Abstract
Background/Objectives: 16S ribosomal RNA sequencing has, for several years, been the main means of identifying bacterial and archaeal species. Low-throughput Sanger sequencing is often used for the detection and identification of microbial species, but this technique has several limitations. The use of high-throughput sequencers may be a good alternative to improve patient identification, especially for polyclonal infections and management. Kraken 2 and KrakenUniq are free, high-throughput tools providing a very rapid and accurate classification for metagenomic analyses. However, Kraken 2 can present false-positive results relative to KrakenUniq, which can be limiting in hospital settings requiring high levels of accuracy. The aim of this study was to establish an alternative next-generation sequencing technique to replace Sanger sequencing and to confirm that KrakenUniq is an excellent analysis tool that does not present false results relative to Kraken 2. Methods: DNA was extracted from reference bacterial samples for Laboratory Quality Controls (QCMDs) and the V2-V3 and V3-V4 regions of the 16S ribosomal gene were amplified. Amplified products were sequenced with the Illumina 16S Metagenomic Sequencing protocol with minor modifications to adapt and sequence an Illumina 16S library with a small 500-cycle nano-flow cell. The raw files (Fastq) were analyzed on a commercial Smartgene platform for comparison with Kraken 2 and KrakenUniq results. KrakenUniq was used with a standard bacterial database and with the 16S-specific Silva138, RDP11.5, and Greengenes 13.5 databases. Results: Seven of the eight (87.5%) QCMDs were correctly sequenced and identified by Sanger sequencing. The remaining QCMD, QCMD6, could not be identified through Sanger sequencing. All QCMDs were correctly sequenced and identified by MiSeq with the commercial Smartgene analysis platform. QCMD6 contained two bacteria, Acinetobacter and Klebsiella. KrakenUniq identification results were identical to those of Smartgene, whereas Kraken 2 yielded 25% false-positive results. Conclusions: If Sanger identification fails, MiSeq with a small nano-flow cell is a very good alternative for the identification of bacterial species. KrakenUniq is a free, fast, and easy-to-use tool for identifying and classifying bacterial infections.
1. Introduction
Bacterial infections are the second leading cause of death worldwide. In 2019, bacterial infections caused 13.7 million deaths around the world. Five of the thirty-three most common bacteria are responsible for half of all deaths: Staphylococcus aureus, Escherichia coli, Streptococcus pneumoniae, Klebsiella pneumoniae, and Pseudomonas aeruginosa. Staphylococcus aureus is the leading bacterial cause of death in 135 countries. In children under the age of five years, it causes the most deadly pneumococcal infection [1].
In France, incidence was significantly higher in 2022 than in 2021 for invasive infections caused by airborne and/or contact-transmitted bacteria: Haemophilus influenzae, Neisseria meningitidis, Streptococcus pneumoniae, and Streptococcus pyogenes. Streptococcus pyogenes was the most common species in children under 10 years of age [2].
Identification is traditionally based on the phenotypic characteristics of the bacterium: staining, morphology, ability to grow on certain culture media, and biochemical characteristics detected with various commercial techniques. However, it is difficult to identify some bacteria on the basis of phenotype, for various reasons [3,4,5,6]. Indeed, some bacteria express few phenotypic traits. In other species, stress may alter phenotypic characteristics [7]. Consequently, identification methods based exclusively on phenotypic characteristics can give erroneous results. Matrix-assisted laser desorption–ionization time of flight mass spectrometry (MALDI-TOF MS), a new technology applied to the problem of bacterial species identification, has been introduced in several microbiology laboratories [8]. However, although this technique offers an accurate identification approach, there may be limitations in the diagnosis of bacteria [9].
In such situations, amplification and sequencing of the 16S ribosomal RNA (rRNA) gene, followed by comparison of the resulting sequence with database sequences, have proven effective for bacterial identification [10,11,12,13]. The 16S rRNA gene encodes the 16S subunit of rRNA and has a structure that is highly conserved in all bacteria. It consists of a succession of conserved domains containing binding sites for the universal primers used to sequence this gene. The advantages of molecular biology techniques over traditional techniques include better sensitivity, significant time savings, and greater specificity for the identification of unusual bacteria in cultures.
