Simple Summary
Insects regulate their physiology and behavior through their circadian clock in response to daily changes in the photoperiod. Parasitoid wasps are well-known biocontrol agents. Therefore, understanding the circadian activities of parasitoid adults may help improve biological control strategies. In the present study, we characterized the circadian patterns of emergence, mating, and oviposition of an ectoparasitoid wasp, Pachycrepoideus vindemmiae. We also identified eight clock candidate genes, most of which showed significant rhythmic expressions. These results serve as a starting point for further functional studies of the clock genes in P. vindemmiae as well as in other parasitoid wasps. The findings of this study also provide information that could contribute to improving biological control strategies using parasitoid wasps.
Abstract
Despite the importance of circadian rhythms in insect behavior, our understanding of circadian activity and the molecular oscillatory mechanism in parasitoid wasp circadian clocks is limited. In this study, behavioral activities expected to be under the control of the endogenous circadian system were characterized in an ectoparasitoid wasp, Pachycrepoideus vindemmiae. Most adults exhibited emergence between late night and early morning, while mating only occurred during the daytime, with a peak at midday. Oviposition had three peaks in the early morning, late day, or early night and late night. Additionally, we identified eight putative clock genes from P. vindemmiae. The quantitative PCR (qPCR) results indicate that most clock genes showed significant rhythmic expressions. Our comparative analysis of clock genes in P. vindemmiae and 43 other parasitoid wasps revealed that none of the wasps possessed the timeless and cry1 genes commonly found in some other insect species, suggesting that the circadian clock system in parasitoid wasps is distinct from that in other non-Hymenoptera insects such as Drosophila. Thus, this study attempted to build the first hypothetical circadian clock model for a parasitoid wasp, thus generating hypotheses and providing a platform for the future functional characterization of P. vindemmiae clock genes as well as those of other parasitoid wasps. Finally, these findings on P. vindemmiae circadian activity will aid the development of effective field release programs for biological control, which can be tested under field conditions.
1. Introduction
Insects regulate their physiology and behavior through their circadian clock in response to daily changes in the photoperiod. In parasitoids, the circadian system controls the timing of activities such as locomotor, emergence, mating, and oviposition activity [1]. For example, the locomotor activity of Brachymeria intermedia only occurs during the photophase [2], while the locomotor activity of female Meteorus pulchricornis wasps peaks just after light off under a photoperiod of 16:8 h (light/dark), indicating a primarily nocturnal pattern [3]. The emergence of Cotesia kariyai larvae occurs solely during the photophase and is not dependent on the developmental duration, and they can emerge in the new photophase under the reversed cycle [4]. The emergence of Trichogramma brassicae male and female adults occurs within three hours of lighting [5]. Nasonia vitripennis males emerge preferentially around light-on when they anticipate the light-on signal [6]. The daily oviposition rhythms of three Drosophila parasitoids were monitored under a light–dark cycle. Leptopilina heterotoma and Asobara tabida mainly parasitize hosts in the morning, and L. boulardi oviposits just before light off [7]. Eretmocerus warrae females oviposit throughout the 24 h period with a peak at 10–14 h into the photophase [8]. The emergence, mating, and oviposition rhythms of Tamarixia triozae have been recorded and analyzed, with most emergence and mating activities taking place in the morning and oviposition exclusively occurring during the daytime with a peak in the mid-morning to mid-afternoon [9].
At the molecular level, the central circadian clock has been most extensively studied in the fruit fly Drosophila melanogaster. The basic mechanism of the clock involves three interlocked autoregulatory transcriptional translational feedback loops consisting of a set of clock genes [10]. In the first major loop, the products of Clock (Clk) and cycle (cyc) genes form heterodimers, which activate the transcription of the period (per) and timeless (tim) during the late day to early night [11,12,13,14]. In the middle of the night, the proteins PER and TIM form heterodimers and enter the nucleus, where they suppress the transcriptional activity of the CLK/CYC complex and, thus, repress their own transcription [12,15]. Light exposure causes the Drosophila-type cryptochrome (CRY-d) to disrupt PER and TIM heterodimers, releasing the inhibition of transcription [1,16]. The rhythmic expression of per and tim is produced by this negative feedback. The second loop involves the genes Clk, cyc, vrille, and Par domain protein 1 (Pdp1) [17]. During the night, CLK/CYC activates the transcription of vrille and Pdp1 through an E-box [18]. The VRILLE protein accumulates earlier than PDP1 and represses Clk transcription through a V/P-box. PDP1 accumulates later than VRILLE and triggers Clk transcription during the day, resulting in a peak of CLK during the day [10,19]. The third loop is made up of the clockwork orange (cwo) gene, a member of the orange superfamily. The protein CWO binds to the E-box, competing with CLK/CYC, to regulate the amplitude of Clk, per, and tim mRNA oscillations [20]. Although other insects have a basic clock mechanism similar to that of Drosophila, some differences have been observed [21]. For example, in Antheraea pernyi and Bombyx mori, PER does not enter the nucleus [22,23]. In the genome of hymenopteran species, tim is absent [24,25]. The involvement of CRY2 in the PER/TIM feedback loop has been reported in Apis mellifera and Danaus plexippus [16,24,26].
The molecular mechanisms of the circadian clock in parasitoid wasps are largely unknown. Most studies on the circadian clocks of parasitoid wasps have been limited to those of N. vitripennis [27]. For instance, in the heads of N. vitripennis females, per and cry mRNA levels were found to oscillate synchronously in L12: D12 and L16: D8 photoperiods, as well as in constant light and darkness following entrainment in the same photoperiods. The light-on signal determined the occurrence of per and cry oscillations [28]. In contrast, other clock genes, including Clk, cwo, and Pdp1, showed no significant rhythmic expression in the heads of N. vitripennis females under any photoperiods [29]. Per RNAi altered the expression of cry2, Clk, and cyc, changed the locomotor activity, and affected male courtship behavior [27,30]. Furthermore, N. vitripennis females injected with dsRNA of per were unable to produce diapause-destined eggs in response to short days, indicating that per plays a crucial role in the photoperiodic perception and the timing of photoperiodic diapause induction [27,29].
Pachycrepoideus vindemmiae (Hymenoptera: Pteromalidae), a solitary and generalist pupal wasp, can successfully parasitize a wide range of fly (Diptera) hosts [31,32,33]. The potential of P. vindemmiae as biological control agents against Diptera pests has been evaluated. For example, P. vindemmiae has been found to be one of only two parasitoid species that can successfully attack and kill Drosophila suzukii in the field in Europe and the Americas [34]. Despite its potential, the circadian activities of P. vindemmiae are not well understood, and research exploring the molecular mechanisms of its circadian clock is limited, as is the case with many other parasitoid wasps. In the present study, we characterized circadian activities under a photoperiod of 12:12 h (light/dark) and identified eight clock candidate genes of P. vindemmiae. The expression profiles of these genes were also determined. These results provide a basis for comparison with the circadian systems of other insect species and serve as a starting point for further functional studies of the clock genes in P. vindemmiae as well as in other parasitoid wasps. The findings of this study can also provide information to improve biological control strategies using parasitoid wasps.
2. Materials and Methods
2.1. Insect Rearing
The D. melanogaster w1118 obtained from the Bloomington Stock Center (Indiana University, Bloomington, IL, USA) was reared on standard medium. The P. vindemmiae colony was kindly provided by Prof. Gongyin Ye (Zhejiang University, Hangzhou, China) and reared by parasitizing the pupae of w1118 as described in [31,35,36]. Adult wasps were held in culture tubes (2.5 cm in diameter and 10 cm in height) (ASOCC507601, Sinopharm, Shanghai, China) and fed with a 20% v/v sucrose solution (A610498, Sangon Biotech, Shanghai, China). Both laboratory cultures were maintained at 25 °C and 60 ± 5% relative humidity under a photoperiod of 12:12 h of light and darkness [31].
