Abstract
SARS-CoV-2 wastewater epidemiology suffers from uncertainties concerning sample storage. We show the effect of the storage of wastewater on the detectable SARS-CoV-2 load. Storage at 4 °C for up to 9 days had no significant effect, while storage at −20 °C led to a significant reduction in gene copy numbers.
1. Introduction, Aims and Methods
In the context of the global COVID-19 pandemic, the quantification of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) fragments in wastewater offers the opportunity to monitor the level of infection in large populations, independent of apparent symptoms [1,2]). With the growing number of SARS-CoV-2 wastewater studies, we need comprehensive knowledge on common storage procedures for raw wastewater to generate valid data from sewage surveillance. Temperature, as a central environmental parameter, is a main driver of microbial decay and significantly alters the persistence of viruses in wastewater. Thereby, storage at lower temperatures (<4 °C) increases the persistence of coronaviruses in wastewater [3]. The aim of this investigation was to compare the effect of the most common storage temperatures (≤4 °C) of wastewater samples, +4 °C and −20 °C [4,5], on the detectability of SARS-CoV-2 gene copy numbers.
Therefore, we analyzed 24 h composite samples of raw influent wastewater from the wastewater treatment plant (WWTP) Zirl, Tyrol, Austria (19 April 2020, 30,000 population equivalents (PE)) and the WWTP Siggerwiesen, Salzburg, Austria (4 October 2020, 680,000 PE). The wastewater from the WWTP Zirl was roughly composed of 1/3 industrial and 2/3 domestic wastewater, with a catchment length (main collector) of 50 km. The sample from this plant was taken during dry weather with a mean ambient temperature of 15.2 °C. Meanwhile, the wastewater from the WWTP in Salzburg was mainly composed of domestic wastewater, with a catchment length (main collector) of 140 km. Sampling for this site was conducted during dry weather with a mean ambient temperature of 11.4 °C. The former samples (Zirl) were pasteurized prior to analysis due to uncertainties of the safety status of the wastewater at this time, while the latter (Salzburg) remained unpasteurized. Pasteurization of the wastewater involved an exposure of the samples to 60 °C for 1.5 h prior to sample processing [6]. The investigated storage conditions are summarized in Table 1.

Table 1.
Experimental design.
For SARS-CoV-2 RNA extraction, we modified the protocol from Wu et al. [2]. In a first step, larger particles were removed to decrease the amount of non-viral RNA and PCR inhibitors. For this purpose, 40–70 mL of wastewater was transferred to centrifugation tubes and centrifuged at 4500× g for 30 min at 4 °C. To precipitate viral fragments, the resulting supernatant was immediately transferred into a fresh tube containing 10% w/v polyethylene glycol (PEG) 8000 (CarlRoth, Karlsruhe, Germany) and 2.25% w/v NaCl. The Reax2™ overhead shaker (Heidolph, Schwabach, Germany) was used until both additives were dissolved within a few minutes. Subsequently, the samples were centrifuged at 12,000× g for 99 min at 4 °C to obtain a pellet containing the viral fragments. The supernatant was removed in two steps. First, most of the supernatant was carefully decanted, and then, after additional centrifugation at 12,000× g for 5 min, a pipette was used to remove the remaining fluid.
Following the precipitation of the viral fragments, pellets from the Zirl samples were resuspended with 800 µL TRIzol® (Invitrogen, Waltham, MA, USA), and TRIzol®-chloroform extraction was performed. For the Salzburg samples, we substituted hazardous TRIzol® with 800 µL lysis buffer (Monarch™ total RNA Miniprep Kit, NewEnglandBiolabs, Ipswich, MA, USA). The aqueous, pale phase from TRIzol®-chloroform extraction or the pellet resuspended in lysis buffer was purified according to the manufacturer protocol of the Monarch™ total RNA Miniprep Kit with non-enzymatic gDNA removal. RNA was eluted in 40 µL RNase-free water.
RNA concentrations of the templates were quantified via a Nanodrop, and extracts with RNA concentrations above 200 ng µL−1 were diluted as needed. RNA copy numbers were determined using the N1 primers/probe according to the CDC protocol [7] targeting the nucleocapsid gene of SARS-CoV-2. RT-qPCR reactions contained the following per 20 µL: 10 µL Luna Universal Probe One-Step Reaction Mix (2X) from NEB, 1 µL Luna WarmStart® RT Enzyme Mix (20×) from NEB, 0.8 µL primer (final concentration 0.4 µM), 0.4 µL probe (final concentration 0.2 µM), 2 µL PCR-grade water, and 5 µL template. Analyses were conducted on a RotorGene cycler (Qiagen, Hilden, Germany). After an initial reverse transcription at 55 °C for 10 min, followed by 95 °C for 1 min of denaturation, 45 cycles of 95 °C for 10 sec and 60 °C for 40 sec were performed. To calculate copy numbers, a plasmid standard containing the N gene of SARS-CoV-2 (2019-nCoV_N_Positive Control, IDT, Leuven, Belgium) was used. All variants were processed in parallel (n ≥ 3) and were tested for significant differences against day 0 using the Mann–Whitney U Test (α = 0.05) in the software package Past 4.03 [8].