Two sequencing techniques are available for bacterial identification: low-throughput analysis (Sanger sequencing) and high-throughput analysis (next-generation sequencing or NGS). Both methods analyze DNA at the single-base level, but they use very different approaches. Sanger sequencing generates fragments of the target sequence, each initiated from a primer. Each fragment ends with a fluorescent marker, with different colors corresponding to the nucleotides A, C, G, and T. These fragments are distinguished after separation via capillary electrophoresis. By contrast, NGS encompasses a wide range of techniques for sequencing DNA libraries. These libraries include vast collections of short DNA fragments, amplicons, or even mixed genome fragments from various species. In this high-throughput method, all of the sequences in a library are sequenced collectively. Unlike Sanger sequencing, NGS requires bioinformatics tools for the interpretation and analysis of the data obtained at the end of sequencing. Kraken was the first tool to introduce rapid taxonomic classification based on exact k-mer matches [14]. Kraken 2 constitutes a significant improvement in speed and memory while maintaining the classification algorithm of the original Kraken tool [15,16]. Kraken 2 uses a compact hash table and a probabilistic data structure, and it applies a spaced seed mask of s spaces to the minimizer and calculates a compact hash code [15]. It is generally superior for standard metagenomic analyses, but can have a low false-positive rate, making it less suitable for diagnosing infections [17,18]. KrakenUniq enhances Kraken by adding counts of unique k-mers for each classification, thereby providing a more accurate estimate of species abundance [19]. KrakenUniq uses the efficient cardinality estimation algorithm HyperLogLog for counting the number of unique k-mers identified for each taxon [20,21].
In this study, we sequenced the 16S gene with Sanger and NGS sequencing techniques and then used Kraken 2 and KrakenUniq to analyze the NGS-generated data. Several studies have shown that Kraken 2 is a very good tool for analyzing microorganisms, but very few studies have described its limitations [22,23,24]. The aim of this study was to set up an NGS technique as an alternative to Sanger sequencing and to confirm that KrakenUniq is an excellent bioinformatics analysis tool with a very low false-positive rate.
2. Materials and Methods
2.1. Validation Samples
Reference bacterial samples for quality control were obtained from the independent international organization Quality Control for Molecular Diagnostics (QCMD) in the form of supernatants from cultured reference bacterial strains. QCMD provides more than 2000 laboratories in more than 120 countries with a quality assessment service, with the aim of evaluating the ability of laboratories to detect, identify, and interpret the bacterial species supplied [25]. In addition to QCMD samples, we also included a positive and a negative control (DNA-free PCR).
2.2. Bacterial DNA Extraction
DNA was extracted with the EZ1 Virus Mini kit v2.0 (Qiagen S.A.S., Courtaboeuf, France) in accordance with the manufacturer’s instructions, but with an additional proteinase K pretreatment at 56 °C for 60 min.
2.3. Sanger PCR Amplification
Targeted PCR was performed on the V2/V3 region of the 16S rRNA with the following primers: 27F and 244R as previously described by Moumile et al. [26]. The PCR mixture contained 10 μL extracted DNA, 1 µL of each primer (12.5 μM), 1 µL dNTPs, 2 µL MgSO4, 5 µL 10 × buffer, and 0.2 µL Taq HIFI enzyme (Invitrogen, Carlsbad, CA, USA). We added 35 µL nuclease-free water to obtain a volume of 50 µL. PCR was performed under the following conditions: initial denaturation for 10 min at 94 °C, followed by 35 cycles of 30 s at 94 °C, 30 s at 58 °C, 30 s at 68 °C, and a final extension for 7 min at 68 °C. The amplified products were subjected to quality control via agarose gel electrophoresis. The amplicon obtained was about 357 base pairs (bp) long.
2.4. MiSeqPCR Amplification
Targeted PCR was performed on the V3/V4 region of the 16S RNA with primers containing Illumina adapter sequences, 341F and 785R, as previously described [27]. The reaction mixture contained 12.5 µL KAPA HiFi DNA polymerase (Reference KK2601, Roche, Cape, South Africa), 0.5 µL of each primer, 1.5 µL nuclease-free water, and 10 µL extracted DNA. The following PCR conditions were used: initial denaturation for 3 min at 95 °C, followed by 45 cycles of 30 s at 95 °C, 30 s at 55 °C, 30 s at 72 °C, and a final extension for 5 min at 72 °C. Amplified products of 550 bp in size were detected on migration in a TapeStation system.