2.2. The Circadian Activity of P. vindemmiae
We recorded sex-dependent emergence rhythms using a method described by Bertossa et al. [6] and Chen et al. [9]. Briefly, 15 two-day-old mated female wasps were placed in Petri dishes (10 cm in diameter) (F611004, Sangon Biotech, Shanghai, China) with 60 host pupae aged two days old and allowed to parasitize for 6 h (Zeitgeber time (ZT) 9–15; ZT0 corresponds to light on and ZT12 corresponds to light off) under a 12:12 light–dark photoperiod. A total of 16 days after oviposition, the emergence rhythms of 30 P. vindemmiae females (5 replicates of 6 females each) and 25 males (5 replicates of five males each) from the parasitized host pupae were observed for 3 successive days under the same photoperiod. During the scotophase, the infrared night vision system connected to a PC was used to record the observations.
For measuring circadian mating rhythm, we individually paired one-day-old virgin females with one-day-old virgin males at the beginning of the photophase under a photoperiod of 12:12 h (light/dark). The mating events of 26 pairs (5 replicates of 5 or 6 pairs each) were observed continuously throughout both the photophase and scotophase. During the scotophase, the infrared night vision system was used, as described above.
The two-day-old mated females were used for recording circadian oviposition rhythm. We paired a one-day-old virgin female with a one-day-old virgin male in a petri dish (3.5 cm in diameter) (F611201, Sangon Biotech, Shanghai, China). The male was removed immediately after a mating event was observed. Then, we obtained a mated female and fed it until this female was two days old. A single two-day-old mated female was held in the Petri dish (3.5 cm in diameter) (F611201, Sangon Biotech, Shanghai, China) with 10 host pupae aged two days old at the beginning of the photophase under a photoperiod of 12:12 h (light/dark). The oviposition rhythms of 32 mated females (5 replicates of 6 or 7 females each) were observed continuously throughout photophase and scotophase. During the scotophase, the infrared night vision system was used. Female wasps were fed on 20% v/v sucrose solution.
2.3. Identification of Clock Systems from Parasitoid Wasps
The P. vindemmiae transcriptome can be obtained from the National Center for Biotechnology Information (NCBI) Sequence Read Archive (SRA) Databases (https://www.ncbi.nlm.nih.gov/sra/PRJNA573955, accessed on 15 December 2022). The genome data of 43 other parasitoid wasps (Table S1) were obtained from InsectBase 2.0 (http://v2.insect-genome.com/, accessed on 15 December 2022). Firstly, candidate clock genes of P. vindemmiae and other parasitoid wasps were identified through TBLASTN searches against transcriptomes and genomes using local BLAST with an E-value cutoff of 1 × 10−5 using known clock protein sequences of D. melanogaster, Anopheles gambiae, A. mellifera, D. plexippus, Tribolium castaneum, and Gryllus bimaculatus. Then, candidate genes were further confirmed manually using online BLASTP versus NCBI non-redundant protein sequences without species limits (E-value: 1 × 10−5).
2.4. Sequence Alignment and Phylogenetic Analysis
Domain analyses of CRY2, VRILLE, PDP1 and CWO proteins were conducted according to a CDD search (https://www.ncbi.nlm.nih.gov/cdd, accessed on 15 December 2022) [37]. The domains of other clock proteins were analyzed based on their corresponding references. Multiple sequence alignments of the amino acid sequences were performed using ClustalX2 [38] and then edited using GeneDoc. Phylogenetic analysis was conducted using MEGA 7 with 1000 bootstrap replicates based on the maximum likelihood method [39].
2.5. Quantitative Real-Time PCR (qPCR)
The mRNA levels of the clock genes from P. vindemmiae were measured using qPCR. One day after emergence, the heads of adult insects were collected into a TRIzol reagent (15596018, Invitrogen, Carlsbad, CA, USA) every 4 h (ZT2, 6, 10, 14, 18, 22; ZT0 corresponds to light-on and ZT12 corresponds to light-off). Total RNA was extracted from the collected heads according to the manufacturer’s protocol. The first-strand complementary DNA (cDNA) was synthesized using TransScript One-Step gDNA Removal and cDNA Synthesis SuperMix (AT311, Transgen, Beijing, China) as previously described [40]. The specific qPCR primers were designed using Primer3 Input (version 0.4.0, https://bioinfo.ut.ee/primer3-0.4.0/, accessed on 15 November 2022) (Table S2). qPCR was carried out for three biological replicates, with 20 adult heads pooled together for each replicate. The experiments were conducted on a CFX96™ Real-Time PCR Detection System (Bio-Rad, Hercules, CA, USA) using the following program: 95 °C for 30 s, followed by 40 cycles of 95 °C for 5 s, and 60 °C for 30 s. We used the 28S rRNA gene as the reference gene [31]. Relative expression levels were calculated using the comparative 2−ΔΔCT method [41], following the guidelines described by Bustin et al. [42].
2.6. Data Analysis
The differences between the means were analyzed using one-way analysis of variance (ANOVA) and Tukey’s test with statistical significance set at p < 0.05 using SPSS version 22 (IBM SPSS Statistics for Windows, Version 22.0). The results of the circadian rhythm observation and qPCR were visualized using GraphPad Prism 7.0 (GraphPad, San Diego, CA, USA).
3. Results
3.1. The Circadian Activity of P. vindemmiae
The emergence of both sexes was highest between the late scotophase and the early photophase and then significantly decreased. All males and females emerged from ZT 22 to ZT 2. One sex difference noted in the emergence of P. vindemmiae was that males started to emerge at ZT 22, half an hour earlier than females, and completed emergence at ZT 0.5, 1.5 h later than females (Figure 1A). Mating was observed only during the daytime, with the highest peak occurring at midday (Figure 1B). Oviposition had three peaks in the early morning, late day or early night, and late night. Nearly 70% of female adults oviposited within six hours into the photophase (Figure 1C).
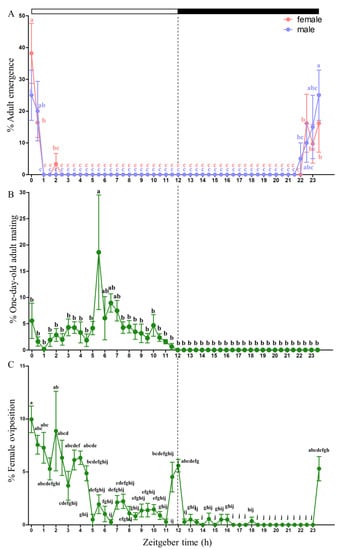
Figure 1.
Pachycrepoideus vindemmiae males and females emergence(A), mating (B) and oviposition (C) patterns under a photoperiod of 12: 12 h (light/dark). Data are presented as the means ± standard deviation. The different letters are significantly different based on one-way analysis of variance (ANOVA) and Tukey’s test with differences considered significant at p < 0.05.
