A Meta-Analysis of Bacterial Communities in Food Processing Facilities: Driving Forces for Assembly of Core and Accessory Microbiomes across Different Food Commodities
Abstract
1. Introduction
2. Data Collection
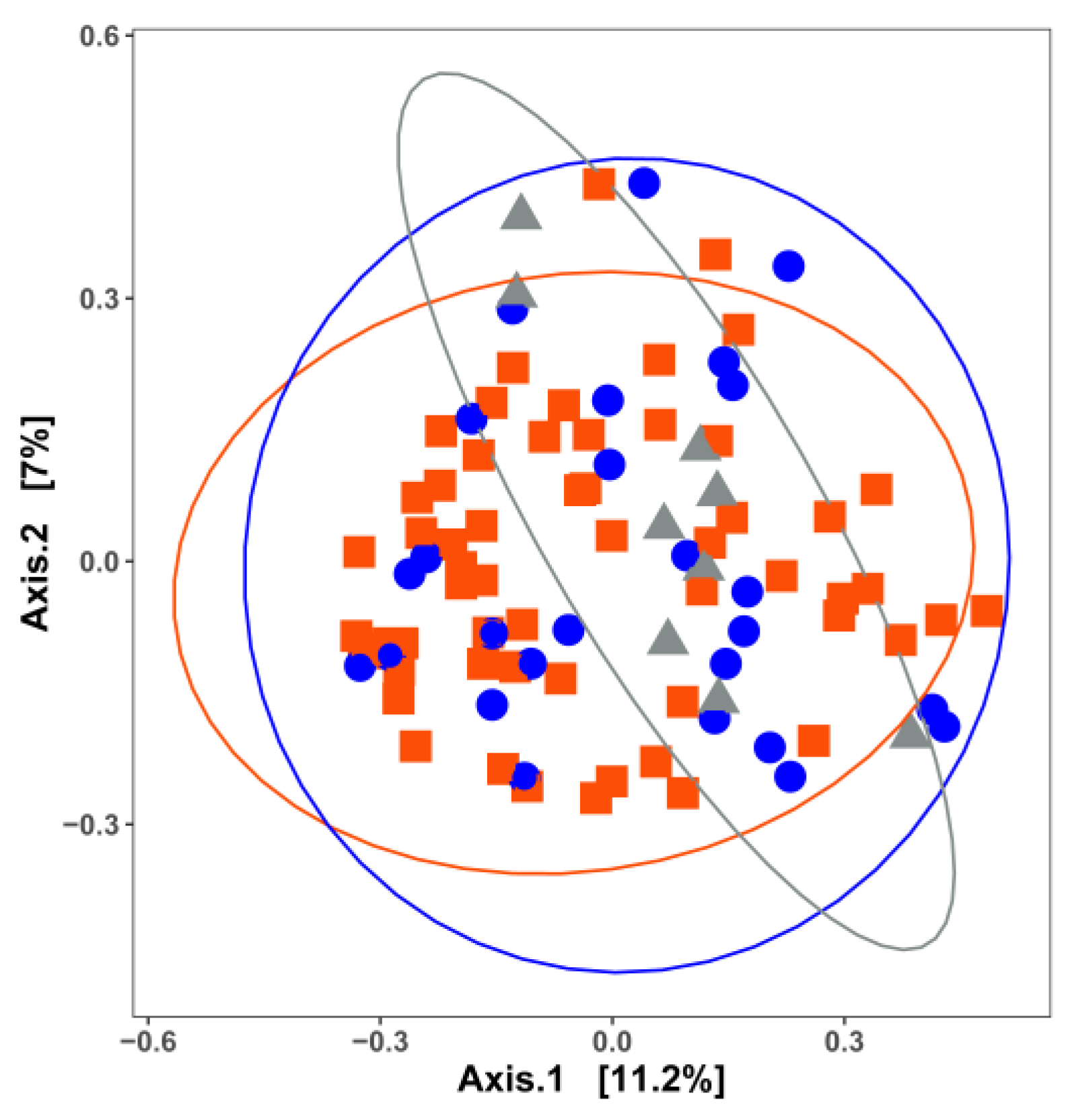
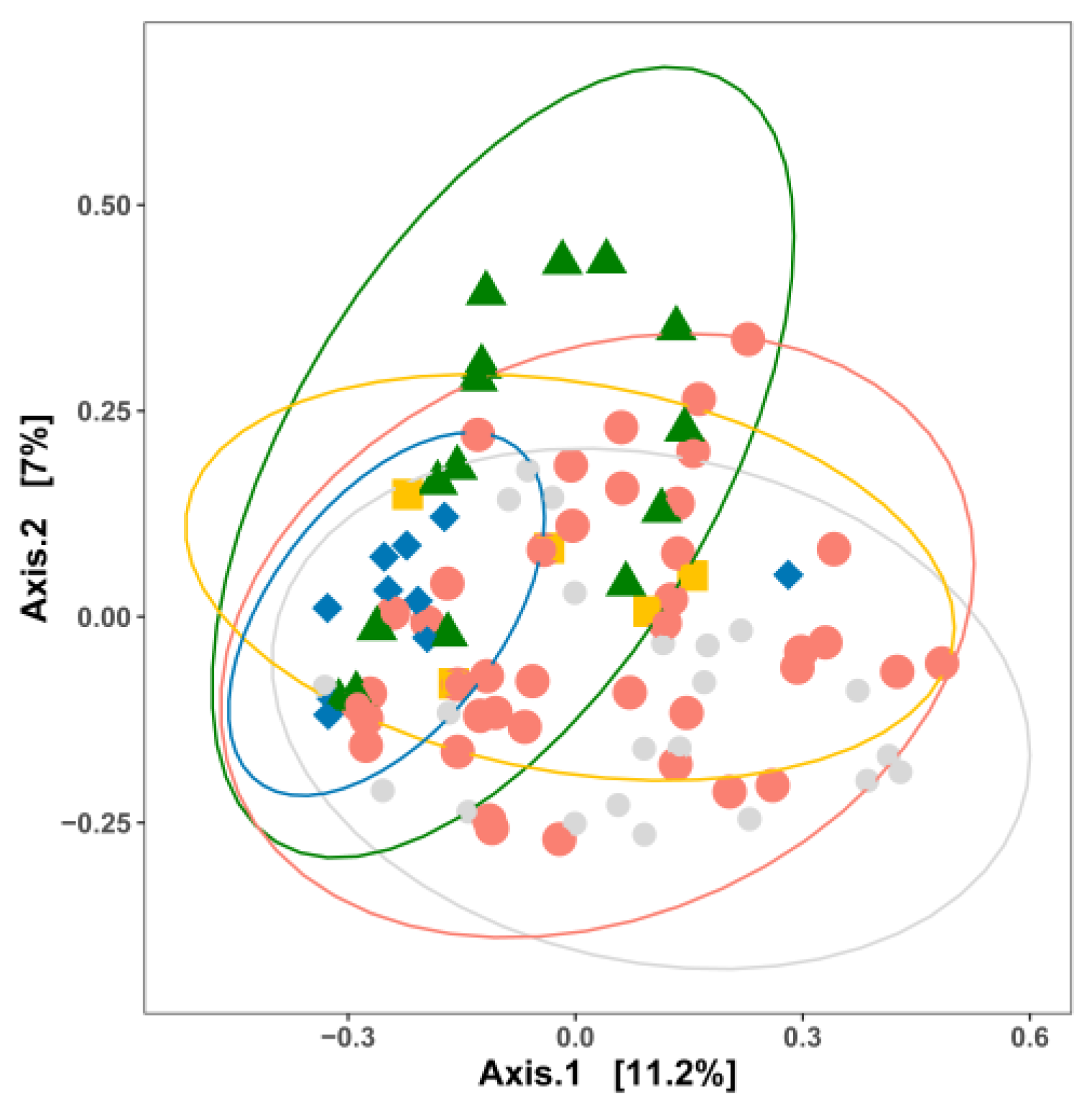
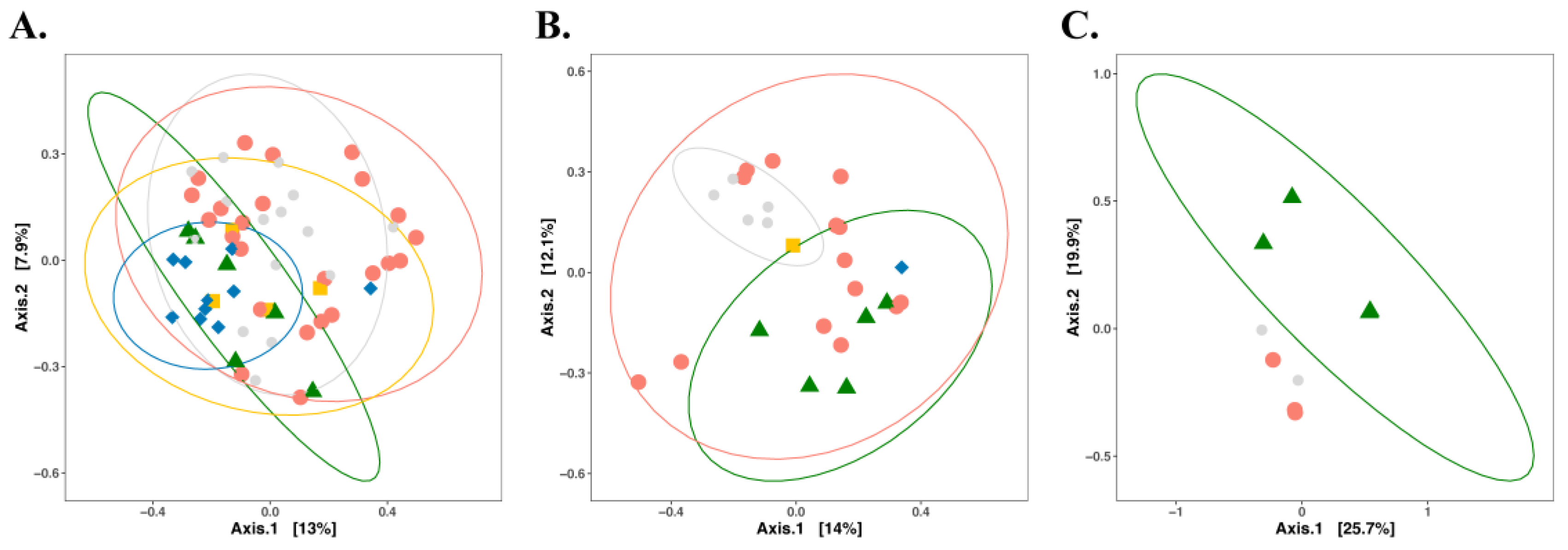
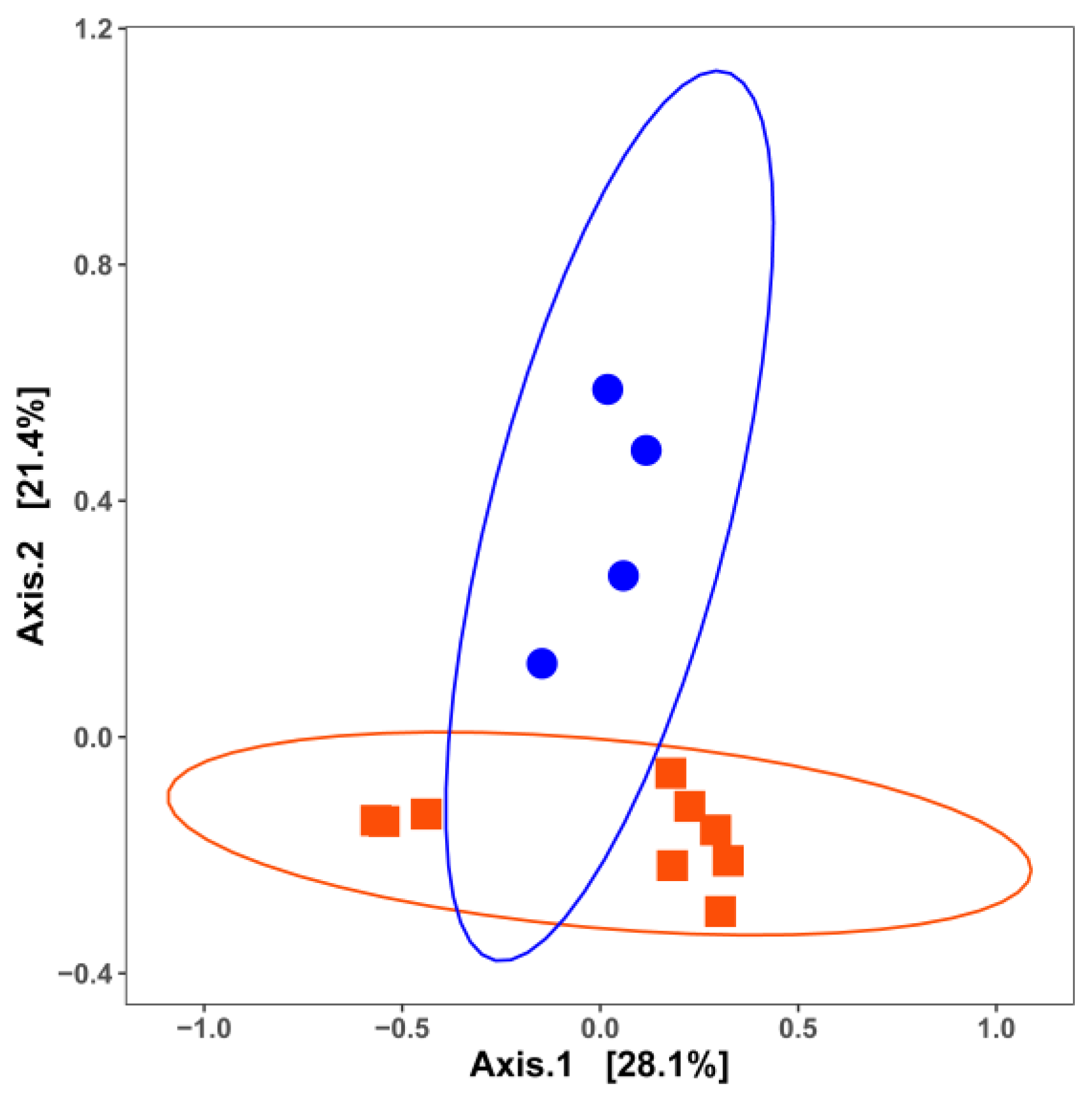
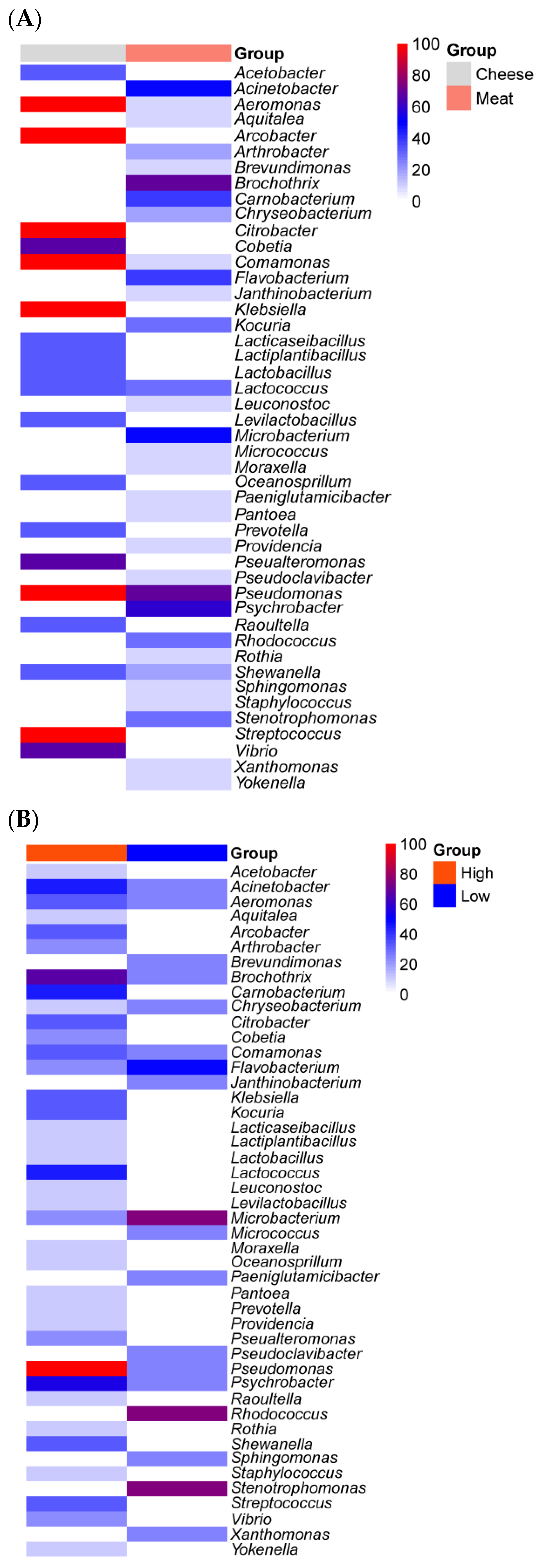
3. Impact of Nutrient Source and Commodity on the Compositions of Bacterial Communities
4. Which Bacteria Are Where?
5. Can a Core Microbiome in Food Processing Facilities Be Identified?
6. The Use of Sanitizers and Selective Ecology
7. Limitations
8. Conclusions and Perspectives
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- FAO. How to Feed the World in 2050; Food and Agriculture Organization: Rome, Italy, 2009; pp. 1–35. [Google Scholar]
- Janet, R.; Richard, W.; Tim, S.; Craig, H. How to Sustainably Feed 10 Billion People by 2050, in 21 Charts. World Resour. Inst. 2018, 1–15. [Google Scholar]
- Holt, R.D. Bringing the Hutchinsonian Niche into the 21st Century: Ecological and Evolutionary Perspectives. Proc. Natl. Acad. Sci. USA 2009, 106, 19659–19665. [Google Scholar] [CrossRef]
