Abstract
Our systematic review aimed to evaluate the effect of periodontal interventions on the diversity and composition of periodontal microbiota assessed by high throughput sequencing (HTS) metagenomics analysis. An electronic search was conducted from database inception to November 2021. All clinical trials that evaluated the effect of periodontal interventions on the gingival microbiota through HTS were selected. The measures of alpha diversity, richness, Shannon diversity index, and the Chao1 index, were used as the primary outcome, whereas relative abundances of bacterial genera were considered as the secondary outcome. Overall, 24 studies were eligible for the systematic review, of which 13 studies were included in the meta-analysis. Periodontal intervention for the test group decreased Shannon diversity, richness, and Chao1 index (alpha diversity), as observed from baseline to post-treatment. The most common genera that increased after periodontal therapy were Rothia, Actinomyces, Streptococcus, Veillonella, and Hemophilus, whilst Porphyromonas, Tannerella, Fusobacterium, and Treponema decreased after periodontal therapy. Periodontal interventions may decrease the bacterial diversity and richness and alter the composition of oral microbiota in the short term. Periodontal microbiota signatures could potentially be used for the assessment of periodontal disease development, progression, and success of the intervention.
1. Introduction
The human body is a superorganism with trillions of associated microorganisms that are essential for maintaining health or eliciting disease. The term microbiota generally refers to all organisms comprising bacteria, fungi, protozoa, and viruses, and a perturbation of the healthy microbiota due to complex interactions of genetic, microbial, host, and environmental factors results in the emergence of pathobionts [1]. These microbiota easily outnumber the number of human cells within the body [2]. The total mass of bacteria in an average human is estimated to be about 0.2 kg, and the total number of bacteria cells is around 3.8 × 1013 [3]. The activity of the microbiota and the expression of their genomic information, known as the microbiome, provides humans with traits that are not usually present within the human genome [4]. In the mouth alone, over 700 bacterial species have been identified in oral samples by DNA-based microbiome analysis, and they form complex mixtures of species in different micro-niches on the teeth, tongue, and soft and hard tissues of the mouth [5].
Periodontal diseases, including gingivitis and periodontitis, result from inflammation caused by ecological disturbances in the periodontal microbiota [6,7]. In periodontal health, the prevalence of potentially virulent species (pathobionts) are present, but in lower abundance than in individuals with the disease. Clinical evidence suggests that increased periodontal inflammation may not be associated with distinct microbiota, but with an increase in abundance of potentially virulent species, such as Porphyromonas gingivalis, Treponema denticola, and Tannerella forsythia, as well as others, which increases the total bacterial biomass [8]. As a result, several authors have reported increased bacterial richness, evenness, and diversity associated with periodontal disease, although this is unusual, as a disease or microbiota dysbiosis is usually associated with a less diverse microbiome and an increase in certain interventions in other parts of the body [8].
Periodontal treatment consists of a broad range of interventions aimed at controlling the infection and arresting the inflammation [9]. The first step in periodontal therapy consists of non-surgical therapy, i.e., scaling and root planning (SRP) and controlling of risk factors (i.e., smoking and uncontrolled Type II diabetes) [10]. Over the years, researchers have shown that SRP led to a clear improvement in periodontal pocket depth, suppressed periodontal bleeding, and reduced microbial dysbiosis [11]. Several authors have suggested the use of adjuncts to SRP such as antibiotics (commonly amoxicillin and/or metronidazole), mouthwashes (e.g., chlorhexidine), local drug delivery (e.g., doxycycline), and host modulating therapy (e.g., sub-antimicrobial dose doxycycline) [12], especially in periodontitis patients. Periodontal interventions, such as SRP without chemical adjuncts, aim at tampering with the periodontal microflora that would eventually lead to the resolution of inflammation. But these procedures can decrease microbial diversity and significantly reduce the relative abundances of both healthy- and gingivitis-associated bacterial species [13,14]. The development of emerging techniques, such as the potential for oral microbiota transplantation, has also sparked an interest in the response of periodontal microbiota to different therapies to re-establish a healthy microbiota without reducing the bacterial richness and diversity [15]. Novel kinds of toothpaste are also available, which claim to shape the oral microbiota via proteins designed to foster species associated with the healthy oral microbial communities [16]. Similarly, mouthwashes are also available in the market to reduce the bacterial load by antimicrobial effects by altering the plaque microbiota [17,18].
Previous studies have used low throughput measurement techniques (microbial culture, checkerboard hybridization, and PCR analysis), which have resulted in the incomplete characterization of the oral microbiome composition [19,20,21]. Little is known about how the periodontal microbiota composition and diversity changes metagenomically in response to the periodontal treatment [22]. Research can now examine differences in the periodontal microbiota between health and disease through high throughput sequencing (HTS) or next-generation sequencing (NGS). The 16S rRNA amplicon sequencing and random shotgun sequencing are the two main approaches to sequencing. The amplicon sequencing of 16S rRNA genes allows thousands of sequences per sample and provides the power to comprehensively study bacterial community diversity and composition within a specific niche. The shotgun approach allows the study of the entire genome based on random fragments of DNA that are then assembled by finding overlapping ends [23]. There are several bioinformatics tools that provide pipelines used for the generation, clustering, and assigning of operational taxonomic unit (OTU) and building OTU tables and analysing microbial communities and diversities. Recently, with the increased use of denoising methods, amplicon sequence variants (ASVs) are produced instead of clusters. The two most used matrices to assess and compare microbial communities are alpha (within-sample) and beta (between-sample) diversity. The measures of alpha diversity include the number of OTUs/ASVs count or richness (total number of organisms within a sample), the evenness (the relative abundance of the organisms), or indices that combine these two dimensions (e.g., Shannon’s diversity; Chao1 index) [24]. The beta diversity is the number of species shared between microbial communities between samples. The measures of beta diversity include UniFrac, Bray Curtis dissimilarity, Jaccard distance, Principal Component Analysis (PCA), and Principal Coordinates Analysis (PCoA) [25] (Figure 1).
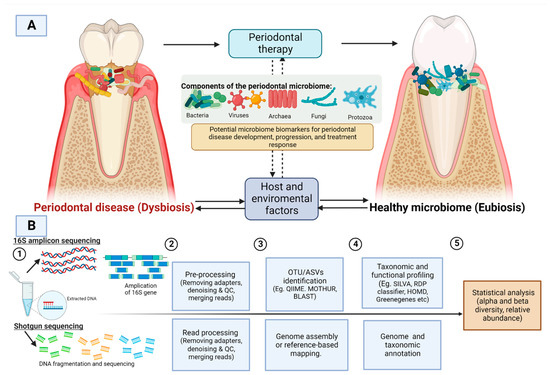
Figure 1.
(A) Dysbiosis caused by the periodontal microbiome is influenced by various host and environmental factors. Periodontal intervention could alter the microbial composition from a state of dysbiosis (periodontal disease) to eubiosis (periodontally healthy) (B) A general workflow for 16S amplicon sequencing and shotgun sequencing. QC: quality control, OTU: operational taxonomic unit; ASV: Amplicon sequence variants. Image created with BioRender.com, accessed om 27 July 2022.
To our knowledge, there has been no meta-analysis conducted on the changes in the periodontal microbiome analyzed through HTS exclusively after the periodontal intervention on periodontal disease patients. This review will be a recent update of the periodontal microbiome literature. The research question was based on the PICOS (Population, Intervention, Comparison, Outcome, and Study) format. In clinical trials (S), we sought to answer: does any form of periodontal treatment (I) compared to intervention without active agent/placebo/SRP (C) affect the composition of the periodontal microbiota (O) involving adults with periodontal disease (P) using HTS methodologies? The authors hypothesized that periodontal intervention could reverse the dysbiotic microbiota associated with periodontal disease towards a balanced state consistent with oral health.
2. Materials and Methods
2.1. Study Registration
The study review protocol was registered with the PROSPERO database (International Prospective Register of Systematic Reviews), registered under CRD42020188531, and can be accessed on https://www.crd.york.ac.uk/prospero/display_record.php?RecordID=188531. This systematic review and meta-analysis has been prepared according to the Preferred Reporting Items for Systematic Reviews and Meta-Analysis (PRISMA) guidelines [26].
2.2. Specific Research Goals
- To find the changes in the composition of the periodontal microbiome community after periodontal treatment. By composition, we intend to find patterns in disease-associated shifts in the periodontal microbiota that differ in their directionality (microbiota pre-intervention vs. microbiota post-intervention);
- To find the magnitude of difference in alpha diversity metrics before and after periodontal treatment. Some of the commonly used measures include several OTU counts (richness), Shannon diversity index (accounts for both abundance and evenness of the species), and Chao1 index (non-parametric method for assessing the number of species in a community). To find the difference in beta diversity before and after treatment. The commonly used measures of beta diversity include UniFrac, PCA, PCoA, Bray Curtis dissimilarity, Jaccard distance, and Principal Coordinates Analysis; and
- To find the predominant bacterial species present in the periodontal microbiome and the total number of bacterial species that differ between the treatment group and control group identified through high throughput sequencing.
2.3. Search Strategy
An electronic search was conducted on the following databases to identify eligible studies: MEDLINE, Scopus, and EBSCOHost (Dentistry & Oral Sciences Sources). For grey literature, we used the Cochrane database of systematic review, OpenGrey database, ProQuest Dissertation, and clinicaltrial.gov. In addition, a manual hand-searching of reference lists of relevant papers was screened to identify articles that might have been missed on the electronic search. We refrained from using Google Scholar; although it is a very powerful tool, it has a low threshold of reproducibility, accepts only very basic Boolean logic, the algorithm by which the search results are ordered has not been disclosed, and it may not be an effective means of identifying grey literature [27]. The articles were searched from database inception to November 2021.
The search strategy used for the MEDLINE database was (“Metagenomics” [MeSH] OR “Metagenome” [MeSH] OR “High-Throughput Nucleotide Sequencing” [MeSH] OR “microbiota” [MeSH] OR “Genes, Bacterial” [MeSH] OR “metagenomics” [tiab] OR “16S rDNA” [tiab] OR “16S rRNA” [tiab] Or Pyrosequencing [tiab] OR “next-generation sequencing” [tiab] OR “Illumina sequencing” [tiab] OR “Functional gene array” [tiab] OR “Oral microbiome” [tiab] OR “Bacteria*” [tiab] OR “Bacterial diversity” [tiab] OR “Bacterial community” [tiab]) AND (“Tooth Diseases” [MeSH] OR “Mouth Diseases” [MeSH] OR “Oral Health” [MeSH] OR “Gingival diseases” [MeSH] OR “Gingivitis” [MeSH] OR “Periodontal diseases” [MeSH] OR “Periodontal debridement” [MeSH] OR “Periodontal index” [MeSH] OR “Periodontal pocket” [MeSH] OR “probing depth” [tiab] OR “periodont*” OR “plaque score”).
For Scopus and EBSCOHost, we used a similar search strategy, but for our grey literature search, we used a truncated search string to maximize the number of results (Table A1). The search string was pilot tested using a combination of MeSH and key terms. Systematic reviews, narrative reviews, and standard textbooks were additionally searched to identify all the other eligible studies. Whenever possible, citation tracking was performed on search engines to keep track of the latest publication.
