Characterization of the Tellurite-Resistance Properties and Identification of the Core Function Genes for Tellurite Resistance in Pseudomonas citronellolis SJTE-3
Abstract
1. Introduction
2. Materials and Methods
2.1. Strains, Plasmids, Media, and Chemicals
2.2. Standard DNA Operation
2.3. Bioinformatics Analysis
2.4. Combination of the Tellurite-Resistance Genes
2.5. Evaluation of Tellurite Resistance Properties
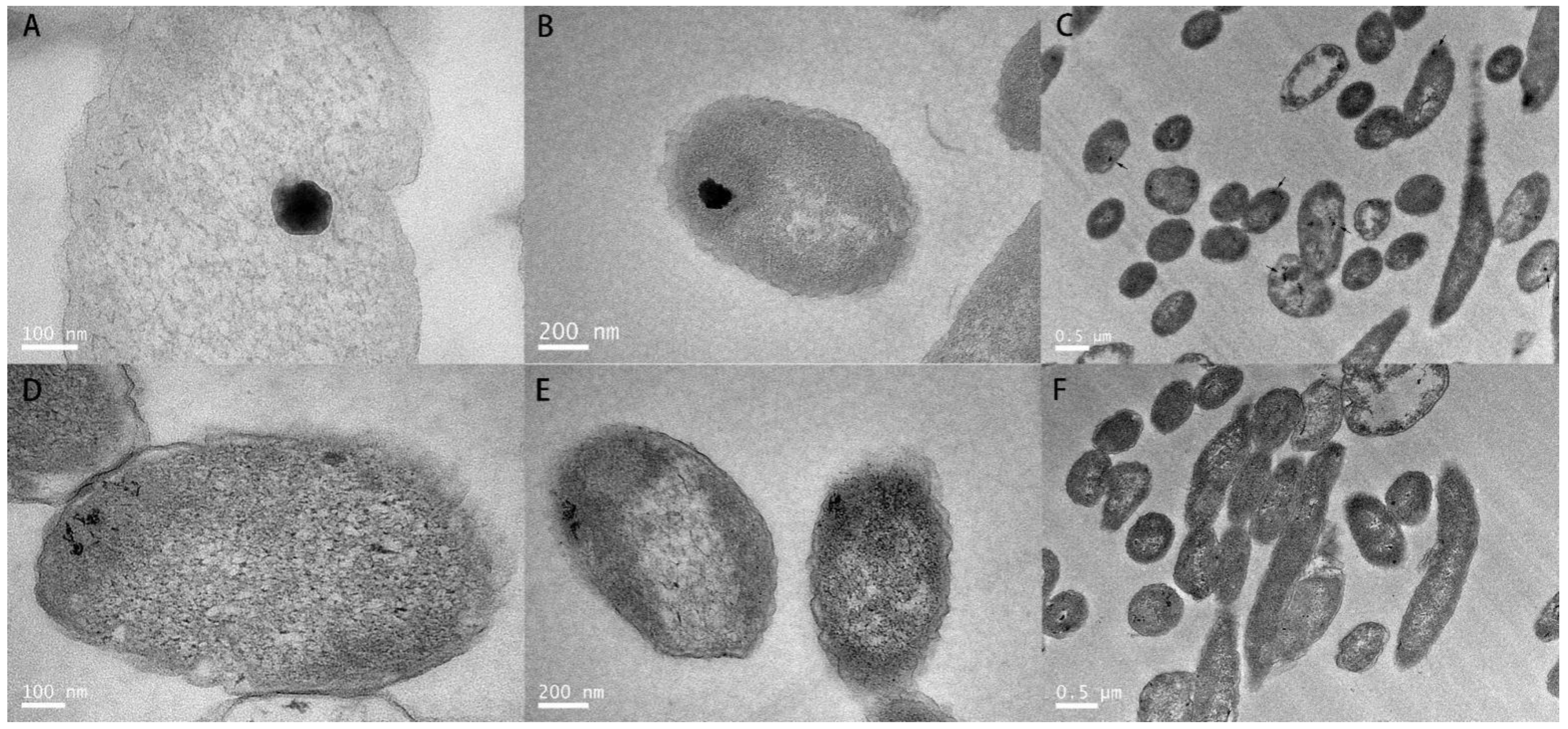
2.6. Transmission Electron Microscope (TEM) Observation
3. Results
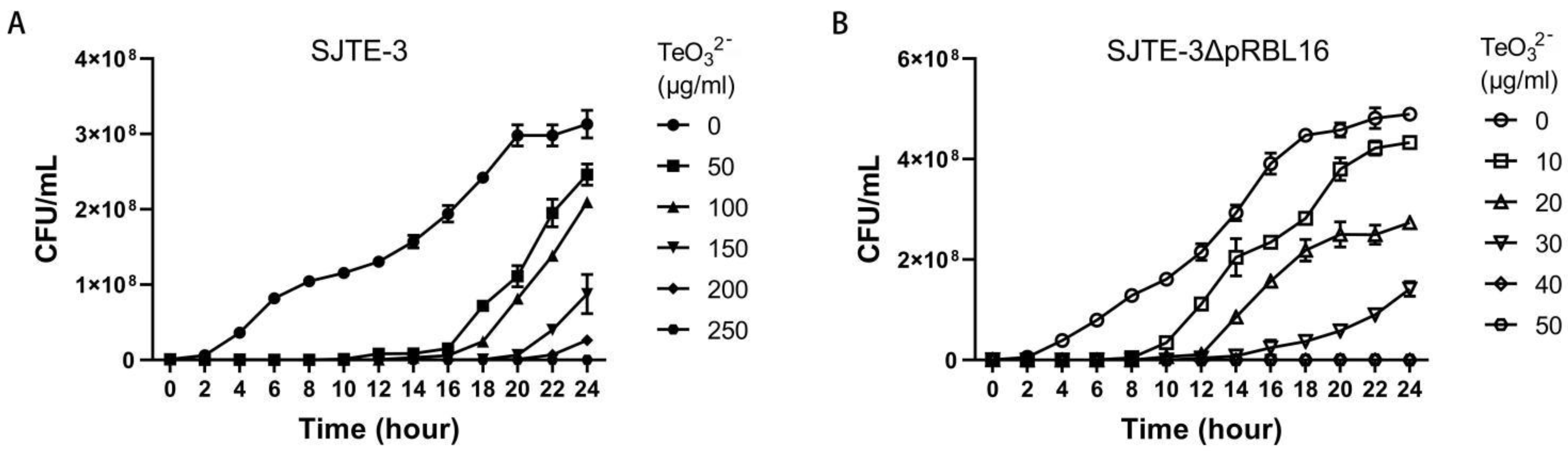
3.1. Plasmid pRBL16 Endowed P. citronellolis SJTE-3 with High-Level Tellurite Resistance
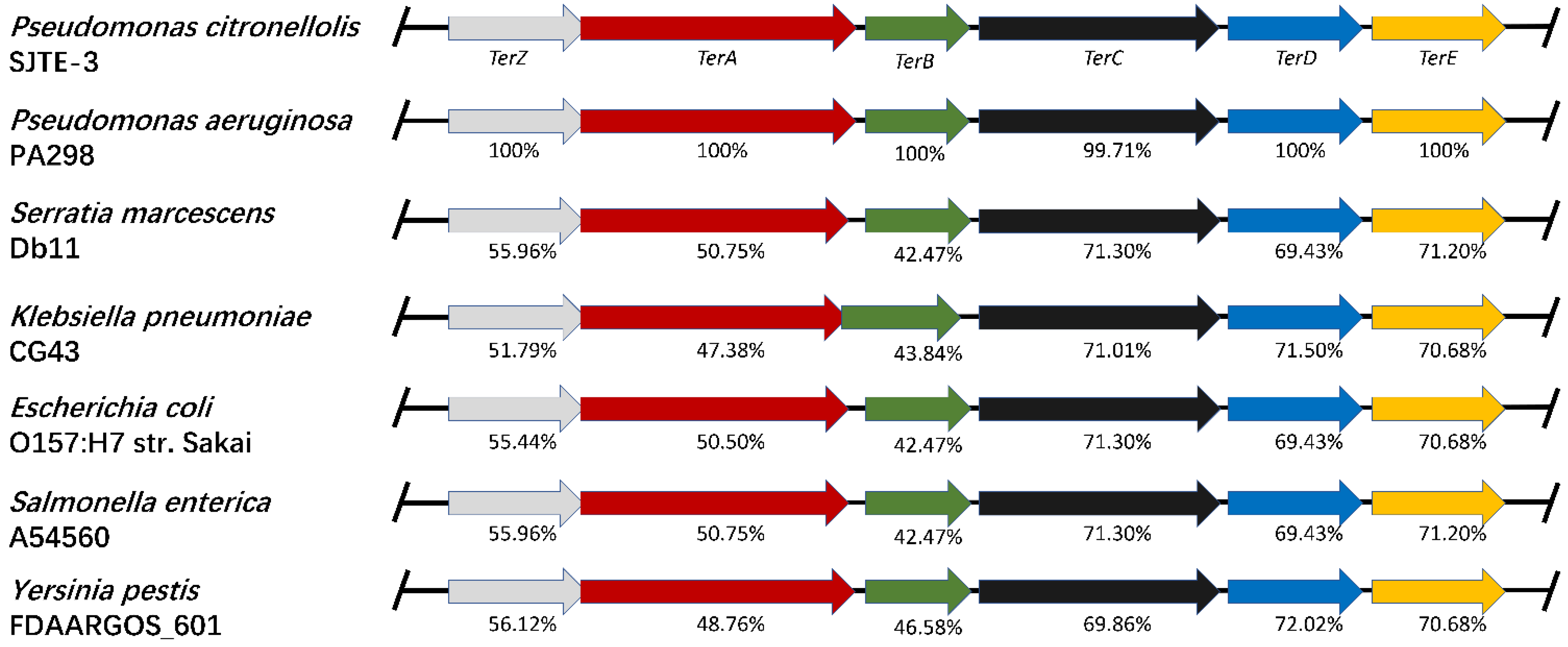
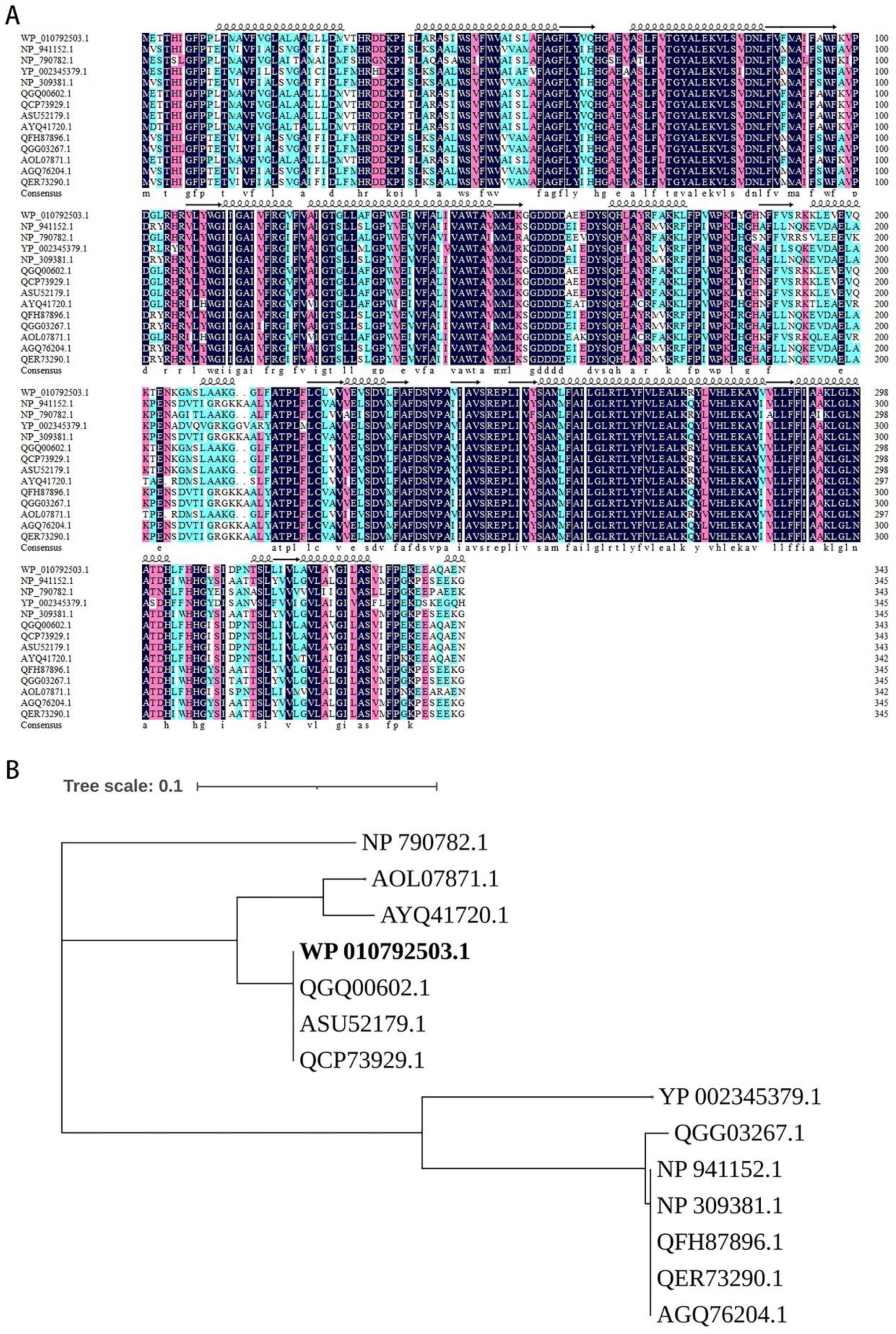
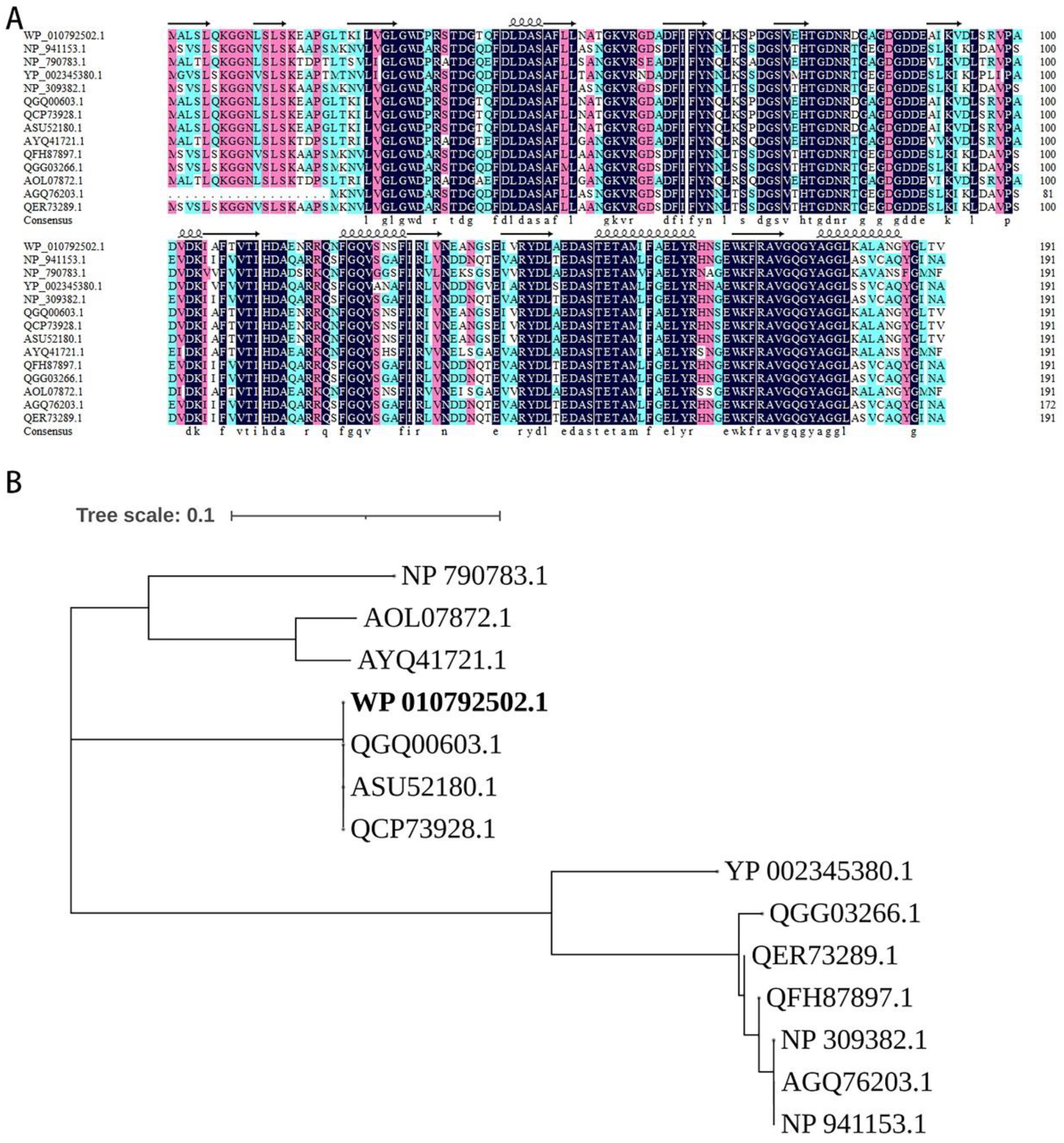
3.2. The terZABCDE Gene Clusters in Different Bacteria May Have the Same Origin
3.3. The terCD Genes Are the Minimal Function Combination for Tellurite Resistance
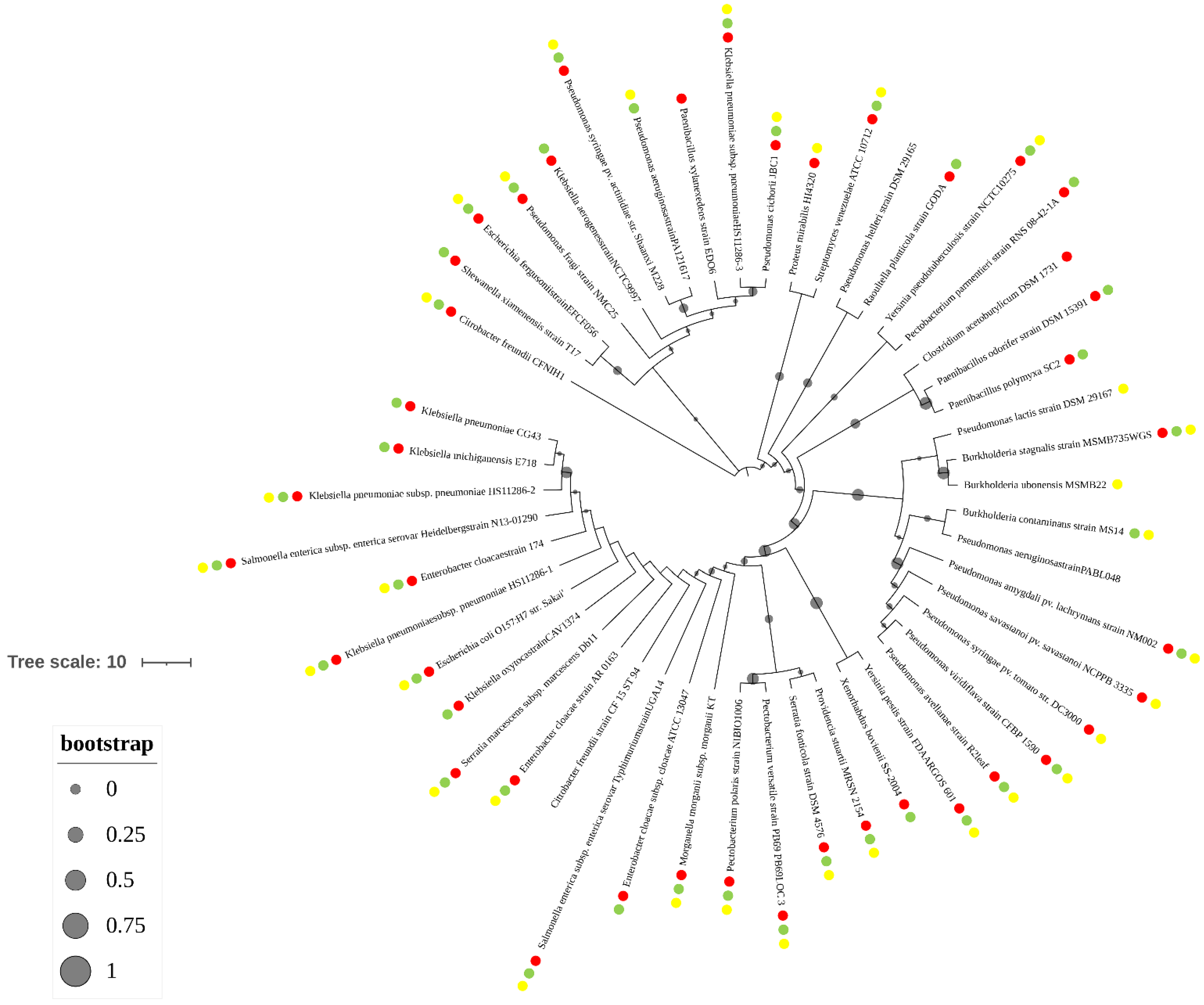
3.4. The terCD Genes and Pathogenic Genes Are in Co-Distribution
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Belzile, N.; Chen, Y. Tellurium in the environment: A critical review focused on natural waters, soils, sediments and airborne particles. Appl. Geochem. 2015, 63, 83–92. [Google Scholar] [CrossRef]
- He, Z.; Yang, Y.; Liu, J.-W.; Yu, S.-H. Emerging tellurium nanostructures: Controllable synthesis and their applications. Chem. Soc. Rev. 2017, 46, 2732–2753. [Google Scholar] [CrossRef]
- Xie, H.G.; Xia, W.; Chen, M.; Wu, L.C.; Tong, J. Isolation and Characterization of the tellurite-reducing photosynthetic bacterium, Rhodopseudomonas palustris strain TX618. Water Air Soil Pollut. 2018, 229, 158. [Google Scholar] [CrossRef]
- Shen, J.; Jia, S.; Shi, N.; Ge, Q.; Gotoh, T.; Lv, S.; Zhu, M. Elemental electrical switch enabling phase segregation-free operation. Science 2021, 374, 1390–1394. [Google Scholar] [CrossRef] [PubMed]
- Ba, L.A.; Döring, M.; Jamier, V.; Jacob, C. Tellurium: An element with great biological potency and potential. Org. Biomol. Chem. 2010, 8, 4203–4216. [Google Scholar] [CrossRef] [PubMed]
- Vaigankar, D.C.; Dubey, S.K.; Mujawar, S.Y.; D’Costa, A.; Shyama, S.K. Tellurite biotransformation and detoxification by Shewanella baltica with simultaneous synthesis of tellurium nanorods exhibiting photo-catalytic and anti-biofilm activity. Ecotoxicol. Environ. Saf. 2018, 165, 516–526. [Google Scholar] [CrossRef] [PubMed]