- Vellend, M. Conceptual Synthesis in Community Ecology. Q. Rev. Biol. 2010, 85, 183–206. [Google Scholar] [CrossRef]
- Gill, C.O. Spoilage Factors, Affecting|Microbiological. In Encyclopedia of Meat Sciences, 2nd ed.; Academic Press: Cambridge, MA, USA, 2014; pp. 388–393. [Google Scholar] [CrossRef]
- Zhu, Y.; Wang, W.; Li, M.; Zhang, J.; Ji, L.; Zhao, Z.; Zhang, R.; Cai, D.; Chen, L. Microbial Diversity of Meat Products under Spoilage and Its Controlling Approaches. Front. Nutr. 2022, 9, 2976. [Google Scholar] [CrossRef] [PubMed]
- Alegbeleye, O.; Odeyemi, O.A.; Strateva, M.; Stratev, D. Microbial Spoilage of Vegetables, Fruits and Cereals. Appl. Food Res. 2022, 2, 100122. [Google Scholar] [CrossRef]
- Bridier, A.; Briandet, R.; Thomas, V.; Dubois-Brissonnet, F. Resistance of Bacterial Biofilms to Disinfectants: A Review. Biofouling 2011, 27, 1017–1032. [Google Scholar] [CrossRef] [PubMed]
- Mertz, A.W.; Koo, O.K.; O’Bryan, C.A.; Morawicki, R.; Sirsat, S.A.; Neal, J.A.; Crandall, P.G.; Ricke, S.C. Microbial Ecology of Meat Slicers as Determined by Denaturing Gradient Gel Electrophoresis. Food Control 2014, 42, 242–247. [Google Scholar] [CrossRef]
- Wang, H.; He, A.; Yang, X. Dynamics of Microflora on Conveyor Belts in a Beef Fabrication Facility during Sanitation. Food Control 2018, 85, 42–47. [Google Scholar] [CrossRef]