2.5. Sanger Library Sequencing
Amplified products were first purified with the Monarch® Spin PCR & DNA Cleanup Kit (Biolabs, Newburyport, MA, USA) according to the manufacturer’s instructions. Sequencing was then performed with the Big Dye Terminator mix. The reaction mixture consisted of 1 µL of the sense and antisense primers used in the first amplification, 13 µL nuclease-free water, 2 µL purified DNA, and 4 µL Big Dye Terminator mix. PCR was performed with 30 cycles of 10 s at 96 °C, 10 s at 50 °C, and 4 min at 60 °C. At the end of the reaction, ddNTPs were removed with rehydrated Sephadex G50 (Cytiva, Uppsala, Sweden), as recommended by the manufacturer. The purified products were then sequenced on a Thermo Fisher ABI 3500 sequencer (Thermo Fisher, Waltham, MA, USA).
2.6. MiSeqLibrary Sequencing
The library was prepared with a slightly modified and adapted version of the Illumina 16S Metagenomic Sequencing protocol (Part # 15044223 Rev. B). PCR products were purified with 0.8× AMPure XP beads and then diluted to 3.5 ng/µL. An index PCR was then performed on a mixture containing 25 µL KAPA HiFi mix (Beckman, CA, USA), 5 µL of each Nextra XT index (Illumina, San Diego, CA, USA), and 15 µL purified DNA. PCR conditions were as follows: initial denaturation for 3 min at 95 °C, followed by 12 cycles of 30 s at 95 °C, 30 s at 55 °C, 30 s at 72 °C, and a final extension for 5 min at 72 °C. After this index PCR, a second purification was performed with 1.2× magnetic beads. Each amplicon was diluted to a final concentration of 2 nM. All amplicons were pooled in a single tube, and 5 µL of this pool was denatured with 0.2 NaOH and diluted to 8 pM in HT1. The library was sequenced with a MiSeq Nano reagent kit v2, 500 cycles (Illumina Inc., San Diego, CA, USA).
2.7. Analysis Results
For Sanger sequencing, we used the sequences for a BLAST+2.16.0 analysis of the NCBI database to identify the various bacteria. By contrast, for MiSeq, we used three methods for bioinformatics analysis: the Advanced Sequencing Platform (ASP) v3.15.0 available from SmartGene (SmartGene, Unterägeri, Switzerland, www.smartgene.com (accessed on 1 May 2025)) [28], Kraken 2, and KrakenUniq.
The SmartGene application was used as a validation method. The SmartGene Bacteria 16S Microbiome App is a commercial CE-IVD-labeled automated cloud application service that can handle base-called sequencing files generated by different sequencing technologies. *.fastq files obtained with the MiSeq platform (Illumina) were uploaded into ASP. The application functions as follows. It first performs the following steps in an automated manner:
- (1)
- Paired-end detection of read files, if uploaded in the same batch;
- (2)
- Technology-specific quality filtering to trim or remove low-quality reads;
- (3)
- Establishment of a work list for the batch.
The user then selects the analysis pipeline to be used, in this case, the Microbiome 16S targeted workflow. This automated analysis pipeline performs the following actions:
- (1)
- Quality filtering: R1 and R2 files are merged for Illumina data, and a sliding window (25 nt) is then applied to reads, enhancing the trimming of poor-quality sections with a low Phred score (<23 for this study) and the filtering of short reads (<20 nt in this study).
- (2)
- Read mapping: Reads passing the quality filters are mapped against the SmartGene 16S Centroid database without prior binning. The SmartGene “16S Centroid” database consists of non-redundant representative bacterial 16S rRNA sequences covering 17,314 species from 3439 genera as of November 2024. It is maintained and updated by AI and algorithms (patent #EP02215578).
