20220703-1-(19) | T. epicoccoides | OR961631 | OR981074 | OR973115 | BAA |
| 13 | Guangxi | CSF25083 | 20220703-3-(1) | T. destructans | OR961632 | OR981075 | OR973116 | AAA |
| 13 | Guangxi | CSF25084 | 20220703-3-(2) | T. destructans | OR961633 | OR981076 | OR973117 | CAA |
| 13 | Guangxi | CSF25085 | 20220703-3-(3) | T. destructans | OR961634 | OR981077 | OR973118 | AAA |
| 13 | Guangxi | CSF25086 | 20220703-3-(4) | T. destructans | OR961635 | OR981078 | OR973119 | CAA |
| 13 | Guangxi | CSF25087 | 20220703-3-(5) | T. destructans | OR961636 | OR981079 | OR973120 | CAA |
| 13 | Guangxi | CSF25088 | 20220703-3-(6) | T. epicoccoides | OR961637 | OR981080 | OR973121 | XBA |
| 13 | Guangxi | CSF25089 | 20220703-3-(7) | T. destructans | OR961638 | OR981081 | OR973122 | AAA |
| 13 | Guangxi | CSF25090 | 20220703-3-(9) | T. destructans | OR961639 | OR981082 | OR973123 | CAA |
| 13 | Guangxi | CSF25091 | 20220703-3-(10) | T. destructans | OR961640 | OR981083 | OR973124 | AAA |
| 13 | Guangxi | CSF25240 c | 20220703-3-(17) | T. epicoccoides | OR961641 | OR981084 | OR973125 | EAA |
| 13 | Guangxi | CSF25241 | 20220703-3-(19) | T. epicoccoides | OR961642 | OR981085 | OR973126 | EAA |
| 13 | Guangxi | CSF25242 | 20220703-3-(20) | T. epicoccoides | OR961643 | OR981086 | OR973127 | EAA |
| 13 | Guangxi | CSF25243 | 20220703-3-(22) | T. epicoccoides | OR961644 | OR981087 | OR973128 | ACA |
| 13 | Guangxi | CSF25244 | 20220703-3-(23) | T. epicoccoides | OR961645 | OR981088 | OR973129 | AAA |
| 13 | Guangxi | CSF25245 | 20220703-3-(24) | T. epicoccoides | OR961646 | OR981089 | OR973130 | ACA |
| 14 | Guangxi | CSF25073 c | 20220703-2-(1) | T. destructans | OR961647 | OR981090 | OR973131 | AAA |
| 14 | Guangxi | CSF25074 | 20220703-2-(2) | T. destructans | OR961648 | OR981091 | OR973132 | AAA |
| 14 | Guangxi | CSF25075 c | 20220703-2-(3) | T. epicoccoides | OR961649 | OR981092 | OR973133 | XBA |
| 14 | Guangxi | CSF25076 | 20220703-2-(4) | T. destructans | OR961650 | OR981093 | OR973134 | AAA |
| 14 | Guangxi | CSF25077 | 20220703-2-(5) | T. destructans | OR961651 | OR981094 | OR973135 | AAA |
| 14 | Guangxi | CSF25078 | 20220703-2-(6) | T. destructans | OR961652 | OR981095 | OR973136 | AAA |
| 14 | Guangxi | CSF25079 | 20220703-2-(7) | T. epicoccoides | OR961653 | OR981096 | OR973137 | ABA |
| 14 | Guangxi | CSF25080 | 20220703-2-(8) | T. destructans | OR961654 | OR981097 | OR973138 | CAA |
| 14 | Guangxi | CSF25081 | 20220703-2-(9) | T. epicoccoides | OR961655 | OR981098 | OR973139 | XBA |
| 14 | Guangxi | CSF25082 | 20220703-2-(10) | T. destructans | OR961656 | OR981099 | OR973140 | CAA |
| 14 | Guangxi | CSF25231 | 20220703-2-(21) | T. epicoccoides | OR961657 | OR981100 | OR973141 | AAA |
| 14 | Guangxi | CSF25232 | 20220703-2-(22) | T. epicoccoides | OR961658 | OR981101 | OR973142 | AAA |
| 14 | Guangxi | CSF25233 | 20220703-2-(25) | T. epicoccoides | OR961659 | OR981102 | OR973143 | EAA |
| 15 | Guangxi | CSF25092 | 20220703-4-(1) | T. destructans | OR961660 | OR981103 | OR973144 | AAA |
| 15 | Guangxi | CSF25093 | 20220703-4-(3) | T. destructans | OR961661 | OR981104 | OR973145 | AAA |
| 15 | Guangxi | CSF25094 | 20220703-4-(4) | T. destructans | OR961662 | OR981105 | OR973146 | CAA |
| 15 | Guangxi | CSF25095 | 20220703-4-(5) | T. destructans | OR961663 | OR981106 | OR973147 | CAA |
| 15 | Guangxi | CSF25096 | 20220703-4-(6) | T. destructans | OR961664 | OR981107 | OR973148 | AAA |
| 15 | Guangxi | CSF25097 | 20220703-4-(7) | T. destructans | OR961665 | OR981108 | OR973149 | AAA |
| 15 | Guangxi | CSF25098 | 20220703-4-(8) | T. destructans | OR961666 | OR981109 | OR973150 | CAA |
| 15 | Guangxi | CSF25099 | 20220703-4-(9) | T. destructans | OR961667 | OR981110 | OR973151 | CAA |
| 15 | Guangxi | CSF25247 c | 20220703-4-(10) | T. epicoccoides | OR961668 | OR981111 | OR973152 | ECA |
| 15 | Guangxi | CSF25248 | 20220703-4-(11) | T. epicoccoides | OR961669 | OR981112 | OR973153 | AAA |
| 16 | Guangxi | CSF25056 | 20220702-3-(1) | T. destructans | OR961670 | OR981113 | OR973154 | CAA |
| 16 | Guangxi | CSF25057 | 20220702-3-(2) | T. destructans | OR961671 | OR981114 | OR973155 | CAA |
| 16 | Guangxi | CSF25058 | 20220702-3-(3) | T. destructans | – f | OR981115 | OR973156 | -AA |
| 16 | Guangxi | CSF25059 | 20220702-3-(4) | T. destructans | OR961672 | OR981116 | OR973157 | CAA |
| 16 | Guangxi | CSF25060 | 20220702-3-(5) | T. destructans | OR961673 | OR981117 | OR973158 | AAA |
| 16 | Guangxi | CSF25061 | 20220702-3-(6) | T. destructans | OR961674 | OR981118 | OR973159 | CAA |
| 16 | Guangxi | CSF25062 | 20220702-3-(7) | T. destructans | OR961675 | OR981119 | OR973160 | AAA |
| 16 | Guangxi | CSF25063 | 20220702-3-(8) | T. destructans | OR961676 | OR981120 | OR973161 | CAA |
| 16 | Guangxi | CSF25064 | 20220702-3-(9) | T. destructans | OR961677 | OR981121 | OR973162 | CAA |
| 16 | Guangxi | CSF25065 | 20220702-3-(10) | T. destructans | OR961678 | OR981122 | OR973163 | CAA |
| 16 | Guangxi | CSF25214 c | 20220702-3-(17) | T. destructans | OR961679 | OR981123 | OR973164 | CBA |
| 16 | Guangxi | CSF25215 | 20220702-3-(18) | T. epicoccoides | OR961680 | OR981124 | OR973165 | ECA |
| 16 | Guangxi | CSF25216 | 20220702-3-(19) | T. epicoccoides | OR961681 | OR981125 | OR973166 | ECA |
| 16 | Guangxi | CSF25217 | 20220702-3-(20) | T. epicoccoides | OR961682 | OR981126 | OR973167 | EAA |
| 17 | Guangxi | CSF25046 | 20220702-2-(1) | T. destructans | OR961683 | OR981127 | OR973168 | CAA |
| 17 | Guangxi | CSF25047 | 20220702-2-(2) | T. destructans | OR961684 | OR981128 | OR973169 | AAA |
| 17 | Guangxi | CSF25048 c | 20220702-2-(3) | T. epicoccoides | OR961685 | OR981129 | OR973170 | CBA |
| 17 | Guangxi | CSF25049 | 20220702-2-(4) | T. destructans | OR961686 | OR981130 | OR973171 | AAA |
| 17 | Guangxi | CSF25050 | 20220702-2-(5) | T. destructans | OR961687 | OR981131 | OR973172 | AAA |
| 17 | Guangxi | CSF25051 | 20220702-2-(6) | T. destructans | OR961688 | OR981132 | OR973173 | CAA |
| 17 | Guangxi | CSF25052 | 20220702-2-(7) | T. destructans | OR961689 | OR981133 | OR973174 | AAA |
| 17 | Guangxi | CSF25053 | 20220702-2-(8) | T. destructans | OR961690 | OR981134 | OR973175 | AAA |
| 17 | Guangxi | CSF25054 c | 20220702-2-(9) | T. destructans | OR961691 | OR981135 | OR973176 | CAA |
| 17 | Guangxi | CSF25055 | 20220702-2-(10) | T. destructans | OR961692 | OR981136 | OR973177 | AAA |
| 17 | Guangxi | CSF25204 | 20220702-2-(17) | T. epicoccoides | OR961693 | OR981137 | OR973178 | EAA |
| 17 | Guangxi | CSF25205 | 20220702-2-(18) | T. epicoccoides | OR961694 | OR981138 | OR973179 | EAA |
| 17 | Guangxi | CSF25206 | 20220702-2-(19) | T. epicoccoides | OR961695 | OR981139 | OR973180 | EAA |
| 17 | Guangxi | CSF25207 | 20220702-2-(20) | T. epicoccoides | OR961696 | OR981140 | OR973181 | ECA |
| 18 | Guangxi | CSF25036 | 20220702-1-(1) | T. destructans | OR961697 | OR981141 | – | AA- |
| 18 | Guangxi | CSF25037 | 20220702-1-(2) | T. destructans | OR961698 | OR981142 | OR973182 | AAA |
| 18 | Guangxi | CSF25038 | 20220702-1-(3) | T. destructans | OR961699 | OR981143 | OR973183 | CAA |
| 18 | Guangxi | CSF25039 | 20220702-1-(4) | T. destructans | OR961700 | OR981144 | OR973184 | AAA |
| 18 | Guangxi | CSF25040 | 20220702-1-(5) | T. destructans | OR961701 | OR981145 | OR973185 | AAA |
| 18 | Guangxi | CSF25041 | 20220702-1-(6) | T. destructans | OR961702 | OR981146 | OR973186 | AAA |
| 18 | Guangxi | CSF25042 c | 20220702-1-(7) | T. epicoccoides | OR961703 | OR981147 | OR973187 | DJA |
| 18 | Guangxi | CSF25043 c | 20220702-1-(8) | T. epicoccoides | OR961704 | OR981148 | OR973188 | AHA |
| 18 | Guangxi | CSF25044 c | 20220702-1-(9) | T. epicoccoides | OR961705 | OR981149 | OR973189 | DJA |
| 18 | Guangxi | CSF25045 c | 20220702-1-(10) | T. epicoccoides | OR961706 | OR981150 | OR973190 | DHA |
| 18 | Guangxi | CSF25188 c | 20220702-1-(11) | T. epicoccoides | OR961707 | OR981151 | OR973191 | ACA |
| 18 | Guangxi | CSF25189 | 20220702-1-(12) | T. epicoccoides | OR961708 | OR981152 | OR973192 | ACA |
| 18 | Guangxi | CSF25192 | 20220702-1-(15) | T. epicoccoides | OR961709 | OR981153 | OR973193 | ACA |
| 18 | Guangxi | CSF25196 | 20220702-1-(19) | T. epicoccoides | OR961710 | OR981154 | OR973194 | AEA |
| 19 | Guangxi | CSF25100 | 20220704-1-(1) | T. destructans | OR961711 | OR981155 | OR973195 | CAA |
| 19 | Guangxi | CSF25101 | 20220704-1-(2) | T. destructans | OR961712 | OR981156 | OR973196 | CAA |
| 19 | Guangxi | CSF25102 | 20220704-1-(3) | T. destructans | OR961713 | OR981157 | OR973197 | CCA |
| 19 | Guangxi | CSF25103 | 20220704-1-(4) | T. destructans | OR961714 | OR981158 | OR973198 | AAA |
| 19 | Guangxi | CSF25104 | 20220704-1-(5) | T. destructans | OR961715 | OR981159 | OR973199 | CAA |
| 19 | Guangxi | CSF25105 | 20220704-1-(6) | T. destructans | OR961716 | OR981160 | OR973200 | CAA |
| 19 | Guangxi | CSF25106 | 20220704-1-(7) | T. destructans | OR961717 | OR981161 | OR973201 | CAA |
| 19 | Guangxi | CSF25107 | 20220704-1-(8) | T. destructans | OR961718 | OR981162 | OR973202 | CAA |
| 19 | Guangxi | CSF25108 | 20220704-1-(12) | T. destructans | OR961719 | OR981163 | OR973203 | CAA |
| 19 | Guangxi | CSF25109 | 20220704-1-(13) | T. destructans | OR961720 | OR981164 | OR973204 | CAA |
| 19 | Guangxi | CSF25250 | 20220704-1-(17) | T. epicoccoides | OR961721 | OR981165 | OR973205 | ECA |
| 19 | Guangxi | CSF25251 c | 20220704-1-(18) | T. epicoccoides | OR961722 | OR981166 | OR973206 | CCA |
| 19 | Guangxi | CSF25252 | 20220704-1-(19) | T. epicoccoides | OR961723 | OR981167 | OR973207 | ACA |
| 19 | Guangxi | CSF25253 c | 20220704-1-(20) | T. epicoccoides | OR961724 | OR981168 | OR973208 | GGA |
| 19 | Guangxi | CSF25254 | 20220704-1-(21) | T. epicoccoides | OR961725 | OR981169 | OR973209 | ACA |
| 20 | Guangxi | CSF25110 | 20220704-2-(1) | T. destructans | OR961726 | OR981170 | OR973210 | CAA |
| 20 | Guangxi | CSF25111 | 20220704-2-(2) | T. destructans | OR961727 | OR981171 | OR973211 | AAA |
| 20 | Guangxi | CSF25112 | 20220704-2-(4) | T. destructans | OR961728 | OR981172 | OR973212 | AAA |