2.4. Study Selection Criteria
2.4.1. Types of Participants
The population selected was any individual ≥18 years of age who underwent any procedure or intervention for restoration of periodontal health. As we get older, the oral microbiome is stabilized compared to that of youth or children. This is due to the establishment of independent oral hygiene, maintenance habits, permanent dentition, and a stable adult diet with defined dietary patterns [28,29]. Having said that, even the adult microbiome can be altered throughout life with changes in dietary habits, increasing age, oral hygiene practices, and tobacco and alcohol use [29,30].
2.4.2. Type of Interventions
Interventions aimed at achieving periodontal health were included. All types of interventions such as standard periodontal therapy (i.e., SRP either as the sole procedure (having controls as no treatment) or combined with mouthwash or antibiotics, toothpaste with active agents, and customized diets for achieving periodontal health) were included. Standard treatment, including SRP, antibiotics (amoxicillin, or metronidazole), toothpaste without active ingredients, placebos, or no treatment, were included as controls. For both test and control groups, changes in microbial communities were noted at baseline and post-intervention. Therefore, we are comparing: (1) test vs. control and (2) each group individually (test group (baseline vs. post-intervention) and control group (baseline vs. post-intervention)). Trials that evaluated the effectiveness as a single interventional trial arm by comparing before and after treatment (baseline vs. post-intervention only) were also included, though there was no control group for comparison.
2.4.3. Types of Outcome Measures
The primary outcomes were measures of (1) alpha diversity of periodontal microbiota at baseline compared to post-treatment values for treatment and control subgroups (mean ± standard deviation) and (2) comparison of alpha diversity among treatment and control group post periodontal intervention (mean ± standard deviation). The relative abundance level (relative prevalence percentage) for each bacterial genera and species and beta diversity were included as secondary outcomes.
2.5. Selection Criteria
2.5.1. Inclusion Criteria
Studies with the following criteria were selected: (1) original studies; (2) evaluation of an intervention compared to control for restoration of periodontal health; (3) having periodontal disease (gingivitis and periodontitis); (4) analysis of periodontal microbiota through bacterial high throughput sequencing; and (5) randomized clinical trial, before and after trials and/or quasi-experimental.
2.5.2. Exclusion Criteria
Studies that analyzed salivary microbiome, tongue scraping, or other parts of oral mucosa instead of supragingival plaque or subgingival plaque for microbial profiling were excluded. Healthy participants without any forms of periodontal disease were excluded. In vitro or animal studies, case reports, case series, retrospective studies, literature reviews, opinion papers, letters to conference proceedings, and abstracts were also excluded.
2.5.3. Selection of Studies
All the articles found through searching of the databases were uploaded in referencing software, EndNote X9 Version 3.3 (Clarivate Analytics, Philadelphia, PA, USA), and the duplicates were removed. All the references were imported into systematic review management software, Covidence, and were used for screening title and abstracts and full texts. Two independent and calibrated reviewers (SN and SJP) assessed the studies by title and abstracts against the eligibility criteria. The full text was uploaded in Covidence, and assessments were performed only for selected articles that followed the inclusion and exclusion criteria. If there was any disagreement and consensus could not be reached among the two reviewers, a third reviewer (LMJ) was referred for the final decision. The reasons for the exclusion of an article have been recorded separately.
2.6. Data Extraction
Data extraction was performed independently from the selected studies by the two reviewers (SN and SJP) using a customized data extraction form containing all information necessary to answer the research question (Table A2). The data extraction form was first pilot tested on a random sample (5% of included studies) and modified until all the reviewers agreed upon the key variables and outcomes. There are various computational diversity matrices used among researchers for assessing alpha diversity such as Pielou’s evenness index, observed ASVs and OTUs count, Faith’s phylogenetic diversity, etc. However, for our review, the three most used (richness (including ASVs/OUT count), Shannon index, and Chao1 index) were selected as outcomes. When the included studies had any missing information, the authors were contacted via email correspondence for further details and information. The data extraction form included details on (1) study characteristics: surname of the first author, year and country of study, study design; (2) patient characteristics: disease type and definition, age, number of participants for test and control group, treatment and control description, and duration of treatment; (3) collection and extraction method: plaque collection method and hypervariable region for sequencing; and (4) outcome measures: Alpha diversity (richness, Shannon diversity, and Chao1 index) and relative abundance levels for genera identified in the study. The method of OTU/ASV generation, databases used taxonomic and functional profiling, and methods used for statistical analysis has been recorded for each included publication.
For pooled data and meta-analysis, data extraction from graphs or charts was performed using the WebPlotDigitizer tool (Version 4.2 GNU Affero General Public License). In studies that reported only the median, the estimates were converted to mean and standard deviation [31].
2.7. Assessment of Risk of Bias and Quality of Evidence Assessment
For quality assessment of clinical trials, the “Risk of Bias Assessment 2” (RoB 2.0) guidelines formed by the Cochrane Collaboration was used [32]. For each study, bias was assessed in five domains: (1) bias due to randomization; (2) bias due to deviations from intended intervention; (3) bias due to missing data; (4) bias due to outcome measurement and (5) bias due to selection of reported results. Each clinical trial was scored as ‘high risk’, ‘some concerns’, or ‘low risk’ and given an overall score. The robvis visualization tool was used for visualizing the risk of bias assessment. No study would be excluded based on the quality of the paper.
The quality of evidence across the included studies was assessed using the Grading of Recommendations Assessment, Development and Evaluation (GRADE) methodology [33]. The risk of bias and quality of evidence assessment was performed independently and in duplicates by two reviewers (SN and SJP). Any disagreements were resolved through discussion between the two reviewers, or a third reviewer was consulted (LMJ).
2.8. Statistical Analysis
All data were extracted and collated in a spreadsheet. All the findings are represented in narrative table form, and where possible, the data was meta-analyzed. Meta-analysis was performed using a random-effects model. We used the random-effects model in contrast to the fixed-effect model, as the former model assumes that the observed estimates of treatment can vary across studies due to real differences in the interventions and sampling variability [34]. In our review, we expected heterogeneity in periodontal interventions, study population, and follow-up length. We used the restricted maximum likelihood (REML) method as the estimation method for meta-analysis, which is a commonly used method when the number of studies is small and produces an unbiased estimate of between studies variability [35]. To compute an effect size, Hedges’s g standardized mean difference and 95% confidence intervals were estimated for measures of alpha diversity (1) within a group test (baseline vs. post-treatment) and control groups (baseline vs. post-treatment) and (2) between groups (test vs. control group). Heterogeneity was measured using Galbraith plots and I2 statistics. A galbraith plot is a scatterplot of the standardized effect size, used as an alternative to a forest plot for assessing heterogeneity and detecting potential outliers. I2 was used to represent the percentage of variation attributable to statistical heterogeneity and was categorized as low (25–50%), moderate (51–75%), or high (>75%) [36]. The leave-one-out meta-analysis performs multiple meta-analyses, omitting one study each time, such as sensitivity analysis. There is a tendency for smaller studies to report larger effect sizes than larger studies, which could be due to between-study heterogeneity and publication bias. Publication bias was assessed using contour-enhanced funnel plots constructed for visualization. Asymmetry in the plots may indicate publication bias. The meta-analysis was performed using STATA 17 software (StataCorp. 2017, Stata Statistical Software: Release 17.0 StataCorp LLC, College Station, TX, USA).
2.9. Power Calculation for Meta-Analysis
We performed a power calculation that allowed us to assess whether the included studies had sufficient statistical power to detect small effect sizes. Power calculation was carried out according to the methods described by Bohrentein et al. (2009) [37]. Since there was no previous meta-analysis performed, we conducted the calculation based on the effect size (d = −0.60) we calculated in our meta-analysis (alpha diversity between treatment and control group) and found that at least 7 studies are required with 200 participants for the test and control group assuming high heterogeneity and statistical power of 80% and alpha of 5%.
3. Results
3.1. Search Strategy and Screening Process
The search details are provided in the PRISMA flow chart (Figure 2). The search strategy resulted in 5019 potential articles: 1020 articles from PubMed (MEDLINE), 2274 articles from Scopus, 1444 articles from the Dentistry and Oral Sciences database, 25 from ProQuest Dissertation, 256 from Cochrane database of systematic reviews, and no articles from OpenGrey. After the removal of 1245 articles as duplicates, 3374 articles were screened. All articles were assessed according to the inclusion and exclusion criteria, and this led to a full-text analysis of 28 articles [6,13,14,16,17,18,23,38,39,40,41,42,43,44,45,46,47,48,49,50,51,52,53,54,55,56,57,58]. Two studies did not report baseline data [6,53], and two studies assessed healthy participants [16,46]; therefore, all four were excluded. Overall, 24 studies were included for descriptive analysis and 13 studies [13,14,17,18,39,41,43,44,49,51,52,57] were meta-analyzed.
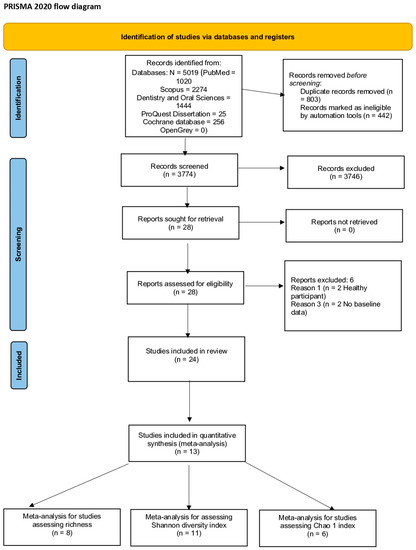
Figure 2.
PRISMA flow chart 2020 of the included trials.
3.2. Characteristics of Included Studies
Characteristics of the included studies were examined in the context of study design, periodontal disease definition, age, description of test and control group, plaque collection method, hypervariable region of 16S rRNA gene, and follow-up period (Table 1).

Table 1.
Characteristics of included studies according to treatment groups.
3.2.1. Study Type and Intervention
Of the 24 studies, 14 were randomized clinical trials (RCTs) [13,17,18,39,41,43,44,47,48,49,51,54,56,57], whereas eight studies [14,23,38,42,45,50,55,58] were pre- and post-intervention studies (before and after SRP) and two studies [40,52]) were clinical trials without clearly defining randomization. For this analysis, one RCT [13] was split into three sub-studies, two prevention sub-studies and one treatment sub-study, while another clinical trial [39] described their results as above or below median responses; thus, we analyzed this data separately. In total, six types of interventions were observed: (a) seven studies [14,23,38,42,45,55,58] were single-arm studies comparing pre- and post-SRP; (b) five studies supplemented SRP with antibiotics [39,43,48,50,51] or probiotics [41]; (c) four studies [13,18,40,56] used mouthwash rinses only; (d) two studies [49,52] compared ultrasonic scaling to air polishing; (e) three studies [17,44,47] used toothpaste with active ingredients; (f) one study used a regenerative material (Beta-tricalcium phosphate/EMD/Hydroxyapatite) for furcation therapy [55]; and (g) one study used an anti-inflammatory diet [57]. The duration of the interventions ranged from immediately after treatment [58] to three weeks [13,18], 4 weeks [17,41,42,45], 6 weeks [14,55,57], 8 weeks [43,48,50], 12 weeks [38,40,44,47,49,52], 6 months [51,54,56], and 12 months [39].