- Chasteen, T.G.; Fuentes, D.E.; Tantaleán, J.C.; Vásquez, C.C. Tellurite: History, oxidative stress, and molecular mechanisms of resistance. FEMS Microbiol. Rev. 2009, 33, 820–832. [Google Scholar] [CrossRef] [PubMed]
- Perez, J.M.; Calderón, I.L.; Arenas, F.A.; Fuentes, D.E.; Pradenas, G.A.; Fuentes, E.L.; Sandoval, J.M.; Castro, M.E.; Elías, A.O.; Vásquez, C.C. Bacterial toxicity of potassium tellurite: Unveiling an ancient enigma. PLoS ONE 2007, 2, e211. [Google Scholar] [CrossRef] [PubMed]
- Calderón, I.L.; Arenas, F.A.; Pérez, J.M.; Fuentes, D.E.; Araya, M.A.; Saavedra, C.P.; Tantaleán, J.C.; Pichuantes, S.E.; Youderian, P.A.; Vásquez, C.C. Catalases are NAD(P)H-dependent tellurite reductases. PLoS ONE 2006, 1, e70. [Google Scholar] [CrossRef] [PubMed]
- Borsetti, F.; Tremaroli, V.; Michelacci, F.; Borghese, R.; Winterstein, C.; Daldal, F.; Zannoni, D. Tellurite effects on Rhodobacter capsulatus cell viability and superoxide dismutase activity under oxidative stress conditions. Res. Microbiol. 2005, 156, 807–813. [Google Scholar] [CrossRef]
- Turner, R.J.; Weiner, J.H.; Taylor, D.E. The tellurite-resistance determinants tehAtehB and klaAklaBtelB have different biochemical requirements. Microbiology 1995, 141, 3133–3140. [Google Scholar] [CrossRef]
- Fuentes, D.E.; Fuentes, E.L.; Castro, M.E.; Pérez, J.M.; Araya, M.A.; Chasteen, T.G.; Pichuantes, S.E.; Vásquez, C.C. Cysteine metabolism-related genes and bacterial resistance to potassium tellurite. J. Bacteriol. 2007, 189, 8953–8960. [Google Scholar] [CrossRef]
- Moore, M.D.; Kaplan, S. Identification of intrinsic high-level resistance to rare-earth oxides and oxyanions in members of the class Proteobacteria: Characterization of tellurite, selenite, and rhodium sesquioxide reduction in Rhodobacter sphaeroides. J. Bacteriol. 1992, 174, 1505–1514. [Google Scholar] [CrossRef]
- Avazéri, C.; Turner, R.J.; Pommier, J.; Weiner, J.H.; Giordano, G.; Verméglio, A. Tellurite reductase activity of nitrate reductase is responsible for the basal resistance of Escherichia coli to tellurite. Microbiology 1997, 143, 1181–1189. [Google Scholar] [CrossRef]
- O’Gara, J.P.; Gomelsky, M.; Kaplan, S. Identification and molecular genetic analysis of multiple loci contributing to high-level tellurite resistance in Rhodobacter sphaeroides 2.4.1. Appl. Environ. Microbiol. 1997, 63, 4713–4720. [Google Scholar] [CrossRef]