| 20 | Guangxi | CSF25113 | 20220704-2-(5) | T. destructans | OR961729 | OR981173 | OR973213 | AAA |
| 20 | Guangxi | CSF25114 | 20220704-2-(6) | T. destructans | OR961730 | OR981174 | OR973214 | CAA |
| 20 | Guangxi | CSF25115 | 20220704-2-(7) | T. destructans | OR961731 | OR981175 | OR973215 | AAA |
| 20 | Guangxi | CSF25116 | 20220704-2-(8) | T. destructans | OR961732 | OR981176 | OR973216 | CAA |
| 20 | Guangxi | CSF25117 | 20220704-2-(10) | T. destructans | OR961733 | OR981177 | OR973217 | AAA |
| 20 | Guangxi | CSF25118 | 20220704-2-(11) | T. destructans | OR961734 | OR981178 | OR973218 | AAA |
| 20 | Guangxi | CSF25119 | 20220704-2-(12) | T. destructans | OR961735 | OR981179 | OR973219 | AAA |
| 20 | Guangxi | CSF25120 | 20220704-2-(13) | T. destructans | OR961736 | OR981180 | OR973220 | CAA |
| 20 | Guangxi | CSF25121 | 20220704-2-(14) | T. destructans | OR961737 | OR981181 | OR973221 | ACA |
| 20 | Guangxi | CSF25122 | 20220704-2-(15) | T. destructans | OR961738 | OR981182 | OR973222 | AAA |
| 20 | Guangxi | CSF25123 | 20220704-2-(16) | T. destructans | OR961739 | OR981183 | OR973223 | CAA |
| 20 | Guangxi | CSF25258 | 20220704-2-(17) | T. epicoccoides | OR961740 | OR981184 | OR973224 | DIA |
| 20 | Guangxi | CSF25259 | 20220704-2-(18) | T. epicoccoides | OR961741 | OR981185 | OR973225 | ACA |
| 20 | Guangxi | CSF25260 | 20220704-2-(19) | T. epicoccoides | OR961742 | OR981186 | OR973226 | AFA |
| 20 | Guangxi | CSF25261 | 20220704-2-(20) | T. epicoccoides | OR961743 | OR981187 | OR973227 | ACA |
| 20 | Guangxi | CSF25262 | 20220704-2-(21) | T. epicoccoides | OR961744 | OR981188 | OR973228 | ACA |
| 20 | Guangxi | CSF25263 | 20220704-2-(22) | T. epicoccoides | OR961745 | OR981189 | OR973229 | ACA |
| 21 | Guangxi | CSF25124 | 20220704-3-(1) | T. destructans | OR961746 | OR981190 | OR973230 | CAA |
| 21 | Guangxi | CSF25125 | 20220704-3-(2) | T. destructans | OR961747 | OR981191 | OR973231 | CAA |
| 21 | Guangxi | CSF25126 | 20220704-3-(3) | T. destructans | OR961748 | OR981192 | OR973232 | CAA |
| 21 | Guangxi | CSF25127 | 20220704-3-(4) | T. destructans | OR961749 | OR981193 | OR973233 | CAA |
| 21 | Guangxi | CSF25128 | 20220704-3-(5) | T. destructans | – | – | OR973234 | --A |
| 21 | Guangxi | CSF25129 | 20220704-3-(6) | T. destructans | OR961750 | OR981194 | OR973235 | CAA |
| 21 | Guangxi | CSF25130 | 20220704-3-(7) | T. destructans | OR961751 | OR981195 | OR973236 | CAA |
| 21 | Guangxi | CSF25131 | 20220704-3-(8) | T. destructans | OR961752 | OR981196 | OR973237 | CAA |
| 21 | Guangxi | CSF25132 | 20220704-3-(9) | T. destructans | OR961753 | OR981197 | OR973238 | AAA |
| 21 | Guangxi | CSF25133 | 20220704-3-(10) | T. destructans | OR961754 | OR981198 | OR973239 | CAA |
| 21 | Guangxi | CSF25270 | 20220704-3-(11) | T. epicoccoides | OR961755 | OR981199 | OR973240 | ACA |
| 21 | Guangxi | CSF25271 | 20220704-3-(13) | T. epicoccoides | OR961756 | OR981200 | OR973241 | EAA |
| 21 | Guangxi | CSF25272 | 20220704-3-(14) | T. epicoccoides | OR961757 | OR981201 | OR973242 | EAA |
| 21 | Guangxi | CSF25273 | 20220704-3-(16) | T. epicoccoides | OR961758 | OR981202 | OR973243 | ACA |
| 21 | Guangxi | CSF25274 | 20220704-3-(18) | T. epicoccoides | OR961759 | – | OR973244 | A-A |
| 21 | Guangxi | CSF25275 | 20220704-3-(20) | T. epicoccoides | OR961760 | OR981203 | OR973245 | EAA |
| 22 | Hainan | CSF25309 | 20221026-6-(1) | T. epicoccoides | OR961761 | OR981204 | OR973246 | ECA |
| 22 | Hainan | CSF25310 | 20221026-6-(2) | T. epicoccoides | OR961762 | OR981205 | OR973247 | ECA |
| 22 | Hainan | CSF25311 | 20221026-6-(3) | T. epicoccoides | OR961763 | OR981206 | OR973248 | EAA |
| 23 | Hainan | CSF25158 | 20221026-7-(1) | T. destructans | OR961764 | OR981207 | OR973249 | CAA |
| 23 | Hainan | CSF25159 | 20221026-7-(2) | T. destructans | OR961765 | OR981208 | OR973250 | CAA |
| 23 | Hainan | CSF25160 | 20221026-7-(3) | T. destructans | OR961766 | OR981209 | OR973251 | CAA |
| 23 | Hainan | CSF25161 | 20221026-7-(4) | T. destructans | OR961767 | OR981210 | OR973252 | AAA |
| 23 | Hainan | CSF25162 | 20221026-7-(5) | T. destructans | OR961768 | OR981211 | OR973253 | CAA |
| 23 | Hainan | CSF25163 | 20221026-7-(6) | T. destructans | OR961769 | OR981212 | OR973254 | AAA |
| 23 | Hainan | CSF25164 | 20221026-7-(7) | T. destructans | OR961770 | OR981213 | OR973255 | AAA |
| 23 | Hainan | CSF25165 | 20221026-7-(8) | T. destructans | OR961771 | OR981214 | OR973256 | AAA |
| 23 | Hainan | CSF25166 | 20221026-7-(9) | T. destructans | OR961772 | OR981215 | OR973257 | AAA |
| 23 | Hainan | CSF25167 | 20221026-7-(10) | T. destructans | OR961773 | OR981216 | OR973258 | CAA |
| 23 | Hainan | CSF25168 | 20221026-7-(11) | T. destructans | OR961774 | OR981217 | OR973259 | CAA |
| 23 | Hainan | CSF25312 | 20221026-7-(12) | T. epicoccoides | OR961775 | OR981218 | OR973260 | ACA |
| 23 | Hainan | CSF25313 | 20221026-7-(13) | T. epicoccoides | OR961776 | OR981219 | OR973261 | ACA |
| 23 | Hainan | CSF25314 | 20221026-7-(14) | T. epicoccoides | OR961777 | OR981220 | OR973262 | ACA |
| 23 | Hainan | CSF25315 | 20221026-7-(15) | T. epicoccoides | OR961778 | OR981221 | OR973263 | DIA |
| 23 | Hainan | CSF25316 | 20221026-7-(16) | T. epicoccoides | OR961779 | OR981222 | OR973264 | ACA |
| 23 | Hainan | CSF25317 c | 20221026-7-(17) | T. epicoccoides | OR961780 | OR981223 | OR973265 | AIA |
| 24 | Hainan | CSF25144 | 20221026-4-(1) | T. destructans | OR961781 | OR981224 | OR973266 | CAA |
| 24 | Hainan | CSF25145 | 20221026-4-(2) | T. destructans | OR961782 | OR981225 | OR973267 | CAA |
| 24 | Hainan | CSF25146 | 20221026-4-(3) | T. destructans | OR961783 | OR981226 | OR973268 | AAA |
| 24 | Hainan | CSF25147 | 20221026-4-(4) | T. destructans | OR961784 | OR981227 | OR973269 | AAA |
| 24 | Hainan | CSF25292 c | 20221026-4-(5) | T. epicoccoides | OR961785 | OR981228 | OR973270 | GGA |
| 24 | Hainan | CSF25293 c | 20221026-4-(6) | T. epicoccoides | OR961786 | OR981229 | OR973271 | AFA |
| 24 | Hainan | CSF25294 | 20221026-4-(7) | T. epicoccoides | – | OR981230 | OR973272 | -EA |
| 24 | Hainan | CSF25295 c | 20221026-4-(8) | T. epicoccoides | OR961787 | OR981231 | OR973273 | AEA |
| 24 | Hainan | CSF25296 | 20221026-4-(9) | T. epicoccoides | OR961788 | OR981232 | OR973274 | EAA |
| 24 | Hainan | CSF25297 | 20221026-4-(10) | T. epicoccoides | OR961789 | OR981233 | OR973275 | DIA |
| 25 | Hainan | CSF25148 | 20221026-5-(1) | T. destructans | OR961790 | OR981234 | OR973276 | CAA |
| 25 | Hainan | CSF25149 | 20221026-5-(2) | T. destructans | OR961791 | OR981235 | OR973277 | AAA |
| 25 | Hainan | CSF25150 | 20221026-5-(3) | T. destructans | OR961792 | OR981236 | OR973278 | AAA |
| 25 | Hainan | CSF25151 | 20221026-5-(4) | T. destructans | OR961793 | OR981237 | OR973279 | CAA |
| 25 | Hainan | CSF25152 | 20221026-5-(5) | T. destructans | OR961794 | OR981238 | OR973280 | CAA |
| 25 | Hainan | CSF25153 | 20221026-5-(6) | T. destructans | OR961795 | OR981239 | OR973281 | AAA |
| 25 | Hainan | CSF25154 | 20221026-5-(7) | T. destructans | OR961796 | OR981240 | OR973282 | ACA |
| 25 | Hainan | CSF25155 | 20221026-5-(8) | T. destructans | OR961797 | OR981241 | OR973283 | CAA |
| 25 | Hainan | CSF25156 | 20221026-5-(9) | T. destructans | OR961798 | – | OR973284 | C-A |
| 25 | Hainan | CSF25157 | 20221026-5-(10) | T. destructans | OR961799 | OR981242 | OR973285 | AAA |
| 25 | Hainan | CSF25299 c | 20221026-5-(11) | T. epicoccoides | OR961800 | OR981243 | OR973286 | AEA |
| 25 | Hainan | CSF25300 | 20221026-5-(12) | T. epicoccoides | OR961801 | OR981244 | OR973287 | AEA |
| 25 | Hainan | CSF25301 | 20221026-5-(13) | T. epicoccoides | OR961802 | OR981245 | OR973288 | AIA |
| 25 | Hainan | CSF25302 c | 20221026-5-(14) | T. epicoccoides | OR961803 | OR981246 | OR973289 | AFA |
| 25 | Hainan | CSF25303 | 20221026-5-(15) | T. epicoccoides | OR961804 | OR981247 | OR973290 | AAA |
| 26 | Hainan | CSF25134 c | 20221026-2-(1) | T. destructans | OR961805 | OR981248 | OR973291 | CCA |
| 26 | Hainan | CSF25135 | 20221026-2-(2) | T. destructans | OR961806 | OR981249 | OR973292 | ACA |
| 26 | Hainan | CSF25136 | 20221026-2-(3) | T. destructans | OR961807 | OR981250 | OR973293 | ACA |
| 26 | Hainan | CSF25286 | 20221026-2-(4) | T. epicoccoides | – | OR981251 | OR973294 | -EA |
| 26 | Hainan | CSF25287 | 20221026-2-(5) | T. epicoccoides | OR961808 | OR981252 | OR973295 | CAA |
| 26 | Hainan | CSF25288 | 20221026-2-(6) | T. epicoccoides | OR961809 | OR981253 | OR973296 | AAA |
| 26 | Hainan | CSF25289 | 20221026-2-(7) | T. epicoccoides | – | OR981254 | OR973297 | -IA |
| 26 | Hainan | CSF25290 | 20221026-2-(8) | T. epicoccoides | OR961810 | OR981255 | – | AE- |
| 26 | Hainan | CSF25291 | 20221026-2-(9) | T. epicoccoides | – | OR981256 | OR973298 | -EA |
| 27 | Hainan | CSF25137 c | 20221026-3-(1) | T. destructans | OR961811 | OR981257 | OR973299 | ACA |
| 27 | Hainan | CSF25138 | 20221026-3-(2) | T. destructans | OR961812 | OR981258 | OR973300 | CAA |
| 27 | Hainan | CSF25139 | 20221026-3-(3) | T. destructans | OR961813 | OR981259 | OR973301 | AAA |
| 27 | Hainan | CSF25140 | 20221026-3-(4) | T. destructans | OR961814 | OR981260 | OR973302 | AAA |
| 27 | Hainan | CSF25141 | 20221026-3-(5) | T. destructans | OR961815 | OR981261 | OR973303 | AAA |
| 27 | Hainan | CSF25142 | 20221026-3-(6) | T. destructans | OR961816 | OR981262 | OR973304 | CAA |
| 27 | Hainan | CSF25143 | 20221026-3-(7) | T. destructans | OR961817 | OR981263 | OR973305 | CAA |
| 28 | Hainan | CSF25276 | 20221026-1-(51) | T. epicoccoides | OR961818 | OR981264 | OR973306 | EAA |
| 28 | Hainan | CSF25277 c | 20221026-1-(52) | T. epicoccoides | OR961819 | OR981265 | OR973307 | DEA |
| 28 | Hainan | CSF25280 | 20221026-1-(55) | T. epicoccoides | OR961820 | OR981266 | OR973308 | BAA |
| 28 | Hainan | CSF25283 | 20221026-1-(58) | T. epicoccoides | OR961821 | OR981267 | OR973309 | AAA |
| 28 | Hainan | CSF25284 | 20221026-1-(59) | T. epicoccoides | OR961822 | OR981268 | OR973310 | CAA |
| 29 | Guangdong | CSF25612 | 20230221-2-(1) | T. destructans | OR961823 | OR981269 | OR973311 | CAA |