3.2.2. Study Participants
All the study subjects were generally systematically healthy patients, and the age ranged from 18–73 years. All the studies included both the male and female sex except for one study [45] that only included female patients. For this review, we included studies on all forms of periodontal diseases, including generalized gingivitis [6,55,57], chronic periodontitis [23,38,39,42,43,44,48,49,54,55,56], aggressive periodontitis [14,45,50], periodontitis (unclassified) [40,41,47,52,58] and experimental gingivitis [13,18].
3.2.3. Study Methodology and Metagenomics Analysis
The plaque collection methods varied across studies; the most popular methods for collection of plaque was sterile paper points [13,39,42,43,44,48,49,50,54,56] and sterile Gracey curettes [17,18,23,38,40,41,45,51,52,57,58] or periodontal scaler [55]. Few authors preferred filter paper [14] and swabs [47] for plaque collection. The most commonly used hypervariable region observed was the V3–V4/ V4 [14,38,42,43,45,50,51,52,56,57] region of 16S rRNA, while some authors used V1–V2 [13,17,18,55,58], V4-V6 [44], V4–V5 [40,47], V5–V7 [39], and V6 [48].
3.2.4. Bioinformatics and Statistical Test
The most popular method for sequencing is 16S rRNA sequencing (Table 2); however, one author used a combination of shotgun and 16S amplicon sequencing [40] and one author used only shotgun sequencing [23]. The OTU-based method for clustering has been commonly used across many of the studies, i.e., more than 97% similarity. All the studies used OTU as the basis of metagenomics analysis, except for two studies [43,44] that used ribosomal sequencing variants (RSVs), and two authors used amplicon sequence variant (ASV) [47,51]. The commonly used platforms for generating OTU/abundance tables from raw sequence reads were MOTHUR or QIIME/QIIME 2 (Quantitative Insights into Microbial Ecology) or pipelines in R programming software (DADA2, Phyloseq) used independently or in comparison with a referencing database such as Human Oral Microbiome database, SILVA 16S rRNA database, Greengenes database, and RDP.

Table 2.
Description of microbial data analysis from the included studies.
Additional statistical analysis was carried out by several authors for hypothesis testing of alpha and beta diversity indices before and after periodontal therapy. Depending on the normality and non-normality of the data, either t-test [23,49,52,58], analysis of variance (ANOVA), or a non-parametric test such as Wilcoxon rank-sum/signed signed-rank test [13,18,41,44,52,56,58] or Mann–Whitney test [13,14,39,41,44] and Kruskal Wallis test [38,57] was performed. Categorical variables were analyzed using the Chi-squared test, and McNemar and Fischer test [41]. For analyzing the association of microbial composition and environmental covariates and outcomes, several multivariate analysis methods were used. The analysis of group similarities (ANOSIM) [14,23,47], analysis of covariance (ANCOVA) [17,39,44], and multivariate analysis of variance was conducted with permutation (PERMANOVA) [39] and the Mantel test [40]. Sample size calculation was only carried out by few clinical trials [17,41,47,51,54,56,57].
3.2.5. Measures of Alpha Diversity
Alpha diversity is a measure of within-sample diversity of the community, described in terms of the number (richness) or distribution (evenness) [24]. The bacterial richness (number of observed species) was assessed commonly by most authors [13,14,18,43,44,47,49,58]. The commonly used indice to measure alpha diversity was Shannon Diversity Index [13,14,17,18,39,41,43,51,52,58]. Some studies used the Chao1 index [13,14,47,51,52,58], Simpson index [14,42,48], Pilous evenness [43,48], and Faith’s phylogenetic diversity [40]. The findings of alpha diversity from the included studies have been described in Table 2.
3.2.6. The Measure of Beta Diversity
Beta diversity is a measure of the between-sample differences between pairs of communities. It can be measured either using principal coordinates analysis (PCoA) or principal component analysis (PCA) [25]. To assess the beta-diversity, PCoA was used in twelve studies [13,14,17,23,40,43,44,45,47,51,52], and only three studies used the PCA [18,39,57]. Meta-analysis could not be conducted for beta diversity. The descriptive finding of each included study is summarized in Table 2. We tried contacting the authors of the papers for raw primary data for beta-diversity for meta-analysis but failed to receive any additional information.
3.2.7. Relative Abundance of Bacterial Genera
One of the main findings of the review was the consistency of the identified bacteria among the studies. We tried to identify the most abundant species before and after periodontal intervention and tried to find the common species that decreased and increased after periodontal intervention (Table A3). There was more prevalence of bacteria of the Red complex in the pre-intervention stage and more health-associated bacteria after an intervention. Post-intervention Porphyromonas, Fusobacterium, Tannerella, Treponema, Selenomonas, Parvimonas, TM7, Fillibactor, Fretibacterium, and Campylobacter decreased in number. While Escherichia, Neisseria, Prevotella, Capnocytophaga, Lautropia, Haemophilus, Veillonella, Actinomyces, Streptococcus, and Rothia increased in number post-intervention (Figure 3).
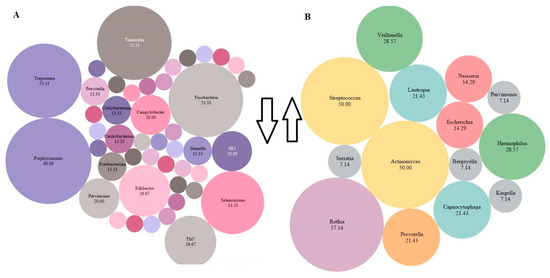
Figure 3.
The most abundant species present across all the studies post-intervention. The percentage of abundance was calculated by cumulative % of all bacterial species present in the included articles. The size of the bubble is related to the abundance (higher abundance = large bubble). The smaller bubbles represent species <7% (Megasphaera, Oribacterium, Peptococcus, Peptostreptococcus, Solobacterium, Actinomyces, Atopobium, Bacteroidacea, Clostridiales, Cornynebacterium, Dialaster, Granulicatella, Kingella, Mogibacterium, Olsnella, Propionibacterium, Spirochates, Stretococcus, Veillonella. (A) Decrease (downward arrow) in species post-intervention (B) Increase (upward arrow) in species post-intervention. The drawing was made in Tableau version 9.1, Seattle, WA, USA.
3.3. Synthesis of Results
3.3.1. Within-Group Alpha Diversity for Treatment and Control Groups (Baseline vs. Treatment)
Meta-analysis could not be conducted for studies that did not include mean and SD. Two studies [14,58] were before and after the trial, did not have control groups, and were excluded from the control sub-group analysis. The difference in alpha diversity from baseline to post-intervention was compared independently for both pooled test and pooled control groups. Post-intervention, the pooled test group showed lowered richness, Shannon’s diversity, and Chao1 index when compared to the control group. Eight trials [13,14,18,43,44,47,49,58] revealed a decrease in richness in the treatment group (SMD = 0.29; 95% CI = −0.10, 0.68) (Figure 4). In contrast, the richness resembled baseline in the control group (SMD = 0.08, 95% CI = −0.73, 0.90). Ten studies for the test group were meta-analyzed for Shannon’s diversity [13,14,17,18,39,41,43,51,52,58] (Figure 5). The test group showed a decrease in Shannon diversity in the post-treatment samples (SMD: 0.35, 95% CI: 0.08, 0.62), whereas the control group (SMD: −0.05, CI: −0.65, 0.54) resembled baseline. Six trials examined the Chao1 of observed species [13,14,47,51,52,58] (Figure 6), and there was a decrease in the Chao1 Index between baseline and post-treatment in the test group (SMD: 0.38, 95% CI: −0.06, 0.82) and the control group (SMD: 0.51, 95% CI: −0.12, 1.14).
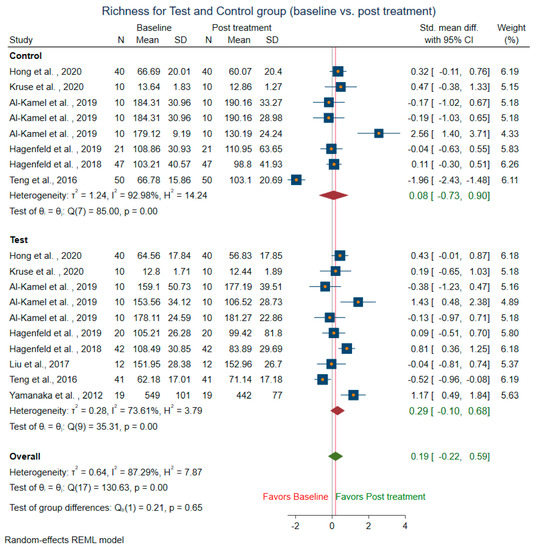
Figure 4.
Subgroup meta-analysis of richness based on test subgroup (baseline vs. post-treatment) and control subgroup (baseline vs. post-treatment).
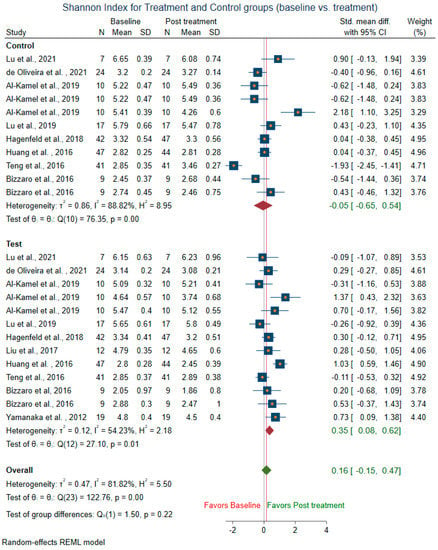
Figure 5.
Subgroup meta-analysis of Shannon index based on test subgroup (baseline vs. post-treatment) and control subgroup (baseline vs. post-treatment).
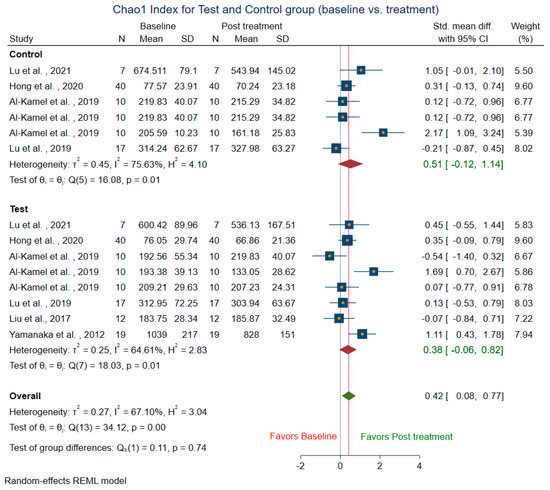
Figure 6.
Subgroup meta-analysis of Chao1 index based on test subgroup (baseline vs. post-treatment) and control subgroup (baseline vs. post-treatment).
3.3.2. Between-Group Alpha Diversity (Test vs. Control)
Two studies [14,58] were dropped from the analysis, as they did not have control groups. The richness was lower after periodontal intervention in the treatment group than in the control group (SMD: −0.45, 95 CI: −1.34, 0.43) (Supplementary Figure S1). Similarly, Shannon index (SMD: −0.61; 95% CI: −1.27, 0.06) (Supplementary Figure S2) and Chao1 index (SMD: −0.18; 95% CI: −1.19, 0.84) (Supplementary Figure S3) was lower among treatment groups when compared against the control group favoring periodontal treatment.