- (3)
- Production of a quality report and display of the results: Results are grouped according to match quality (e.g., % coverage, number of mismatches, etc.), match specificity (matching a single species or not), and match consistency (close matches belonging to the same genus). The system produces a confidence score for the matching taxon, and it is not possible to provide a call at the species level; a call is made at the next taxon up, indicating all possible matching species and genera. Results are displayed in table format, along with the number of reads and relative abundance, with the possibility of consolidation at species, genus, and family levels, and the generation of a dynamic Krona diagram. Abundances are evaluated by counting the reads mapped to a particular species/genus/family.
Kraken 2 and KrakenUniq were installed on a 64-bit Linux computer with 8 GB RAM. The Kraken 2 package was downloaded as a Zip file from https://github.com/DerrickWood/kraken2.git (accessed on 1 May 2025) (Figure 1).
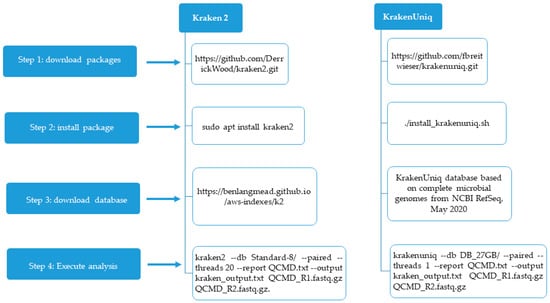
Figure 1.
The different steps for configuring Kraken 2 and KrakenUniq.
This file was unzipped and the following command line was used to install Kraken 2: sudo apt install kraken 2. For the running of Kraken 2, we downloaded an 8 GB (Standard-8) database from https://benlangmead.github.io/aws-indexes/k2. (accessed on 2 May 2025). We used the following command line to analyze each sample: kraken2 --db Standard-8/ --paired --threads 20 --report QCMD.txt --output kraken_output.txt QCMD_R1.fastq.gz QCMD_R2.fastq.gz. In this command line, Standard-8 is the database, and QCMD.txt is the file containing the classification results from the fastq.gz files.
We also used specific 16S databases—the SILVA database, Greengenes 13.5, and the RDP 11.5 database—for the Kraken2 analysis. These databases are all available from https://benlangmead.github.io/aws-indexes/k2. (accessed on 2 May 2025).
KrakenUniq was downloaded from https://github.com/fbreitwieser/krakenuniq.git (accessed on 2 May 2025) and installed with the following command line: ./install_krakenuniq.sh.
We downloaded an indexed 27 G database by clicking on the following link: KrakenUniq database based on complete microbial genomes from NCBI RefSeq, May 2020.
We used the following command line for the analysis of each sample: krakenuniq --db DB_27 GB/ --paired --threads 1 --report QCMD.txt --output kraken_output.txt QCMD_R1.fastq.gz QCMD_R2.fastq.gz.
3. Results
In total, eight QCMDs were subjected to Sanger sequencing and sequencing on a MiSeq system. Seven of these QCMDs (87.5%) were correctly sequenced through Sanger sequencing and were analyzed with NCBI Nucleotide Blast (Table 1). Total score and percent identity (Per. Indent) ranged from 963% to 11,245% and from 99.55% to 100%, respectively. Each QCMD was identified with a single species different from the others. The results of QCMD6 were not interpretable through Sanger sequencing, as this QCMD is a mixture of two microbes.

Table 1.
Sanger sequencing results.
All eight QCMDs (100%) were correctly sequenced by MiSeq. The number of reads mapped with the Smartgene analysis application ranged from 116,822 (95.77%) to 188,722 (95%) (Table 2). Two genera were identified for QCMD6—Acinetobacter and Klebsiella—accounting for 52.02% and 40.09% of reads, respectively (Figure 2A). The mean analysis time for the eight QCMDs was 4080 s.

Table 2.
Results of Smartgene, KrakenUniq, and Kraken 2 analyses with the standard database.
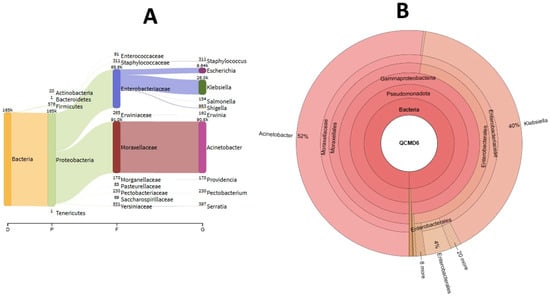
Figure 2.