3.2. Core Clock Genes in P. vindemmiae and Other Parasitoid Wasps
Eight core clock genes were identified in P. vindemmiae, including Clock (Pv_Clk), cycle (Pv_cyc), period (Pv_per), timeout (Pv_timout), cryptochrome2 (Pv_cry2), vrille (Pv_vrille), Par domain protein 1 (Pv_Pdp1) and clockwork orange gene (Pv_cwo) (Table 1 and Table 2). BLASTP (https://blast.ncbi.nlm.nih.gov/Blast.cgi, accessed on 5 May 2023) analysis revealed that six of the eight clock proteins exhibited the highest sequence homology with those of N. vitripennis, with the exception of Pv_VRILLE, which showed the highest sequence homology with that of Melipona quadrifasciata, and Pv_CWO, which showed the highest sequence homology with that of Colletes gigas (Table 1). tim and cry1 were not identified in all the wasps analyzed in this study, and cyc, per, cry2, and vrille were not identified in eight, one, two, and one species of these wasps, respectively. However, Clk, timeout, Pdp1, and cwo were identified in all the parasitoid wasps (Table 2 and Table S1).

Table 1.
Clock genes found in Pachycrepoideus vindemmiae.

Table 2.
Clock genes identified from parasitoid wasps.
3.3. CLK
Pv_CLK shared 79% identity (BlastP, E-value = 0) with the CLK protein from N. vitripennis (Table 1). Pv_CLK contains an N-terminal basic helix loop helix (bHLH) domain for binding to DNA [43]. Two PER-ARNT-SIM (PAS) domains mediating the binding to the heterodimeric partner CYC were also detected in Pv_CLK. In the region immediately carboxy-terminal to the PAS-B domain, a PAS-associated C terminal (PAC) domain was detected, which was proposed to be necessary for dimer formation (Figure S1) [44]. Phylogenetic analysis of CLKs from parasitoid wasps showed all the proteins could be divided into two groups (Figure S2). Group 1 CLKs formed three branches with CLKs from Figitidae, Ichneumonidae, and Pteromalidae. Group 2 CLKs from five families of parasitoids are separated into five clusters to be family-specific. Pv_CLK shared a much closer evolutionary relationship with CLKs from Pteromalidae in group 1.
3.4. CYC
CYC is also termed aryl hydrocarbon receptor nuclear translocator (ARNT) or brain and muscle ARNT-like protein (BMAL) [44,45]. Pv_CYC shared about 78% identity (BlastP, E-value = 0) with ARNT-like protein 1 isoform X2 from N. vitripennis (Table 1). Pv_CYC contains three highly conserved regions that are characteristic of known CYC proteins, bHLH, PAS-A, and PAS-B domains (Figure S3). The evolutionary tree showed that Pv_CYC clusters with corresponding proteins from other Pteromalidae parasitoids. The other CYCs, deriving from five families of parasitoid wasps, were separated into five clusters to be family-specific (Figure S4).
3.5. PER
Pv_PER had 63% sequence similarity (BlastP, E-value = 0) to PER in N. vitripennis (Table 1). Pv_PER contains PAS-A and PAS-B regions that mediate the binding of PER to its heterodimeric partner TIM [15]. A cytoplasmic localization domain (CLD) involved in the retention of PER in the cytoplasm [46] and nuclear localization signal (NLS) mediating nuclear entry of the PER-TIM complex [47] were also detected in Pv_PER (Figure S5). The phylogenetic tree indicated Pv_PER clusters with corresponding proteins from other Pteromalidae parasitoids. The other PERs, deriving from five families of parasitoid wasps, were separated into five clusters to be family-specific (Figure S6).
3.6. TIMEOUT
Pv_TIMEOUT exhibited 89% sequence similarity (BlastP, E-value = 0) to the TIMELESS homolog from N. vitripennis (Table 1). Pv_TIMEOUT contains the conserved TIMELESS domain and TIMELESS-C domain as found in other species (Figure S7) [48]. A phylogenetic tree was constructed based on TIMEOUT and TIMELESS sequences for 34 parasitoid species and nine other insect species, respectively. The analysis suggested TIMEOUT and TIMELESS proteins fall out into their respective clades (Figure 2). Pv_TIMEOUT was located in the same clade as TIMEOUT proteins from other Pteromalidae parasitoids, and the other TIMEOUTs, deriving from four families of parasitoid wasps, were separated into four family-specific clusters (Figure 2).
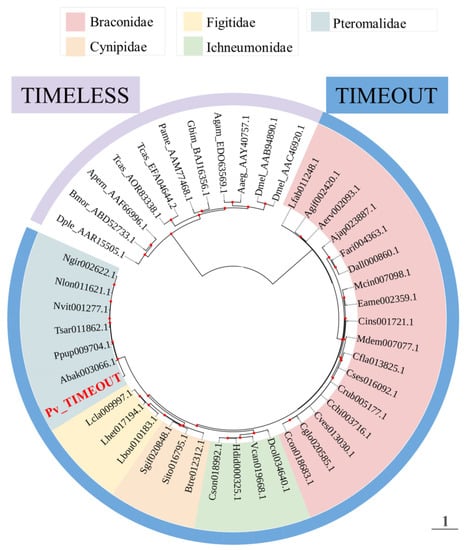
Figure 2.
Phylogenetic relationships of TIMEOUT proteins from parasitoid wasps including Pachycrepoideus vindemmiae and TIMELESS proteins from Aedes aegypti (Aaeg), Anopheles gambiae (Agam), Antheraea pernyi (Aper), Bombyx mori (Bmor), Drosophila melanogaster (Dmel), Danaus plexippus (Dple), Gryllus bimaculatus (Gbim), Periplaneta americana (Pame) and Tribolium castaneum (Tcas). GenBank numbers of TIMELESS proteins are listed after the abbreviations of species names. For TIMEOUT proteins, tip labels show the protein names in InsectBase 2.0 and the species information is listed in Table S1. The phylogenetic tree is constructed using the maximum likelihood method. The best models is JTT+G4. Red dots at the nodes denote bootstrap values greater than 500 from 1000 trials.
3.7. CRY2
Pv_CRY2 shared 86% identity (BlastP, E-value = 0) with the CRY1 isoform X3 identified from N. vitripennis (Table 1). However, this protein is listed as ‘CRY1′ in NCBI despite clearly being CRY2 [28]. Pv_CRY2 contains a DNA photolyase and a flavin adenine dinucleotide (FAD) binding domain (Figure S8). The CLK: BMAL interaction domains (RD-1, RD-2a, and RD-2b) present in mice are also found in P. vindemmiae [49], as well as a coiled-coil domain. A conserved NLS, which is necessary for CRY nuclear localization, is also found within the RD-2b domain [50] (Figure S8). For the phylogenetic analysis of insect CRY1 and CRY2 proteins, we chose 6-4 photolyases and vertebrate CRY4 since they are most closely related to insect CRY1 and CRY2 phylogenetically [1]. Consistent with the previous results [1], the phylogenetic tree of CRY/DNA photolyase proteins mapped with the functional character revealed that all vertebrate CRY and insect CRY2 proteins had repressive transcriptional activity except for CRY3 from Danio rerio. All CRY2 proteins from parasitoid wasps clustered within the insect CRY2 group, which have been reported to repress CLK: CYC transcription in cell culture [1]. Pv_CRY2 clusters with corresponding proteins from other Pteromalidae parasitoids and the other CRY2 proteins from five families of parasitoid wasps were separated into five clusters to be family-specific (Figure 3). The evolutionary tree showed that the repressive transcriptional ability of insect CRY2 evolved from a photolyase-like ancestral gene lacking the ability to function as a transcriptional suppressor, and Pv_CRY2 might function as a transcriptional suppressor of CLK: CYC-mediated transcription like other insect CRY2 proteins.