| 29 | Guangdong | CSF25613 | 20230221-2-(2) | T. destructans | OR961824 | OR981270 | OR973312 | CAA |
| 29 | Guangdong | CSF25614 | 20230221-2-(3) | T. destructans | OR961825 | OR981271 | OR973313 | ACA |
| 29 | Guangdong | CSF25615 | 20230221-2-(4) | T. destructans | OR961826 | OR981272 | OR973314 | CAA |
| 29 | Guangdong | CSF25616 | 20230221-2-(5) | T. destructans | OR961827 | OR981273 | OR973315 | AAA |
| 29 | Guangdong | CSF25617 | 20230221-2-(6) | T. destructans | OR961828 | OR981274 | OR973316 | AAA |
| 29 | Guangdong | CSF25618 | 20230221-2-(7) | T. destructans | OR961829 | OR981275 | OR973317 | CAA |
| 29 | Guangdong | CSF25619 | 20230221-2-(8) | T. destructans | OR961830 | OR981276 | OR973318 | CAA |
| 29 | Guangdong | CSF25620 | 20230221-2-(9) | T. destructans | OR961831 | OR981277 | OR973319 | CAA |
| 29 | Guangdong | CSF25621 | 20230221-2-(10) | T. destructans | OR961832 | OR981278 | OR973320 | ACA |
| 29 | Guangdong | CSF25622 | 20230221-2-(11) | T. destructans | OR961833 | OR981279 | OR973321 | CAA |
| 29 | Guangdong | CSF25623 | 20230221-2-(12) | T. destructans | OR961834 | OR981280 | OR973322 | AAA |
| 29 | Guangdong | CSF25520 | 20230221-2-(20) | T. epicoccoides | OR961835 | OR981281 | OR973323 | EAA |
| 29 | Guangdong | CSF25521 | 20230221-2-(21) | T. epicoccoides | OR961836 | OR981282 | OR973324 | ACA |
| 29 | Guangdong | CSF25522 | 20230221-2-(22) | T. epicoccoides | OR961837 | OR981283 | OR973325 | AAA |
| 29 | Guangdong | CSF25523 | 20230221-2-(23) | T. epicoccoides | OR961838 | OR981284 | OR973326 | EAA |
| 29 | Guangdong | CSF25524 | 20230221-2-(25) | T. epicoccoides | OR961839 | OR981285 | OR973327 | EAA |
| 29 | Guangdong | CSF25525 | 20230221-2-(26) | T. epicoccoides | OR961840 | OR981286 | OR973328 | EAA |
| 30 | Guangdong | CSF25599 | 20230221-1-(1) | T. destructans | OR961841 | OR981287 | OR973329 | CAA |
| 30 | Guangdong | CSF25600 | 20230221-1-(2) | T. destructans | OR961842 | OR981288 | OR973330 | AAA |
| 30 | Guangdong | CSF25601 c | 20230221-1-(3) | T. destructans | OR961843 | OR981289 | OR973331 | CCA |
| 30 | Guangdong | CSF25602 | 20230221-1-(4) | T. destructans | OR961844 | OR981290 | OR973332 | AAA |
| 30 | Guangdong | CSF25603 | 20230221-1-(5) | T. destructans | OR961845 | OR981291 | OR973333 | AAA |
| 30 | Guangdong | CSF25604 | 20230221-1-(6) | T. destructans | OR961846 | OR981292 | OR973334 | AAA |
| 30 | Guangdong | CSF25605 | 20230221-1-(7) | T. destructans | OR961847 | OR981293 | OR973335 | CAA |
| 30 | Guangdong | CSF25606 | 20230221-1-(8) | T. destructans | OR961848 | OR981294 | OR973336 | AAA |
| 30 | Guangdong | CSF25607 | 20230221-1-(9) | T. destructans | OR961849 | OR981295 | OR973337 | ACA |
| 30 | Guangdong | CSF25608 | 20230221-1-(10) | T. destructans | OR961850 | OR981296 | OR973338 | AAA |
| 30 | Guangdong | CSF25609 | 20230221-1-(11) | T. destructans | OR961851 | OR981297 | OR973339 | CAA |
| 30 | Guangdong | CSF25610 | 20230221-1-(12) | T. destructans | OR961852 | OR981298 | OR973340 | AAA |
| 30 | Guangdong | CSF25611 | 20230221-1-(14) | T. destructans | OR961853 | OR981299 | OR973341 | AAA |
| 30 | Guangdong | CSF25512 | 20230221-1-(24) | T. epicoccoides | OR961854 | OR981300 | OR973342 | ACA |
| 30 | Guangdong | CSF25513 c | 20230221-1-(26) | T. epicoccoides | OR961855 | OR981301 | OR973343 | AIA |
| 30 | Guangdong | CSF25514 | 20230221-1-(27) | T. epicoccoides | OR961856 | OR981302 | OR973344 | ACA |
| 30 | Guangdong | CSF25515 | 20230221-1-(28) | T. epicoccoides | OR961857 | OR981303 | OR973345 | ACA |
| 30 | Guangdong | CSF25516 | 20230221-1-(29) | T. epicoccoides | OR961858 | OR981304 | OR973346 | AAA |
| 30 | Guangdong | CSF25517 | 20230221-1-(30) | T. epicoccoides | OR961859 | OR981305 | OR973347 | ACA |
| 31 | Guangdong | CSF25587 | 20230220-3-(1) | T. destructans | OR961860 | OR981306 | OR973348 | AAA |
| 31 | Guangdong | CSF25588 | 20230220-3-(2) | T. destructans | OR961861 | OR981307 | OR973349 | AAA |
| 31 | Guangdong | CSF25589 | 20230220-3-(3) | T. destructans | OR961862 | OR981308 | OR973350 | AAA |
| 31 | Guangdong | CSF25590 | 20230220-3-(4) | T. destructans | OR961863 | OR981309 | OR973351 | AAA |
| 31 | Guangdong | CSF25591 | 20230220-3-(5) | T. destructans | OR961864 | OR981310 | OR973352 | AAA |
| 31 | Guangdong | CSF25592 | 20230220-3-(6) | T. destructans | OR961865 | OR981311 | OR973353 | AAA |
| 31 | Guangdong | CSF25593 | 20230220-3-(7) | T. destructans | OR961866 | OR981312 | OR973354 | AAA |
| 31 | Guangdong | CSF25594 | 20230220-3-(8) | T. destructans | OR961867 | OR981313 | OR973355 | AAA |
| 31 | Guangdong | CSF25595 | 20230220-3-(10) | T. destructans | OR961868 | OR981314 | OR973356 | AAA |
| 31 | Guangdong | CSF25596 | 20230220-3-(11) | T. destructans | OR961869 | OR981315 | OR973357 | AAA |
| 31 | Guangdong | CSF25597 | 20230220-3-(12) | T. destructans | OR961870 | OR981316 | OR973358 | CAA |
| 31 | Guangdong | CSF25598 | 20230220-3-(13) | T. destructans | OR961871 | OR981317 | OR973359 | CAA |
| 31 | Guangdong | CSF25500 | 20230220-3-(25) | T. epicoccoides | OR961872 | OR981318 | OR973360 | ACA |
| 31 | Guangdong | CSF25501 | 20230220-3-(26) | T. epicoccoides | OR961873 | OR981319 | OR973361 | ACA |
| 31 | Guangdong | CSF25502 | 20230220-3-(27) | T. epicoccoides | OR961874 | OR981320 | OR973362 | AAA |
| 31 | Guangdong | CSF25503 | 20230220-3-(28) | T. epicoccoides | OR961875 | OR981321 | OR973363 | ACA |
| 31 | Guangdong | CSF25504 | 20230220-3-(29) | T. epicoccoides | OR961876 | OR981322 | OR973364 | ACA |
| 31 | Guangdong | CSF25505 | 20230220-3-(30) | T. epicoccoides | – | OR981323 | OR973365 | -EA |
| 32 | Guangdong | CSF25624 | 20230221-3-(3) | T. destructans | OR961877 | OR981324 | OR973366 | CCA |
| 32 | Guangdong | CSF25625 | 20230221-3-(4) | T. destructans | OR961878 | OR981325 | OR973367 | ACA |
| 32 | Guangdong | CSF25626 | 20230221-3-(5) | T. destructans | OR961879 | OR981326 | OR973368 | AAA |
| 32 | Guangdong | CSF25627 | 20230221-3-(6) | T. destructans | OR961880 | OR981327 | OR973369 | CAA |
| 32 | Guangdong | CSF25628 | 20230221-3-(7) | T. destructans | OR961881 | OR981328 | OR973370 | AAA |
| 32 | Guangdong | CSF25629 c | 20230221-3-(8) | T. destructans | OR961882 | OR981329 | OR973371 | ACA |
| 32 | Guangdong | CSF25630 | 20230221-3-(9) | T. destructans | OR961883 | OR981330 | OR973372 | AAA |
| 32 | Guangdong | CSF25631 | 20230221-3-(10) | T. destructans | OR961884 | OR981331 | OR973373 | AAA |
| 32 | Guangdong | CSF25632 | 20230221-3-(11) | T. destructans | OR961885 | OR981332 | OR973374 | AAA |
| 32 | Guangdong | CSF25633 | 20230221-3-(12) | T. destructans | OR961886 | OR981333 | OR973375 | AAA |
| 32 | Guangdong | CSF25634 | 20230221-3-(13) | T. destructans | OR961887 | OR981334 | OR973376 | CCA |
| 32 | Guangdong | CSF25635 | 20230221-3-(14) | T. destructans | OR961888 | OR981335 | OR973377 | CAA |
| 32 | Guangdong | CSF25636 | 20230221-3-(15) | T. destructans | OR961889 | OR981336 | OR973378 | AAA |
| 32 | Guangdong | CSF25531 | 20230221-3-(18) | T. epicoccoides | OR961890 | OR981337 | OR973379 | ACA |
| 32 | Guangdong | CSF25532 | 20230221-3-(19) | T. epicoccoides | – | OR981338 | OR973380 | -IA |
| 32 | Guangdong | CSF25533 | 20230221-3-(20) | T. epicoccoides | OR961891 | OR981339 | OR973381 | ACA |
| 32 | Guangdong | CSF25534 | 20230221-3-(21) | T. epicoccoides | – | OR981340 | OR973382 | -AA |
| 32 | Guangdong | CSF25535 | 20230221-3-(22) | T. epicoccoides | OR961892 | OR981341 | OR973383 | ACA |
| 32 | Guangdong | CSF25536 | 20230221-3-(23) | T. epicoccoides | – | OR981342 | OR973384 | -AA |
| 33 | Guangdong | CSF25576 | 20230220-2-(1) | T. destructans | OR961893 | OR981343 | OR973385 | AAA |
| 33 | Guangdong | CSF25577 | 20230220-2-(2) | T. destructans | OR961894 | OR981344 | OR973386 | AAA |
| 33 | Guangdong | CSF25578 | 20230220-2-(3) | T. destructans | OR961895 | OR981345 | OR973387 | AAA |
| 33 | Guangdong | CSF25579 | 20230220-2-(4) | T. destructans | OR961896 | OR981346 | OR973388 | AAA |
| 33 | Guangdong | CSF25580 | 20230220-2-(5) | T. destructans | OR961897 | OR981347 | OR973389 | AAA |
| 33 | Guangdong | CSF25581 | 20230220-2-(6) | T. destructans | OR961898 | OR981348 | OR973390 | CCA |
| 33 | Guangdong | CSF25582 | 20230220-2-(7) | T. destructans | OR961899 | OR981349 | OR973391 | AAA |
| 33 | Guangdong | CSF25583 | 20230220-2-(8) | T. destructans | OR961900 | OR981350 | OR973392 | CCA |
| 33 | Guangdong | CSF25584 | 20230220-2-(9) | T. destructans | OR961901 | OR981351 | OR973393 | AAA |
| 33 | Guangdong | CSF25585 | 20230220-2-(11) | T. destructans | OR961902 | OR981352 | OR973394 | AAA |
| 33 | Guangdong | CSF25586 | 20230220-2-(12) | T. destructans | OR961903 | OR981353 | OR973395 | CAA |
| 33 | Guangdong | CSF25497 | 20230220-2-(18) | T. epicoccoides | OR961904 | OR981354 | OR973396 | EAA |
| 33 | Guangdong | CSF25498 | 20230220-2-(19) | T. epicoccoides | OR961905 | OR981355 | OR973397 | ECA |
| 33 | Guangdong | CSF25499 | 20230220-2-(20) | T. epicoccoides | OR961906 | OR981356 | OR973398 | EAA |
| 34 | Guangdong | CSF25568 | 20230220-1-(2) | T. destructans | OR961907 | OR981357 | OR973399 | AAA |
| 34 | Guangdong | CSF25569 | 20230220-1-(5) | T. destructans | OR961908 | OR981358 | OR973400 | CCA |
| 34 | Guangdong | CSF25570 | 20230220-1-(7) | T. destructans | OR961909 | OR981359 | OR973401 | CAA |
| 34 | Guangdong | CSF25571 | 20230220-1-(9) | T. destructans | OR961910 | OR981360 | OR973402 | AAA |
| 34 | Guangdong | CSF25572 | 20230220-1-(10) | T. destructans | OR961911 | OR981361 | OR973403 | CCA |
| 34 | Guangdong | CSF25573 | 20230220-1-(11) | T. destructans | OR961912 | OR981362 | OR973404 | CAA |
| 34 | Guangdong | CSF25574 | 20230220-1-(14) | T. destructans | OR961913 | OR981363 | OR973405 | AAA |
| 34 | Guangdong | CSF25575 c | 20230220-1-(16) | T. destructans | OR961914 | OR981364 | OR973406 | AAA |
| 34 | Guangdong | CSF25495 | 20230220-1-(17) | T. epicoccoides | – | OR981365 | OR973407 | -EA |
| 34 | Guangdong | CSF25496 | 20230220-1-(18) | T. epicoccoides | – | OR981366 | OR973408 | -IA |
| 35 | Guangdong | CSF25371 | 20230216-5-(20) | T. epicoccoides | OR961915 | OR981367 | OR973409 | CAA |