KrakenUniq (A) and Smartgene (B) analyses.
The number of reads mapped with KrakenUniq ranged from 117,091 (99.28%) to 189,218 (99.5%). Acinetobacter and Klebsiella were also identified in QCMD6 and these two genera accounted for 69.87% and 20% of reads, respectively, according to KrakenUniq (Figure 2B). The mean analysis time for the eight QCMDs was 27 s using KrakenUniq. No false positives were observed with Smartgene and KrakenUniq.
By contrast, Kraken 2 analysis with the standard database presented 25% (2 of 8 QCMDs) false positives, including QCMD1, identified as Pseudomonas with 158,379 reads (99.26%), and QCMD5, identified as Enterococcus with a total of 165,385 reads (99.83%) (Figure 3). Conversely, the mixture of bacteria in QCMD6 was correctly identified with 21.21% Acinetobacter and 25.14% Klebsiella reads. The mean analysis time was 27 s.

Figure 3.
Kraken 2 analysis for QCMD1 (A) and QCMD5 (B).
An analysis of the Kraken 2 results obtained with the 16S-specific databases also revealed several false-positive results (Table 3). In the Silva138 database, QCMD2 was identified as Streptococcus with a total of 13,598 reads (34.97%), and QCMD7 was identified as a mixture of two bacteria: Enterobacter with 7798 (25.6%) reads and Klebsiella with 7060 (23.5%) reads. Only one of the microbes present in QCMD6, Acinetobacter, was correctly identified, with 85,118 reads (83.59%), despite the mixed nature of the infection, with both Acinetobacter and Klebsiella present.

Table 3.
Kraken 2 results with 16S-specific databases.
The Greengenes database gave incorrect results for QCMD1 and QCMD5, both of which were identified as Serratia, with 17,029 (72.03%) and 1694 (11.23%) reads, respectively. The mixed infection in QCMD6 was not correctly detected, as only Acinetobacter was identified, with a total of 55,180 reads (86.58%).
For the RDP11.5 database, QCMD1 was also identified as Serratia, with 90,122 (93.03%) reads. QCMD7 was identified as a mixture of two bacteria, Klebsiella with 5054 (29.05%) reads and Enterobacter with 3287 (18.9%) reads. The QCMD6 was identified as a single bacterium, Acinetobacter, with 23,369 (69.72%) reads.
The total time for library preparation was 3 h 8 min for MiSeq and 4 h 45 min for Sanger. The sequencing time was 8 h for MiSeq and 1 h 30 min for Sanger.
4. Discussion
For over 35 years, clinical microbiology laboratories have used targeted sequencing for the definitive identification of bacterial pathogens of humans [29,30,31,32]. The 16S rRNA gene is a slowly evolving, highly conserved gene found in all microorganisms, features that have led to it becoming the most widely used target for bacterial and archaeal pathogen identification studies [33,34,35,36,37]. In this study, we used Sanger 16S sequencing technology to detect and identify various pathogens. This technique is often used when attempts to identify or detect bacteria in clinical samples are unsuccessful. However, this technique is unsuitable for the simultaneous detection of more than one species in a single sample and is, therefore, of limited value for use in cases of polymicrobial infection [38]. This limitation is illustrated by our results for QCMD6, corresponding to a polymicrobial infection with two species, Acinetobacter baumannii and Klebsiella pneumoniae. Despite this limitation, the rest of the samples (85.7%) were successfully identified through Sanger sequencing. The Sanger technique remains a rapid, cost-effective, high-performance method for detecting monomicrobial infections.