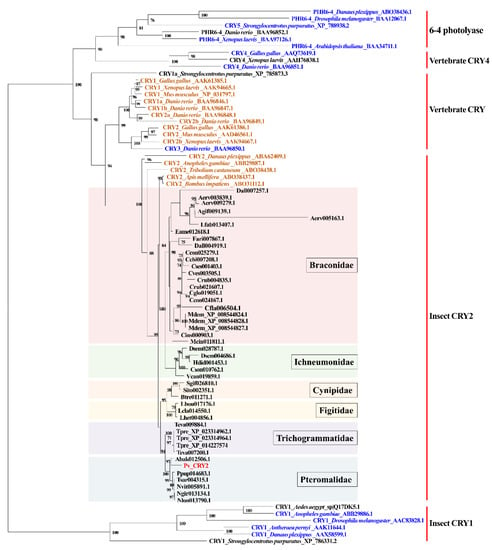
Figure 3.
Phylogenetic relationships of DNA photolyases and CRYs. The phylogenetic tree of amino acid sequences was constructed using the maximum likelihood method. The best model is LG+I+G4. Bootstrap values below 70% were removed from the phylogenetic tree. The functional character is mapped onto this phylogenetic tree based on [1]. Orange letters indicate the proteins that can repress CLK: CYC (BMAL)-mediated transcription in cell culture. Blue letters indicate the proteins that lack this ability in cell culture. Black and red letters indicate the proteins whose transcriptional repressive activity remain unknown. The tip labels of parasitoid wasps show the protein names in InsectBase 2.0. Species information for parasitoid wasps is listed in Table S1 and other species are selected based on [1], whose tip labels consist of protein names, species names and Genbank numbers.
3.8. VRILLE and PDP1
Both Pv_VRILLE and Pv_PDP1 are members of the basic leucine zipper (bZIP) transcription factors [18]. Pv_VRILLE and Pv_PDP1 shared 77.95% identity (BlastP, E-value = 1 × 10−163) with nuclear factor interleukin-3-regulated protein (a vertebrate homolog of VRILLE protein) from Melipona quadrifasciata, and 99% identity (BlastP, E-value = 0) with the hepatic leukemia factor (a vertebrate homolog of PDP1 protein) isoform X9 from N. vitripennis (Table 1). Both Pv_VRILLE and Pv_PDP1 contain bZIP domains, which are essential for their DNA-binding function in the Clk gene promoter to regulate Clk transcription [18,51] (Figures S9 and S10). The evolutionary trees show that both Pv_VRILLE and Pv_PDP1 clustered with corresponding proteins from other Pteromalidae parasitoids. The other VRILLE and PDP1 proteins from different families of parasitoid wasps were separated into their respective clusters to be family-specific, with the exception of a PDP1 protein (Lcla007363.1) from Leptopilina clavipes (Hymenoptera: Figitidae), which clustered into the Pteromalidae group (Figures S11 and S12).
3.9. CWO
Pv_CWO displayed 64% identity (BlastP, E-value = 1 × 10−153) with the transcription factor cwo isoform X2 from Colletes gigas (Table 1). Pv_CWO contains two highly conserved regions that are characteristic of known CWO proteins, namely, the bHLH and Hairy Orange domains (Figure S13) [52]. The bHLH domain is responsible for binding to DNA [43]. The Hairy Orange is an important functional domain in the Drosophila proteins Hesr-1, Hairy, and Enhancer of Split, which play a key role in inhibiting specific transcriptional activators [53]. According to the evolutionary tree, all CWO proteins were grouped into six major clusters to be family-specific and Pv_CWO clusters with corresponding proteins from other Pteromalidae parasitoids (Figure S14).
3.10. Expression Profiles of Clock Genes in P. vindemmiae Females and Males
We obtained daily expression profiles of eight core clock genes in the heads of P. vindemmiae females and males. In the heads of female adults, Pv_Clk and Pv_cyc mRNA showed similar daily oscillations, with a peak in the early morning (ZT 2) and a trough at midnight (ZT 18) (Figure 4A,B). In contrast, Pv_per, Pv_timeout, Pv_cry2, Pv_Pdp1, and Pv_cwo mRNA levels were in an almost anti-phase to those of Pv_clk and Pv_cyc. Pv_per and Pv_timeout mRNA levels varied over time, with a peak at ZT 18 (Figure 4C,D). Pv_vrille showed a similar expression pattern to Pv_per and Pv_timeout, with no significant circadian changes detected (Figure 4F). Pv_cry2 and Pv_Pdp1 mRNA levels cycled in the heads of female adults with a peak at early night (ZT 14) and a trough at late night (ZT 22) and early morning (ZT 2), respectively (Figure 4E,G). Pv_cwo mRNA levels peaked between ZT 10 and 14 and declined to low levels between ZT 18 and 8 (Figure 4H).
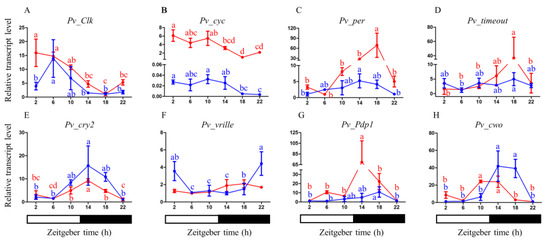
Figure 4.
qPCR results showing the mRNA abundance levels of the eight clock genes in females (solid red lines) and males (solid blue lines) (A–H). Data are presented as the means ± standard deviation. The different letters are significantly different based on one-way analysis of variance (ANOVA) and Tukey’s test with differences considered significant at p < 0.05.
The expression profiles of the same core clock genes were also measured In the heads of male adults. Pv_Clk and Pv_cyc mRNA levels were low during the night and reached their maxima at ZT 6, ZT 2, and ZT10 during the day, respectively (Figure 4A,B). Pv_per, Pv_timeout, Pv_cry2, Pv_Pdp1, and Pv_cwo mRNA levels were almost in anti-phase with those of Pv_clk and Pv_cyc. Pv_per and Pv_cry2 transcript levels were low during the day, started to increase in the late day, and reached their maxima at ZT 14 (Figure 4C,E). Pv_timeout and Pv_Pdp1 mRNA levels peaked at midnight (ZT 18) (Figure 4D,G). The peak levels of Pv_vrille mRNA from males occurred late at night or early morning (ZT 22-2) and decreased during the day (Figure 4F). Pv_cwo mRNA levels peaked between ZT 14 and 18 and declined to low levels between ZT 22 and 10 (Figure 4H). The qPCR results show differences in the clock gene expression patterns between female and male individuals. In males, the expression peaks of Pv_Clk, Pv_Pdp1, and Pv_cwo were delayed for several hours compared to females. Pv_vrille showed no significant rhythmic expression in females, while it peaked at late night and early morning in males.
4. Discussion
In parasitoid wasps, different circadian activities are performed at specific times to maximize fitness gains. P. vindemmiae oviposition had three peaks in the early morning, late day or early night, and late night (Figure 1C), while mating peaked at midday with no overlap with the oviposition activity (Figure 1B). In insects, repeated mating attempts by males can result in harassment of females and cause physical injuries, reduced foraging efficiency, and energy expenditure. Harassment by males can also interfere with the process of oviposition and reduce the longevity and fecundity of females [54]. No overlap between oviposition and mating peaks suggested that males were not able to disturb females during oviposition, indicating that P. vindemmiae has developed strategies to perform different life functions at specific times for maximum fitness gain. A similar situation has also been observed in T. triozae [9] and E. warrae [8].
Sex differences in circadian behavior have been observed in parasitoids. Firstly, a distinct difference between males and females in circadian activity is the time at which activity begins, which has been observed in N. giraulti [55], Trichogramma species [5,56], and Encarsia formosa [57]. Secondly, there is a difference in activity duration and intensity between males and females. For example, T. brassicae males have been observed to be less active than females since their phase of activity is shorter and their hourly activities fewer [58]. The daily rhythm of emergence also varied according to sex. It has been reported that males emerge before females (protandry) in some parasitoids [5]. In this study, the results show that P. vindemmiae males started to emerge half an hour earlier than females at ZT 22 (Figure 1A). The advantage of parasitoid wasp protandry is that the males emerge first, so they have a greater opportunity to mate with females. For females, protandry can reduce the time between emergence and mating, thereby reducing the risk of death before reproduction.