3.3.3. Heterogeneity among Studies and Publication Bias
For detection of heterogeneity, we used I2 statistics and Galbraith’s plot. The I2 was high (>80%) for all the forest plots of alpha diversity. The Galbraith scatter plot for richness showed all the studies except one lowered the richness after periodontal treatment (all the dot points are below the no-effect line) (Supplementary Figure S4) and heterogeneity was detected; three studies were outliers out of seven studies. For the Shannon index, there were four outliers out of ten groups (Supplementary Figure S5), and for the Chao1 index there were two outliers (Supplementary Figure S6), which may be the reason for heterogeneity.
The leave one out sensitivity analysis showed that one study [13] might have influenced the outcome for richness (Supplementary Figure S7), Shannon index (Supplementary Figure S8), and Chao1 (Supplementary Figure S9).
The contour-enhanced funnel plots were asymmetrical for all the measures of alpha diversity (Supplementary Figures S10–S12), but no small study effect was detected, and no publication bias was detected, as there were studies present in the non-significant region.
3.4. Risk of Bias Assessment and Quality of Evidence
The robvis tool was used for visualizing the risk of bias assessment (Supplementary Figure S13). The risk of bias for each study was assessed for all domains, as described in the Cochrane Handbook [32]. From the 24 studies, the overall risk of bias was high for all but six trials [13,41,44,47,49,51]. The main reason for high-risk bias was unclear reporting about random sequence generation and allocation concealment.
The GRADE analysis assessed the evidence of the outcome for alpha diversity (richness, Shannon, and Chao1) between the test and control groups, and they were of low quality (Supplementary Figure S14). This could have been due to a lack of randomization, high heterogeneity among studies, and lack of consistency in the methodology.
4. Discussion
To the best of our knowledge, this is the first systematic review that assessed the effects of periodontal interventions on the diversity of periodontal microbiota meta-analytically. The effects of periodontal interventions on the diversity and abundance of certain genera were assessed to test if periodontal interventions could alter the microbial composition of the periodontal plaque to a resolved or healthier state. The findings from our review suggest that periodontal intervention could lead to low diversity, richness, and community evenness, as we observed a decrease in the alpha diversity.
When interpreting the results of this systematic review, the following limitations should be considered. The heterogeneity could be primarily due to methodological variation (study design, disease levels, interventions, and study population) and sequencing techniques. There were a limited number of clinical trials reporting on the periodontal microbiome. We used six databases for our search strategy and could not expand our search strategy to other databases (e.g., Web of Science, Embase), and this could have led to publication bias; publications in the English language were selected, and this could have led to additional bias. For the meta-analysis, we have combined all forms of periodontal intervention which were directed toward treating periodontal disease. The main goal of this review was to observe changes in microbial communities (alpha and beta diversity) before and after periodontal therapy, regardless of the intervention. Therefore, we grouped studies based on the outcome and not the treatment. Previously, authors had similarly combined interventions in meta-analysis for the gut microbiome [59,60]. Periodontal disease includes both conditions, gingivitis, and periodontitis, and for this review, they were not separated. We included studies that have assessed the periodontal microbiome through a sampling of supra and subgingival plaque. Periodontal pathogens found in saliva may not be reflective of periodontal microbiota [38] and were excluded from this review. As observed from our review, the method of plaque collection varied from the use of paper points and sterile Gracey curettes, but both methods are equally effective for the collection of samples [61]. The included studies also used a wide range of different interventions to restore periodontal health from common procedures such as SRP to diet. The patient population also differed in geographic and ethnic variability.
The composition and diversity of the microbiome could be dependent on factors such as the hypervariable region selected, DNA extraction methods, sampling, the sequencing platform used, and the database for taxonomy [62]. The OTUs, ribosomal sequence variants (RSV) or ASVs, the number of reads per sample, the quality of filtering, and the normalization of data could also be the reason for heterogeneity. The authors of this review found methodological inconsistencies for HTS in all the included studies. For example, there were inconsistencies in DNA extraction and library preparation method and variability in marker selection (e.g., V1–V3 vs V3–V4). The laboratory environment should be standardized and applied across studies. Our systematic review and meta-analysis included studies with similar methods and outcome measures for microbial profiling. Systematic and comparable methodologies and rigorous statistical analysis are required for a more accurate and precise estimation of these effects. The latest use of HTS of the 16S rRNA gene allows a deeper analysis of the subgingival microbiota, but also has limited our ability to interpret data sets with wide ranges in methodologies, techniques, and analysis methods [63]. We also found most authors used the traditional statistical methods for microbial data analysis. Sparsity with many zeroes, overinflation, and over-dispersed data often poses a challenge for accurate statistical analysis for microbiome researchers. Handling rare taxa, and large p and small n [64] pose additional statistical issues. Due to these problems, the traditional parametric and non-parametric methods might not be suitable to analyze the microbiome data with many excess zeroes, and failure to account for this may result in misleading inference and estimation.
Periodontal disease leads to an oral microbiota with higher richness, evenness, and diversity. There is a shift in the microbial communities at the species and even at the strain/clone level. A diverse combination of species has been reported, but oral health-associated species are usually lower in frequency [8,65], whereas periodontal health is related to lowered diversity and evenness [23]. Our findings suggest that the richness and diversity were significantly lower in the pooled (all studies) treatment group compared to baseline. Periodontal interventions resulted in a less rich and diverse microbiota, which is a normal characteristic seen in a periodontally healthy individual. Therefore, periodontal interventions might have resulted in a shift in oral microbiota with fewer variations in the microbial community. Our findings are in contrast to those of Galimanas et al. (2014) [66] and Kirst et al. (2015) [67], who failed to capture any differences in microbial diversity and overall composition between healthy (no periodontitis) and periodontitis patients. While an overriding host defence could limit the community composition to non-pathogenic commensals, technical and methodological implications also need to be further investigated when comparing studies [65]. Overall, the diversity depends on the mean number of reads. A lot of information might be lost if the number of reads is too low due to extraction methods, and low abundance OTU might be discarded or the reads might be too short and might not provide a good identification of the microbial community [68,69]. The low abundance of organisms may make all the difference among the studies. NGS is moving towards producing longer reads (>500–700 bp for one single 16S rRNA) and therefore better identification. However, the targeting of the variable regions to the most appropriate region of 16S rRNA gene even, with larger reads, is more crucial. The microbiome is resilient due to richness, kinds of interactions, and functional redundancy.
While there is considerable debate about the presence of a core human microbiota, many microbes are generally common among most individuals and comprise dominant species that exist under healthy conditions [1]. The variable sets of species that are exclusive to an individual are often linked to lifestyle and environmental changes. It additionally depends on phenotypic and genotypic determinants, as well as one’s evolutionary history [70,71]. This signal is as unique across individuals as a human fingerprint [4]. In our review, we found several species increased and decreased post-intervention, and these species could be used as periodontal-based signatures for assessment of the efficacy of the periodontal intervention. The species Porphyromonas, Tannerella, Fusobacterium, Treponema, and Selenomonas were found consistently to decrease post-intervention among all the pooled studies. Collectively, these species are associated with gingivitis and chronic periodontitis [1,6,13,67]. A decrease in the abundance of these species has been associated with recovery from periodontal disease. Streptococcus, Actinomyces, Rothia, Veillonella, Prevotella, and Veillonella increased post-intervention. Among these, the classes Firmicutes (e.g., Streptococcus) and Actinobacteria (e.g., Actinomyces) were most commonly found (Figure 2). Previous studies have shown that Firmicutes and Actinobacteria are routinely found in dental plaque from healthy individuals [72] and Streptococcus have also been associated with healthy oral conditions and are an indicator of periodontal stability [63]. Similarly, Actinomyces are generally associated with health [1,6,13]. Due to a limited number of eligible studies, the abundance could not be calculated at a species level, which is a more robust technique to discriminate the role of specific community members in driving the ecological shift [16]. A combination of both amplicon and shotgun sequencing could identify bacteria more in-depth to the species level. The main advantage of HTS is the ability to discriminate species at the clonal level.
The current interventions for periodontal treatment aim at reducing the bacterial biomass but do not necessarily return it to a state of periodontal health [8]. Our review highlights the fact there is not a single composition of bacteria that represents either a healthy state or a diseased state, e.g., the genus Prevotella and Streptococcus are detected in health and some species are detected in disease. There has been an increase in interest in therapeutic interventions that can restore periodontal health by modulating microbial ecology [73]. Oral microbiome transplantation (OMTs) could be potentially beneficial in replacing diseased microbiota with healthy microbes [15]. This could ensure that the diseased site is repopulated with microbiota associated with health.
5. Conclusions
There is a need for more standardized clinical trial and NGS methods and validated reference databases for better taxonomy. Here, we conduct a preliminary, qualitative comparison to begin to understand the overarching trends of how periodontal microbiota respond to treatment.
Decreased alpha diversity indicates a shift of the periodontal microbiota towards periodontal health following a periodontal intervention. Few genera were consistently present in many of the included studies and could be part of the core microbiota. There was consistency in the microbiota species that increased and decreased post-intervention and could serve as microbiota-based signatures for understanding and comparing different periodontal interventions in the future.
Future Research
Future research in this area should be oriented towards examining different treatments while adopting the same methodologies for oral microbial profiling. Our findings highlight the importance of complete characterization of the periodontal microbiota to accurately evaluate the effectiveness of new therapeutic and diagnostic methods for periodontal disease treatment. The periodontal microbiota could be an alternative target for new therapies and should be monitored to better understand the efficacy of periodontal treatment outcomes.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/microorganisms10081582/s1, Figure S1: Meta-analysis of richness post-treatment (treatment vs. control); Figure S2. Meta-analysis of Shannon index post-treatment (treatment vs. control); Figure S3. Meta-analysis of Chao1 index post-treatment (treatment vs. control); Figure S4. Galbraith scatter plot of richness; Figure S5. Galbraith scatter plot of Chao1 index; Figure S6. Galbraith scatter plot of Shannon index; Figure S7. Leave one out analysis richness; Figure S8. Leave one out analysis Shannon index; Figure S9. Leave one out analysis Chao1 index; Figure S10. Funnel plot richness; Figure S11. Funnel plot Shannon index; Figure S12. Funnel plot Chao1 index; Figure S13. Risk of bias assessment; Figure S14. GRADE assessment.
Author Contributions
S.N., study conception and design, search and selection, statistical evaluation, data interpretation, and manuscript writing; S.J.P., study conception, search and selection, analysis, data interpretation, manuscript preparation, and writing; L.W., study concept and design, critically reading the manuscript; P.Z., data interpretation, critically reading the manuscript; K.K., statistical analyses, data interpretation, critically reading the manuscript; L.J., statistical analyses, data interpretation, critically reading the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
Sonia Nath is supported by the University of Adelaide, Research Training Program Stipend Scholarship 2020.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
All data generated or analyzed during this study will be included in a published article (and its supplementary information file).
Acknowledgments
We wish to acknowledge the University of Adelaide.
Conflicts of Interest
The authors declare no conflict of interest.
Appendix A

Table A1.
Search strategy.
Table A1.