- Wagner, E.M.; Pracser, N.; Thalguter, S.; Fischel, K.; Rammer, N.; Pospíšilová, L.; Alispahic, M.; Wagner, M.; Rychli, K. Identification of Biofilm Hotspots in a Meat Processing Environment: Detection of Spoilage Bacteria in Multi-Species Biofilms. Int. J. Food Microbiol. 2020, 328, 108668. [Google Scholar] [CrossRef]
- Liu, N.T.; Bauchan, G.R.; Francoeur, C.B.; Shelton, D.R.; Lo, Y.M.; Nou, X. Ralstonia insidiosa Serves as Bridges in Biofilm Formation by Foodborne Pathogens Listeria monocytogenes, Salmonella enterica, and Enterohemorrhagic Escherichia coli. Food Control 2016, 65, 14–20. [Google Scholar] [CrossRef]
- Abdallah, M.; Benoliel, C.; Drider, D.; Dhulster, P.; Chihib, N.E. Biofilm Formation and Persistence on Abiotic Surfaces in the Context of Food and Medical Environments. Arch. Microbiol. 2014, 196, 453–472. [Google Scholar] [CrossRef]
- Mcglynn, W. Guidelines for the Use of Chlorine Bleach as a Sanitizer in Food Processing Operations. Handling Articles. Food Technology Fact Sheet. Oklahoma State University. Available online: https://shareok.org/bitstream/handle/11244/50164/oksd_fapc_116_2013-12.pdf?sequence=1 (accessed on 13 June 2023).