Knowledge of the circadian activity of parasitoid wasps can contribute to decisions on field release. For diurnal parasitoids, it is better to release the parasitoid wasps in the morning [9,59]. The circadian oviposition patterns of Diachasmimorpha longicaudata and Doryctobracon crawfordi overlap. The control efficacy is reduced when both parasitoid wasps are simultaneously released as a result of substantial competition [60]. Our study revealed the circadian activity of P. vindemmiae and provides information for the enhancement of its biological control. For instance, we may achieve better results if we carry out field release of mated P. vindemmiae females instead of newly emerged females in the early morning when environmental conditions are more favorable to avoid affecting the sex ratio of offspring due to insufficient mating because most P. vindemmiae female oviposition occurs in the early morning earlier than the peak they mate at noon.
The absence of the timeless and cry1 genes in parasitoid wasps suggests that their circadian clock system is distinct from that of other non-Hymenoptera insects such as Drosophila. While CRY1 is light-sensitive and does not exhibit transcriptional repressive activity, CRY2, a vertebrate-like protein, is a potent transcriptional repressor of CLK: CYC-mediated transcription but is not light-sensitive. These two cry genes give rise to different types of circadian clocks. Light-sensing in different types of circadian clocks is mediated by different mechanisms, leading to the synchronization of the clock via TIMELESS degradation [1]. However, Hymenoptera species, including parasitoid wasps, have lost both CRY1 and TIMELESS proteins, implying a completely novel light input mechanism for their circadian clocks [16,25,61]. Figure 5 presents a circadian clock model for P. vindemmiae that comprises three interlocked autoregulatory transcriptional-translational feedback loops. In the first major loop, heterodimers formed by the products of Clk and cyc genes activate the transcription of per and cry2. Subsequently, PER/CRY2 enter the nucleus to inhibit their own transcription by repressing CLK/CYC transcriptional activity. Because of the absence of CRY1, the clock uses a new light input pathway for entrainment. The second loop consists of the genes Clk, cyc, vrille, and Pdp1. CLK/CYC activates the transcription of vrille and Pdp1 leading VRILLE to accumulate earlier than PDP1, which then suppresses Clk transcription. PDP1 accumulates later and triggers Clk transcription. Finally, CWO modulates the amplitude of the clock in the third loop.
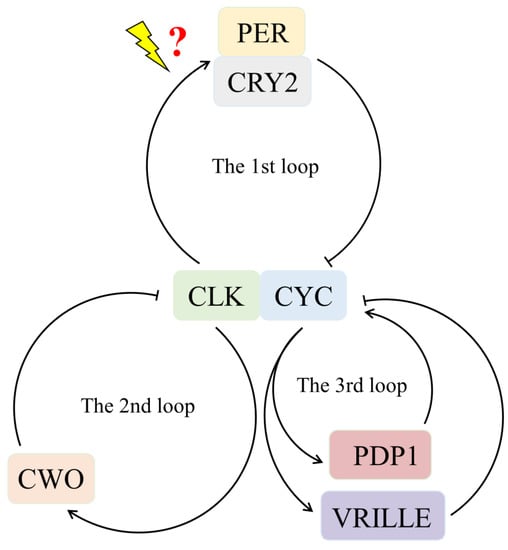
Figure 5.
A hypothetical circadian clock model for Pachycrepoideus vindemmiae.
Studies examining the expression patterns of core clock genes have demonstrated the diversity of the molecular clock machinery in insects. Pv_Clk mRNA in females exhibits a similar daily oscillation to Clk in D. melanogaster, peaking in the early morning [62] (Figure 4A), while Clk shows no significant rhythmic expression in Solenopsis invicta [19], A. mellifera [24] or Gryllus bimaculatus [18]. Pv_cyc mRNA levels are low during the night and reach their maxima in the early morning (Figure 4B), while cyc shows no significant rhythmic expression in D. melanogaster males [62] or N. vitripennis females [29]. In hymenopteran insects, timeout mRNA levels in both P. vindemmiae (Figure 4D) and A. mellifera [24] peak at midnight, while timeout mRNA levels in S. invicta are almost in anti-phase to these [19]. Pv_cry2 mRNA cycles in the heads of female adults peak in the early night (Figure 4E), similar to those of A. mellifera females, while cry2 mRNA levels peak at late night in S. invicta [19] and N. vitripennis [29]. However, Pv_per mRNA levels vary over time, with a peak at midnight (Figure 4C), similar to those of D. melanogaster [62], S. invicta [19], A. mellifera [24], and G. bimaculatus [18]. Overall, the expression profiles of core clock genes in P. vindemmiae, such as Pv_Clk, Pv_cyc, Pv_per, Pv_timeout, and Pv_cry2, are more similar to those of A. mellifera than other insects. To further investigate the diversity of circadian clock mechanisms, the expression patterns of clock genes need to be analyzed in more insect species, and the functions of the clock genes should be studied using molecular genetic techniques.
Although there have been only a few comparative studies on the expression patterns of female and male clock genes, previous research has shown that some clock genes are expressed consistently in both sexes [13,18,51], while others are quite variable [63,64]. Sex differences in circadian clock gene expression may arise from environmental variability and gene functions. In Ceratosolen solmsi, a pollinating wasp that has an obligate mutualism with fig trees, timeout is only rhythmically expressed in females that are dispersing from the fig syconium, while it is arrhythmic in males and females inside the syconium [63]. Among the core clock genes, per shows the largest divergence between the sexes in C. solmsi. Moreover, the mRNA levels of per are closely correlated with emergence rates at specific time intervals in both male and female wasps. The present study detected sexual dimorphism in the clock gene expression in P. vindemmiae, which suggests that these clock genes with different expression patterns in females and males may play a key role in protandry in P. vindemmiae (Figure 1A). More research on sexual dimorphism in clock gene expression is needed to reveal the extensive diversity of gene functions under various environmental conditions.
Although the molecular oscillatory mechanisms consisting of a set of clock genes have been extensively studied in some model insects [16,21,65,66], how the circadian clocks control daily rhythms in physiology and behavior via mechanisms that regulate gene expressions remains unknown. In Anopheles species, clock genes affect swarming and mating behavior by regulating the gene desat1, a dehydrogenase gene involved in the synthesis of a hydrocarbon on the body surface of male mosquitoes, which is a component of the sex pheromone of Anopheles [67]. Our present study characterized the circadian activity and putative clock genes in P. vindemmiae; however, the mechanisms by which the clock genes regulate circadian behaviors such as emergence, mating, and oviposition require further research.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/insects14050486/s1, Table S1: Circadian clock proteins in parasitoid wasps; Table S2: Primers used in qPCR analysis; Figure S1: Sequence alignment of CLK amino acid sequences; Figure S2: Phylogenetic relationships of CLKs from parasitoid wasps; Figure S3: Sequence alignment of alignment of of CYC amino acid sequences; Figure S4: Phylogenetic relationships of CYCs from parasitoid wasps; Figure S5: Sequence alignment of PER amino acid sequences; Figure S6: Phylogenetic relationships of PERs from parasitoid wasps; Figure S7: Sequence alignment of TIMEOUT amino acid sequences; Figure S8: (A) Structure and organization of Pv_CRY2; (B) Sequence alignment of CRY amino acid sequences; Figure S9: Sequence alignment of VRILLE amino acid sequences; Figure S10: Sequence alignment of PDP1 amino acid sequences; Figure S11: Phylogenetic relationships of VRILLE from parasitoid wasps; Figure S12: Phylogenetic relationships of PDP1 from parasitoid wasps; Figure S13: Sequence alignment of CWO amino acid sequences; Figure S14: Phylogenetic relationships of CWO proteins from parasitoid wasps. References [14,15,37,49,68,69] are cited in the supplementary materials.