- Cournoyer, B.; Watanabe, S.; Vivian, A. A tellurite-resistance genetic determinant from phytopathogenic pseudomonads encodes a thiopurine methyltransferase: Evidence of a widelyconserved family of methyltransferases. Biochim. Biophys. Acta 1998, 1397, 161–168. [Google Scholar] [CrossRef]
- Tantaleán, J.C.; Araya, M.A.; Saavedra, C.P.; Fuentes, D.E.; Pérez, J.M.; Calderón, I.L.; Youderian, P.; Vásquez, C.C. The Geobacillus stearothermophilus V iscS gene, encoding cysteine desulfurase, confers resistance to potassium tellurite in Escherichia coli K-12. J. Bacteriol. 2003, 185, 5831–5837. [Google Scholar] [CrossRef] [PubMed]
- Taylor, D.E. Bacterial tellurite resistance. Trends Microbiol. 1999, 7, 111–115. [Google Scholar] [CrossRef]
- Zeinert, R.; Martinez, E.; Schmitz, J.; Senn, K.; Usman, B.; Anantharaman, V.; Aravind, L.; Waters, L.S. Structure-function analysis of manganese exporter proteins across bacteria. J. Biol. Chem. 2018, 293, 5715–5730. [Google Scholar] [CrossRef] [PubMed]
- Dambach, M.; Sandoval, M.; Updegrove, T.B.; Anantharaman, V.; Aravind, L.; Waters, L.S.; Storz, G. The ubiquitous yybP-ykoY riboswitch is a manganese-responsive regulatory element. Mol. Cell 2015, 57, 1099–1109. [Google Scholar] [CrossRef]
- Guzzo, J.; Dubow, M.S. A novel selenite- and tellurite-inducible gene in Escherichia coli. Appl. Environ. Microbiol. 2000, 66, 4972–4978. [Google Scholar] [CrossRef]
- Chiang, S.K.; Lou, Y.C.; Chen, C. NMR solution structure of KP-TerB, a tellurite-resistance protein from Klebsiella pneumoniae. Protein Sci. 2008, 17, 785–789. [Google Scholar] [CrossRef]
- Pan, Y.-R.; Lou, Y.-C.; Seven, A.B.; Rizo, J.; Chen, C. NMR structure and calcium-binding properties of the tellurite resistance protein TerD from Klebsiella pneumoniae. J. Mol. Biol. 2011, 405, 1188–1201. [Google Scholar] [CrossRef]
- Bhatia, M.; Girdhar, A.; Tiwari, A.; Nayarisseri, A. Implications of a novel Pseudomonas species on low density polyethylene biodegradation: An in vitro to in silico approach. Springerplus 2014, 3, 497. [Google Scholar] [CrossRef]
- Zheng, D.; Wang, X.; Wang, P.; Peng, W.; Ji, N.; Liang, R. Genome sequence of Pseudomonas citronellolis SJTE-3, an estrogen- and polycyclic aromatic hydrocarbon-degrading bacterium. Genome Announc. 2016, 4, e01373-16. [Google Scholar] [CrossRef]