| 35 | Guangdong | CSF25372 c | 20230216-5-(22) | T. epicoccoides | OR961916 | OR981368 | OR973410 | ECA |
| 35 | Guangdong | CSF25373 | 20230216-5-(24) | T. epicoccoides | OR961917 | OR981369 | OR973411 | CAA |
| 35 | Guangdong | CSF25374 c | 20230216-5-(27) | T. epicoccoides | OR961918 | OR981370 | OR973412 | EAA |
| 35 | Guangdong | CSF25375 | 20230216-5-(30) | T. epicoccoides | OR961919 | OR981371 | OR973413 | EAA |
| 35 | Guangdong | CSF25376 | 20230216-5-(32) | T. epicoccoides | OR961920 | OR981372 | OR973414 | ECA |
| 36 | Guangdong | CSF25356 | 20230216-3-(9) | T. epicoccoides | OR961921 | OR981373 | OR973415 | AAA |
| 36 | Guangdong | CSF25357 | 20230216-3-(10) | T. epicoccoides | OR961922 | OR981374 | OR973416 | EAA |
| 36 | Guangdong | CSF25358 | 20230216-3-(12) | T. epicoccoides | OR961923 | OR981375 | OR973417 | ECA |
| 36 | Guangdong | CSF25359 | 20230216-3-(14) | T. epicoccoides | OR961924 | OR981376 | OR973418 | EAA |
| 36 | Guangdong | CSF25360 | 20230216-3-(15) | T. epicoccoides | OR961925 | OR981377 | OR973419 | CAA |
| 36 | Guangdong | CSF25361 | 20230216-3-(16) | T. epicoccoides | OR961926 | OR981378 | OR973420 | ECA |
| 37 | Guangdong | CSF25367 c | 20230216-4-(1) | T. epicoccoides | OR961927 | OR981379 | OR973421 | CCA |
| 37 | Guangdong | CSF25368 c | 20230216-4-(3) | T. epicoccoides | OR961928 | OR981380 | OR973422 | BAA |
| 37 | Guangdong | CSF25369 | 20230216-4-(4) | T. epicoccoides | OR961929 | OR981381 | OR973423 | EAA |
| 37 | Guangdong | CSF25370 | 20230216-4-(5) | T. epicoccoides | OR961930 | OR981382 | OR973424 | EAA |
| 38 | Guangdong | CSF25347 | 20230216-2-(10) | T. epicoccoides | OR961931 | OR981383 | OR973425 | EAA |
| 38 | Guangdong | CSF25348 | 20230216-2-(11) | T. epicoccoides | OR961932 | OR981384 | OR973426 | AAA |
| 38 | Guangdong | CSF25349 | 20230216-2-(12) | T. epicoccoides | OR961933 | OR981385 | OR973427 | CAA |
| 38 | Guangdong | CSF25350 | 20230216-2-(13) | T. epicoccoides | OR961934 | OR981386 | OR973428 | AAA |
| 38 | Guangdong | CSF25351 c | 20230216-2-(14) | T. epicoccoides | OR961935 | OR981387 | OR973429 | HAA |
| 38 | Guangdong | CSF25352 | 20230216-2-(15) | T. epicoccoides | OR961936 | OR981388 | OR973430 | AAA |
| 39 | Guangdong | CSF25325 | 20230215-1-(1) | T. epicoccoides | OR961937 | OR981389 | OR973431 | CCA |
| 39 | Guangdong | CSF25326 c | 20230215-1-(2) | T. epicoccoides | OR961938 | OR981390 | OR973432 | CAA |
| 39 | Guangdong | CSF25327 | 20230215-1-(3) | T. epicoccoides | OR961939 | OR981391 | OR973433 | EAA |
| 39 | Guangdong | CSF25328 | 20230215-1-(6) | T. epicoccoides | OR961940 | OR981392 | OR973434 | ACA |
| 39 | Guangdong | CSF25329 | 20230215-1-(7) | T. epicoccoides | OR961941 | OR981393 | OR973435 | EAA |
| 39 | Guangdong | CSF25330 | 20230215-1-(8) | T. epicoccoides | OR961942 | OR981394 | OR973436 | EAA |
| 40 | Guangdong | CSF25336 | 20230216-1-(11) | T. epicoccoides | OR961943 | OR981395 | OR973437 | ECA |
| 40 | Guangdong | CSF25337 | 20230216-1-(13) | T. epicoccoides | OR961944 | OR981396 | OR973438 | ECA |
| 40 | Guangdong | CSF25338 | 20230216-1-(15) | T. epicoccoides | OR961945 | OR981397 | OR973439 | EAA |
| 40 | Guangdong | CSF25339 | 20230216-1-(16) | T. epicoccoides | OR961946 | OR981398 | OR973440 | AAA |
| 40 | Guangdong | CSF25340 c | 20230216-1-(17) | T. epicoccoides | OR961947 | OR981399 | OR973441 | BCA |
| 40 | Guangdong | CSF25341 | 20230216-1-(18) | T. epicoccoides | OR961948 | OR981400 | OR973442 | EAA |
| 41 | Guangdong | CSF25546 | 20230217-3-(1) | T. destructans | OR961949 | OR981401 | OR973443 | CCA |
| 41 | Guangdong | CSF25547 c | 20230217-3-(2) | T. epicoccoides | OR961950 | OR981402 | OR973444 | XBA |
| 41 | Guangdong | CSF25548 | 20230217-3-(6) | T. destructans | OR961951 | OR981403 | OR973445 | AAA |
| 41 | Guangdong | CSF25549 | 20230217-3-(8) | T. destructans | OR961952 | OR981404 | OR973446 | CAA |
| 41 | Guangdong | CSF25550 | 20230217-3-(13) | T. destructans | OR961953 | OR981405 | OR973447 | AAA |
| 41 | Guangdong | CSF25551 | 20230217-3-(16) | T. destructans | OR961954 | OR981406 | OR973448 | AAA |
| 41 | Guangdong | CSF25552 | 20230217-3-(17) | T. destructans | OR961955 | OR981407 | OR973449 | AAA |
| 41 | Guangdong | CSF25553 | 20230217-3-(21) | T. destructans | OR961956 | OR981408 | OR973450 | CAA |
| 41 | Guangdong | CSF25554 | 20230217-3-(24) | T. destructans | OR961957 | OR981409 | OR973451 | CCA |
| 41 | Guangdong | CSF25555 | 20230217-3-(25) | T. destructans | OR961958 | OR981410 | OR973452 | CAA |
| 41 | Guangdong | CSF25556 | 20230217-3-(30) | T. destructans | OR961959 | OR981411 | OR973453 | CAA |
| 41 | Guangdong | CSF25557 c | 20230217-3-(37) | T. destructans | OR961960 | OR981412 | OR973454 | CAA |
| 41 | Guangdong | CSF25399 | 20230217-3-(51) | T. epicoccoides | OR961961 | OR981413 | OR973455 | AAA |
| 41 | Guangdong | CSF25400 | 20230217-3-(52) | T. epicoccoides | OR961962 | OR981414 | OR973456 | AAA |
| 41 | Guangdong | CSF25401 | 20230217-3-(53) | T. epicoccoides | OR961963 | OR981415 | OR973457 | CAA |
| 41 | Guangdong | CSF25402 | 20230217-3-(54) | T. epicoccoides | OR961964 | OR981416 | OR973458 | ECA |
| 41 | Guangdong | CSF25403 | 20230217-3-(55) | T. epicoccoides | OR961965 | OR981417 | OR973459 | AAA |
| 41 | Guangdong | CSF25404 | 20230217-3-(56) | T. epicoccoides | OR961966 | OR981418 | OR973460 | ECA |
| 42 | Guangdong | CSF25558 | 20230219-1-(5) | T. destructans | OR961967 | OR981419 | OR973461 | AAA |
| 42 | Guangdong | CSF25559 | 20230219-1-(8) | T. destructans | OR961968 | OR981420 | OR973462 | AAA |
| 42 | Guangdong | CSF25560 | 20230219-1-(9) | T. destructans | OR961969 | OR981421 | OR973463 | CAA |
| 42 | Guangdong | CSF25561 | 20230219-1-(15) | T. destructans | OR961970 | OR981422 | OR973464 | AAA |
| 42 | Guangdong | CSF25562 | 20230219-1-(17) | T. destructans | OR961971 | OR981423 | OR973465 | AAA |
| 42 | Guangdong | CSF25563 | 20230219-1-(23) | T. destructans | OR961972 | OR981424 | OR973466 | CAA |
| 42 | Guangdong | CSF25564 | 20230219-1-(25) | T. destructans | OR961973 | OR981425 | OR973467 | CAA |
| 42 | Guangdong | CSF25565 | 20230219-1-(26) | T. destructans | OR961974 | OR981426 | OR973468 | CAA |
| 42 | Guangdong | CSF25566 | 20230219-1-(27) | T. destructans | OR961975 | OR981427 | OR973469 | AAA |
| 42 | Guangdong | CSF25567 | 20230219-1-(29) | T. destructans | OR961976 | OR981428 | OR973470 | CAA |
| 43 | Guangdong | CSF25390 | 20230217-2-(18) | T. epicoccoides | OR961977 | OR981429 | OR973471 | ECA |
| 43 | Guangdong | CSF25391 | 20230217-2-(20) | T. epicoccoides | OR961978 | OR981430 | OR973472 | AAA |
| 43 | Guangdong | CSF25392 | 20230217-2-(21) | T. epicoccoides | OR961979 | OR981431 | OR973473 | AAA |
| 43 | Guangdong | CSF25393 | 20230217-2-(24) | T. epicoccoides | OR961980 | OR981432 | OR973474 | EAA |
| 43 | Guangdong | CSF25394 | 20230217-2-(25) | T. epicoccoides | OR961981 | OR981433 | OR973475 | EAA |
| 43 | Guangdong | CSF25395 | 20230217-2-(26) | T. epicoccoides | OR961982 | OR981434 | OR973476 | EAA |
| 44 | Guangdong | CSF25378 | 20230217-1-(13) | T. epicoccoides | OR961983 | OR981435 | OR973477 | ECA |
| 44 | Guangdong | CSF25379 | 20230217-1-(14) | T. epicoccoides | OR961984 | OR981436 | OR973478 | CAA |
| 44 | Guangdong | CSF25380 | 20230217-1-(15) | T. epicoccoides | OR961985 | OR981437 | OR973479 | ECA |
| 44 | Guangdong | CSF25381 | 20230217-1-(17) | T. epicoccoides | OR961986 | OR981438 | OR973480 | EAA |
| 44 | Guangdong | CSF25382 | 20230217-1-(18) | T. epicoccoides | OR961987 | OR981439 | OR973481 | EAA |
| 44 | Guangdong | CSF25383 | 20230217-1-(21) | T. epicoccoides | OR961988 | OR981440 | OR973482 | EAA |
| 45 | Guangdong | CSF25436 c | 20230218-1-(6) | T. epicoccoides | OR961989 | OR981441 | OR973483 | ACA |
| 45 | Guangdong | CSF25437 | 20230218-1-(7) | T. epicoccoides | OR961990 | OR981442 | OR973484 | EAA |
| 45 | Guangdong | CSF25438 | 20230218-1-(8) | T. epicoccoides | OR961991 | OR981443 | OR973485 | AAA |
| 45 | Guangdong | CSF25439 | 20230218-1-(9) | T. epicoccoides | OR961992 | OR981444 | OR973486 | EAA |
| 45 | Guangdong | CSF25440 | 20230218-1-(10) | T. epicoccoides | OR961993 | OR981445 | OR973487 | EAA |
| 45 | Guangdong | CSF25441 | 20230218-1-(11) | T. epicoccoides | OR961994 | OR981446 | OR973488 | BAA |
| 46 | Guangdong | CSF25419 | 20230217-4-(3) | T. epicoccoides | OR961995 | OR981447 | OR973489 | EAA |
| 46 | Guangdong | CSF25420 | 20230217-4-(4) | T. epicoccoides | OR961996 | OR981448 | OR973490 | AAA |
| 46 | Guangdong | CSF25421 | 20230217-4-(5) | T. epicoccoides | OR961997 | OR981449 | OR973491 | AAA |
| 46 | Guangdong | CSF25422 | 20230217-4-(6) | T. epicoccoides | OR961998 | OR981450 | OR973492 | ECA |
| 46 | Guangdong | CSF25423 | 20230217-4-(7) | T. epicoccoides | – | OR981451 | OR973493 | -CA |
| 46 | Guangdong | CSF25424 | 20230217-4-(8) | T. epicoccoides | OR961999 | OR981452 | OR973494 | EAA |
| 47 | Guangdong | CSF25451 | 20230218-2-(7) | T. epicoccoides | OR962000 | OR981453 | OR973495 | AAA |
| 47 | Guangdong | CSF25452 | 20230218-2-(8) | T. epicoccoides | OR962001 | OR981454 | OR973496 | EAA |
| 47 | Guangdong | CSF25453 | 20230218-2-(9) | T. epicoccoides | OR962002 | OR981455 | OR973497 | BAA |
| 47 | Guangdong | CSF25454 | 20230218-2-(10) | T. epicoccoides | OR962003 | OR981456 | OR973498 | AAA |
| 47 | Guangdong | CSF25455 | 20230218-2-(11) | T. epicoccoides | OR962004 | OR981457 | OR973499 | ECA |
| 47 | Guangdong | CSF25456 | 20230218-2-(13) | T. epicoccoides | OR962005 | OR981458 | OR973500 | ACA |
| 48 | Guangdong | CSF25468 | 20230218-3-(12) | T. epicoccoides | OR962006 | OR981459 | OR973501 | AAA |
| 48 | Guangdong | CSF25469 | 20230218-3-(13) | T. epicoccoides | OR962007 | OR981460 | OR973502 | ACA |
| 48 | Guangdong | CSF25470 | 20230218-3-(14) | T. epicoccoides | OR962008 | OR981461 | OR973503 | AAA |
| 48 | Guangdong | CSF25471 | 20230218-3-(15) | T. epicoccoides | OR962009 | OR981462 | OR973504 | ACA |
| 48 | Guangdong | CSF25472 | 20230218-3-(16) | T. epicoccoides | OR962010 | OR981463 | OR973505 | EAA |