Within all samples stored at 4 °C, variation coefficients spanned from 2% to 51%, with a median of 37%. This variance inhomogeneity may be explained by the heterogeneity of the influent wastewater and by the accumulation of inaccuracies during the multi-step extraction protocol. Large variance and inhomogeneity were reported earlier by Wu et al. [9] and seemed to be independent of the method of viral fragment concentration as reported by Ahmed et al. [2]. Pasteurization of the former samples (Zirl) may have had an impact on the recovery and analysis of SARS-CoV-2 from wastewater. Pecson et al. [10] showed that pasteurization may lead to a slight increase in the recovery of SARS-CoV-2 fragments. Nevertheless, samples were pasteurized right before sample processing, and storage conditions were equal for all variants.
2. Results and Discussion
Short-term storage of wastewater for up to 9 days at 4 °C had no significant effect on the number of detectable SARS-CoV-2 fragments (Figure 1 and Figure 2). These findings are in accordance with earlier studies on enveloped viruses [11,12]. In contrast to our storage experiments, Ahmed et al. [3] chose a spike-in approach, using high loads of gamma-irradiated SARS-CoV-2 virions (approximately 6.7 × 105 gc mL−1), and stated a decay rate of approximately 8% per day at 4 °C.
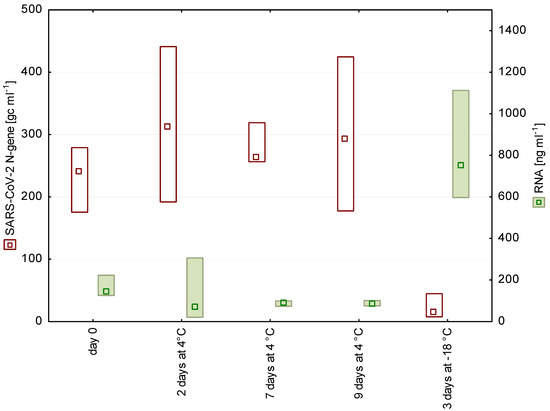
Figure 1.
N gene copy numbers and RNA concentrations detected in wastewater from a WWTP in Salzburg after 0, 2, 7, and 9 days of storage at 4 °C as well as after freezing (n = 4, median, box: min-max).
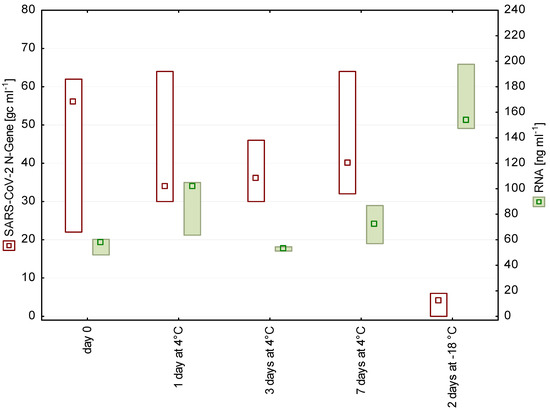
Figure 2.
N gene copy numbers and RNA concentrations detected in wastewater from a WWTP in Tyrol after 0, 1, 3, and 7 days of storage at 4 °C as well as after freezing (n = 3, median, box: min-max).
Freezing–thawing of samples led to a significant loss of the signal. A possible reason is that the freeze–thaw cycle disrupts cells, which is also reflected in the increased RNA concentration in the frozen samples (Figure 1 and Figure 2). The release of cell contents possibly includes also proteases and RNases, which may impair the subsequent SARS-CoV-2 detection.
In conclusion, we recommend storing wastewater samples for SARS-CoV-2 analysis at 4 °C upon analysis and not freezing them.
Author Contributions
Conceptualization, R.M., N.L. and A.O.W.; methodology, R.M. and N.L.; software, M.M.; validation, E.P. and R.M.; formal analysis, R.M. and E.P.; investigation, R.M. and N.L.; resources, H.I.; data curation, R.M. and M.M.; writing—original draft preparation, R.M.; writing—review and editing, R.M., N.L., A.O.W. and H.I.; visualization, R.M.; supervision, A.O.W.; project administration, H.I.; funding acquisition, H.I. All authors have read and agreed to the published version of the manuscript.