- Peng, W.; Fu, Y.; Jia, B.; Sun, X.; Wang, Y.; Deng, Z.; Lin, S.; Liang, R. Metabolism analysis of 17α-ethynylestradiol by Pseudomonas citronellolis SJTE-3 and identification of the functional genes. J. Hazard. Mater. 2022, 423, 127045. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Davison, H.C.; Woolhouse, M.E.; Low, J.C. What is antibiotic resistance and how can we measure it? Trends Microbiol. 2000, 8, 554–559. [Google Scholar] [CrossRef]
- Wiegand, I.; Hilpert, K.; Hancock, R.E.W. Agar and broth dilution methods to determine the minimal inhibitory concentration (MIC) of antimicrobial substances. Nat. Protoc. 2008, 3, 163–175. [Google Scholar] [CrossRef] [PubMed]
- Morales, E.H.; Pinto, C.A.; Luraschi, R.; Muñoz-Villagrán, C.M.; Cornejo, F.A.; Simpkins, S.W.; Nelson, J.; Arenas, F.A.; Piotrowski, J.S.; Myers, C.L.; et al. Accumulation of heme biosynthetic intermediates contributes to the antibacterial action of the metalloid tellurite. Nat. Commun. 2017, 8, 15320. [Google Scholar] [CrossRef]
- Cazares, A.; Moore, M.P.; Hall, J.P.J.; Wright, L.L.; Grimes, M.; Emond-Rhéault, J.-G.; Pongchaikul, P.; Santanirand, P.; Levesque, R.C.; Fothergill, J.L.; et al. A megaplasmid family driving dissemination of multidrug resistance in Pseudomonas. Nat. Commun. 2020, 11, 1370. [Google Scholar] [CrossRef]
- Turner, R.J.; Hou, Y.; Weiner, J.H.; Taylor, D.E. The arsenical ATPase efflux pump mediates tellurite resistance. J. Bacteriol. 1992, 174, 3092–3094. [Google Scholar] [CrossRef] [PubMed]
- Alonso, G.; Gomes, C.; González, C.; Rodríguez Lemoine, V. On the mechanism of resistance to channel-forming colicins (PacB) and tellurite, encoded by plasmid Mip233 (IncHI3). FEMS Microbiol. Lett. 2000, 192, 257–261. [Google Scholar] [CrossRef][Green Version]
- Berks, B.C.; Richardson, D.J.; Robinson, C.; Reilly, A.; Aplin, R.T.; Ferguson, S.J. Purification and characterization of the periplasmic nitrate reductase from Thiosphaera pantotropha. Eur. J. Biochem. 1994, 220, 117–124. [Google Scholar] [CrossRef]
- Whelan, K.F.; Sherburne, R.K.; Taylor, D.E. Characterization of a region of the IncHI2 plasmid R478 which protects Escherichia coli from toxic effects specified by components of the tellurite, phage, and colicin resistance cluster. J. Bacteriol. 1997, 179, 63–71. [Google Scholar] [CrossRef] [PubMed]
- Taylor, D.E.; Rooker, M.; Keelan, M.; Ng, L.K.; Martin, I.; Perna, N.T.; Burland, N.V.; Blattner, F.R. Genomic variability of O islands encoding tellurite resistance in enterohemorrhagic Escherichia coli O157:H7 isolates. J. Bacteriol. 2002, 184, 4690–4698. [Google Scholar] [CrossRef]