As a means of overcoming the problem of bacterial identification in polymicrobial infections, we optimized and adapted the Illumina 16S protocol for the rapid sequencing of samples and bacterial identification within 24 h. The recent arrival of an Illumina MiSeq sequencer in our CHV laboratory [39] has opened up new possibilities for 16S sequencing, including the use of a nano-flow cell for the sequencing of a small number of samples with about one million reads. This new technique is highly suitable for use in clinical practice, in which results for individual patients must be obtained rapidly to optimize treatment. The results obtained with the Smartgene analysis platform were entirely concordant with those obtained through Sanger sequencing for the identification of bacteria in monomicrobial infections. Polymicrobial infection was correctly identified and differentiated. We chose to use this commercial Smartgene analysis platform because it was specifically developed to describe the composition of microbiota [28]. It is, thus, suitable for use to identify mixed (polymicrobial) infections and minority species. With this commercial platform, we were able to validate the quality of reads (Q30 ≥ 89% for all samples) obtained through sequencing and to validate the other analysis tools (Kraken 2 and KrakenUniq) assessed in this study. An analysis performed after sequencing yielded a Q30 value ≥ 89% and a Clusters Passing Filter rate ≥ 92%, demonstrating successful sequencing with a very good read quality. The technique used for library preparation is faster (libraries for eight samples prepared in about 3 h and 10 min, Figure 4) than other technologies requiring an additional fragmentation step. Furthermore, this protocol does not require the purchase of a specific library kit (just the KAPA HiFi HotStart ReadyMix with Illumina indices). This technique therefore remains a very good alternative approach to the identification of bacteria based on 16S RNA sequences for NGS.
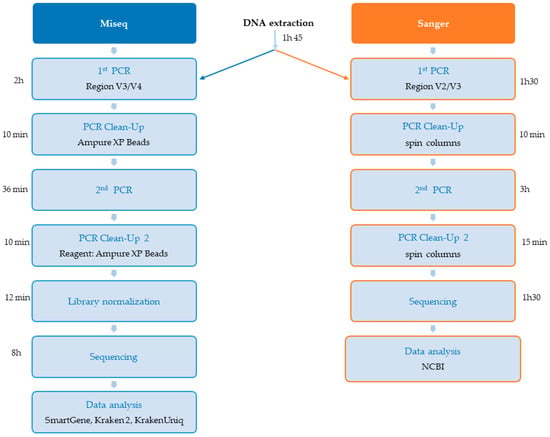
Figure 4.
Sequencing times with MiSeq and Sanger.
Kraken 2 and KrakenUniq are free analysis tools from Kraken, the first tool for very fast taxonomic classification based on exact k-mer matches. The number of reads mapped for all samples was similar for Smartgene and Kraken 2 (standard database), but false positive results were obtained for QCMD1 and QCMD5. We also tested Kraken 2 with all of the available 16S-specific databases (Silva, Greengenes, and RDP), as in several other studies [40,41,42,43,44,45], with the aim of identifying a database giving no false positive detections. Unfortunately, all Kraken 2 databases gave at least 14.2% false positives for monomicrobial infections, and polymicrobial infection was not correctly identified with any of the 16S-specific databases. Thus, regardless of the database used, Kraken 2 is less suitable for the diagnosis of infections in hospital settings, in which a high degree of precision is required.
The KrakenUniq analysis yielded results comparable to those obtained with Smartgene. No false positives were observed, and the polymicrobial infection (QCMD6) was correctly identified. This study thus provides the first demonstration of practical cases of false positives with Kraken 2 analysis being corrected by KrakenUniq analysis. In addition to its simplicity of execution and speed of analysis (mean of 27 s per sample), KrakenUniq is clearly highly suitable for use in mono- and polymicrobial infections. We show herein how it is possible to install and run bioinformatics analyses without any specific prerequisite knowledge, on a low-capacity 8G RAM computer, as reported by C. Pockrandt et al. 2022 [45], to obtain reliable and robust analysis results. Furthermore, these analyses with KrakenUniq can be performed locally, without the need for Internet or a specific network.
Encouragingly, MiSeq sequencing successfully resolved all of the cases for which Sanger sequencing had yielded uninterpretable results in the last five years. In total, 32 clinical samples were sequenced on the MiSeq system and analyzed with KrakenUniq (unpublished data). Eleven of these samples (35%) were identified as polymicrobial infections, with the remainder being identified as monomicrobial. These results indicate that there are very few polymicrobial infections in our study (7% per year on average). The inability of Sanger sequencing to identify certain monomicrobial infections may be due to a low DNA concentration [46] following prior antibiotic treatment. As a means of ensuring better patient care, we will, from now on, systematically subject samples for which Sanger sequencing is unsuccessful to sequencing on the MiSeq sequencer. We strongly recommend the use of the 16S Illumina library preparation technique and KrakenUniq in all laboratories involved in microbial diagnosis.