Search strategy.
| Database | Search String | Results |
|---|---|---|
| Medline(PubMed) | ((((“Metagenomics” [MeSH Terms] OR “Metagenome” [MeSH Terms] OR “High-Throughput Nucleotide Sequencing” [MeSH Terms] OR “microbiota” [MeSH Terms] OR “genes, bacterial” [MeSH Terms] OR “Metagenomics” [Title/Abstract] OR “16S rDNA” [Title/Abstract] OR “16S rRNA” [Title/Abstract]) AND (“Or” [All Fields] AND “Pyrosequencing” [Title/Abstract])) OR “next-generation sequencing” [Title/Abstract] OR “Illumina sequencing” [Title/Abstract] OR “Functional gene array” [Title/Abstract] OR “Oral microbiome” [Title/Abstract] OR “Bacterial diversity” [Title/Abstract] OR “Bacterial community” [Title/Abstract]) AND (“Tooth Diseases” [MeSH Terms] OR “Mouth Diseases” [MeSH Terms] OR “Gingival diseases” [MeSH Terms] OR “Gingivitis” [MeSH Terms] OR “Periodontal diseases” [MeSH Terms] OR “Periodontal debridement” [MeSH Terms] OR “Periodontal index” [MeSH Terms] OR “Periodontal pocket” [MeSH Terms] OR “probing depth” [Title/Abstract] OR “periodont*” [All Fields] OR “plaque score” [All Fields]) AND “humans” [MeSH Terms]) AND (humans [Filter]) | 1020 |
| Scopus | (TITLE-ABS-KEY (“Metagenomics” OR “Metagenome” OR “High-Throughput Nucleotide Sequencing” OR “microbiota” OR “Genes, Bacterial” OR “metagenomics” OR “16S rDNA” OR “16S rRNA” OR pyrosequencing OR “next-generation sequencing” OR “Illumina sequencing” OR “Functional gene array” OR “Oral microbiome” OR “Bacterial diversity” OR “Bacterial community”) AND TITLE-ABS-KEY (“Tooth Diseases” OR “Mouth Diseases” OR “Gingival diseases” OR “Gingivitis” OR “Periodontal diseases” OR “Periodontal debridement” OR “Periodontal index” OR “Periodontal pocket” OR “probing depth” OR “periodont*” OR “plaque score”)) AND (LIMIT-TO (EXACTKEYWORD, “Human”)) AND (EXCLUDE (DOCTYPE, “re”) OR EXCLUDE (DOCTYPE, “ed”) OR EXCLUDE (DOCTYPE, “le”) OR EXCLUDE (DOCTYPE, “no”) OR EXCLUDE (DOCTYPE, “cp”) OR EXCLUDE (DOCTYPE, “ch”) OR EXCLUDE (DOCTYPE, “sh”) OR EXCLUDE (DOCTYPE, “tb”) OR EXCLUDE (DOCTYPE, “Undefined”)) | 2274 |
| Dentistry & Oral Sciences source | (“Metagenomics” OR “Metagenome” OR “High-Throughput Nucleotide Sequencing” OR “microbiota” OR “Genes, Bacterial” OR “metagenomics” OR “16S rDNA” OR “16S rRNA” OR pyrosequencing OR “next-generation sequencing” OR “Illumina sequencing” OR “Functional gene array” OR “Oral microbiome” OR “Bacterial diversity” OR “Bacterial community”) AND (“Tooth Diseases” OR “Mouth Diseases” OR “Gingival diseases” OR “Gingivitis” OR “Periodontal diseases” OR “Periodontal debridement” OR “Periodontal index” OR “Periodontal pocket” OR “probing depth” OR “periodont*” OR “plaque score”) | 1444 |
| ProQuest Dissertation | “Oral microbiome” OR “16S rDNA” OR “16S rRNA” AND (“Gingival diseases” OR “Gingivitis” OR “Periodontal diseases”) AND “clinical trial” AND filter (dissertations) | 25 |
| OpenGrey database | “Oral microbiome” OR “16S rDNA” OR “16S rRNA”AND (“Gingival diseases” OR “Gingivitis” OR “Periodontal diseases”) AND “clinical trial” | 0 |
| Cochrane database of systematic reviews | “Oral microbiome” OR “16S rDNA” OR “16S rRNA” AND (“Gingival diseases” OR “Gingivitis” OR “Periodontal diseases”) AND “clinical trial” | 256 |

Table A2.
Data extraction form.
Table A2.
Data extraction form.
| Variable | Definition | |
|---|---|---|
| Study Characteristics | ||
| 1. | SID | Unique identification number of study |
| 2. | Author | Last name of first author |
| 3. | Year | Year of publication |
| 4. | Country | Country of study conducted |
| 5. | Study design | Design of the study (e.g., parallel or split) |
| 6. | Randomization | Describes if randomization was performed (Yes = 1, No = 2, Not clear = 99) |
| 7. | Blinding | Describes if blinding was done and level of blinding (Single = 1, double = 2, triple = 3, none = 4, not clear = 99) |
| 8. | Sample size | Did the researchers calculate the sample size (Yes = 1, No = 2, Not clear = 99) |
| Participant Characteristics | ||
| 9. | Cases_n | Total number of cases |
| 10. | Control_n | Total number of control |
| 11. | age_mean, age_sd | Age of included participants [mean (SD), median (IQR), or categorical age, as reported] |
| 12. | Periodontal disease type | Describe the periodontal condition |
| 13. | Periodontal definition | Describe the definition used for classifying periodontal disease |
| 14. | Test description | Describes the intervention used for test group |
| 15. | Control description | Describes the intervention/placebo/no treatment for the control group |
| 16. | Treatment duration | Describes the recalls, and total duration of intervention |
| Collection and extraction method | ||
| 17. | Plaque_collection | Describes the site for plaque collection (supra or subgingival) and the instrument used for collection (e.g., paper point or sterile curette) |
| 18. | Hypervariable region | Describes the variable region used for DNA extraction |
| Measures of alpha diversity | ||
| 19. | OTU count (richness) | The OTU count scores at baseline and post-intervention; mean and SD |
| 20. | Chao1 index | The Chao1 scores at baseline and post-intervention; mean and SD |
| 21. | Shannon diversity index | The Shannon diversity scores at baseline and post-intervention; mean and SD |
| Outcome | Overall results of the study. | |

Table A3.
The relative abundances of bacteria.
Table A3.
The relative abundances of bacteria.
| Author and Year | Pre-Intervention Abundant Taxa | Post-Intervention Abundant Taxa | Post-Intervention Increase | Post-Intervention Decrease | Overall Abundant Taxa |
|---|---|---|---|---|---|
| Lu et al., 2021 | Porphyromonas, Treponema, Prevotella, Fusobacterium, Filifactor, Saccharibacteria TM7 G-5, and Peptostreptococcaceae XIG-6, | Actinomyces and Capnocytophaga | Actinomyces, Rothia, Neisseria, Capnocytophaga, Lautropia, and Cardiobacterium | Porphyromonas, Treponema, Filifactor, TM7 G-5, Peptostreptococcaceae XI G-6, Fretibacterium, Dialister, and Peptococcus. | Porphyromonas, Treponema, Prevotella, Fusobacterium, Filifactor, Saccharibacteria, Peptostreptococcaceae XIG-6, Actinomyces and Capnocytophaga |
| de Oliveira et al., 2021 | Capnocytophaga spp., Enterobactereacea, P.acnes, Staphylococus CN. | Actinomyces spp. Aggregatibacter actinomycetemcomitans, Acinetobacter baumannii, Clostridium difficile, Candida albicans, Campylobacter spp., Corynebacterium matruchotii, Dialister pneumosintes, Eikenella corrodens, Eubacterium spp., Enterococcus faecalis, Fusobacterium periodonticum, Fusobacterium nucleatum, Filifactor alocis, Gemella spp., Hafnia alvei, Helicobacter pylori, L.buccalis, Lactobacillus spp. Olsenella uli, Parvimonas micra, Paeriginosa, Porphyromonas gingivalis, Prevotella spp., streptococci spp. Staphylococcus aureus, Treponema socranskii, Treponema denticola, Tannerella forsythia, Veillonella parvula | N/A | N/A | Prevotella spp., Tannerella forsythia, Treponema denticola, Pseudomonas aeruginosa, Porphyromonas gingivalis, Actinomyces spp. |
| Wang et al., 2021 | Red complex bacteria | N/A | N/A | Porphyromonas gingivalis and Treponema denticola, Tannerella forsythia | Porphyromonas gingivalis and Treponema denticola, Tannerella forsythia |
| Hong et al., 2020 | Firmicutes, Proteobacteria | Streptococcus | Firmicutes | Phylum: Spirochaetes, Proteobacteria, Fusobacteria. Genus: Haemophilus, Fusobacterium and Capnocytophaga | Phylum: Firmicutes, Proteobacteria, Actinobacteria Genus: Streptococcus, Actinomyces, Lautropia, Actinomyces, Haemophilus, f_Neisseriaceae, Neisseria, Rothia, Corynebacterium, Capnocytophaga |
| Kruse et al., 2020 | Gram-positive aerobic cocci, Gram-positive aerobic rods, Gram-positive anaerobic rods, and Gram-negative anaerobic rods. | Gram-positive aerobic cocci, Gram-positive anaerobic cocci, Gram-positive anaerobic rods | Slackia exigua, Eubacterium yurii, Atopobium rimae, Filifactor alocis, Bifidobacterium dentium, Solobacterium moorei, Olsenella uli | Actinomyces meyeri, Actinomyces oris, Actinomyces odontolyticus, Actinomyces naeslundii, Actinomyces gerencseriae, Corynebacterium matruchotii, Rothia mucilaginosa, Rothia aeria, Streptococcus oralis, Streptococcus mitis, Streptococcus sanguinis, Streptococcus cristatus, Streptococcus sinensis, Streptococcus salivarius, Streptococcus anginosus, Streptococcus constellatus, Streptococcus intermedius, Streptococcus mutans, Gemella morbillorum, Streptococcus spp. | Porphyromonas gingivalis, Prevotella intermedia, Prevotella nigrescens, Prevotella tannerae, Prevotella buccae, Fusobacterium nucleatum, Campylobacter rectus, Tannerella forsythia, Selenomonas spp., parvula, Dialister pneumosintes, Anaeroglobus geminaus, Anaeroglobus geminatus, Parvimonas micra, Capnocytophaga ochracea, Capnocytophaga gingivalis, Aggregatibacter aphrophilus, Cardiobacterium hominis, Eikenella corrodens, Neisseria macacae/mucosa, Neisseria elongate, Neisseria flavescens, Neisseria bacilliformis, Nesseria spp., Lautropia mirabilis, Actinomyces meyeri, Actinomyces oris, Actinomyces odontolyticus, Actinomyces naeslundii, Actinomyces gerencseriae, Corynebacterium matruchotii, Rothia mucilaginosa, Rothia aeria, Streptococcus oralis, Streptococcus mitis, Streptococcus sanguinis, Streptococcus cristatus, Streptococcus sinensis, Streptococcus salivarius, Streptococcus anginosus, Streptococcus constellatus, Streptococcus intermedius, Streptococcus mutans, Gemella morbillorum, Streptococcus |