- Nguyen, T.T.H.; Kikuchi, T.; Tokunaga, T.; Iyoda, S.; Iguchi, A. Diversity of the tellurite resistance gene operon in Escherichia coli. Front. Microbiol. 2021, 12, 681175. [Google Scholar] [CrossRef] [PubMed]
- Whelan, K.F.; Colleran, E.; Taylor, D.E. Phage inhibition, colicin resistance, and tellurite resistance are encoded by a single cluster of genes on the IncHI2 plasmid R478. J. Bacteriol. 1995, 177, 5016–5027. [Google Scholar] [CrossRef]
- Liu, M.; Turner, R.J.; Winstone, T.L.; Saetre, A.; Dyllick-Brenzinger, M.; Jickling, G.; Tari, L.W.; Weiner, J.H.; Taylor, D.E. Escherichia coli TehB requires S-adenosylmethionine as a cofactor to mediate tellurite resistance. J. Bacteriol. 2000, 182, 6509–6513. [Google Scholar] [CrossRef]
- Borsetti, F.; Borghese, R.; Cappelletti, M.; Zannoni, D. Tellurite processing by cells of Rhodobacter capsulatus involves a periplasmic step where the oxyanion causes a malfunction of the cytochrome C maturation system. Int. Biodeterior. Biodegrad. 2018, 130, 84–90. [Google Scholar] [CrossRef]
- SVávrová, S.; Struhárňanská, E.; Turňa, J.; Stuchlík, S. Tellurium: A rare element with influence on prokaryotic and eukaryotic biological systems. Int. J. Mol. Sci. 2021, 22, 5924. [Google Scholar] [CrossRef]
- Vornhagen, J.; Bassis, C.M.; Ramakrishnan, S.; Hein, R.; Mason, S.; Bergman, Y.; Sunshine, N.; Fan, Y.; Holmes, C.L.; Timp, W.; et al. A plasmid locus associated with Klebsiella clinical infections encodes a microbiome-dependent gut fitness factor. PLoS Pathog. 2021, 17, e1009537. [Google Scholar] [CrossRef]
- Vávrová, S.; Valkova, D.; Drahovská, H.; Kokavec, J.; Mravec, J.; Turna, J. Analysis of the tellurite resistance determinant on the pNT3B derivative of the pTE53 plasmid from uropathogenic Escherichia coli. Biometals 2006, 19, 453–460. [Google Scholar] [CrossRef]
- Turkovicova, L.; Smidak, R.; Jung, G.; Turna, J.; Lubec, G.; Aradska, J. Proteomic analysis of the TerC interactome: Novel links to tellurite resistance and pathogenicity. J. Proteom. 2016, 136, 167–173. [Google Scholar] [CrossRef]
- Kwon, K.C.; Cho, M.H. Deletion of the chloroplast-localized at TerC gene product in Arabidopsis thaliana leads to loss of the thylakoid membrane and to seedling lethality. Plant J. 2008, 55, 428–442. [Google Scholar] [CrossRef]
- Schneider, A.; Steinberger, I.; Strissel, H.; Kunz, H.H.; Manavski, N.; Meurer, J.; Burkhard, G.; Jarzombski, S.; Schünemann, D.; Geimer, S.; et al. The Arabidopsis Tellurite resistance C protein together with ALB3 is involved in photosystem II protein synthesis. Plant. J. 2014, 78, 344–356. [Google Scholar] [CrossRef] [PubMed]