- Wheeler, T.L.; Kalchayanand, N.; Bosilevac, J.M. Pre- and Post-Harvest Interventions to Reduce Pathogen Contamination in the U.S. Beef Industry. Meat Sci. 2014, 98, 372–382. [Google Scholar] [CrossRef]
- Dickson, J.S.; Anderson, M.E. Microbiological Decontamination of Food Animal Carcasses by Washing and Sanitizing Systems: A Review. J. Food Prot. 1992, 55, 360–366. [Google Scholar] [CrossRef]
- United States of Food and Drug Administration Raw Milk Misconceptions and the Danger of Raw Milk Consumption. Available online: http://www.fda.gov/food/foodborneillnesscontaminants/buystoreservesafefood/ucm247991.htm (accessed on 3 May 2023).
- Ryan, R.P.; Germaine, K.; Franks, A.; Ryan, D.J.; Dowling, D.N. Bacterial Endophytes: Recent Developments and Applications. FEMS Microbiol. Lett. 2008, 278, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Kandel, S.L.; Joubert, P.M.; Doty, S.L. Bacterial Endophyte Colonization and Distribution within Plants. Microorganisms 2017, 5, 77. [Google Scholar] [CrossRef]
- Lodewyckx, C.; Vangronsveld, J.; Porteous, F.; Moore, E.R.B.; Taghavi, S.; Mezgeay, M.; Van Der Lelie, D. Critical Reviews in Plant Sciences Endophytic Bacteria and Their Potential Applications Endophytic Bacteria and Their Potential Applications. CRC Crit. Rev. Plant Sci. 2002, 21, 583–606. [Google Scholar] [CrossRef]
- Valentino, V.; Sequino, G.; Cobo-Díaz, J.F.; Álvarez-Ordóñez, A.; De Filippis, F.; Ercolini, D. Evidence of Virulence and Antibiotic Resistance Genes from the Microbiome Mapping in Minimally Processed Vegetables Producing Facilities. Food Res. Int. 2022, 162, 112202. [Google Scholar] [CrossRef] [PubMed]
- De Filippis, F.; La Storia, A.; Villani, F.; Ercolini, D. Exploring the Sources of Bacterial Spoilers in Beefsteaks by Culture-Independent High-Throughput Sequencing. PLoS ONE 2013, 8, 70222. [Google Scholar] [CrossRef]
- Falardeau, J.; Keeney, K.; Trmčić, A.; Kitts, D.; Wang, S. Farm-to-Fork Profiling of Bacterial Communities Associated with an Artisan Cheese Production Facility. Food Microbiol. 2019, 83, 48–58. [Google Scholar] [CrossRef]
- Hultman, J.; Rahkila, R.; Ali, J.; Rousu, J.; Björkroth, K.J. Meat Processing Plant Microbiome and Contamination Patterns of Cold-Tolerant Bacteria Causing Food Safety and Spoilage Risks in the Manufacture of Vacuum-Packaged Cooked Sausages. Appl. Environ. Microbiol. 2015, 81, 7088–7097. [Google Scholar] [CrossRef]
- De Filippis, F.; Valentino, V.; Alvarez-Ordóñez, A.; Cotter, P.D.; Ercolini, D. Environmental Microbiome Mapping as a Strategy to Improve Quality and Safety in the Food Industry. Curr. Opin. Food Sci. 2021, 38, 168–176. [Google Scholar] [CrossRef]
- Fagerlund, A.; Langsrud, S.; Møretrø, T. Microbial Diversity and Ecology of Biofilms in Food Industry Environments Associated with Listeria monocytogenes Persistence. Curr. Opin. Food Sci. 2021, 37, 171–178. [Google Scholar] [CrossRef]
- Yuan, L.; Hansen, M.F.; Røder, H.L.; Wang, N.; Burmølle, M.; He, G. Mixed-Species Biofilms in the Food Industry: Current Knowledge and Novel Control Strategies. Crit. Rev. Food Sci. Nutr. 2019, 60, 2277–2293. [Google Scholar] [CrossRef]
- Alvarez-Ordóñez, A.; Coughlan, L.M.; Briandet, R.; Cotter, P.D. Annual Review of Food Science and Technology Biofilms in Food Processing Environments: Challenges and Opportunities. Annu. Rev. Food Sci. Technol 2019, 10, 173–195. [Google Scholar] [CrossRef]
- Møretrø, T.; Langsrud, S.; Heir, E. Bacteria on Meat Abattoir Process Surfaces after Sanitation: Characterisation of Survival Properties of Listeria monocytogenes and the Commensal Bacterial Flora. Adv. Microbiol. 2013, 3, 255–264. [Google Scholar] [CrossRef]
- Cobo-Díaz, J.F.; Alvarez-Molina, A.; Alexa, E.A.; Walsh, C.J.; Mencía-Ares, O.; Puente-Gómez, P.; Likotrafiti, E.; Fernández-Gómez, P.; Prieto, B.; Crispie, F.; et al. Microbial Colonization and Resistome Dynamics in Food Processing Environments of a Newly Opened Pork Cutting Industry during 1.5 Years of Activity. Microbiome 2021, 9, 204. [Google Scholar] [CrossRef]
- Brightwell, G.; Boerema, J.; Mills, J.; Mowat, E.; Pulford, D. Identifying the Bacterial Community on the Surface of IntraloxTM Belting in a Meat Boning Room by Culture-Dependent and Culture-Independent 16S RDNA Sequence Analysis. Int. J. Food Microbiol. 2006, 109, 47–53. [Google Scholar] [CrossRef]
- Marouani-Gadri, N.; Augier, G.; Carpentier, B. Characterization of Bacterial Strains Isolated from a Beef-Processing Plant Following Cleaning and Disinfection—Influence of Isolated Strains on Biofilm Formation by Sakaï and EDL 933 E. coli O157:H7. Int. J. Food Microbiol. 2009, 133, 62–67. [Google Scholar] [CrossRef] [PubMed]
- Røder, H.L.; Raghupathi, P.K.; Herschend, J.; Brejnrod, A.; Knøchel, S.; Sørensen, S.J.; Burmølle, M. Interspecies Interactions Result in Enhanced Biofilm Formation by Co-Cultures of Bacteria Isolated from a Food Processing Environment. Food Microbiol. 2015, 51, 18–24. [Google Scholar] [CrossRef] [PubMed]
- Zwirzitz, B.; Wetzels, S.U.; Dixon, E.D.; Stessl, B.; Zaiser, A.; Rabanser, I.; Thalguter, S.; Pinior, B.; Roch, F.F.; Strachan, C.; et al. The Sources and Transmission Routes of Microbial Populations throughout a Meat Processing Facility. Npj Biofilms Microbiomes 2020, 6, 26. [Google Scholar] [CrossRef] [PubMed]
- Zwirzitz, B.; Wetzels, S.U.; Dixon, E.D.; Fleischmann, S.; Selberherr, E.; Thalguter, S.; Quijada, N.M.; Dzieciol, M.; Wagner, M.; Stessl, B. Co-Occurrence of Listeria spp. and Spoilage Associated Microbiota during Meat Processing Due to Cross-Contamination Events. Front. Microbiol. 2021, 12, 632935. [Google Scholar] [CrossRef] [PubMed]
- Fox, E.M.; Solomon, K.; Moore, J.E.; Wall, P.G.; Fanning, S. Phylogenetic Profiles of In-House Microflora in Drains at a Food Production Facility: Comparison and Biocontrol Implications of Listeria-Positive and -Negative Bacterial Populations. Appl. Environ. Microbiol. 2014, 80, 3369–3374. [Google Scholar] [CrossRef]
- Mettler, E.; Carpentier, B. Variations over Time of Microbial Load and Physicochemical Properties of Floor Materials after Cleaning in Food Industry Premises. J. Food Prot. 1998, 61, 57–65. [Google Scholar] [CrossRef]
- Stellato, G.; La Storia, A.; De Filippis, F.; Borriello, G.; Villani, F.; Ercolini, D. Overlap of Spoilage-Associated Microbiota between Meat and the Meat Processing Environment in Small-Scale and Large-Scale Retail Distributions. Appl. Environ. Microbiol. 2016, 82, 4045. [Google Scholar] [CrossRef] [PubMed]