Funding
This work was financially supported by the “Förderkreis 1669–Wissenschaft Gesellschaft” and by the Austrian Federal Ministry of Education, Science and Research.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Acknowledgments
We would like to thank the WWTPs Zirl and Siggerwiesen very much for their collaboration and for supplying the samples.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Medema, G.; Heijnen, L.; Elsinga, G.; Italiaander, R.; Brouwer, A. Presence of SARS-Coronavirus-2 RNA in Sewage and Correlation with Reported COVID-19 Prevalence in the Early Stage of the Epidemic in The Netherlands. Environ. Sci. Technol. Lett. 2020, 7, 511–516. [Google Scholar] [CrossRef]
- Wu, F.; Zhang, J.; Xiao, A.; Gu, X.; Lee, W.L.; Armas, F.; Kauffman, K.; Hanage, W.; Matus, M.; Ghaeli, N.; et al. SARS-CoV-2 Titers in wastewater are higher than expected from clinically confirmed cases. MSystems 2020, 5, e00614-20. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, W.; Bertsch, P.M.; Bibby, K.; Haramoto, E.; Hewitt, J.; Huygens, F.; Gyawali, P.; Korajkic, A.; Riddell, S.; Sherchan, S.P.; et al. Decay of SARS-CoV-2 and surrogate murine hepatitis virus RNA in untreated wastewater to inform application in wastewater-based epidemiology. Environ. Res. 2020, 191, 110092. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, W.; Bivins, A.; Bertsch, P.M.; Bibby, K.; Choi, P.M.; Farkas, K.; Gyawali, P.; Hamilton, K.A.; Haramoto, E.; Kitajima, M.; et al. Surveillance of SARS-CoV-2 RNA in wastewater: Methods optimisation and quality control are crucial for generating reliable public health information. Curr. Opin. Environ. Sci. Health 2020. [Google Scholar] [CrossRef] [PubMed]
- Michael-Kordatou, I.; Karaolia, P.; Fatta-Kassinos, D. Sewage analysis as a tool for the COVID-19 pandemic response and management: The urgent need for optimised protocols for SARS-CoV-2 detection and quantification. J. Environ. Chem. Eng. 2020, 8, 104306. [Google Scholar] [CrossRef] [PubMed]
- Darnell, M.E.R.; Subbarao, K.; Feinstone, S.M.; Taylor, D.R. Inactivation of the coronavirus that induces severe acute respiratory syndrome, SARS-CoV. J. Virol. Meth. 2004, 121, 85–91. [Google Scholar] [CrossRef] [PubMed]
- CDC. 2019-Novel Coronavirus (2019-nCoV) Real-Time rRT-PCR Panel Primers and Probes. 2020. Available online: https://www.cdc.gov/coronavirus/2019-ncov/lab/rt-pcr-panel-primer-probes.html (accessed on 1 April 2020).
- Hammer, Ø.; Harper, D.A.T.; Ryan, P.D. Past: Paleontological Statistics Software Package for Education and Data Analysis. Palaeont. Electron. 2001, 4, 9. [Google Scholar]
- Wu, F.; Xiao, A.; Zhang, J.; Moniz, K.; Endo, N.; Armas, F.; Bonneau, R.; Brown, M.A.; Bushman, M.; Chai, P.R.; et al. SARS-CoV-2 titers in wastewater foreshadow dynamics and clinical presentation of new COVID-19 cases. MedRxiv 2020. [Google Scholar] [CrossRef]
- Pecson, B.M.; Darby, E.; Haas, C.N.; Amha, Y.M.; Bartolo, M.; Danielson, R.; Dearborn, Y.; Giovanni, G.D.; Ferguson, C.; Fevig, S.; et al. Reproducibility and sensitivity of 36 methods to quantify the SARS-CoV-2 genetic signal in raw wastewater: Findings from an interlaboratory methods evaluation in the U.S. Environ. Sci. Water Res. Technol. 2021, 7, 504–520. [Google Scholar] [CrossRef] [PubMed]
- Casanova, L.; Rutala, W.A.; Weber, D.J.; Sobsey, M.D. Survival of surrogate coronaviruses in water. Water Res. 2009, 43, 1893–1898. [Google Scholar] [CrossRef] [PubMed]
- Muirhead, A.; Zhu, K.; Brown, J.; Basu, M.; Brinton, M.A.; Costa, F.; Hayat, M.J.; Stauber, C.E. Zika Virus RNA Persistence in Sewage. Environ. Sci. Technol. Lett. 2020, 7, 659–664. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).