- Sanssouci, E.; Lerat, S.; Grondin, G.; Shareck, F.; Beaulieu, C. tdd8: A TerD domain-encoding gene involved in Streptomyces coelicolor differentiation. Antonie Leeuwenhoek 2011, 100, 385–398. [Google Scholar] [CrossRef]
- Valkova, D.; Valkovičová, L.; Vávrová, S.; Kováčová, E.; Mravec, J.; Turna, J. The contribution of tellurite resistance genes to the fitness of Escherichia coli uropathogenic strains. Open Life Sci. 2007, 2, 182–191. [Google Scholar] [CrossRef]
- Ponnusamy, D.; Clinkenbeard, K.D. Role of tellurite resistance operon in filamentous growth of Yersinia pestis in macrophages. PLoS ONE 2015, 10, e0141984. [Google Scholar] [CrossRef] [PubMed]
- Yin, X.; Wheatcroft, R.; Chambers, J.R.; Liu, B.; Zhu, J.; Gyles, C.L. Contributions of O island 48 to adherence of enterohemorrhagic Escherichia coli O157:H7 to epithelial cells in vitro and in ligated pig ileal loops. Appl. Environ. Microbiol. 2009, 75, 5779–5786. [Google Scholar] [CrossRef] [PubMed]
- Tarr, P.I.; Bilge, S.S.; Vary, J.C.; Jelacic, S.; Habeeb, R.L.; Ward, T.R.; Baylor, M.R.; Besser, T.E. Iha: A novel Escherichia coli O157:H7 adherence-conferring molecule encoded on a recently acquired chromosomal island of conserved structure. Infect. Immun. 2000, 68, 1400–1407. [Google Scholar] [CrossRef] [PubMed]
- Azeddoug, H.; Reysset, G. Cloning and sequencing of a chromosomal fragment from Clostridium acetobutylicum strain ABKn8 conferring chemical-damaging agents and UV resistance to E. coli recA strains. Curr. Microbiol. 1994, 29, 229–235. [Google Scholar] [CrossRef] [PubMed]
- Ponnusamy, D.; Hartson, S.D.; Clinkenbeard, K.D. Intracellular Yersinia pestis expresses general stress response and tellurite resistance proteins in mouse macrophages. Vet. Microbiol. 2011, 150, 146–151. [Google Scholar] [CrossRef]
- Poirier, K.; Faucher, S.; Béland, M.; Brousseau, R.; Gannon, V.; Martin, C.; Harel, J.; Daigle, F. Escherichia coli O157:H7 survives within human macrophages: Global gene expression profile and involvement of the Shiga toxins. Infect. Immun. 2008, 76, 4814–4822. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.; Li, T.; Xia, X.; Zhou, Z.; Zheng, S.; Wang, G. Reduction of tellurite in Shinella sp. WSJ-2 and adsorption removal of multiple dyes and metals by biogenic tellurium nanorods. Int. Biodeterior. Biodegrad. 2019, 144, 104751. [Google Scholar] [CrossRef]