- Ellerbroek, L. Airborne Microflora in Poultry Slaughtering Establishments. Food Microbiol. 1997, 14, 527–531. [Google Scholar] [CrossRef]
- Yang, X.; Wang, H.; Hrycauk, S.; Holman, D.B.; Ells, T.C. Microbial Dynamics in Mixed-Culture Biofilms of Salmonella Typhimurium and Escherichia coli O157:H7 and Bacteria Surviving Sanitation of Conveyor Belts of Meat Processing Plants. Microorganisms 2023, 11, 421. [Google Scholar] [CrossRef]
- Rodríguez-López, P.; Saá-Ibusquiza, P.; Mosquera-Fernández, M.; López-Cabo, M. Listeria monocytogenes—Carrying Consortia in Food Industry. Composition, Subtyping and Numerical Characterisation of Mono-Species Biofilm Dynamics on Stainless Steel. Int. J. Food Microbiol. 2015, 206, 84–95. [Google Scholar] [CrossRef] [PubMed]
- Langsrud, S.; Moen, B.; Møretrø, T.; Løype, M.; Heir, E. Microbial Dynamics in Mixed Culture Biofilms of Bacteria Surviving Sanitation of Conveyor Belts in Salmon-Processing Plants. J. Appl. Microbiol. 2016, 120, 366–378. [Google Scholar] [CrossRef]
- Guobjörnsdóttir, B.; Einarsson, H.; Thorkelsson, G. Microbial Adhesion to Processing Lines for Fish Fillets and Cooked Shrimp: Influence of Stainless Steel Surface Finish and Presence of Gram-Negative Bacteria on the Attachment of Listeria Monocytogenes. Available online: https://hrcak.srce.hr/file/162696 (accessed on 26 January 2023).
- Møretrø, T.; Moen, B.; Heir, E.; Hansen, A.; Langsrud, S. Contamination of Salmon Fillets and Processing Plants with Spoilage Bacteria. Int. J. Food Microbiol. 2016, 237, 98–108. [Google Scholar] [CrossRef]
- Maillet, A.; Bouju-Albert, A.; Roblin, S.; Vaissié, P.; Leuillet, S.; Dousset, X.; Jaffrès, E.; Combrisson, J.; Prévost, H. Impact of DNA Extraction and Sampling Methods on Bacterial Communities Monitored by 16S RDNA Metabarcoding in Cold-Smoked Salmon and Processing Plant Surfaces. Food Microbiol. 2021, 95, 103705. [Google Scholar] [CrossRef]
- Bjørge Thomassen, G.M.; Krych, L.; Knøchel, S.; Mehli, L. Bacterial Community Development and Diversity during the First Year of Production in a New Salmon Processing Plant. Food Microbiol. 2023, 109, 104138. [Google Scholar] [CrossRef] [PubMed]
- Pothakos, V.; Stellato, G.; Ercolini, D.; Devlieghere, F. Processing Environment and Ingredients Are Both Sources of Leuconostoc gelidum, Which Emerges as a Major Spoiler in Ready-to-Eat Meals. Appl. Environ. Microbiol. 2015, 81, 3529–3541. [Google Scholar] [CrossRef] [PubMed]
- Fagerlund, A.; Møretrø, T.; Heir, E.; Briandet, R.; Langsrud Nofima, S.; Christopher Elkins, E.A. Cleaning and Disinfection of Biofilms Composed of Listeria monocytogenes and Background Microbiota from Meat Processing Surfaces. Appl. Environ. Microbiol. 2017, 83, e01046-17. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.G.; Huang, X.N.; Meng, J.; Chen, J.Y.; Han, B.Z. Characterization and Comparison of the Bacterial Community on Environmental Surfaces through a Fresh-Cut Vegetables Processing Line in China. Food Res. Int. 2022, 155, 111075. [Google Scholar] [CrossRef] [PubMed]
- Liu, N.T.; Lefcourt, A.M.; Nou, X.; Shelton, D.R.; Zhang, G.; Lo, Y.M. Native Microflora in Fresh-Cut Produce Processing Plants and Their Potentials for Biofilm Formation. J. Food Prot. 2013, 76, 827–832. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.G.; Meng, J.; Bao, W.J.; Kang, J.M.; Chen, J.Y.; Han, B.Z. Occurrence of Disinfectant-Resistant Bacteria in a Fresh-Cut Vegetables Processing Facility and Their Role in Protecting Salmonella Enteritidis. RSC Adv. 2021, 11, 10291–10299. [Google Scholar] [CrossRef]
- Tan, X.; Chung, T.; Chen, Y.; Macarisin, D.; Laborde, L.; Kovac, J. The Occurrence of Listeria Monocytogenes Is Associated with Built Environment Microbiota in Three Tree Fruit Processing Facilities. Microbiome 2019, 7, 115. [Google Scholar] [CrossRef]
- Gu, G.; Ottesen, A.; Bolten, S.; Wang, L.; Luo, Y.; Rideout, S.; Lyu, S.; Nou, X. Impact of Routine Sanitation on the Microbiomes in a Fresh Produce Processing Facility. Int. J. Food Microbiol. 2019, 294, 31–41. [Google Scholar] [CrossRef]
- Dzieciol, M.; Schornsteiner, E.; Muhterem-Uyar, M.; Stessl, B.; Wagner, M.; Schmitz-Esser, S. Bacterial Diversity of Floor Drain Biofilms and Drain Waters in a Listeria monocytogenes Contaminated Food Processing Environment. Int. J. Food Microbiol. 2016, 223, 33–40. [Google Scholar] [CrossRef]
- Bokulich, N.A.; Mills, D.A. Facility-Specific “House” Microbiome Drives Microbial Landscapes of Artisan Cheesemaking Plants. Appl. Environ. Microbiol. 2013, 79, 5214–5223. [Google Scholar] [CrossRef]
- Calasso, M.; Ercolini, D.; Mancini, L.; Stellato, G.; Minervini, F.; Di Cagno, R.; De Angelis, M.; Gobbetti, M. Relationships among House, Rind and Core Microbiotas during Manufacture of Traditional Italian Cheeses at the Same Dairy Plant. Food Microbiol. 2016, 54, 115–126. [Google Scholar] [CrossRef]
- Guzzon, R.; Carafa, I.; Tuohy, K.; Cervantes, G.; Vernetti, L.; Barmaz, A.; Larcher, R.; Franciosi, E. Exploring the Microbiota of the Red-Brown Defect in Smear-Ripened Cheese by 454-Pyrosequencing and Its Prevention Using Different Cleaning Systems. Food Microbiol. 2017, 62, 160–168. [Google Scholar] [CrossRef] [PubMed]
- Quijada, N.M.; Mann, E.; Wagner, M.; Rodríguez-Lázaro, D.; Hernández, M.; Schmitz-Esser, S. Autochthonous Facility-Specific Microbiota Dominates Washed-Rind Austrian Hard Cheese Surfaces and Its Production Environment. Int. J. Food Microbiol. 2018, 267, 54–61. [Google Scholar] [CrossRef]
- Schön, K.; Schornsteiner, E.; Dzieciol, M.; Wagner, M.; Müller, M.; Schmitz-Esser, S. Microbial Communities in Dairy Processing Environment Floor-Drains Are Dominated by Product-Associated Bacteria and Yeasts. Food Control 2016, 70, 210–215. [Google Scholar] [CrossRef]
- Stellato, G.; De Filippis, F.; La Storia, A.; Ercolini, D. Coexistence of Lactic Acid Bacteria and Potential Spoilage Microbiota in a Dairy Processing Environment. Appl. Environ. Microbiol. 2015, 81, 7893–7904. [Google Scholar] [CrossRef] [PubMed]
- Marsha, A. Echols Food Safety Regulation in the European Union and the United States: Different Cultures, Different Laws. Columbia J. Eur. Law 1998, 4, 525. [Google Scholar]
- Johnson, R. The U.S.—EU Beef Hormone Dispute; Library of Congress, Congressional Research Service: Washington, DC, USA, 2015. Available online: https://sgp.fas.org/crs/row/R40449.pdf (accessed on 13 June 2023).