| Al-Kamel et al., 2019 Prevention sub-study | Streptococcus, Fusobacterium, Leptotrichia, Veillonella, Haemophilus, Actinomyces, Lautropia, Rothia, Capnocytophaga | NAC: Fusobacterium, Streptococcus, Leptotrichia, Cardiobacterium, Campylobacter, TM7_G_1_. CHX: Fusobacterium. Streptococcus, Granulicatella, Neisseria, Capnocytophaga. | NAC: Haemophilus, Lautropia, Rothia, Kingella, Brevundimonas, Escherichia. CHX: Rothia, Actinomyces, Cardiobacterium, Porphyromonas, Peptostreptococcus, Actinobaculum, Lachnospiraceae_G_3_, Lachnoanaerobaculum. | NAC: SR1_G_1_, Selenomonas, Olsenella, Parvimonas, Dialister, Oribacterium, TM7_G_1_, Cardiobacterium, Campylobacter, Fusobacterium. CHX: Granulicatella | Fusobacterium, Streptococcus, Leptotrichia, Veillonella, Propionibacterium, Actinomyces |
| Al-Kamel et al., 2019 Treatment sub-study | Fusobacterium, Streptococcus, TM7_G_1_, Leptotrichia, Prevotella, Veillonella, Porphyromonas | CHX: Fusobacterium, Streptococcus, Capnocytophaga, Bacteroidales_G_2, Prevotella, Veillonella NAC: Fusobacterium, Streptococcus, TM7_G_1, Leptotrichia, Propionibacterium, Cardiobacterium | Capnocytophaga | Corynebacterium, Stomatobaculum, Selenomonas, SR1-G_1, Lachnospiraceae_G1, TM7_G_3_, Lachnoanaerobaculum, Gemella, Propionibacterium, Veillonella, Tannerella, Cardiobacterium, Actinomyces, TM7_G_1_ | Fusobacterium, Streptococcus, TM7_G_1_, Leptotrichia, Prevotella, Veillonella, Propionibacterium. |
| Hagenfeld et al., 2019 | Fusobacterium, Prevotella, Veillonella | N/A | N/A | N/A | Fusobacterium, Prevotella, Veillonella, Porphyromonas, Alloprevotella, Campylobacter, Treponema_2, Streptococcus |
| Lu et al., 2019 | Streptococcus, and Saccharibacteria | N/A | Streptococcus, Escherichia, Acidopropionibacterium, Serratia | Treponema, Bacteroidacea | Actinomyces, Streptococcus, Leptotrichia, Capnocytophaga, Lautropia, Fusobacterium, Neisseria. |
| Woelber et al., 2019 | Streptococcus, Veillonella, Fusobacterium, Actinomyces, Prevotella | Fusobacterium, Veillonella, Streptococcus, Actinomyces, Prevotella. | Prevotella, Veillonella, Rothia | Streptococcus, Fusobacterium, Campylobacter, Cornybacterium | Fusobacterium, Veillonella, Streptococcus, Actinomyces, Rothia, Prevotella. |
| Hagenfeld et al., 2018 | Fusobacterium, Porphyromonas, Tannerella, Fretibacterium. | N/A | Streptococcus, Veillonella. | Porphyromonas, Tannerella, Treponema, Prevotella, Campylobacter, Fusobacterium, Parvimonas, Fretibacterium, Fillibactor, Oceanivirga. | N/A |
| Chen et al., 2018 | Fillifactor, Desulfobulbus, Eubacterium, Hallella, Porphyromonas, Phocaeicola, Tanerella, Bacteroidetes, Alloprevotella, Johnsonella, Treponema, Leptotrichia, Mogibacterium | N/A | N/A | N/A | Fusobacterium, Prevotella, Porphyromonas, Treponema, Corynebacterium, Leptotrichia, Selenomonas, Actinomyces, Campylobacter, Tanerella (total 27 genus) |
| Belstrom et al., 2018 | Prevotella, Treponema, Porphyromonas, Fusobacterium | Rothia, Prevotella, Streptococcus | Streptococcus, Rothia, Actinomyces | Porphyromonas, Treponema | Prevotella, Treponema, Porphyromonas, Fusobacterium, Rothia, Streptococcus, Cornyebacterium, Actinomyces. |
| Quieiroz et al., 2017 | Fusobacterium, Pseudomonas, Streptococcus, Fillifactor, Parvimonas. | N/A | N/A | Selenomonas, Fillifactor, Fusobacterium | Actinomyces, Campylobacter, Fillifactor, Fusobacterium, Gemella, Parvimonas, Psedomonas, Propionibacterium, Selenomonas, Streptococcus, Haemophilus, Veillonella |
| Han et al., 2017 | Sharpea, Moryella, Fusobacterium, Johnsonella, Peptostreptococcus, Peptococcus, Treponema, TG5, Desulfobulbus, Fillifactor, Tannerella, Porphyromonas, Megamonas, Esherichia, Selemonas, Dialister, Megasphaera, Prevotella, Leptotrichia, Hylemonella, Campylobacter, Bacteroides, Syntrophomonas | Kingella, Sphingopyxis, Lautropia, Capnocytophagam Neisseria, Aggregatibacter, Cornybacterium, Actinomyces, Parasscardovia, Veillonella, Rothia, Streptococcus | Actinobacteria, Proteobacteria | Bacteroidetes, Spirochaetes, Fusobacteria | Phyla: Bacteroidetes, Actinobacteria, Proteobacteria, Firmicutes, Fusobacteria, Spirochaetes, Synergistetes |
| Liu et al., 2017 | Porphyromonas, Treponema, Fretibacterium | Streptococcus, Lautropia, Haemophilus, Actinomyces | Lautropia, Actinomyces, Haemophilus | Treponema, Porphyromonas, Fretibacterium | Neisseria, Streptococcus, Fusobacterium |
| Califf et al., 2017 | Porphyromonas, Desulfovibrio, SHD-231, Treponema, Haemophilus, Acholeplasma, TG5, Mycoplasma, Eikenella, Desulfobulbus, Pseudoramibacter_Eubacterium, Methylobacterium Mogibacterium, Scardovia | Desulfovibrio, Streptococcus, Methanobrevibacter, PedobacterBE24, Butyrivibrio Peptococcus, Rhizobium, Aerococcus, Filifactor, Slackia | N/A | N/A | Desulfovibrio, Butyrivibrio, Methanobrevibacter, Pedobacter, Peptococcus, Filifactortreptococcus, Aerococcus, Slackia |
| Bizarro et al., 2016 | Porphyromonas, Treponema, Fusobacterium, Fillifactor. | Actinomyces, Streptococcus, Veillonella, Neisseria, Haemophilus | Both groups: Neisseria, Rothia, Capnocytophaga, Streptococcus Test group: Veillonella, Haemophilus Control group: Parvimonas, Actinomyces | Both groups: Fillifactor, Tannerella, uncultured Clostridiales family xiii incertae sedis, Porphyromonas, Treponema, uncultured Synergistaceae Test group: Paludibacter, Fusobacterium, Parvimonas | Fusobacterium, Prevotella, Treponema, Parvimonas, Porphyromonas, Paludibacter, Neisseria, Rothia, Fillifactor, Actinomyces, Streptococcus, Veillonella, Tannerella, Uncult. Clostridiales, Capnocytophaga, Haemophilus, Campylobacter |
| Teng et al., 2016 | Streptococcus, Actinomyces, Rothia, Veillonella | Leptotrichia, Neisseria, Capnocytophaga, Prevotella, Fusobacterium, Haemophilus, Lautropia, Abiotropia | Haemophilus, Lautropia, Neisseria, Capnocytophaga, Propioni-bacterium | Porphyromonas, Peptostreptococcus, Prevotella, Peptococcus, Selenomonas, Solobacterium, SR1, Tannerella, TM7 genus, Uncultured_Lachnospiraceae, Atopobium, Gemella, Megasphaera, Mogibacterium, Moraxella, Oribacterium and Shuttleworthia | Strptococcus, Leptotrichia, Actinomyces, Neisseria, Capnocytophaga, Rothia, Prevotella, Fusobacterium, Haemophilus, Lautropia, Porphyromonas, Cornyebacterium, Abiotropia |
| Huang et al., 2016 | Prevotella, Leptotrichia, Selenomonas, uncultured Lachnospiraceae, TM7, Tannerella, Peptococcus, and unclassified Veillonellaceae | Rothia, Granulicatella, Bergeyella and Lautropia | Actinomyces | T-Actinobaculum, TM7 and Leptotrichia C-Actinobaculum, TM7 and Leptotrichia | Rothia, Bergeyella, Lautropia, Granulicatella, Prevotella, Leptotrichia, Selenomonas, uncultured Lachnospiraceae, TM7, Tannerella, Peptococcus, and unclassified Veillonellaceae. |
| Shi et al., 2015 | Porphyromonas, Treponema, Tannerella, Olsnella, Peptostreptococcus, Synergistes, Fillifactor, Mycoplasma | Actinomyces, Streptococcus, Rothia, Bergeyella | Actinomyces, Streptococcus, Rothia, Bergeyella. | Porphyromonas, Treponema, Tannerella | Prevotella, Fusobacterium |
| Schwarzberg et al., 2014 | N/A | N/A | Streptococcus, Veillonella | Prevotella, Fusobacterium, Leptotrichia | Prevotella, Fusobacterium, Streptococcus |
| Laksmana et al., 2012 | Fusobacterium, Porphyromonas, Prevotella, Synergistetes spp., Fillifactor, Actinomyces, Treponema | Fusobacterium, Porphyromonas, Prevotella, Streptococcus, Veillonella | Streptococcus, Rothia, Actinomyces, Veillonella | Fusobacterium, Porphyromonas, Treponema, Tannerella | N/A |
| Junemann et al., 2012 | Porphyromonas, Prevotella, Treponema, Fusobacterium, Tannerella. | Prevotella, Streptococcus, Fusobacterium | Prevotella, Selenomonas, Streptococcus, Actinomyces, Rothia | Test group: Treponema, Fillifactor, Porphyromonas, Tannerella Control group: Porphyromonas, Tannerella | Phyla: Bacteroidetes, Firmicutes, Fusobacteria, Proteobacteria, Actinobacteria, Spirochaetes, and Synergistetes |
| Yamanaka et al., 2012 | Streptococcus, Leptotrichia, Actinomyces, Rothia, Fusobacterium | Streptococcus, Leptotrichia, Actinomyces, Rothia, Corynebacterium | Corynebacterium | Fusobacterium, Kingella | Streptococcus, Prevotella, Veillonella, Rothia, Actinomyces, Neisseria, Porphyromonas, Gemelia, Fusobacterium, Leptotrichia, Granulicatella, Capnocytophaga, Corynebacterium |
References
- Verma, D.; Garg, P.K.; Dubey, A.K. Insights into the human oral microbiome. Arch. Microbiol. 2018, 200, 525–540. [Google Scholar] [CrossRef] [PubMed]
- Dewhirst, F.E.; Chen, T.; Izard, J.; Paster, B.J.; Tanner, A.C.; Yu, W.H.; Lakshmanan, A.; Wade, W.G. The human oral microbiome. J. Bacteriol. 2010, 192, 5002–5017. [Google Scholar] [CrossRef] [PubMed]
- Sender, R.; Fuchs, S.; Milo, R. Revised Estimates for the Number of Human and Bacteria Cells in the Body. PLoS Biol. 2016, 14, e1002533. [Google Scholar] [CrossRef] [PubMed]