| Features | Values |
|---|---|
| Length (bp) | 370,338 |
| ORF number | 512 |
| Longest ORF (bp) | 5766 |
| Average ORF length (bp) | 649.18 |
| Coding Region (bp) | 332,382 |
| GC content (%) | 56.57 |
| % of genome | 89.75 |
| Tellurite Concentration (μg/mL) | 1.0 | 2.0 | 5.0 | 10.0 | 20.0 | 50.0 | 100 | |
|---|---|---|---|---|---|---|---|---|
| Recombinant Strains | ||||||||
| MG1655 (pBS-terZABCDE) | + | + | + | + | + | + | + | |
| MG1655 (pBS-terZABCE) | + | W | W | W | W | W | W | |
| MG1655 (pBS-terZBCDE) | + | + | + | + | + | + | + | |
| MG1655 (pBS-terZABDE) | W | − | − | − | − | − | − | |
| MG1655 (pBS-terZACDE) | + | + | + | + | + | + | + | |
| MG1655 (pBS-terZABCD) | + | + | + | + | + | + | + | |
| MG1655 (pBS-terABCDE) | + | + | + | + | + | + | + | |
| MG1655 (pBS-terABCD) | + | + | + | + | + | + | + | |
| MG1655 (pBS-terBCDE) | + | + | + | + | + | + | + | |
| MG1655 (pBS-terABCE) | + | + | − | − | − | − | − | |
| MG1655 (pBS-terZACE) | + | + | + | + | + | + | − | |
| MG1655 (pBS-terZACD) | + | + | + | + | + | + | + | |
| MG1655 (pBS-terZABD) | − | − | − | − | − | − | − | |
| MG1655 (pBS-terZADE) | W | W | − | − | − | − | − | |
| MG1655 (pBS-terZCDE) | + | + | + | + | + | + | + | |
| MG1655 (pBS-terZABE) | − | − | − | − | − | − | − | |
| MG1655 (pBS-terZBDE) | + | W | W | − | − | − | − | |
| MG1655 (pBS-terABDE) | W | − | − | − | − | − | − | |
| MG1655 (pBS-terACDE) | + | + | + | + | + | + | + | |
| MG1655 (pBS-terZBCD) | + | + | + | + | + | + | + | |
| MG1655 (pBS-terZBCE) | + | + | W | W | W | W | W | |
| MG1655 (pBS-terZABC) | − | − | − | − | − | − | − | |
| MG1655 (pBS-terZAB) | − | − | − | − | − | − | − | |
| MG1655 (pBS-terZAC) | − | − | − | − | − | − | − | |
| MG1655 (pBS-terZAD) | − | − | − | − | − | − | − | |
| MG1655 (pBS-terZAE) | − | − | − | − | − | − | − | |
| MG1655 (pBS-terZBC) | − | − | − | − | − | − | − | |
| MG1655 (pBS-terZBD) | − | − | − | − | − | − | − | |
| MG1655 (pBS-terZBE) | − | − | − | − | − | − | − | |
| MG1655 (pBS-terZCD) | + | + | + | + | + | + | W | |
| MG1655 (pBS-terZCE) | W | W | W | W | W | − | − | |
| MG1655 (pBS-terZDE) | − | − | − | − | − | − | − | |
| MG1655 (pBS-terABC) | − | − | − | − | − | − | − | |
| MG1655 (pBS-terABD) | − | − | − | − | − | − | − | |
| MG1655 (pBS-terABE) | − | − | − | − | − | − | − | |
| MG1655 (pBS-terACD) | + | + | + | + | + | + | + | |
| MG1655 (pBS-terACE) | + | + | + | + | W | − | − | |