- Beardsley, E. In Europe, A Cow over Hormone-Treated U.S. Beef. Available online: https://www.npr.org/templates/story/story.php?storyId=113314725 (accessed on 30 March 2023).
- Zhang, Z.; Yang, L.; He, Y.; Luo, X.; Zhao, S.; Jia, X. Composition of Fecal Microbiota in Grazing and Feedlot Angus Beef Cattle. Animals 2021, 11, 3167. [Google Scholar] [CrossRef]
- Bell, A.W.; Charmley, E.; Hunter, R.A.; Archer, J.A. The Australasian Beef Industries—Challenges and Opportunities in the 21st Century. Anim. Front. 2011, 1, 10–19. [Google Scholar] [CrossRef][Green Version]
- Canadian Food Inspection Agency Handling of Meat Products—Archived—Chapter 4—Meat Processing Controls and Procedures—Food Safety for Industry—Canadian Food Inspection Agency. Available online: https://inspection.canada.ca/food-safety-for-industry/archived-food-guidance/meat-and-poultry-products/manual-of-procedures/chapter-4/eng/1367622697439/1367622787568?chap=4 (accessed on 30 March 2023).
- Zheng, J.; Wittouck, S.; Salvetti, E.; Franz, C.M.A.P.; Harris, H.M.B.; Mattarelli, P.; O’toole, P.W.; Pot, B.; Vandamme, P.; Walter, J.; et al. A Taxonomic Note on the Genus Lactobacillus: Description of 23 Novel Genera, Emended Description of the Genus Lactobacillus beijerinck 1901, and Union of Lactobacillaceae and Leuconostocaceae. Int. J. Syst. Evol. Microbiol. 2020, 70, 2782–2858. [Google Scholar] [CrossRef]
- Wolfe, B.E.; Button, J.E.; Santarelli, M.; Dutton, R.J. Cheese Rind Communities Provide Tractable Systems for in Situ and in Vitro Studies of Microbial Diversity. Cell 2014, 158, 422–433. [Google Scholar] [CrossRef]
- Hassan, A.N.; Frank, J.F. Microorganisms Associated with Milk. In Encyclopedia of Dairy Sciences, 2nd ed.; Academic Press: Cambridge, MA, USA, 2011; pp. 447–457. [Google Scholar] [CrossRef]
- Kable, M.E.; Srisengfa, Y.; Laird, M.; Zaragoza, J.; McLeod, J.; Heidenreich, J.; Marco, M.L. The Core and Seasonal Microbiota of Raw Bovine Milk in Tanker Trucks and the Impact of Transfer to a Milk Processing Facility. mBio 2016, 7, e00836-16. [Google Scholar] [CrossRef]
- Xue, T.; Chen, X.; Shang, F. Short Communication: Effects of Lactose and Milk on the Expression of Biofilm-Associated Genes in Staphylococcus aureus Strains Isolated from a Dairy Cow with Mastitis. J. Dairy Sci. 2014, 97, 6129–6134. [Google Scholar] [CrossRef] [PubMed]
- Illikoud, N.; Gohier, R.; Werner, D.; Barrachina, C.; Roche, D.; Jaffrès, E.; Zagorec, M. Transcriptome and Volatilome Analysis During Growth of Brochothrix thermosphacta in Food: Role of Food Substrate and Strain Specificity for the Expression of Spoilage Functions. Front. Microbiol. 2019, 10, 2527. [Google Scholar] [CrossRef] [PubMed]
- Brightwell, G.; Broda, D.M.; Boerema, J.A. Sources of Psychrophilic and Psychrotolerant Clostridia Causing Spoilage of Vacuum-packed Chilled Meats, as Determined by PCR Amplification Procedure. J. Appl. Microbiol. 2009, 107, 178–186. [Google Scholar] [CrossRef]
- Canadian Food Inspection Agency Food Safety Facts on Scombroid Poisoning—Canadian Food Inspection Agency. Available online: http://www.inspection.gc.ca/food/information-for-consumers/fact-sheets-and-infographics/food-poisoning/scombroid/eng/1332280657698/1332280735024 (accessed on 17 April 2023).