| 48 | Guangdong | CSF25473 | 20230218-3-(17) | T. epicoccoides | OR962011 | OR981464 | OR973506 | EAA |
| 49 | Guangdong | CSF25482 | 20230218-4-(2) | T. epicoccoides | OR962012 | OR981465 | OR973507 | AAA |
| 49 | Guangdong | CSF25483 | 20230218-4-(4) | T. epicoccoides | OR962013 | OR981466 | OR973508 | EAA |
| 49 | Guangdong | CSF25484 | 20230218-4-(5) | T. epicoccoides | OR962014 | OR981467 | OR973509 | BAA |
| 49 | Guangdong | CSF25485 | 20230218-4-(6) | T. epicoccoides | OR962015 | OR981468 | OR973510 | EAA |
| 49 | Guangdong | CSF25486 | 20230218-4-(7) | T. epicoccoides | OR962016 | OR981469 | OR973511 | ACA |
| 49 | Guangdong | CSF25487 | 20230218-4-(8) | T. epicoccoides | OR962017 | OR981470 | OR973512 | EAA |
| 50 | Fujian | CSF25801 | 20230330-1-(21) | T. epicoccoides | OR962018 | OR981471 | OR973513 | ACA |
| 50 | Fujian | CSF25802 | 20230330-1-(22) | T. epicoccoides | OR962019 | OR981472 | OR973514 | EAA |
| 50 | Fujian | CSF25803 | 20230330-1-(23) | T. epicoccoides | OR962020 | OR981473 | OR973515 | AAA |
| 50 | Fujian | CSF25804 | 20230330-1-(24) | T. epicoccoides | OR962021 | OR981474 | OR973516 | AAA |
| 50 | Fujian | CSF25805 | 20230330-1-(25) | T. epicoccoides | OR962022 | OR981475 | OR973517 | ECA |
| 51 | Fujian | CSF25773 c | 20230328-1-(2) | T. epicoccoides | OR962023 | OR981476 | OR973518 | DIA |
| 51 | Fujian | CSF25774 | 20230328-1-(5) | T. epicoccoides | OR962024 | OR981477 | OR973519 | DIA |
| 51 | Fujian | CSF25775 | 20230328-1-(7) | T. epicoccoides | OR962025 | OR981478 | OR973520 | DIA |
| 51 | Fujian | CSF25776 | 20230328-1-(8) | T. epicoccoides | OR962026 | OR981479 | OR973521 | DIA |
| 51 | Fujian | CSF25777 | 20230328-1-(9) | T. epicoccoides | OR962027 | OR981480 | OR973522 | ECA |
| 51 | Fujian | CSF25778 | 20230328-1-(11) | T. epicoccoides | OR962028 | OR981481 | OR973523 | AAA |
| 52 | Fujian | CSF25782 | 20230329-1-(1) | T. epicoccoides | – | OR981482 | OR973524 | -EA |
| 52 | Fujian | CSF25783 | 20230329-1-(2) | T. epicoccoides | OR962029 | OR981483 | OR973525 | EAA |
| 52 | Fujian | CSF25784 | 20230329-1-(3) | T. epicoccoides | OR962030 | OR981484 | OR973526 | EAA |
| 52 | Fujian | CSF25785 | 20230329-1-(4) | T. epicoccoides | OR962031 | OR981485 | OR973527 | ACA |
| 52 | Fujian | CSF25786 | 20230329-1-(5) | T. epicoccoides | – | OR981486 | OR973528 | -CA |
| 52 | Fujian | CSF25787 | 20230329-1-(6) | T. epicoccoides | OR962032 | OR981487 | OR973529 | EAA |
| 53 | Fujian | CSF25790 | 20230329-2-(1) | T. epicoccoides | OR962033 | OR981488 | OR973530 | DIA |
| 53 | Fujian | CSF25791 | 20230329-2-(4) | T. epicoccoides | OR962034 | OR981489 | OR973531 | DIA |
| 53 | Fujian | CSF25792 | 20230329-2-(5) | T. epicoccoides | OR962035 | OR981490 | OR973532 | DIA |
| 53 | Fujian | CSF25793 | 20230329-2-(6) | T. epicoccoides | OR962036 | OR981491 | OR973533 | DIA |
| 53 | Fujian | CSF25794 | 20230329-2-(7) | T. epicoccoides | – | OR981492 | OR973534 | -IA |
| 53 | Fujian | CSF25795 | 20230329-2-(8) | T. epicoccoides | OR962037 | OR981493 | OR973535 | DIA |
| 54 | Fujian | CSF25850 | 20230331-3-(1) | T. epicoccoides | OR962038 | OR981494 | OR973536 | CAA |
| 54 | Fujian | CSF25851 | 20230331-3-(2) | T. epicoccoides | OR962039 | OR981495 | OR973537 | ECA |
| 54 | Fujian | CSF25852 | 20230331-3-(4) | T. epicoccoides | OR962040 | OR981496 | OR973538 | EAA |
| 54 | Fujian | CSF25853 | 20230331-3-(5) | T. epicoccoides | OR962041 | OR981497 | OR973539 | CAA |
| 54 | Fujian | CSF25854 | 20230331-3-(6) | T. epicoccoides | OR962042 | OR981498 | OR973540 | CAA |
| 54 | Fujian | CSF25855 | 20230331-3-(7) | T. epicoccoides | OR962043 | OR981499 | OR973541 | EAA |
| 55 | Fujian | CSF25840 c | 20230331-2-(2) | T. epicoccoides | OR962044 | OR981500 | OR973542 | BAA |
| 55 | Fujian | CSF25841 | 20230331-2-(3) | T. epicoccoides | OR962045 | OR981501 | OR973543 | ECA |
| 55 | Fujian | CSF25842 | 20230331-2-(4) | T. epicoccoides | OR962046 | OR981502 | OR973544 | EAA |
| 55 | Fujian | CSF25843 | 20230331-2-(5) | T. epicoccoides | OR962047 | OR981503 | OR973545 | AAA |
| 55 | Fujian | CSF25844 | 20230331-2-(6) | T. epicoccoides | OR962048 | OR981504 | OR973546 | AAA |
| 55 | Fujian | CSF25845 | 20230331-2-(9) | T. epicoccoides | OR962049 | OR981505 | OR973547 | EAA |
| 56 | Fujian | CSF25830 | 20230331-1-(1) | T. epicoccoides | OR962050 | OR981506 | OR973548 | ACA |
| 56 | Fujian | CSF25831 | 20230331-1-(2) | T. epicoccoides | OR962051 | OR981507 | OR973549 | EAA |
| 56 | Fujian | CSF25832 | 20230331-1-(3) | T. epicoccoides | OR962052 | OR981508 | OR973550 | BAA |
| 56 | Fujian | CSF25833 | 20230331-1-(5) | T. epicoccoides | OR962053 | OR981509 | OR973551 | CCA |
| 56 | Fujian | CSF25834 | 20230331-1-(6) | T. epicoccoides | OR962054 | OR981510 | OR973552 | EAA |
| 56 | Fujian | CSF25835 | 20230331-1-(9) | T. epicoccoides | OR962055 | OR981511 | OR973553 | AAA |
| 57 | Fujian | CSF25820 | 20230330-3-(1) | T. epicoccoides | OR962056 | OR981512 | OR973554 | EAA |
| 57 | Fujian | CSF25821 | 20230330-3-(2) | T. epicoccoides | OR962057 | OR981513 | OR973555 | ECA |
| 57 | Fujian | CSF25822 | 20230330-3-(3) | T. epicoccoides | OR962058 | OR981514 | OR973556 | CAA |
| 57 | Fujian | CSF25823 | 20230330-3-(4) | T. epicoccoides | OR962059 | OR981515 | OR973557 | ECA |
| 57 | Fujian | CSF25824 | 20230330-3-(5) | T. epicoccoides | OR962060 | OR981516 | OR973558 | EAA |
| 57 | Fujian | CSF25825 | 20230330-3-(6) | T. epicoccoides | OR962061 | OR981517 | OR973559 | ECA |
| 58 | Fujian | CSF25810 | 20230330-2-(1) | T. epicoccoides | OR962062 | OR981518 | OR973560 | AAA |
| 58 | Fujian | CSF25811 | 20230330-2-(3) | T. epicoccoides | OR962063 | OR981519 | OR973561 | CCA |
| 58 | Fujian | CSF25812 | 20230330-2-(4) | T. epicoccoides | OR962064 | OR981520 | OR973562 | CAA |
| 58 | Fujian | CSF25813 | 20230330-2-(5) | T. epicoccoides | OR962065 | OR981521 | OR973563 | CCA |
| 58 | Fujian | CSF25814 | 20230330-2-(6) | T. epicoccoides | OR962066 | OR981522 | OR973564 | EAA |
| 58 | Fujian | CSF25815 | 20230330-2-(8) | T. epicoccoides | OR962067 | OR981523 | OR973565 | ACA |
| 59 | Fujian | CSF25860 | 20230401-1-(1) | T. epicoccoides | OR962068 | OR981524 | OR973566 | AAA |
| 59 | Fujian | CSF25861 | 20230401-1-(2) | T. epicoccoides | OR962069 | OR981525 | OR973567 | EAA |
| 59 | Fujian | CSF25862 | 20230401-1-(3) | T. epicoccoides | OR962070 | OR981526 | OR973568 | AAA |
| 59 | Fujian | CSF25863 | 20230401-1-(4) | T. epicoccoides | OR962071 | OR981527 | OR973569 | EAA |
| 59 | Fujian | CSF25864 | 20230401-1-(5) | T. epicoccoides | OR962072 | OR981528 | OR973570 | EAA |
| 59 | Fujian | CSF25865 | 20230401-1-(7) | T. epicoccoides | OR962073 | OR981529 | OR973571 | EAA |
a Code of 59 sampling sites connecting to “Sampling Site No.” in Table 1. b CSF: Culture Collection located at Research Institute of Fasting-growing Trees (RIFT), Chinese Academy of Forestry, Zhanjiang, Guangdong Province, China. c Isolates used for phylogenetic analyses in this study. d Information associated with sample site and isolate, for example, “20230315-2-(2)” indicates sample number “20230315-2-(2)” and isolate from this sample; the sample number connecting to “Sample and Isolate Information” in Table 1. e ITS = internal transcribed spacer regions and intervening 5.8S nrRNA gene; tef1 = translation elongation factor 1-alpha; tub2 = β-tubulin. f “–” represents the relative locus that was not successfully amplified in this study. g Genotype within each Teratosphaeria species, determined by sequences of the ITS, tef1 and tub2 regions. The same letter among isolates from each species means they shared the same genotype; “-” means not available.
Sequences downloaded from the NCBI database and sequences generated in this study were aligned using MAFFT online v. 7 (http://mafft.cbrc.jp/alignment/server/ (accessed on 2 November 2023)) [50], with the iterative refinement method (FFT-NS-i setting). The alignments were further edited manually with MEGA v. 7.0 software [49] when necessary. Sequences of each of the ITS, tef1, and tub2 gene regions, as well as the combination of these three gene regions, were analyzed.
Maximum likelihood (ML) analyses were conducted for the three individual gene sequences of ITS, tef1, and tub2, as well as for a concatenated dataset of all three genes. ML analyses were performed with RaxML v. 8.2.4 on the CIPRES Science Gateway v. 3.3 [51], with default GTR substitution matrix and 1000 bootstrap replicates [52]. Phylogenetic trees were viewed using MEGA v. 7.0 [49]. Sequence data of one isolate of Staninwardia suttonii (CBS 120061) were used as the outgroup [30].
3. Results
3.1. Fungal Isolations
Diseased leaves with conidiomata of Mycosphaerellaceae and Teratosphaeriaceae were collected from 59 sites in five provinces in southern China (Table 1; Figure 2). Single conidia from the conidiomata on the diseased leaves were transferred to fresh 2% MEA for fungal isolation. These conidia exhibited the typical morphological characteristics of Teratosphaeria species. Two groups of fungi corresponding to the disease symptoms were observed. The conidia of the first group of fungi were generally brown, and straight to slightly curved in shape. The number of septa in the conidia ranged from one to seven; the majority of conidia had three to five septa (Figure 3). The conidia of the second group of fungi were light brown, variously curved, and rarely straight. The conidia had one to three septa; the majority of conidia had three septa (Figure 4). The conidia of both groups of fungi were base truncate and apex obtuse. The conidia of the first group of fungi were wider, straighter, and darker than those of the second group (Figure 3 and Figure 4). These two groups of fungi had the typical morphological characteristics of T. epicoccoides and T. destructans, respectively [43,53]. In total, 558 isolates were obtained from the 59 sites. Three to twenty isolates were obtained from each sampled site, depending on the number of samples collected and the morphological variations in the conidia on the diseased leaves (Table 1).