| MG1655 (pBS-terADE) | − | − | − | − | − | − | − | |
| MG1655 (pBS-terBCD) | + | + | + | W | W | − | − | |
| MG1655 (pBS-terBCE) | W | W | W | W | − | − | − | |
| MG1655 (pBS-terBDE) | − | − | − | − | − | − | − | |
| MG1655 (pBS-terCDE) | W | − | − | − | − | − | − | |
| MG1655 (pBS-terZA) | − | − | − | − | − | − | − | |
| MG1655 (pBS-terZB) | − | − | − | − | − | − | − | |
| MG1655 (pBS-terZC) | − | − | − | − | − | − | − | |
| MG1655 (pBS-terZD) | − | − | − | − | − | − | − | |
| MG1655 (pBS-terZE) | − | − | − | − | − | − | − | |
| MG1655 (pBS-terAB) | − | − | − | − | − | − | − | |
| MG1655 (pBS-terAC) | − | − | − | − | − | − | − | |
| MG1655 (pBS-terAD) | − | − | − | − | − | − | − | |
| MG1655 (pBS-terAE) | − | − | − | − | − | − | − | |
| MG1655 (pBS-terBC) | − | − | − | − | − | − | − | |
| MG1655 (pBS-terBD) | − | − | − | − | − | − | − | |
| MG1655 (pBS-terBE) | − | − | − | − | − | − | − | |
| MG1655 (pBS-terCD) | W | W | − | − | − | − | − | |
| MG1655 (pBS-terCE) | W | − | − | − | − | − | − | |
| MG1655 (pBS-terDE) | − | − | − | − | − | − | − | |
| MG1655 (pBSPPc) | − | − | − | − | − | − | − | |
| MG1655 | − | − | − | − | − | − | − | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Peng, W.; Wang, Y.; Fu, Y.; Deng, Z.; Lin, S.; Liang, R. Characterization of the Tellurite-Resistance Properties and Identification of the Core Function Genes for Tellurite Resistance in Pseudomonas citronellolis SJTE-3. Microorganisms 2022, 10, 95. https://doi.org/10.3390/microorganisms10010095
Peng W, Wang Y, Fu Y, Deng Z, Lin S, Liang R. Characterization of the Tellurite-Resistance Properties and Identification of the Core Function Genes for Tellurite Resistance in Pseudomonas citronellolis SJTE-3. Microorganisms. 2022; 10(1):95. https://doi.org/10.3390/microorganisms10010095
Chicago/Turabian StylePeng, Wanli, Yanqiu Wang, Yali Fu, Zixin Deng, Shuangjun Lin, and Rubing Liang. 2022. "Characterization of the Tellurite-Resistance Properties and Identification of the Core Function Genes for Tellurite Resistance in Pseudomonas citronellolis SJTE-3" Microorganisms 10, no. 1: 95. https://doi.org/10.3390/microorganisms10010095
APA StylePeng, W., Wang, Y., Fu, Y., Deng, Z., Lin, S., & Liang, R. (2022). Characterization of the Tellurite-Resistance Properties and Identification of the Core Function Genes for Tellurite Resistance in Pseudomonas citronellolis SJTE-3. Microorganisms, 10(1), 95. https://doi.org/10.3390/microorganisms10010095