Author Contributions
Conceptualization, Z.T.; Data curation, Z.T.; Formal analysis, Z.T., M.H., Y.Z. (Yanan Zhou), Y.Z. (Yuqi Zhou), Y.L. (Yunjie Liu), Y.L. (Yan Lin), Q.Z. and Z.Z.; Funding acquisition, Z.T. and H.Z.; Investigation, M.H., Y.Z. (Yanan Zhou), Y.Z. (Yuqi Zhou), Y.L. (Yunjie Liu), Y.L. (Yan Lin), Q.Z. and Z.Z.; Methodology, M.H., Y.Z. (Yanan Zhou) and Y.Z. (Yuqi Zhou); Project administration, Z.T., F.W. and H.Z.; Resources, F.W. and H.Z.; Software, Z.T.; Supervision, F.W. and H.Z.; Validation, Z.T., Y.L. (Yunjie Liu), Y.L. (Yan Lin), Q.Z. and Z.Z.; Visualization, Z.T.; Writing—original draft, Z.T.; Writing—review and editing, Z.T., F.W. and H.Z. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Major Scientific and Technological Innovation Projects in Shandong Province (2019JZZY010711), National Natural Science Foundation of China (Grant No. 31801796) and the Scientific Research Fund for High-level Talents in Qingdao Agricultural University (663/1121019).
Data Availability Statement
The data presented in this study are openly available in NCBI SRA database (https://www.ncbi.nlm.nih.gov/sra/PRJNA573955, accessed on 15 December 2022) and InsectBase 2.0 (http://v2.insect-genome.com/, accessed on 15 December 2022).
Acknowledgments
We extend our sincerest appreciation to Gongyin Ye (Zhejiang University, Hangzhou, China) for giving us the P. vindemmiae colony used in this study.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Yuan, Q.; Metterville, D.; Briscoe, A.D.; Reppert, S.M. Insect cryptochromes: Gene duplication and loss define diverse ways to construct insect circadian clocks. Mol. Biol. Evol. 2007, 24, 948–955. [Google Scholar] [CrossRef]
- Barbosa, P.; Frongillo, E.A. Influence of light-intensity and temperature on locomotory and flight activity of Brachymeria intermedia [Hym.: Chalcididae] a pupal parasitoid of gypsy moth. Entomophaga 1977, 22, 405–411. [Google Scholar] [CrossRef]
- Nishimura, T.; Fujii, T.; Sakamoto, K.; Maeto, K. Daily locomotor activity of the parasitoid wasp Meteorus pulchricornis (Hymenoptera: Braconidae) that attacks exposed lepidopteran larvae. Appl. Entomol. Zool. 2015, 50, 525–531. [Google Scholar] [CrossRef]
- Kawaguchi, M.; Tanaka, T. Time and location of larval emergence of the endoparasitoid Cotesia kariyai (Hymenopter: Braconidae) from the lepidopteran host Pseudaletia separata (Lepidoptera: Noctuidae). Ann. Entomol. Soc. Am. 1999, 92, 101–107. [Google Scholar] [CrossRef]
- Pompanon, F.; Fouillet, P.; Bouletreau, M. Emergence rhythms and protandry in relation to daily patterns of locomotor activity in Trichogramma species. Evol. Ecol. 1995, 9, 467–477. [Google Scholar] [CrossRef]
- Bertossa, R.C.; van Dijk, J.; Beersma, D.G.; Beukeboom, L.W. Circadian rhythms of adult emergence and activity but not eclosion in males of the parasitic wasp Nasonia vitripennis. J. Insect Physiol. 2010, 56, 805–812. [Google Scholar] [CrossRef] [PubMed]
- Fleury, F.; Allemand, R.; Vavre, F.; Fouillet, P.; Bouletreau, M. Adaptive significance of a circadian clock: Temporal segregation of activities reduces intrinsic competitive inferiority in Drosophila parasitoids. Proc. Biol. Sci. 2000, 267, 1005–1010. [Google Scholar] [CrossRef] [PubMed]
- Hanan, A.; He, X.Z.; Shakeel, M.; Wang, Q. Diurnal rhythms of emergence, host feeding and oviposition of Eretmocerus warrae (Hymenoptera: Aphelinidae). New Zealand Plant Prot. 2009, 62, 156–160. [Google Scholar] [CrossRef]
- Chen, C.; He, X.Z.; Zhou, P.; Wang, Q. Tamarixia triozae, an important parasitoid of Bactericera cockerelli: Circadian rhythms and their implications in pest management. Biocontrol 2020, 65, 537–546. [Google Scholar] [CrossRef]
- Tomioka, K.; Matsumoto, A. The circadian system in insects: Cellular, molecular, and functional organization. Adv. Insect Physiol. 2019, 56, 73–115. [Google Scholar]
- Ikeda, K.; Daimon, T.; Sezutsu, H.; Udaka, H.; Numata, H. Involvement of the clock gene period in the circadian rhythm of the silkmoth Bombyx mori. J. Biol. Rhythm 2019, 34, 283–292. [Google Scholar] [CrossRef] [PubMed]
- Tobback, J.; Boerjan, B.; Vandersmissen, H.P.; Huybrechts, R. Male reproduction is affected by RNA interference of period and timeless in the desert locust Schistocerca gregaria. Insect. Biochem. Molec. Biol. 2012, 42, 109–115. [Google Scholar] [CrossRef]
- Moriyama, Y.; Kamae, Y.; Uryu, O.; Tomioka, K. Gb’Clock is expressed in the optic lobe and is required for the circadian clock in the cricket Gryllus bimaculatus. J. Biol. Rhythm 2012, 27, 467–477. [Google Scholar] [CrossRef]
- Uryu, O.; Karpova, S.G.; Tomioka, K. The clock gene cycle plays an important role in the circadian clock of the cricket Gryllus bimaculatus. J. Insect Physiol. 2013, 59, 697–704. [Google Scholar] [CrossRef] [PubMed]
- Moriyama, Y.; Sakamoto, T.; Karpova, S.G.; Matsumoto, A.; Noji, S.; Tomioka, K. RNA interference of the clock gene period disrupts circadian rhythms in the cricket Gryllus bimaculatus. J. Biol. Rhythm 2008, 23, 308–318. [Google Scholar] [CrossRef]
- Zhan, S.; Merlin, C.; Boore, J.L.; Reppert, S.M. The monarch butterfly genome yields insights into long-distance migration. Cell 2011, 147, 1171–1185. [Google Scholar] [CrossRef]
- Cyran, S.A.; Buchsbaum, A.M.; Reddy, K.L.; Lin, M.C.; Glossop, N.R.J.; Hardin, P.E.; Young, M.W.; Storti, R.V.; Blau, J. vrille, Pdp1, and dClock form a second feedback loop in the Drosophila circadian clock. Cell 2003, 112, 329–341. [Google Scholar] [CrossRef]
- Narasaki-Funo, Y.; Tomiyama, Y.; Nose, M.; Bando, T.; Tomioka, K. Functional analysis of Pdp1 and vrille in the circadian system of a cricket. J. Insect Physiol. 2020, 127, 104156. [Google Scholar] [CrossRef] [PubMed]
- Ingram, K.K.; Kutowoi, A.; Wurm, Y.; Shoemaker, D.; Meier, R.; Bloch, G. The molecular clockwork of the fire ant Solenopsis invicta. PLoS ONE 2012, 7, e45715. [Google Scholar] [CrossRef]