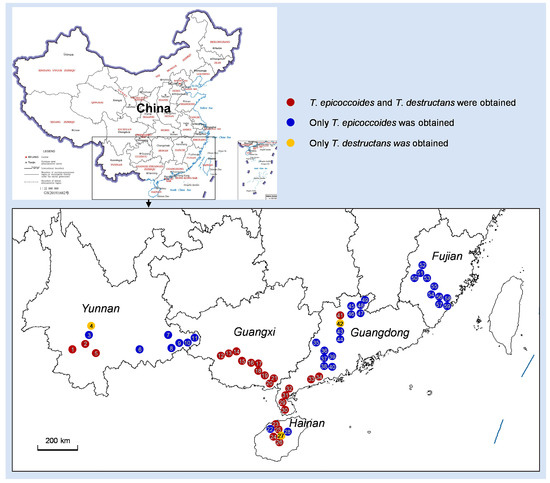
Figure 2.
Map showing the 59 sites in the five provinces in China where the diseased leaf samples were collected, and the Teratosphaeria species identified in each site.
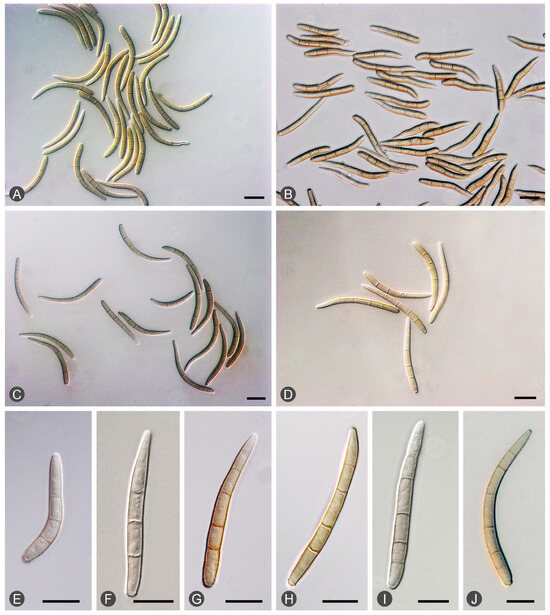
Figure 3.
Morphological features of Teratosphaeria epicoccoides: (A–D) slightly curved conidia with septa from different Eucalyptus trees. (E–J) Conidia with three to six septa: three septa (E,F); four septa (G,H); five septa (I); and six septa (J).
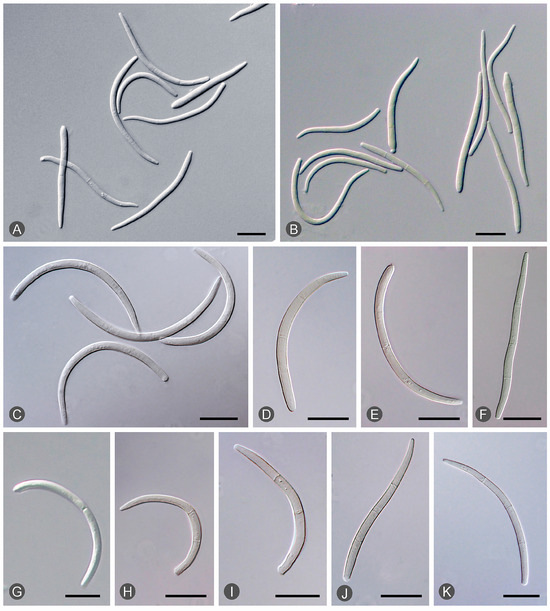
Figure 4.
Morphological features of Teratosphaeria destructans: (A–F) curved and occasionally straight conidia with septa from different Eucalyptus trees. (G–K) Conidia with one to three septa: one septum (G); two septa (H,I); and three septa (J,K).
Both groups of fungi were isolated from plantations of E. urophylla, E. urophylla × E. grandis, E. urophylla × E. pellita and E. urophylla × E. tereticomis, and most especially from multiple genotypes of E. urophylla × E. grandis, which were widely planted in the sampled regions. In addition, the first group of fungi were also frequently observed on the majority of other Eucalyptus genotypes in the plantations, including species of E. camaldulensis, E. grandis, E. pellita and their hybrids with E. urophylla. T. destructans is limited to certain species of E. grandis, E. urophylla, E. tereticornis and their hybrids.
3.2. Multi-Gene Phylogenetic Analyses and Species Identification
The BLAST results indicated that the 558 isolates obtained and sequenced in this study were mainly categorized into two groups. These two groups of fungi were similar to T. epicoccoides (Group A) and T. destructans (Group B), respectively. Amplicons generated for the ITS, tef1, and tub2 gene regions of fungi similar to T. epicoccoides were approximately 500, 350, and 350 bp, respectively. The sequences of ITS, tef1, and tub2 gene regions of fungi similar to T. destructans were approximately 290, 250, and 350 bp, respectively. Sequences of two isolates for each genotype generated by the three genes were used in the analyses. Sequences of ex-type specimen strains and other strains of 29 Teratosphaeria species, including T. epicoccoides and T. destructans, closely related to isolates obtained in the current study, were downloaded from GenBank for sequence comparisons and phylogenetic analyses (Table 3).
Based on the phylogenetic analyses of the ITS, tef1, tub2, and the combined datasets, the isolates of two groups of fungi, Group A and Group B in this study, were most closely related to T. epicoccoides and T. destructans, respectively. The isolates of Group A were consistently grouped with, or close to, the ex-type isolate CMW 5348 of T. epicoccoides in each of the ITS, tef1, tub2, and the combined dataset trees (Figure 5, Figure 6, Figure 7 and Figure 8). Additionally, some isolates of Group A formed independent clades in each of the four phylogenetic trees (Figure 5, Figure 6, Figure 7 and Figure 8). This is consistent with the phylogenetic analysis results presented by Taole et al. [43]. Analysis of the sequence data of the three regions resulted in incongruent genealogies. There was no evidence of distinct species boundaries. Isolates of Group A were identified as T. epicoccoides.
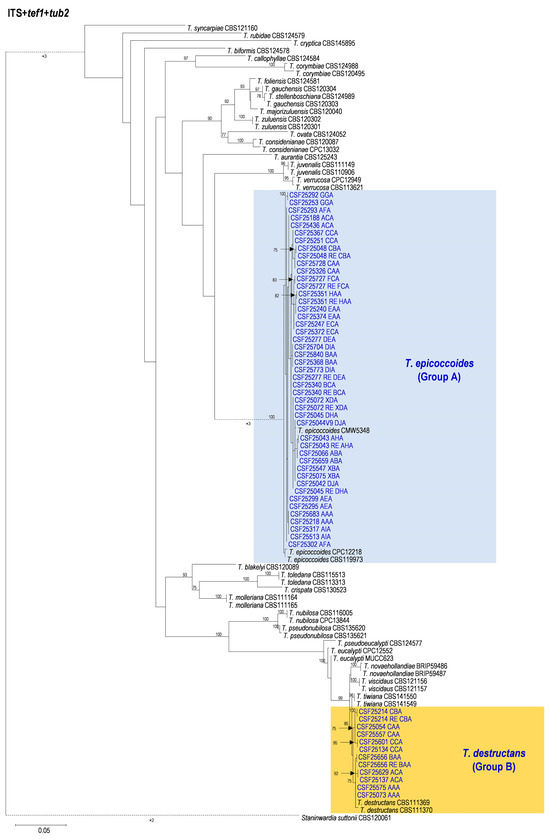
Figure 5.
Phylogenetic tree obtained from the maximum likelihood (ML) analysis of the combined dataset of ITS, tef1, and tub2 in these three gene regions. Bootstrap values ≥ 75% from the ML analysis are indicated at nodes. Isolates reported in this study are highlighted in blue.
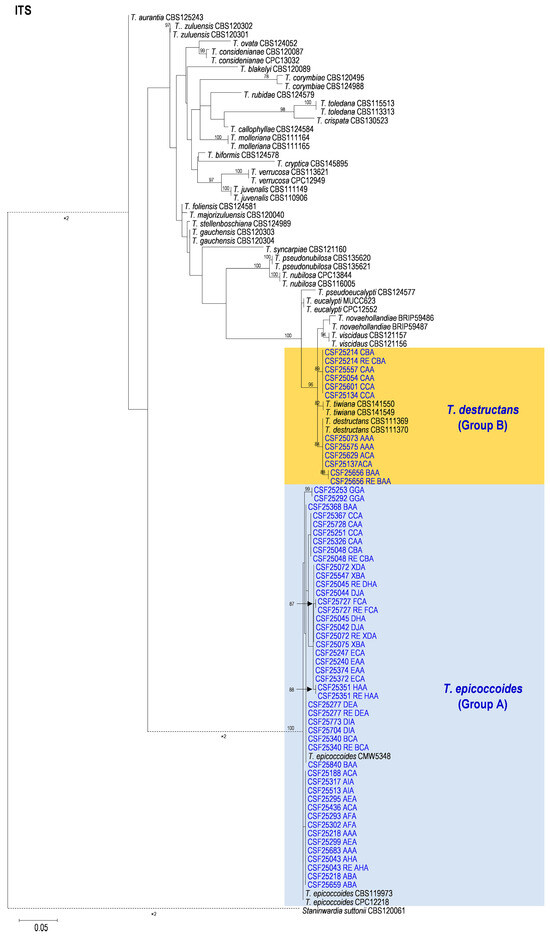
Figure 6.
Phylogenetic tree obtained from the maximum likelihood (ML) analysis of the dataset of the ITS region. Bootstrap values ≥ 75% from the ML analysis are indicated at nodes. Isolates reported in this study are highlighted in blue.
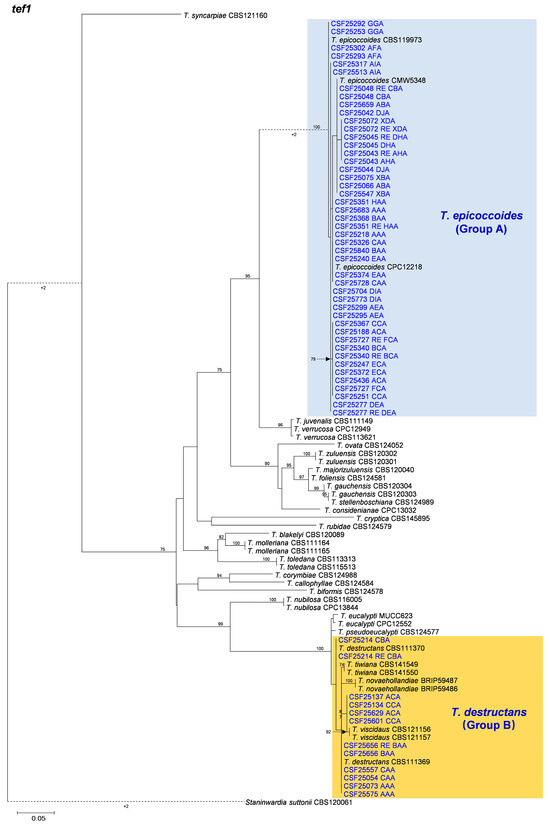
Figure 7.
Phylogenetic tree obtained from the maximum likelihood (ML) analysis of the dataset of the tef1 region. Bootstrap values ≥ 75% from the ML analysis are indicated at nodes. Isolates reported in this study are highlighted in blue.
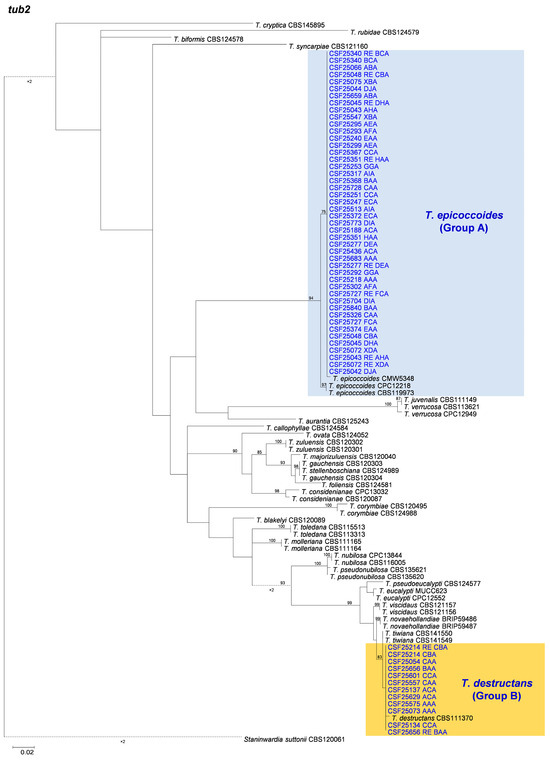
Figure 8.
Phylogenetic tree obtained from the maximum likelihood (ML) analysis of the dataset of the tub2 region. Bootstrap values ≥ 75% from the ML analysis are indicated at nodes. Isolates reported in this study are highlighted in blue.
In Group B, all isolates were grouped with ex-type isolate CBS 111370 of T. destructans in the tub2 tree (Figure 8). Additionally, some isolates of Group B formed independent clades in each of the ITS and tef1 phylogenetic trees. However, these isolates did not form consistently independent clades, nor were they supported by high bootstrap values in both the ITS and tef1 phylogenetic trees (Figure 6 and Figure 7). For example, isolates of genotypes CAA and CBA were grouped together and formed one independent clade with high bootstrap value (89%) in the ITS tree (Figure 6). These isolates were grouped with ex-type isolates CBS 111369 and CBS 111370 of T. destructans in the tef1 tree (Figure 7). The results suggest that these reflect intraspecific sequence differences rather than interspecies variation. Combining the phylogenetic analysis results of ITS, tef1, tub2 and the datasets (Figure 5, Figure 6, Figure 7 and Figure 8), the isolates in Group B were identified as T. destructans.