- Zarco, M.F.; Vess, T.J.; Ginsburg, G.S. The oral microbiome in health and disease and the potential impact on personalized dental medicine. Oral Dis. 2012, 18, 109–120. [Google Scholar] [CrossRef]
- Kilian, M. The oral microbiome—friend or foe? Eur. J. Oral Sci. 2018, 126 (Suppl. S1), 5–12. [Google Scholar] [CrossRef]
- Huang, S.; Li, R.; Zeng, X.; He, T.; Zhao, H.; Chang, A.; Bo, C.; Chen, J.; Yang, F.; Knight, R.; et al. Predictive modeling of gingivitis severity and susceptibility via oral microbiota. ISME J. 2014, 8, 1768–1780. [Google Scholar] [CrossRef]
- Diaz, P.I. Microbial diversity and interactions in subgingival biofilm communities. Front. Oral Biol. 2012, 15, 17–40. [Google Scholar]
- Abusleme, L.; Dupuy, A.K.; Dutzan, N.; Silva, N.; Burleson, J.A.; Strausbaugh, L.D.; Gamonal, J.; Diaz, P.I. The subgingival microbiome in health and periodontitis and its relationship with community biomass and inflammation. ISME J. 2013, 7, 1016–1025. [Google Scholar] [CrossRef]
- Haas, A.N.; Furlaneto, F.; Gaio, E.J.; Gomes, S.C.; Palioto, D.B.; Castilho, R.M.; Sanz, M.; Messora, M.R. New tendencies in non-surgical periodontal therapy. Braz. Oral Res. 2021, 35 (Suppl. S2), e095. [Google Scholar] [CrossRef]
- Bouchard, P.; Carra, M.C.; Boillot, A.; Mora, F.; Rangé, H. Risk factors in periodontology: A conceptual framework. J. Clin. Periodontol. 2017, 44, 125–131. [Google Scholar] [CrossRef]
- Zhang, Y.; Qi, Y.; Lo, E.C.M.; McGrath, C.; Mei, M.L.; Dai, R. Using next-generation sequencing to detect oral microbiome change following periodontal interventions: A systematic review. Oral Dis. 2020, 27, 1073–1089. [Google Scholar] [CrossRef] [PubMed]
- Nath, S.; Pulikkotil, S.J.; Dharmarajan, L.; Arunachalam, M.; Jing, K.T. Effect of locally delivered doxycycline as an adjunct to scaling and root planing in the treatment of periodontitis in smokers: A systematic review of randomized controlled trials with meta-analysis and trial sequential analysis. Dent. Res. J. 2020, 17, 235–243. [Google Scholar] [CrossRef]
- Al-Kamel, A.; Baraniya, D.; Al-Hajj, W.A.; Halboub, E.; Abdulrab, S.; Chen, T.; Al-Hebshi, N.N. Subgingival microbiome of experimental gingivitis: Shifts associated with the use of chlorhexidine and N-acetyl cysteine mouthwashes. J. Oral Microbiol. 2019, 11, 1608141. [Google Scholar] [CrossRef] [PubMed]
- Liu, G.; Luan, Q.; Chen, F.; Chen, Z.; Zhang, Q.; Yu, X. Shift in the subgingival microbiome following scaling and root planing in generalized aggressive periodontitis. J. Clin. Periodontol. 2018, 45, 440–452. [Google Scholar] [CrossRef] [PubMed]
- Nath, S.; Zilm, P.; Jamieson, L.; Kapellas, K.; Goswami, N.; Ketagoda, K.; Weyrich, L.S. Development and characterization of an oral microbiome transplant among Australians for the treatment of dental caries and periodontal disease: A study protocol. PLoS ONE 2021, 16, e0260433. [Google Scholar] [CrossRef]
- Adams, S.E.; Arnold, D.; Murphy, B.; Carroll, P.; Green, A.K.; Smith, A.M.; Marsh, P.D.; Chen, T.; Marriott, R.E.; Brading, M.G. A randomised clinical study to determine the effect of a toothpaste containing enzymes and proteins on plaque oral microbiome ecology. Sci. Rep. 2017, 7, 43344. [Google Scholar] [CrossRef]
- Huang, S.; Li, Z.; He, T.; Bo, C.; Chang, J.; Li, L.; He, Y.; Liu, J.; Charbonneau, D.; Li, R.; et al. Microbiota-based Signature of Gingivitis Treatments: A Randomized Study. Sci. Rep. 2016, 6, 24705. [Google Scholar] [CrossRef]
- Teng, F.; He, T.; Huang, S.; Bo, C.P.; Li, Z.; Chang, J.L.; Liu, J.Q.; Charbonneau, D.; Xu, J.; Li, R.; et al. Cetylpyridinium Chloride Mouth Rinses Alleviate Experimental Gingivitis by Inhibiting Dental Plaque Maturation. Int. J. Oral Sci. 2016, 8, 182–190. [Google Scholar] [CrossRef]
- Haffajee, A.D.; Roberts, C.; Murray, L.; Veiga, N.; Martin, L.; Teles, R.P.; Letteri, M.; Socransky, S.S. Effect of herbal, essential oil, and chlorhexidine mouthrinses on the composition of the subgingival microbiota and clinical periodontal parameters. J. Clin. Dent. 2009, 20, 211–217. [Google Scholar]
- Iniesta, M.; Herrera, D.; Montero, E.; Zurbriggen, M.; Matos, A.R.; Marín, M.J.; Sánchez-Beltrán, M.C.; Llama-Palacio, A.; Sanz, M. Probiotic effects of orally administered Lactobacillus reuteri-containing tablets on the subgingival and salivary microbiota in patients with gingivitis. A randomized clinical trial. J. Clin. Periodontol. 2012, 39, 736–744. [Google Scholar] [CrossRef]
- Morales, A.; Gandolfo, A.; Bravo, J.; Carvajal, P.; Silva, N.; Godoy, C.; Garcia-Sesnich, J.; Hoare, A.; Diaz, P.; Gamonal, J. Microbiological and clinical effects of probiotics and antibiotics on nonsurgical treatment of chronic periodontitis: A randomized placebocontrolled trial with 9-month follow-up. J. Appl. Oral Sci. 2018, 26, e20170075. [Google Scholar] [CrossRef] [PubMed]
- Guerra, F.; Mazur, M.; Ndokaj, A.; Corridore, D.; La Torre, G.; Polimeni, A.; Ottolenghi, L. Periodontitis and the microbiome: A systematic review and meta-analysis. Minerva Stomatol. 2018, 67, 250–258. [Google Scholar] [CrossRef] [PubMed]
- Shi, B.; Chang, M.; Martin, J.; Mitreva, M.; Lux, R.; Klokkevold, P.; Sodergren, E.; Weinstock, G.M.; Haak, S.K.; Lia, H. Dynamic changes in the subgingival microbiome and their potential for diagnosis and prognosis of periodontitis. MBio 2015, 6, e01926-14. [Google Scholar] [CrossRef] [PubMed]
- Moon, J.H.; Lee, J.H.; Lee, J.Y. Subgingival microbiome in smokers and non-smokers in Korean chronic periodontitis patients. Mol. Oral Microbiol. 2015, 30, 227–241. [Google Scholar] [CrossRef]
- Moon, J.-H.; Lee, J.-H. Probing the diversity of healthy oral microbiome with bioinformatics approaches. BMB Rep. 2016, 49, 662–670. [Google Scholar] [CrossRef]
- Page, M.J.; McKenzie, J.E.; Bossuyt, P.M.; Boutron, I.; Hoffmann, T.C.; Mulrow, C.D.; Shamseer, L.; Tetzlaff, J.M.; Akl, E.A.; Brennan, S.E.; et al. The PRISMA 2020 statement: An updated guideline for reporting systematic reviews. BMJ 2021, 372, n71. [Google Scholar] [CrossRef]
- Haddaway, N.R.; Collins, A.M.; Coughlin, D.; Kirk, S. The Role of Google Scholar in Evidence Reviews and Its Applicability to Grey Literature Searching. PLoS ONE 2015, 10, e0138237. [Google Scholar] [CrossRef]
- Burcham, Z.M.; Garneau, N.L.; Comstock, S.S.; Tucker, R.M.; Knight, R.; Metcalf, J.L.; Miranda, A.; Reinhart, B.; Meyers, D.; Woltkamp, D.; et al. Patterns of Oral Microbiota Diversity in Adults and Children: A Crowdsourced Population Study. Sci. Rep. 2020, 10, 2133. [Google Scholar] [CrossRef]
- Sedghi, L.; DiMassa, V.; Harrington, A.; Lynch, S.V.; Kapila, Y.L. The oral microbiome: Role of key organisms and complex networks in oral health and disease. Periodontology 2000 2021, 87, 107–131. [Google Scholar] [CrossRef]
- Gomez, A.; Espinoza, J.L.; Harkins, D.M.; Leong, P.; Saffery, R.; Bockmann, M.; Torralba, M.; Kuelbs, C.; Kodukula, R.; Inman, J.; et al. Host Genetic Control of the Oral Microbiome in Health and Disease. Cell Host Microbe 2017, 22, 269–278.e3. [Google Scholar] [CrossRef]
- Hozo, S.P.; Djulbegovic, B.; Hozo, I. Estimating the mean and variance from the median, range, and the size of a sample. BMC Med. Res. Methodol. 2005, 5, 13. [Google Scholar] [CrossRef]
- Sterne, J.A.C.; Savović, J.; Page, M.J.; Elbers, R.G.; Blencowe, N.S.; Boutron, I.; Cates, C.J.; Cheng, H.Y.; Corbett, M.S.; Eldridge, S.M.; et al. RoB 2: A revised tool for assessing risk of bias in randomised trials. BMJ 2019, 366, l4898. [Google Scholar] [CrossRef] [PubMed]
- Guyatt, G.H.; Oxman, A.D.; Vist, G.E.; Kunz, R.; Falck-Ytter, Y.; Alonso-Coello, P.; Schünemann, H.J. GRADE: An emerging consensus on rating quality of evidence and strength of recommendations. BMJ 2008, 336, 924–926. [Google Scholar] [CrossRef] [PubMed]
- Higgins, J.P.T.; Altman, D.G.; Gøtzsche, P.C.; Jüni, P.; Moher, D.; Oxman, A.D.; Savović, J.; Schulz, K.F.; Weeks, L.; Sterne, J.A.C. The Cochrane Collaboration’s tool for assessing risk of bias in randomised trials. BMJ 2011, 343, d5928. [Google Scholar] [CrossRef] [PubMed]
- Raudenbush, S.W.; Becker, B.J.; Kalaian, H. Modeling multivariate effect sizes. Psychol. Bull. 1988, 103, 111. [Google Scholar] [CrossRef]
- Higgins, J.P.T.; Thompson, S.G.; Deeks, J.J.; Altman, D.G. Measuring inconsistency in meta-analyses. BMJ 2003, 327, 557–560. [Google Scholar] [CrossRef]
- Rosenblad, A. Introduction to Meta-Analysis by Michael Borenstein, Larry V. Hedges, Julian P.T. Higgins, Hannah R. Rothstein. Int. Stat. Rev. 2009, 77, 478–479. [Google Scholar] [CrossRef]
- Belstrøm, D.; Grande, M.A.; Sembler-Møller, M.L.; Kirkby, N.; Cotton, S.L.; Paster, B.J.; Holmstrup, P. Influence of periodontal treatment on subgingival and salivary microbiotas. J. Periodontol. 2018, 89, 531–539. [Google Scholar] [CrossRef]
- Bizzarro, S.; Laine, M.L.; Buijs, M.J.; Brandt, B.W.; Crielaard, W.; Loos, B.G.; Zaura, E. Microbial profiles at baseline and not the use of antibiotics determine the clinical outcome of the treatment of chronic periodontitis. Sci. Rep. 2016, 6, 20205. [Google Scholar] [CrossRef]