- Rivas, G.B.S.; Zhou, J.; Merlin, C.; Hardin, P.E. CLOCKWORK ORANGE promotes CLOCK-CYCLE activation via the putative Drosophila ortholog of CLOCK INTERACTING PROTEIN CIRCADIAN. Curr. Biol. 2021, 31, 4207–4218. [Google Scholar] [CrossRef]
- Tomioka, K.; Matsumoto, A. Circadian molecular clockworks in non-model insects. Curr. Opin. Insect Sci. 2015, 7, 58–64. [Google Scholar] [CrossRef] [PubMed]
- Sauman, I.; Reppert, S.M. Circadian clock neurons in the silkmoth Antheraea pernyi: Novel mechanisms of Period protein regulation. Neuron 1996, 17, 889–900. [Google Scholar] [CrossRef] [PubMed]
- Sehadová, H.; Markova, E.P.; Sehnal, F.; Takeda, M. Distribution of circadian clock-related proteins in the cephalic nervous system of the silkworm, Bombyx mori. J. Biol. Rhythm 2004, 19, 466–482. [Google Scholar] [CrossRef]
- Rubin, E.B.; Shemesh, Y.; Cohen, M.; Elgavish, S.; Robertson, H.M.; Bloch, G. Molecular and phylogenetic analyses reveal mammalian-like clockwork in the honey bee (Apis mellifera) and shed new light on the molecular evolution of the circadian clock. Genome Res. 2006, 16, 1352–1365. [Google Scholar] [CrossRef] [PubMed]
- Kotwica-Rolinska, J.; Chodakova, L.; Smykal, V.; Damulewicz, M.; Provaznik, J.; Wu, B.C.H.; Hejnikova, M.; Chvalova, D.; Dolezel, D. Loss of timeless underlies an evolutionary transition within the circadian clock. Mol. Biol. Evol. 2022, 39, msab346. [Google Scholar] [CrossRef]
- Zhu, H.; Sauman, I.; Yuan, Q.; Casselman, A.; Emery-Le, M.; Emery, P.; Reppert, S.M. Cryptochromes define a novel circadian clock mechanism in monarch butterflies that may underlie sun compass navigation. PLoS Biol. 2008, 6, 138–155. [Google Scholar] [CrossRef]
- Benetta, E.D.; Beukeboom, L.W.; van de Zande, L. Adaptive differences in circadian clock gene expression patterns and photoperiodic diapause induction in Nasonia vitripennis. Am. Nat. 2019, 193, 881–896. [Google Scholar] [CrossRef]
- Bertossa, R.C.; van de Zande, L.; Beukeboom, L.W.; Beersma, D.G.M. Phylogeny and oscillating expression of period and cryptochrome in short and long photoperiods suggest a conserved function in Nasonia vitripennis. Chronobiol. Int. 2014, 31, 749–760. [Google Scholar] [CrossRef]
- Mukai, A.; Goto, S.G. The clock gene period is essential for the photoperiodic response in the jewel wasp Nasonia vitripennis (Hymenoptera: Pteromalidae). Appl. Entomol. Zool. 2016, 51, 185–194. [Google Scholar] [CrossRef]
- Benetta, E.D.; van de Zande, L.; Beukeboom, L.W. Courtship rhythm in Nasonia vitripennis is affected by the clock gene period. Behaviour 2021, 158, 685–704. [Google Scholar] [CrossRef]
- Yang, L.; Wang, B.B.; Qiu, L.M.; Wan, B.; Yang, Y.; Liu, M.M.; Wang, F.; Fang, Q.; Stanley, D.W.; Ye, G.Y. Functional characterization of a venom protein calreticulin in the ectoparasitoid Pachycrepoideus vindemiae. Insects 2020, 11, 29. [Google Scholar] [CrossRef] [PubMed]
- Tormos, J.; Beitia, F.; Bockmann, E.A.; Asis, J.D.; Fernandez, S. The preimaginal phases and development of Pachycrepoideus vindemmiae (Hymenoptera, Pteromalidae) on mediterranean fruit fly, Ceratitis capitata (Diptera, Tephritidae). Microsc. Microanal. 2009, 15, 422–434. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.G.; Hogg, B.N.; Biondi, A.; Daane, K.M. Plasticity of body growth and development in two cosmopolitan pupal parasitoids. Biol. Control 2021, 163, 104738. [Google Scholar] [CrossRef]
- Da Silva, C.S.B.; Price, B.E.; Soohoo-Hui, A.; Walton, V.M. Factors affecting the biology of Pachycrepoideus vindemmiae (Hymenoptera: Pteromalidae), a parasitoid of spotted-wing drosophila (Drosophila suzukii). PLoS ONE 2019, 14, e0218301. [Google Scholar]
- Sadanandappa, M.K.; Sathyanarayana, S.H.; Bosco, G. Parasitoid wasp culturing and assay to study parasitoid-induced reproductive modifications in Drosophila. Bio. Protoc. 2023, 13, e4582. [Google Scholar] [CrossRef] [PubMed]
- Small, C.; Paddibhatla, I.; Rajwani, R.; Govind, S. An introduction to parasitic wasps of Drosophila and the antiparasite immune response. J. Vis. Exp. 2012, e3347. [Google Scholar] [CrossRef]
- Marchler-Bauer, A.; Bo, Y.; Han, L.; He, J.; Lanczycki, C.J.; Lu, S.; Chitsaz, F.; Derbyshire, M.K.; Geer, R.C.; Gonzales, N.R.; et al. CDD/SPARCLE: Functional classification of proteins via subfamily domain architectures. Nucleic Acids Res. 2017, 45, D200–D203. [Google Scholar] [CrossRef]
- Larkin, M.A.; Blackshields, G.; Brown, N.P.; Chenna, R.; McGettigan, P.A.; McWilliam, H.; Valentin, F.; Wallace, I.M.; Wilm, A.; Lopez, R.; et al. Clustal W and clustal X version 2.0. Bioinformatics 2007, 23, 2947–2948. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Teng, Z.W.; Wu, H.Z.; Ye, X.H.; Fang, Q.; Zhou, H.X.; Ye, G.Y. An endoparasitoid uses its egg surface proteins to regulate its host immune response. Insect Sci. 2022, 29, 1030–1046. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−∆∆CT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Bustin, S.A.; Benes, V.; Garson, J.A.; Hellemans, J.; Huggett, J.; Kubista, M.; Mueller, R.; Nolan, T.; Pfaffl, M.W.; Shipley, G.L.; et al. The MIQE guidelines: Minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 2009, 55, 611–622. [Google Scholar] [CrossRef] [PubMed]
- Allada, R.; White, N.E.; So, W.V.; Hall, J.C.; Rosbash, M. A mutant Drosophila homolog of mammalian Clock disrupts circadian rhythms and transcription of period and timeless. Cell 1998, 93, 791–804. [Google Scholar] [CrossRef]
- Takahata, S.; Ozaki, T.; Mimura, J.; Kikuchi, Y.; Sogawa, K.; Fujii-Kuriyama, Y. Transactivation mechanisms of mouse clock transcription factors, mClock and mArnt3. Genes Cells 2000, 5, 739–747. [Google Scholar] [CrossRef]
- Chang, D.C.; McWatters, H.G.; Williams, J.A.; Gotter, A.L.; Levine, J.D.; Reppert, S.M. Constructing a feedback loop with circadian clock molecules from the silkmoth, Antheraea pernyi. J. Biol. Chem. 2003, 278, 38149–38158. [Google Scholar] [CrossRef] [PubMed]