3.3. Distribution of Teratosphaeria Species
Based on the DNA sequence comparisons of ITS, tef1, and tub2 sequences, the 558 isolates obtained from 59 sampling sites in this study were identified as T. epicoccoides (312 isolates; 55.9%) and T. destructans (246 isolates, 44.1%). T. epicoccoides was isolated and identified in 56 (accounting 95%) sampling sites, and T. destructans was obtained from 27 (accounting 46%) sampling sites. These two species were all obtained from 24 (accounting for 41%) sampling sites (Figure 2).
Determined by ITS, tef1, and tub2 sequences, 21 genotypes were generated for the 291 T. epicoccoides isolates (the genotypes of 21 isolates were not clear because the sequences of three gene regions were not all obtained) (Table 4). Six genotypes were generated for the 242 T. destructans isolates (the genotypes of four isolates were not clear because the sequences of three gene regions were not all obtained) (Table 5). The ratios of the genotype number to the isolate number of T. epicoccoides and T. destructans were all highest in Hainan Province.

Table 4.
Isolate numbers of each genotype of T. epicoccoides in the Eucalyptus plantations in each of the five provinces.

Table 5.
Isolate numbers of each genotype of T. destructans in the Eucalyptus plantations in each of the five provinces.
4. Discussion
In this study, systematical disease surveys were conducted to collect diseased Eucalyptus leaves with typical fruiting structures of Mycosphaerellaceae and Teratosphaeriaceae in the core Eucalyptus plantation regions in China. In total, 558 isolates were identified according to disease symptoms and morphological characteristics and were mainly based on DNA sequence comparisons of three gene regions. These fungi were identified as T. epicoccoides and T. destructans. The results of this study indicate that T. epicoccoides and T. destructans are widely distributed in the core Eucalyptus planting regions in southern China.
This study has clarified and expanded the geographic distribution of Teratosphaeria species associated with diseased Eucalyptus leaves in southern China. Teratosphaeria epicoccoides was initially described in 1992. It was indicated that this species is distributed in many countries and regions worldwide. The leaf specimens used for the initial description included a total of 25 species of Eucalyptus and Corymbia in Australia, Brazil, Argentina, Zambia, India, and Ethiopia [54]. Currently, T. epicoccoides is widely reported on Eucalyptus leaves globally [41,43]. Teratosphaeria destructans was initially reported and described in 1996 on the leaves of E. grandis in Indonesia, along with some other unknown Eucalyptus species [53]. Currently, T. destructans is reported in a global range, including countries in Africa such as South Africa, and countries in Asia such as China, Thailand, and East Timor [41,55]. In China, T. epicoccoides and T. destructans were both first isolated from leaves of E. urophylla in Guangdong Province in 2006 [13]. Prior to this study, the distribution reports of these two species in China were both limited to Guangdong Province [13,26]. The results of this study suggest that T. epicoccoides is distributed in areas that are majority-planted with Eucalyptus in southern China, and T. destructans is also widely distributed throughout large regions in this country.
DNA sequence comparisons were considered a reliable method for the classification and identification of Mycosphaerellaceae and Teratosphaeriaceae [23,30]. For species of Mycosphaerellaceae and Teratosphaeriaceae, traditional classification has primarily relied on morphological characteristics. When classifying based on morphological characteristics, the identification of species of Mycosphaerellaceae and Teratosphaeriaceae is influenced by the conserved morphological features of their respective sexual morphs [56,57,58]. Scientists have therefore shifted the focus of classifying and identifying these species mostly towards their asexual morphs [59,60,61]. However, similar asexual morphologies have also independently evolved from different taxa, adding further complexity to the taxonomy of these species [62]. In 2014, Quaedvlieg et al. proposed that the accurate classification of genera and species within Mycosphaerellaceae and Teratosphaeriaceae cannot be achieved solely through morphological comparisons. Instead, it requires the integration of molecular data [30]. The research results indicated that the ITS gene serves as a primary barcode locus to distinguish taxa in Mycosphaerella and Teratosphaeria associated with Eucalyptus leaf disease. The ITS gene is easily generated and has the most extensive dataset available; therefore, tef1, tub, or rpb2 represent useful secondary barcode loci [30]. In this study, species identification was primarily based on the comparison of three gene sequences of ITS, tef1, and tub2. These three gene sequences have been widely employed to differentiate both intra- and inter-specific variations among species in Teratosphaeriaceae [25,43,55].
To date, eight species of Mycosphaerellaceae and Teratosphaeriaceae isolated from Eucalyptus leaves in China have been identified based on DNA sequence comparisons. These species include Neoceratosperma yunnanensis, Pallidocercospora crystallina, Paramycosphaerella marksii, Pseudocercospora flavomarginata, Pse. gracilis, Pse. haiweiensis, Teratosphaeria destructans, and T. epicoccoides [26,27,29,30,63]. With the exception of two species of Teratosphaeria, the other six species reside in Mycosphaerellaceae. It is still unknown whether these eight species are all pathogenic to Eucalyptus in China because no pathogenicity tests have been conducted.
One shortcoming of this study is that the pathogenicity of the two identified Teratosphaeria species was not investigated. At present, on a global scale, methods for assessing the pathogenicity of Mycosphaerellaceae and Teratosphaeriaceae species isolated from diseased Eucalyptus leaves include spraying ascospore suspensions, conidial suspensions, hyphal fragment suspensions, or a mixture of spores and hyphal fragment suspensions onto Eucalyptus seedlings in controlled environments. Additional inoculation methods involve applying ascospore masses or lesions from diseased leaf areas onto Eucalyptus seedling leaves [44,57,64]. Conidial suspensions are considered to be a typical method for evaluating the pathogenicity of Mycosphaerellaceae and Teratosphaeriaceae species; however, this is not possible for species that do not sporulate sufficiently in culture. Very limited research has been conducted on the pathogenicity of T. destructans and T. epicoccoides on Eucalyptus. The Teratosphaeria isolates obtained in the current study failed to produce conidia on media. In order to test the pathogenicity of T. destructans and T. epicoccoides, it is vital to find a way to sufficiently induce the sporulation of these fungi in culture or find a new method to test their pathogenicity. For example, the hyphal fragment suspensions were replaced with conidial suspensions to test the pathogenicity of Calonectria pseudoreteaudii [65]. Hyphal fragment suspension may also be used to assess the pathogenicity of Teratosphaeria species, although this need to be evaluated. Previous research results indicated that T. epicoccoides should be indigenous to eastern Australia, and spread to western Australia and other regions of the world [41,66]. T. destructans is recognized as absent from Australia. The origin of T. destructans remains unknown, but it is likely to have originated in Indonesia or East Timor [41,67,68,69]. Research results support the hypothesis that T. epicoccoides and T. destructans are spread via the human-mediated movement of infected plants and seeds [41,66,70]. This suggests the need for much more stringent and more sophisticated biosecurity measures, such as the use of metabarcoding approaches to test seedlots and plant materials under quarantine [41,67,68].
The results of this study confirmed that both T. epicoccoides and T. destructans are dominant species on diseased Eucalyptus leaves in China. Both species are widely distributed in Eucalyptus plantations that are mainly distributed across regions of southern China. Although previous studies have indicated that T. epicoccoides and T. destructans are widely distributed on Eucalyptus leaves globally [41,44], research on these species on diseased Eucalyptus leaves specifically in China is limited. During the sampling process in the current study, we found that the severity caused by each species of T. epicoccoides and T. destructans on different genotypes of E. urophylla × E. grandis were different. Research results showed that significant differences in resistance exist among the six tested E. grandis × E. urophylla genotypes to the inoculated T. destructans [71]. It is suggested that breeding and selection of Eucalyptus tolerant/resistant species, hybrids and clones are needed for the future management of leaf diseases caused by Teratosphaeria species [41]. Future research should focus on exploring pathogenicity testing methods for T. epicoccoides and T. destructans and clarifying their pathogenic characteristics. This will guide the selection of disease-resistant Eucalyptus genotypes for these two widely distributed species.
5. Conclusions
This study has proven that both T. epicoccoides and T. destructans are dominant species and widely distributed on diseased Eucalyptus leaves in southern China; both T. epicoccoides and T. destructans were present in each of the five sampled provinces with multiple genotypes. These two species are not clonal in China. The genetic diversities of T. epicoccoides and T. destructans were high in the southern sampled province. The wider distribution, and higher genetic diversity than has previously been considered indicates that both T. epicoccoides and T. destructans should be considered seriously in future disease management schemes.
Author Contributions
Conceptualization, S.C.; Data curation, B.C., W.W. and S.C.; Formal analysis, W.W. and S.C.; Funding acquisition, S.C.; Investigation, B.C., W.W. and S.C.; Methodology, W.W. and S.C.; Project administration, S.C.; Resources, S.C.; Supervision, S.C.; Validation, W.W. and S.C.; Visualization, S.C.; Writing—original draft, S.C.; Writing—review and editing, B.C., W.W. and S.C. All authors have read and agreed to the published version of the manuscript.
Funding
This study was supported by the National Key R and D Program of China (China-South Africa Forestry Joint Research Centre Project; project No. 2018YFE0120900), the National Ten-Thousand Talents Program (Project No. W03070115), and the Guangdong Top Young Talents Program in China (Project No. 20171172).
Data Availability Statement
Data are contained within the article.
Acknowledgments
We thank Linqin Lu, Xueying Liang, Simin Xiang and Ying Liu (both at Research Institute of Fast-growing Trees, Chinese Academy of Forestry) for their assistance in collecting samples.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Xie, Y.J. Scientific innovation leads to fast development of eucalypt research and industry in China. Eucalypt Sci. Technol. 2022, 39, 35–42. [Google Scholar]
- Xie, Y.J.; Du, A.P. Eucalypts in Southern China; CCP Central Party School Press: Beijing, China, 2019. [Google Scholar]
- Xie, Y.J.; Arnold, R.J.; Wu, Z.H.; Chen, S.F.; Du, A.P.; Luo, J.Z. Advances in eucalypt research in China. Front. Agric. Sci. Eng. 2017, 4, 380–390. [Google Scholar] [CrossRef]
- Chen, S.F. Summary of fungal pathogens from eucalypts in China: 2006–2013. Eucalypt Sci. Technol. 2014, 31, 37–65. [Google Scholar]
- Turnbull, J.W. Development of Sustainable Forestry Plantations in China: A Review; Australian Centre for International Agricultural Research: Canberra, Australia, 2007.