- Califf, K.J.; Schwarzberg-Lipson, K.; Garg, N.; Gibbons, S.M.; Caporaso, J.G.; Slots, J.; Cohen, C.; Dorrestein, P.C.; Kelley, S.T. Multi-omics Analysis of Periodontal Pocket Microbial Communities Pre- and Posttreatment. Msystems 2017, 2, e00016-17. [Google Scholar] [CrossRef]
- de Oliveira, A.M.; Lourenço, T.G.B.; Colombo, A.P.V. Impact of systemic probiotics as adjuncts to subgingival instrumentation on the oral-gut microbiota associated with periodontitis: A randomized controlled clinical trial. J. Periodontol. 2021, 93, 31–44. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Hemme, C.; Beleno, J.; Shi, Z.J.; Ning, D.; Qin, Y.; Tu, Q.; Jorgensen, M.; He, Z.; Wu, L.; et al. Oral microbiota of periodontal health and disease and their changes after nonsurgical periodontal therapy. ISME J. 2018, 12, 1210–1224. [Google Scholar] [CrossRef]
- Hagenfeld, D.; Koch, R.; Jünemann, S.; Prior, K.; Harks, I.; Eickholz, P.; Hoffmann, T.; Kim, T.S.; Kocher, T.; Meyle, J.; et al. Do we treat our patients or rather periodontal microbes with adjunctive antibiotics in periodontal therapy? A 16S rDNA microbial community analysis. PLoS ONE 2018, 13, e0195534. [Google Scholar] [CrossRef]
- Hagenfeld; Prior, K.; Harks, I.; Jockel-Schneider, Y.; May, T.; Harmsen, D.; Schlagenhauf, U.; Ehmke, B. No differences in microbiome changes between anti-adhesive and antibacterial ingredients in toothpastes during periodontal therapy. J. Periodontal. Res. 2019, 54, 435–443. [Google Scholar] [CrossRef] [PubMed]
- Han, J.; Wang, P.; Ge, S. The microbial community shifts of subgingival plaque in patients with generalized aggressive periodontitis following non-surgical periodontal therapy: A pilot study. Oncotarget 2017, 8, 10609–10619. [Google Scholar] [CrossRef] [PubMed]
- He, T.; Huang, S.; Yue, F.; Wang, L.; Liu, J.; Xu, J. A randomized, controlled comparison of a stannous-containing dentifrice for reducing gingival bleeding and balancing the oral microbiome relative to a positive control. Am. J. Dent. 2021, 34, 222–227. [Google Scholar] [PubMed]
- Hong, I.; Lee, H.G.; Keum, H.L.; Kim, M.J.; Jung, U.W.; Kim, K.; Kim, S.Y.; Park, T.; Kim, H.J.; Kim, J.J.; et al. Clinical and Microbiological Efficacy of Pyrophosphate Containing Toothpaste: A Double-Blinded Placebo-Controlled Randomized Clinical Trial. Microorganisms 2020, 8, 1806. [Google Scholar] [CrossRef]
- Jünemann, S.; Prior, K.; Szczepanowski, R.; Harks, I.; Ehmke, B.; Goesmann, A.; Stoye, J.; Harmsen, D. Bacterial community shift in treated periodontitis patients revealed by ion torrent 16S rRNA gene amplicon sequencing. PLoS ONE 2012, 7, e41606. [Google Scholar] [CrossRef]
- Kruse, A.B.; Maamar, R.; Akakpo, D.L.; Woelber, J.P.; Wittmer, A.; Vach, K.; Ratka-Krüger, P.; Al-Ahmad, A. Effects of subgingival air-polishing with trehalose powder on oral biofilm during periodontal maintenance therapy: A randomized-controlled pilot study. BMC Oral Health 2020, 20, 123. [Google Scholar] [CrossRef]
- Laksmana, T.; Kittichotirat, W.; Huang, Y.; Chen, W.; Jorgensen, M.; Bumgarner, R.; Chen, C. Metagenomic analysis of subgingival microbiota following non-surgical periodontal therapy: A pilot study. Open Dent. J. 2012, 6, 255–261. [Google Scholar] [CrossRef]
- Lu, H.; He, L.; Jin, D.; Zhu, Y.; Meng, H. Effect of adjunctive systemic antibiotics on microbial populations compared with scaling and root planing alone for the treatment of periodontitis: A pilot randomized clinical trial. J. Periodontol. 2021, 93, 570–583. [Google Scholar]
- Lu, H.; Zhao, Y.; Feng, X.; He, L.; Meng, H. Microbiome in maintained periodontitis and its shift over a single maintenance interval of 3 months. J. Clin. Periodontol. 2019, 46, 1094–1104. [Google Scholar]
- Nibali, L.; Sousa, V.; Davrandi, M.; Spratt, D.; Alyahya, Q.; Dopico, J.; Donos, N. Differences in the periodontal microbiome of successfully treated and persistent aggressive periodontitis. J. Clin. Periodontol. 2020, 47, 980–990. [Google Scholar] [CrossRef]
- Queiroz, L.A.; Casarin, R.C.V.; Dabdoub, S.M.; Tatakis, D.N.; Sallum, E.A.; Kumar, P.S. Furcation Therapy With Enamel Matrix Derivative: Effects on the Subgingival Microbiome. J. Periodontol. 2017, 88, 617–625. [Google Scholar] [CrossRef] [PubMed]
- Schwarzberg, K.; Le, R.; Bharti, B.; Lindsay, S.; Casaburi, G.; Salvatore, F.; Saber, M.H.; Alonaizan, F.; Slots, J.; Gottlieb, R.A.; et al. The personal human oral microbiome obscures the effects of treatment on periodontal disease. PLoS ONE 2014, 9, e86708. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zeng, J.; Yuan, Q.; Luan, Q. Efficacy of (-)-epigallocatechin gallate delivered by a new-type scaler tip during scaling and root planing on chronic periodontitis: A split-mouth, randomized clinical trial. BMC Oral Health 2021, 21, 79. [Google Scholar] [CrossRef] [PubMed]
- Woelber, J.P.; Gärtner, M.; Breuninger, L.; Anderson, A.; König, D.; Hellwig, E.; Al-Ahmad, A.; Vach, K.; Dötsch, A.; Ratka-Krüger, P.; et al. The influence of an anti-inflammatory diet on gingivitis. A randomized controlled trial. J. Clin. Periodontol. 2019, 46, 481–490. [Google Scholar] [CrossRef]
- Yamanaka, W.; Takeshita, T.; Shibata, Y.; Matsuo, K.; Eshima, N.; Yokoyama, T.; Yamashita, Y. Compositional stability of a salivary bacterial population against supragingival microbiota shift following periodontal therapy. PLoS ONE 2012, 7, e42806. [Google Scholar] [CrossRef]
- Minichino, A.; Brondino, N.; Solmi, M.; Del Giovane, C.; Fusar-Poli, P.; Burnet, P.; Cipriani, A.; Lennox, B.R. The gut-microbiome as a target for the treatment of schizophrenia: A systematic review and meta-analysis of randomised controlled trials of add-on strategies. Schizophr. Res. 2021, 234, 58–70. [Google Scholar] [CrossRef]
- Sharpton, S.R.; Maraj, B.; Harding-Theobald, E.; Vittinghoff, E.; Terrault, N.A. Gut microbiome–targeted therapies in nonalcoholic fatty liver disease: A systematic review, meta-analysis, and meta-regression. Am. J. Clin. Nutr. 2019, 110, 139–149. [Google Scholar] [CrossRef]
- Jervøe-Storm, P.M.; Alahdab, H.; Koltzscher, M.; Fimmers, R.; Jepsen, S. Comparison of curet and paper point sampling of subgingival bacteria as analyzed by real-time polymerase chain reaction. J. Periodontol. 2007, 78, 909–917. [Google Scholar] [CrossRef]
- Quince, C.; Walker, A.W.; Simpson, J.T.; Loman, N.J.; Segata, N. Shotgun metagenomics, from sampling to analysis. Nat. Biotechnol. 2017, 35, 833–844. [Google Scholar] [CrossRef]
- Pérez-Chaparro, P.J.; McCulloch, J.A.; Mamizuka, E.M.; Moraes, A.; Faveri, M.; Figueiredo, L.C.; Duarte, P.M.; Feres, M. Do different probing depths exhibit striking differences in microbial profiles? J. Clin. Periodontol. 2018, 45, 26–37. [Google Scholar] [CrossRef] [PubMed]
- Xia, Y.; Sun, J. Hypothesis Testing and Statistical Analysis of Microbiome. Genes Dis. 2017, 4, 138–148. [Google Scholar] [CrossRef] [PubMed]
- Griffen, A.L.; Beall, C.J.; Campbell, J.H.; Firestone, N.D.; Kumar, P.S.; Yang, Z.K.; Podar, M.; Leys, E.J. Distinct and complex bacterial profiles in human periodontitis and health revealed by 16S pyrosequencing. ISME J. 2012, 6, 1176–1185. [Google Scholar] [CrossRef] [PubMed]
- Galimanas, V.; Hall, M.W.; Singh, N.; Lynch, M.D.; Goldberg, M.; Tenenbaum, H.; Cvitkovitch, D.G.; Neufeld, J.D.; Senadheera, D.B. Bacterial community composition of chronic periodontitis and novel oral sampling sites for detecting disease indicators. Microbiome 2014, 2, 32. [Google Scholar] [CrossRef]
- Kirst, M.E.; Li, E.C.; Alfant, B.; Chi, Y.Y.; Walker, C.; Magnusson, I.; Wang, G.P. Dysbiosis and alterations in predicted functions of the subgingival microbiome in chronic periodontitis. Appl. Environ. Microbiol. 2015, 81, 783–793. [Google Scholar] [CrossRef]
- Pfeiffer, F.; Gröber, C.; Blank, M.; Händler, K.; Beyer, M.; Schultze, J.L.; Mayer, G. Systematic evaluation of error rates and causes in short samples in next-generation sequencing. Sci. Rep. 2018, 8, 10950. [Google Scholar] [CrossRef]
- Wang, V.X.; Blades, N.; Ding, J.; Sultana, R.G.P. Estimation of sequencing error rates in short reads. BMC Bioinform. 2012, 13, 185. [Google Scholar]
- Weyrich, L.S.; Duchene, S.; Soubrier, J.; Arriola, L.; Llamas, B.; Breen, J.; Morris, A.G.; Alt, K.W.; Caramelli, D.; Dresely, V.; et al. Neanderthal behaviour, diet, and disease inferred from ancient DNA in dental calculus. Nature 2017, 544, 357–361. [Google Scholar] [CrossRef]
- Weyrich, L.S. The evolutionary history of the human oral microbiota and its implications for modern health. Periodontology 2000 2021, 85, 90–100. [Google Scholar] [CrossRef] [PubMed]
- Keijser, B.J.; Zaura, E.; Huse, S.M.; van der Vossen, J.M.; Schuren, F.H.; Montijn, R.C.; ten Cate, J.M.; Crielaard, W. Pyrosequencing analysis of the oral microflora of healthy adults. J. Dent. Res. 2008, 87, 1016–1020. [Google Scholar] [CrossRef] [PubMed]
- Turnbaugh, P.J.; Ley, R.E.; Hamady, M.; Fraser-Liggett, C.M.; Knight, R.; Gordon, J.I. The human microbiome project. Nature 2007, 449, 804–810. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).