- Saez, L.; Young, M.W. Regulation of nuclear entry of the Drosophila clock proteins period and timeless. Neuron 1996, 17, 911–920. [Google Scholar] [CrossRef] [PubMed]
- Chang, D.C.; Reppert, S.M. A novel C-terminal domain of Drosophila PERIOD inhibits dCLOCK: CYCLE-mediated transcription. Curr. Biol. 2003, 13, 758–762. [Google Scholar] [CrossRef]
- Zhu, L.; Feng, S.; Gao, Q.; Liu, W.; Ma, W.H.; Wang, X.P. Host population related variations in circadian clock gene sequences and expression patterns in Chilo suppressalis. Chronobiol. Int. 2019, 36, 969–978. [Google Scholar] [CrossRef]
- Hirayama, J.; Nakamura, H.; Ishikawa, T.; Kobayashi, Y.; Todo, T. Functional and structural analyses of cryptochrome- Vertebrate cry regions responsible for interaction with the CLOCK: BMAL1 heterodimer and its nuclear localization. J. Biol. Chem. 2003, 278, 35620–35628. [Google Scholar] [CrossRef]
- Werckenthin, A.; Derst, C.; Stengl, M. Sequence and expression of per, tim1, and cry2 genes in the madeira cockroach Rhyparobia maderae. J. Biol. Rhythm 2012, 27, 453–466. [Google Scholar] [CrossRef]
- Rego, N.D.C.; Chahad-Ehlers, S.; Campanini, E.B.; Torres, F.R.; de Brito, R.A. VRILLE shows high divergence among Higher Diptera flies but may retain role as transcriptional repressor of clock. J. Insect Physiol. 2021, 133, 104284. [Google Scholar] [CrossRef] [PubMed]
- Nesbit, K.T.; Christie, A.E. Identification of the molecular components of a Tigriopus californicus (Crustacea, Copepoda) circadian clock. Comp. Biochem. Physiol. Part D 2014, 12, 16–44. [Google Scholar] [CrossRef] [PubMed]
- Dawson, S.R.; Turner, D.L.; Weintraub, H.; Parkhurst, S.M. Specificity for the hairy/enhancer of split basic helix-loop-helix (bHLH) proteins maps outside the bHLH domain and suggests two separable modes of transcriptional repression. Mol. Cell. Biol. 1995, 15, 6923–6931. [Google Scholar] [CrossRef] [PubMed]
- Li, X.W.; Jiang, H.X.; Zhang, X.C.; Shelton, A.M.; Feng, J.N. Post-mating interactions and their effects on fitness of female and male Echinothrips americanus (Thysanoptera: Thripidae), a new insect pest in China. PLoS ONE 2014, 9, e87725. [Google Scholar] [CrossRef] [PubMed]
- Bertossa, R.C.; van Dijk, J.; Diao, W.; Saunders, D.; Beukeboom, L.W.; Beersma, D.G.M. Circadian rhythms differ between sexes and closely related species of Nasonia wasps. PLoS ONE 2013, 8, e60167. [Google Scholar] [CrossRef] [PubMed]
- Forsse, E.; Smith, S.M.; Bourchier, R.S. Flight initiation in the egg parasitoid Trichogramma minutum: Effects of ambient temperature, mates, food, and host eggs. Entomol. Exp. Appl. 1992, 62, 147–154. [Google Scholar] [CrossRef]
- Vanlenteren, J.C.; Szabo, P.; Huisman, P.W.T. The parasite-host relationship between Encarsia formosa gahan (Hymenoptera, Aphelinidae) and Trialeurodes vaporariorum (Westwood) (Homoptera, Aleyrodidae). XXXVII. Adult emergence and initial dispersal pattern of E. formosa. J. Appl. Entomol. 1992, 114, 392–399. [Google Scholar] [CrossRef]
- Pompanon, F.; Fouillet, P.; Bouletreau, M. Physiological and genetic factors as sources of variation in locomotion and activity rhythm in a parasitoid wasp (Trichogramma brassicae). Physiol. Entomol. 1999, 24, 346–357. [Google Scholar] [CrossRef]
- Vogt, E.A.; Nechols, J.R. Diel activity patterns of the squash bug egg parasitoid Gryon pennsylvanicum (Hymenoptera: Scelionidae). Ann. Entomol. Soc. Am. 1991, 84, 303–308. [Google Scholar] [CrossRef]
- Miranda, M.; Sivinski, J.; Rull, J.; Cicero, L.; Aluja, M. Niche breadth and interspecific competition between Doryctobracon crawfordi and Diachasmimorpha longicaudata (Hymenoptera: Braconidae), native and introduced parasitoids of Anastrepha spp. fruit flies (Diptera: Tephritidae). Biol. Control 2015, 82, 86–95. [Google Scholar] [CrossRef]
- Merlin, C.; Beaver, L.E.; Taylor, O.R.; Wolfe, S.A.; Reppert, S.M. Efficient targeted mutagenesis in the monarch butterfly using zinc-finger nucleases. Genome Res. 2013, 23, 159–168. [Google Scholar] [CrossRef] [PubMed]
- Rakshit, K.; Krishnan, N.; Guzik, E.M.; Pyza, E.; Giebultowicz, J.M. Effects of aging on the molecular circadian oscillations in Drosophila. Chronobiol. Int. 2012, 29, 5–14. [Google Scholar] [CrossRef]
- Gu, H.F.; Xiao, J.H.; Niu, L.M.; Wang, B.; Ma, G.C.; Dunn, D.W.; Huang, D.W. Adaptive evolution of the circadian gene timeout in insects. Sci. Rep. 2014, 4, 4212. [Google Scholar] [CrossRef] [PubMed]
- Gu, H.F.; Xiao, J.H.; Dunn, D.W.; Niu, L.-M.; Wang, B.; Jia, L.Y.; Huang, D.W. Evidence for the circadian gene period as a proximate mechanism of protandry in a pollinating fig wasp. Biol. Lett. 2014, 10, 20130914. [Google Scholar] [CrossRef]
- Li, C.J.; Yun, X.P.; Yu, X.J.; Li, B. Functional analysis of the circadian clock gene timeless in Tribolium castaneum. Insect Sci. 2018, 25, 418–428. [Google Scholar] [CrossRef] [PubMed]
- Rund, S.S.C.; Hou, T.Y.; Ward, S.M.; Collins, F.H.; Duffield, G.E. Genome-wide profiling of diel and circadian gene expression in the malaria vector Anopheles gambiae. Proc. Natl. Acad. Sci. USA 2011, 108, E421–E430. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.D.; Vega-Rodriguez, J.; Diabate, A.; Liu, J.N.; Cui, C.L.; Nignan, C.; Dong, L.; Li, F.; Ouedrago, C.O.; Bandaogo, A.M.; et al. Clock genes and environmental cues coordinate Anopheles pheromone synthesis, swarming, and mating. Science 2021, 371, 411–415. [Google Scholar] [CrossRef]
- Takekata, H.; Numata, H.; Shiga, S.; Goto, S.G. Silencing the circadian clock gene Clock using RNAi reveals dissociation of the circatidal clock from the circadian clock in the mangrove cricket. J. Insect Physiol. 2014, 68, 16–22. [Google Scholar] [CrossRef]
- Fergus, D.J.; Shaw, K.L. Circadian rhythms and period expression in the Hawaiian cricket genus Laupala. Behav. Genet. 2013, 43, 241–253. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).