- Zhou, X.D.; Wingfield, M.J. Eucalypt diseases and their management in China. Australas. Plant Pathol. 2011, 40, 339–345. [Google Scholar] [CrossRef]
- Old, K.M.; Wingfield, M.J.; Yuan, Z.Q. A Manual of Diseases of Eucalypts in South–East Asia; Center for International Forestry Research: Jakarta, Indonesia, 2003. [Google Scholar]
- Carstensen, G.D.; Venter, S.N.; Wingfield, M.J.; Coutinho, T.A. Two Ralstonia species associated with bacterial wilt of Eucalyptus. Plant Pathol. 2017, 66, 393–403. [Google Scholar] [CrossRef]
- Chen, S.F.; Gryzenhout, M.; Roux, J.; Xie, Y.J.; Wingfield, M.J.; Zhou, X.D. Identification and pathogenicity of Chrysoporthe cubensis on Eucalyptus and Syzygium spp. in South China. Plant Dis. 2010, 94, 1143–1150. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.F.; Barnes, I.; Chungu, D.; Roux, J.; Wingfield, M.J.; Xie, Y.J.; Zhou, X.D. High population diversity and increasing importance of the Eucalyptus stem canker pathogen, Teratosphaeria zuluensis, in South China. Australas. Plant Pathol. 2011, 40, 407–415. [Google Scholar] [CrossRef]
- Li, G.Q.; Liu, F.F.; Li, J.Q.; Liu, Q.L. Botryosphaeriaceae from Eucalyptus plantations and adjacent plants in China. Persoonia 2018, 40, 63–95. [Google Scholar] [CrossRef]
- Wang, W.; Li, G.Q.; Liu, Q.L.; Chen, S.F. Cryphonectriaceae on Myrtales in China: Phylogeny, host range, and pathogenicity. Persoonia 2020, 45, 101–131. [Google Scholar] [CrossRef]
- Burgess, T.I.; Andjic, V.; Hardy, G.E.S.; Dell, B.; Xu, D. First report of Phaeophleospora destructans in China. J. Trop. For. Sci. 2006, 18, 144–146. [Google Scholar]
- Chen, S.F.; Lombard, L.; Roux, J.; Xie, Y.J.; Wingfield, M.J.; Zhou, X.D. Novel species of Calonectria associated with Eucalyptus leaf blight in Southeast China. Persoonia 2011, 26, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.C.; Chen, S.F. Calonectria pentaseptata causes severe leaf disease of cultivated Eucalyptus on the Leizhou Peninsula of southern China. Plant Dis. 2020, 104, 493–509. [Google Scholar] [CrossRef] [PubMed]
- Wu, W.X.; Chen, S.F. Species diversity, mating strategy and pathogenicity of Calonectria species from diseased leaves and soils in the Eucalyptus plantation in southern China. J. Fungi 2021, 7, 73. [Google Scholar] [CrossRef]
- Liang, X.Y.; Wang, Q.C.; Chen, S.F. Phylogeny, morphology, distribution, and pathogenicity of seven Calonectria species from leaf blighted Eucalyptus in HaiNai Island, China. Plant Dis. 2023, 107, 2579–2605. [Google Scholar] [CrossRef]
- Zhou, X.D.; de Beer, Z.W.; Xie, Y.J.; Pegg, G.S.; Wingfield, M.J. DNA based identification of Quambalaria pitereka causing severe leaf blight of Corymbia citriodora in China. Fungal Divers. 2007, 25, 245–254. [Google Scholar]
- Chen, S.F.; Liu, Q.L.; Li, G.Q.; Wingfield, M.J. Quambalaria species associated with eucalypt diseases in southern China. Front. Agric. Sci. Eng. 2017, 4, 433–447. [Google Scholar] [CrossRef]
- Hunter, G.C.; Wingfield, B.D.; Crous, P.W.; Wingfield, M.J. A multi–gene phylogeny for species of Mycosphaerella occurring on Eucalyptus leaves. Stud. Mycol. 2006, 55, 147–161. [Google Scholar] [CrossRef]
- Pérez, C.A.; Wingfield, M.J.; Altier, N.A.; Blanchette, R.A. Mycosphaerellaceae and Teratosphaeriaceae associated with Eucalyptus leaf diseases and stem cankers in Uruguay. For. Pathol. 2009, 39, 349–360. [Google Scholar] [CrossRef]
- Carnegie, A.J.; Pegg, G.S.; White, D.; Burgess, T.I. Species within Mycosphaerellaceae and Teratosphaeriaceae from eucalypts in eastern Australia. Australas. Plant Pathol. 2011, 40, 366–384. [Google Scholar] [CrossRef]
- Crous, P.W.; Wingfield, M.J.; Cheewangkoon, R.; Carnegie, A.J.; Burgess, T.I.; Summerell, B.A.; Edwards, J.; Taylor, P.W.J.; Groenewald, J.Z. Foliar pathogens of eucalypts. Stud. Mycol. 2019, 94, 125–298. [Google Scholar] [CrossRef]
- Pérez, G.; Burgess, T.I.; Slippers, B.; Carnegie, A.J.; Wingfield, B.D.; Wingfield, M.J. Teratosphaeria pseudonubilosa sp. nov., a serious Eucalyptus leaf pathogen in the Teratosphaeria nubilosa species complex. Australas. Plant Pathol. 2014, 43, 67–77. [Google Scholar] [CrossRef]
- Andjic, V.; Maxwell, A.; Hardy, G.E.S.; Burgess, T.I. New cryptic species of Teratosphaeria on Eucalyptus in Australia. IMA Fungus 2016, 7, 253–263. [Google Scholar] [CrossRef] [PubMed]
- Andjic, V.; Hardy, G.E.S.; Cortinas, M.N.; Wingfield, M.J.; Burgess, T.I. Multiple gene genealogies reveal important relationships between species of Phaeophleospora infecting Eucalyptus leaves. FEMS Microbiol. Lett. 2007, 268, 22–33. [Google Scholar] [CrossRef] [PubMed]
- Burgess, T.I.; Barber, P.A.; Sufaati, S.; Xu, D.P.; Hardy, G.E.S.; Dell, B. Mycosphaerella spp. on Eucalyptus in Asia: New species, new hosts and new records. Fungal Divers. 2007, 24, 135–157. [Google Scholar]
- Huang, C.L.; Chen, W.W.; Luo, J.T.; Tan, X.F.; Yang, X.H.; Wei, J.G. Identification of fungal pathogens of fast–growing Eucalyptus leaf disease in Guangxi. For. Pest Dis. 2012, 31, 1–6. [Google Scholar]
- Crous, P.W.; Braun, U.; Hunter, G.C.; Wingfield, M.J.; Verkley, G.J.M.; Shin, H.D.; Nakashima, C.; Groenewald, J.Z. Phylogenetic lineages in Pseudocercospora. Stud. Mycol. 2013, 75, 37–114. [Google Scholar] [CrossRef]
- Quaedvlieg, W.; Binder, M.; Groenewald, J.Z.; Summerell, B.A.; Carnegie, A.J.; Burgess, T.I.; Crous, P.W. Introducing the Consolidated species concept to resolve species in the Teratosphaeriaceae. Persoonia 2014, 33, 1–40. [Google Scholar] [CrossRef]
- Cheewangkoon, R.; Crous, P.W.; Hyde, K.D.; Groenewald, J.Z.; To-anan, C. Species of Mycosphaerell and related anamorphs on Eucalyptus leaves from Thailand. Persoonia 2008, 21, 77–91. [Google Scholar] [CrossRef]
- Liang, Z.C. Forest Diseases in China; China Forestry Publishing House: Beijing, China, 1984. [Google Scholar]
- Zhou, T.S. A preliminary study on pathogenic fungi of the leaf purple spot blotch of Eucalyptus spp. J. Southwest For. Coll. 1990, 10, 180–183. [Google Scholar]
- Wu, X.Y. Occurrence and control of forest diseases in Hainan Island. Trop. For. 2001, 29, 162–166. [Google Scholar]
- Luo, J.T.; Xue, Z.N.; Liao, W.J.; Yuan, C.L.; Ouyang, J.Y.; Shan, Y. Investigation on the diseases in fast growing Eucalyptus spp. in Guangxi. For. Pest Dis. 2012, 31, 21–24. [Google Scholar]
- Zhang, G.H.; Song, S.Y.; Liu, D.B.; Fan, C.M.; Hu, Z.M. Pathogen identification for brown spot disease and anthracnose of Eucalyptus robusta and its analysis. China Plant Prot. 2014, 34, 10–13. [Google Scholar]
- Yang, Y.J. Occurrence and control of main diseases and insect pests of Eucalyptus plantation in Hainan. Mod. Agric. Sci. Technol. 2015, 24, 134–138. [Google Scholar]
- Brooker, M.I.H.; Kleinig, D.A. Field Guide to Eucalypts Volume 2: South-Western and Southern Australia; Inkata Press: Melbourne, Australia, 1990. [Google Scholar]
- Potts, B.M.; Pederick, L.A. Morphology, phylogeny, origin, distribution and genetic diversity of eucalypts. In Diseases and Pathogens of Eucalypts; Keane, P.J., Kile, G.A., Podger, F.D., Brown, B.N., Eds.; CSIRO Publishing: Melbourne, Australia, 2000. [Google Scholar]
- Sankaran, K.V.; Sutton, B.C.; Minter, D. A Checklist of Fungi Recorded on Eucalyptus; CAB International: Oxon, UK, 1995. [Google Scholar]
- Andjic, V.; Carnegie, A.J.; Pegg, G.S.; Hardy, G.E.S.J.; Maxwell, A.; Crous, P.W.; Pérez, C.; Wingfield, M.J.; Burgess, T.I. 23 years of research on Teratosphaeria leaf blight of Eucalyptus. For. Ecol. Manag. 2019, 443, 19–27. [Google Scholar] [CrossRef]
- van Burik, J.A.H.; Schreckhise, R.W.; White, T.C.; Bowden, R.A.; Myerson, D. Comparison of six extraction techniques for isolation of DNA from filamentous fungi. Med. Mycol. 1998, 36, 299–303. [Google Scholar] [CrossRef] [PubMed]
- Taole, M.M.; Burgess, T.I.; Gryzenhout, M.; Wingfield, B.D.; Wingfield, M.J. DNA sequence incongruence and inconsistent morphology obscure species boundaries in the Teratosphaeria suttonii species complex. Mycoscience 2012, 53, 270–283. [Google Scholar] [CrossRef]
- Solís, M.; Wingfield, M.J.; Hammerbacher, A.; Naidoo, S. Comparison of the infection biology of Teratosphaeria destructans and Teratosphaeria epicoccoides on Eucalyptus. Plant Dis. 2022, 106, 1944–1951. [Google Scholar] [CrossRef]
- White, T.; Bruns, T.; Lee, S.; Taylor, J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In PCR Protocols: A Guide to Methods and Applications; Michael, I., Thomas, W.G., John, J.S., Eds.; Academic Press: San Diego, CA, USA, 1990. [Google Scholar]
- Carbone, I.; Kohn, L.M. A method for designing primer sets for speciation studies in filamentous Ascomycetes. Mycologia 1999, 91, 553–556. [Google Scholar] [CrossRef]
- O’Donnell, K.; Cigelnik, E. Two divergent intragenomic rDNA ITS2 types within a monophyletic lineage of the fungus Fusarium are nonorthologous. Mol. Phylogenet. Evol. 1997, 7, 103–116. [Google Scholar] [CrossRef]
- Glass, N.L.; Donaldson, G.C. Development of primer sets designed for use with the PCR to amplify conserved genes from filamentous ascomycetes. Appl. Environ. Microbiol. 1995, 31, 1323–1330. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetic analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT Multiple sequence alignment software Version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Miller, M.A.; Pfeiffer, W.; Schwartz, T. Creating the CIPRES Science Gateway for inference of large phylogenetic trees. In Proceedings of the 2010 Gateway Computing Environments Workshop (GCE), New Orleans, LA, USA, 14 November 2010. [Google Scholar]
- Stamatakis, A. RAxML version 8: A tool for phylogenetic analysis and post analysis of large phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef]
- Wingfield, M.J.; Crous, P.W.; Boden, D. Kirramyces destructans sp. nov., a serious leaf pathogen of Eucalyptus in Indonesia. S. Afr. J. Bot. 1996, 62, 325–327. [Google Scholar] [CrossRef]
- Walker, J.; Sutton, B.C.; Pascoe, I.G. Phaeoseptoria eucalypti and similar fungi on Eucalyptus, with description of Kirramyces gen. nov. (Coelomycetes). Mycol. Res. 1992, 96, 911–924. [Google Scholar] [CrossRef]
- Greyling, I.; Wingfield, M.J.; Coetzee, M.P.; Marincowitz, S.; Roux, J. The Eucalyptus shoot and leaf pathogen Teratosphaeria destructans recorded in South Africa. South. For. 2016, 78, 123–129. [Google Scholar] [CrossRef]
- Park, R.F.; Keane, P.J. Three Mycosphaerella species from leaf diseases of Eucalyptus. Trans. Br. Mycol. Soc. 1982, 79, 95–100. [Google Scholar] [CrossRef]
- Park, R.F.; Keane, P.J. Leaf diseases of Eucalyptus associated with Mycosphaerella species. Trans. Br. Mycol. Soc. 1982, 79, 101–115. [Google Scholar] [CrossRef]
- Aptroot, A. Mycosphaerella and its anamorphs: 2. Conspectus of Mycosphaerella. CBS Biodivers. Ser. 2006, 5, 1–231. [Google Scholar]
- Crous, P.W.; Aptroot, A.; Kang, J.; Braun, U. The genus Mycosphaerella and its anamorphs. Stud. Mycol. 2000, 45, 107–121. [Google Scholar]
- Crous, P.W.; Wingfield, M.J.; Mansilla, J.P.; Alfenas, A.C.; Groenewald, J.Z. Phylogenetic reassessment of Mycosphaerella spp. and their anamorphs occurring on Eucalyptus. II. Stud. Mycol. 2006, 55, 99–131. [Google Scholar] [CrossRef] [PubMed]
- Verkley, G.J.M.; Quaedvlieg, W.; Shin, H.; Crous, P.W. A new approach to species delimitation in Septoria. Stud. Mycol. 2013, 75, 213–305. [Google Scholar] [CrossRef] [PubMed]
- Crous, P.W.; Braun, U.; Groenewald, J.Z. Mycosphaerella is polyphyletic. Stud. Mycol. 2007, 58, 1–32. [Google Scholar] [CrossRef] [PubMed]
- Hunter, G.C.; Crous, P.W.; Wingfield, B.D.; Pongpanich, K.; Wingfield, M.J. Pseudocercospora flavomarginata sp. nov., from Eucalyptus leaves in Thailand. Fungal Divers. 2006, 22, 71–90. [Google Scholar]
- Heather, W.A. Some Aspects of the Ecology and Pathology of Phaeoseptoria eucalypti Hans. Emend. Walker on Some Species of the Genus Eucalyptus. Ph.D. Thesis, The Australian National University, Canberra, Australia, 1965. [Google Scholar]
- Wang, Q.C.; Liu, F.F.; Liu, Q.L.; Wu, W.X.; Wingfield, M.J.; Chen, S.F. Comparison of hyphal fragments and spores to evaluate the pathogenicity of the Eucalyptus leaf and shoot pathogen Calonectria pseudoreteaudii. Plant Dis. 2022, 106, 3145–3153. [Google Scholar] [CrossRef]
- Taole, M.; Bihon, W.; Wingfield, B.D.; Wingfield, M.J.; Burgess, T.I. Multiple introductions from multiple sources: Invasion patterns for an important Eucalyptus leaf pathogen. Ecol. Evol. 2015, 5, 4210–4220. [Google Scholar] [CrossRef]
- Havenga, M.; Wingfield, B.D.; Wingfield, M.J.; Dreyer, L.L.; Roets, F.; Aylward, J. Low genetic diversity and strong geographic structure in six introduced populations of the Eucalyptus foliar pathogen Teratosphaeria destructans. Plant Pathol. 2020, 69, 1540–1550. [Google Scholar] [CrossRef]
- Havenga, M.; Wingfield, B.D.; Wingfield, M.J.; Roets, F.; Dreyer, L.L.; Tatham, C.T.; Duong, D.A.; Wilken, P.M.; Chen, S.; Aylward, J. Mating strategy and mating type distribution in six global populations of the Eucalyptus foliar pathogen Teratosphaeria destructans. Fungal Genet. Biol. 2020, 137, 103350. [Google Scholar] [CrossRef]
- Havenga, M.; Wingfield, B.D.; Wingfield, M.J.; Marincowitz, S.; Dreyer, L.L.; Roets, F.; Japarudin, Y.; Aylward, J. Genetic recombination in Teratosphaeria destructans causing a new disease outbreak in Malaysia. For. Pathol. 2021, 51, e12683. [Google Scholar] [CrossRef]
- Andjic, V.; Dell, B.; Barber, P.; Hardy, G.E.S.; Wingfield, M.J.; Burgess, T.I. Plants for planting; indirect evidence for the movement of a serious forest pathogen, Teratosphaeria destructans, in Asia. Eur. J. Plant Pathol. 2011, 131, 49–58. [Google Scholar] [CrossRef]
- Solís, M.; Hammerbacher, A.; Wingfield, M.J.; Naidoo, S. A Robust Disease Scoring Method to Screen Eucalyptus for Resistance Against the Aggressive Leaf Pathogen Teratosphaeria destructans. Plant Dis. 2023, 107, 1418–1424. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).