- Leisner, J.J.; Laursen, B.G.; Prévost, H.; Drider, D.; Dalgaard, P. Carnobacterium: Positive and Negative Effects in the Environment and in Foods. FEMS Microbiol. Rev. 2007, 31, 592–613. [Google Scholar] [CrossRef] [PubMed]
- Visvalingam, J.; Wang, H.; Ells, T.C.; Yang, X. Facultative Anaerobes Shape Multispecies Biofilms Composed of Meat Processing Surface Bacteria and Escherichia coli O157:H7 or Salmonella enterica Serovar Typhimurium. Appl. Environ. Microbiol. 2019, 85, e01123-19. [Google Scholar] [CrossRef] [PubMed]
- Martin-Visscher, L.A.; Van Belkum, M.J.; Garneau-Tsodikova, S.; Whittal, R.M.; Zheng, J.; McMullen, L.M.; Vederas, J.C. Isolation and Characterization of Carnocyclin A, a Novel Circular Bacteriocin Produced by Carnobacterium maltaromaticum UAL307. Appl. Environ. Microbiol. 2008, 74, 4756–4763. [Google Scholar] [CrossRef] [PubMed]
- Chaillou, S.; Chaulot-Talmon, A.; Caekebeke, H.; Cardinal, M.; Christieans, S.; Denis, C.; Hélène Desmonts, M.; Dousset, X.; Feurer, C.; Hamon, E.; et al. Origin and Ecological Selection of Core and Food-Specific Bacterial Communities Associated with Meat and Seafood Spoilage. ISME J. 2015, 9, 1105–1118. [Google Scholar] [CrossRef]
- Wiernasz, N.; Gigout, F.; Cardinal, M.; Cornet, J.; Rohloff, J.; Courcoux, P.; Vigneau, E.; Skírnisdottír, S.; Passerini, D.; Pilet, M.F.; et al. Effect of the Manufacturing Process on the Microbiota, Organoleptic Properties and Volatilome of Three Salmon-Based Products. Foods 2021, 10, 2517. [Google Scholar] [CrossRef]
- Leff, J.W.; Fierer, N. Bacterial Communities Associated with the Surfaces of Fresh Fruits and Vegetables. PLoS ONE 2013, 8, e59310. [Google Scholar] [CrossRef]
- Verma, S.; Kumar, M.; Kumar, A.; Das, S.; Chakdar, H.; Varma, A.; Saxena, A.K. Diversity of Bacterial Endophytes of Maize (Zea mays) and Their Functional Potential for Micronutrient Biofortification. Curr. Microbiol. 2021, 79, 6. [Google Scholar] [CrossRef]
- Xu, L.; Zhang, Y.; Read, N.; Liu, S.; Friman, V.P. Devosia nitraria sp. nov., a Novel Species Isolated from the Roots of Nitraria sibirica in China. Antonie Van Leeuwenhoek 2017, 110, 1475–1483. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Wei, J.; Qiu, Y.; Niu, C.; Song, Z.; Yuan, Y.; Yue, T. Structure-Dependent Inhibition of Stenotrophomonas maltophilia by Polyphenol and Its Impact on Cell Membrane. Front. Microbiol. 2019, 10, 2646. [Google Scholar] [CrossRef]
- Maes, S.; Heyndrickx, M.; Vackier, T.; Steenackers, H.; Verplaetse, A.; De Reu, K. Identification and Spoilage Potential of the Remaining Dominant Microbiota on Food Contact Surfaces after Cleaning and Disinfection in Different Food Industries. J. Food Prot. 2019, 82, 262–275. [Google Scholar] [CrossRef]
- Gutiérrez, D.; Delgado, S.; Vázquez-Sánchez, D.; Martínez, B.; Cabo, M.L.; Rodríguez, A.; Herrera, J.J.; García, P. Incidence of Staphylococcus aureus and Analysis of Associated Bacterial Communities on Food Industry Surfaces. Appl. Environ. Microbiol. 2012, 78, 8547–8554. [Google Scholar] [CrossRef] [PubMed]
- Dodd, C.E.R. Pseudomonas|Introduction. In Encyclopedia of Food Microbiology, 2nd ed.; Academic Press: Cambridge, MA, USA, 2014; pp. 244–247. [Google Scholar] [CrossRef]
- García-López, M.L.; Santos, J.A.; Otero, A.; Rodríguez-Calleja, J.M. Psychrobacter. In Encyclopedia of Food Microbiology, 2nd ed.; Academic Press: Cambridge, MA, USA, 2014; pp. 261–268. [Google Scholar] [CrossRef]
- Rafii, F. Serratia. In Encyclopedia of Food Microbiology, 2nd ed.; Academic Press: Cambridge, MA, USA, 2014; pp. 371–375. [Google Scholar] [CrossRef]
- Kämpfer, P. Acinetobacter. In Encyclopedia of Food Microbiology, 2nd ed.; Academic Press: Cambridge, MA, USA, 2014; pp. 11–17. [Google Scholar] [CrossRef]
- Audrain, B.; Létoffé, S.; Ghigo, J.M. Airborne Bacterial Interactions: Functions out of Thin Air? Front. Microbiol. 2015, 6, 1476. [Google Scholar] [CrossRef]
- Schulz-Bohm, K.; Martín-Sánchez, L.; Garbeva, P. Microbial Volatiles: Small Molecules with an Important Role in Intra- and Inter-Kingdom Interactions. Front. Microbiol. 2017, 8, 2484. [Google Scholar] [CrossRef]
- Stoddard, S.F.; Smith, B.J.; Hein, R.; Roller, B.R.K.; Schmidt, T.M. RrnDB: Improved Tools for Interpreting RRNA Gene Abundance in Bacteria and Archaea and a New Foundation for Future Development. Nucleic Acids Res. 2015, 43, D593–D598. [Google Scholar] [CrossRef]
- Roller, B.R.K.; Stoddard, S.F.; Schmidt, T.M. Exploiting RRNA Operon Copy Number to Investigate Bacterial Reproductive Strategies. Nat. Microbiol. 2016, 1, 16160. [Google Scholar] [CrossRef] [PubMed]
- Gobbetti, M.; Rizzello, C.G. Arthrobacter. In Encyclopedia of Food Microbiology, 2nd ed.; Academic Press: Cambridge, MA, USA, 2014; pp. 69–76. [Google Scholar] [CrossRef]
- Forquin, M.P.; Weimer, B.C. Brevibacterium. In Encyclopedia of Food Microbiology, 2nd ed.; Academic Press: Cambridge, MA, USA, 2014; pp. 324–330. [Google Scholar] [CrossRef]
- Waśkiewicz, A.; Irzykowska, L. Flavobacterium spp.—Characteristics, Occurrence, and Toxicity. In Encyclopedia of Food Microbiology, 2nd ed.; Academic Press: Cambridge, MA, USA, 2014; pp. 938–942. [Google Scholar] [CrossRef]
- García, G.; Girón, J.A.; Yañez, J.A.; Cedillo, M.L. Stenotrophomonas Maltophilia and Its Ability to Form Biofilms. Microbiol. Res. 2022, 14, 1. [Google Scholar] [CrossRef]
- Wagner, E.M.; Fischel, K.; Rammer, N.; Beer, C.; Palmetzhofer, A.L.; Conrady, B.; Roch, F.F.; Hanson, B.T.; Wagner, M.; Rychli, K. Bacteria of Eleven Different Species Isolated from Biofilms in a Meat Processing Environment Have Diverse Biofilm Forming Abilities. Int. J. Food Microbiol. 2021, 349, 109232. [Google Scholar] [CrossRef] [PubMed]
- Daeschel, D.; Pettengill, J.B.; Wang, Y.; Chen, Y.; Allard, M.; Snyder, A.B. Genomic Analysis of Listeria Monocytogenes from US Food Processing Environments Reveals a High Prevalence of QAC Efflux Genes but Limited Evidence of Their Contribution to Environmental Persistence. BMC Genom. 2022, 23, 488. [Google Scholar] [CrossRef] [PubMed]
- Alvarez-Molina, A.; Cobo-Díaz, J.F.; López, M.; Prieto, M.; de Toro, M.; Alvarez-Ordóñez, A. Unraveling the Emergence and Population Diversity of Listeria monocytogenes in a Newly Built Meat Facility through Whole Genome Sequencing. Int. J. Food Microbiol. 2021, 340, 109043. [Google Scholar] [CrossRef] [PubMed]
- Harrand, A.S.; Jagadeesan, B.; Baert, L.; Wiedmann, M.; Orsi, R.H.; Dudley, E.G. Evolution of Listeria monocytogenes in a Food Processing Plant Involves Limited Single-Nucleotide Substitutions but Considerable Diversification by Gain and Loss of Prophages. Appl. Environ. Microbiol. 2020, 86, e02493-19. [Google Scholar] [CrossRef] [PubMed]
- Arthur, T.M.; Bono, J.L.; Kalchayanand, N. Characterization of Escherichia coli O157:H7 Strains from Contaminated Raw Beef Trim during “High Event Periods”. Appl. Environ. Microbiol. 2014, 80, 506–514. [Google Scholar] [CrossRef]
- Yang, X.; He, A.; Badoni, M.; Tran, F.; Wang, H. Mapping Sources of Contamination of Escherichia Coli on Beef in the Fabrication Facility of a Commercial Beef Packing Plant. Food Control 2017, 75, 153–159. [Google Scholar] [CrossRef]
- Yang, X.; Tran, F.; Youssef, M.K.; Gill, C.O. Determination of Sources of Escherichia coli on Beef by Multiple-Locus Variable-Number Tandem Repeat Analysis. J. Food Prot. 2015, 78, 1296–1302. [Google Scholar] [CrossRef]
- Yang, X.; Wang, H.; He, A.; Tran, F. Microbial Efficacy and Impact on the Population of Escherichia coli of a Routine Sanitation Process for the Fabrication Facility of a Beef Packing Plant. Food Control 2017, 71, 353–357. [Google Scholar] [CrossRef]
- Yang, X.; Tran, F.; Zhang, P. Comparative Genomic Analyses of Escherichia coli from Meat Processing Environment in Relation to Their Biolm Formation and Persistence. Microbiol. Spectr. 2022; e00183-23, e-pub ahead of print. [Google Scholar] [CrossRef]
- Zhang, P.; Essendoubi, S.; Keenliside, J.; Reuter, T.; Stanford, K.; King, R.; Lu, P.; Yang, X. Genomic Analysis of Shiga Toxin-Producing Escherichia coli O157:H7 from Cattle and Pork-Production Related Environments. Npj Sci. food 2021, 5, 15. [Google Scholar] [CrossRef]
- Maes, S.; Vackier, T.; Nguyen Huu, S.; Heyndrickx, M.; Steenackers, H.; Sampers, I.; Raes, K.; Verplaetse, A.; De Reu, K. Occurrence and Characterisation of Biofilms in Drinking Water Systems of Broiler Houses. BMC Microbiol. 2019, 19, 77. [Google Scholar] [CrossRef]
- Yano, T.; Kubota, H.; Hanai, J.; Hitomi, J.; Tokuda, H. Stress Tolerance of Methylobacterium Biofilms in Bathrooms. Microbes Environ. 2013, 28, 87. [Google Scholar] [CrossRef]
- Yam, K.C.; Okamoto, S.; Roberts, J.N.; Eltis, L.D. Adventures in Rhodococcus—From Steroids to Explosives. Can. J. Microbiol. 2011, 57, 155–168. [Google Scholar] [CrossRef]
- Chatterjee, D.; Cooley, R.B.; Boyd, C.D.; Mehl, R.A.; O’Toole, G.A.; Sondermann, H. Mechanistic Insight into the Conserved Allosteric Regulation of Periplasmic Proteolysis by the Signaling Molecule Cyclic-Di-GMP. eLife 2014, 3, e03650. [Google Scholar] [CrossRef]
- Lelieveld, H.L.M.; Mostert, M.A.; Curiel, G.J. Hygienic Design of Food Processing Equipment. In Hygiene in Food Processing Principles and Practice, 2nd ed.; Woodhead Publishing: Sawston, UK, 2014; pp. 91–141. [Google Scholar] [CrossRef]
- Wang, Z.; Fang, Y.; Zhi, S.; Simpson, D.J.; Gill, A.; McMullen, L.M.; Neumann, N.F.; Gänzle, M.G. The Locus of Heat Resistance Confers Resistance to Chlorine and Other Oxidizing Chemicals in Escherichia coli. Appl. Environ. Microbiol. 2020, 86, e02123-19. [Google Scholar] [CrossRef]
- Xu, Z.S.; Yang, X.; Gänzle, M.G. Resistance of Biofilm-and Pellicle-Embedded Strains of Escherichia coli Encoding the Transmissible Locus of Stress Tolerance (TLST) to Oxidative Sanitation Chemicals. Int. J. Food Microbiol. 2021, 359, 109425. [Google Scholar] [CrossRef]
- Møretrø, T.; Schirmer, B.C.T.; Heir, E.; Fagerlund, A.; Hjemli, P.; Langsrud, S. Tolerance to Quaternary Ammonium Compound Disinfectants May Enhance Growth of Listeria monocytogenes in the Food Industry. Int. J. Food Microbiol. 2017, 241, 215–224. [Google Scholar] [CrossRef]
- Dutta, V.; Elhanafi, D.; Kathariou, S. Conservation and Distribution of the Benzalkonium Chloride Resistance Cassette BcrABC in Listeria monocytogenes. Appl. Environ. Microbiol. 2013, 79, 6067–6074. [Google Scholar] [CrossRef] [PubMed]
- Mangalappalli-Illathu, A.K.; Vidović, S.; Korber, D.R. Differential Adaptive Response and Survival of Salmonella Enterica Serovar Enteritidis Planktonic and Biofilm Cells Exposed to Benzalkonium Chloride. Antimicrob. Agents Chemother. 2008, 52, 3669–3680. [Google Scholar] [CrossRef] [PubMed]
- Vestby, L.K.; Grønseth, T.; Simm, R.; Nesse, L.L. Bacterial Biofilm and Its Role in the Pathogenesis of Disease. Antibiotics 2020, 9, 59. [Google Scholar] [CrossRef]
- Saini, J.K.; Marsden, J.L.; Fung, D.Y.C.; Crozier-Dodson, B.A. Evaluation of Potential for Translocation of Listeria monocytogenes from Floor Drains to Food Contact Surfaces in the Surrounding Environment Using Listeria Innocua as a Surrogate. Adv. Microbiol. 2012, 2, 565–570. [Google Scholar] [CrossRef][Green Version]
- Jacques, M.; Malouin, F. One Health-One Biofilm. Vet. Res. 2022, 53, 51. [Google Scholar] [CrossRef] [PubMed]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xu, Z.S.; Ju, T.; Yang, X.; Gänzle, M. A Meta-Analysis of Bacterial Communities in Food Processing Facilities: Driving Forces for Assembly of Core and Accessory Microbiomes across Different Food Commodities. Microorganisms 2023, 11, 1575. https://doi.org/10.3390/microorganisms11061575
Xu ZS, Ju T, Yang X, Gänzle M. A Meta-Analysis of Bacterial Communities in Food Processing Facilities: Driving Forces for Assembly of Core and Accessory Microbiomes across Different Food Commodities. Microorganisms. 2023; 11(6):1575. https://doi.org/10.3390/microorganisms11061575
Chicago/Turabian StyleXu, Zhaohui S., Tingting Ju, Xianqin Yang, and Michael Gänzle. 2023. "A Meta-Analysis of Bacterial Communities in Food Processing Facilities: Driving Forces for Assembly of Core and Accessory Microbiomes across Different Food Commodities" Microorganisms 11, no. 6: 1575. https://doi.org/10.3390/microorganisms11061575
APA StyleXu, Z. S., Ju, T., Yang, X., & Gänzle, M. (2023). A Meta-Analysis of Bacterial Communities in Food Processing Facilities: Driving Forces for Assembly of Core and Accessory Microbiomes across Different Food Commodities. Microorganisms, 11(6), 1575. https://doi.org/10.3390/microorganisms11061575