5. Conclusions
This study confirms that KrakenUniq is a good bioinformatics analysis tool, similar to Kraken 2 in that it yields no false positives. Furthermore, it is free, easy to use, and very fast. Sanger sequencing remains cheaper and faster than NGS, but the use of a nano-flow cell on a MiSeq system remains a good alternative and the best solution for identifying polymicrobial infections while maintaining a reasonable turnaround time to ensure optimal patient care.
Author Contributions
Conceptualization, S.M.J. and N.P.M.; methodology, N.P.M.; validation, N.P.M.; writing—original draft preparation, N.P.M.; writing—review and editing. C.F.-L., S.D., and O.Z. performed DNA extraction and Sanger sequencing. S.M. contributed to the installation and use of KrakenUniq. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Centre Hospitalier de Versailles (grant number: O2024/04).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The original data are included in the article. Further inquiries can be directed to the corresponding author.
Acknowledgments
We thank the personnel of the Versailles Hospital laboratory, especially Benjamin Maneglier and Franck Mausoleo for their technical assistance. We are also very grateful to the SmartGene team, especially Jean Ruelle, for their assistance with the interpretation of sequence data analyses.
Conflicts of Interest
The authors declare no conflicts of interest.
Abbreviations
rRNA, ribosomal RNA; NGS, next-generation sequencing; QCMD, Quality Control for Molecular Diagnostics; NCBI, National Center for Biotechnology Information; ASP, Advanced Sequencing Platform; USD, United States dollars.
References
- GBD 2019 Antimicrobial Resistance Collaborators. Global mortality associated with 33 bacterial pathogens in 2019: A systematic analysis for the Global Burden of Disease Study 2019. Lancet 2022, 400, 2221–2248. [Google Scholar] [CrossRef] [PubMed]
- Santé Publique France. Bulletin Epibac: Surveillance des Infections Invasives Bactériennes. Édition Nationale. 5 Mars 2024. Available online: https://www.santepubliquefrance.fr/content/download/610779/4219986?version=2 (accessed on 1 May 2025).
- Oethinger, M.; Warner, D.K.; Schindler, S.A.; Kobayashi, H.; Bauer, T.W. Diagnosing periprosthetic infection: False-positive intraoperative Gram stains. Clin. Orthop. Relat. Res. 2011, 469, 954–960. [Google Scholar] [CrossRef]
- Tang, Y.-W.; Ellis, N.M.; Hopkins, M.K.; Smith, D.H.; Dodge, D.E.; Persing, D.H. Comparison of phenotypic and genotypic techniques for identification of unusual aerobic pathogenic gram-negative bacilli. J. Clin. Microbiol. 1998, 36, 3674–3679. [Google Scholar] [CrossRef] [PubMed]
- Tang, Y.W.; Hopkins, M.K.; Kolbert, C.P.; Hartley, P.A.; Severance, P.J.; Persing, D.H. Bordetella holmesii-like organisms associated with septicemia, endocarditis, and respiratory failure. Clin. Infect. Dis. 1998, 26, 389–392. [Google Scholar] [CrossRef] [PubMed]
- Hollis, D.G.; Moss, C.W.; Daneshvar, M.I.; Meadows, L.; Jordan, J.; Hill, B. Characterization of Centers for Disease Control group NO-1, a fastidious, nonoxidative, gram-negative organism associated with dog and cat bites. J. Clin. Microbiol. 1993, 31, 746–748. [Google Scholar] [CrossRef]
- Ochman, H.; Lerat, E.; Daubin, V. Examining bacterial species under the specter of gene transfer and exchange. Proc. Natl. Acad. Sci. USA 2005, 102, 6595–6599. [Google Scholar] [CrossRef]
- Seng, P.; Drancourt, M.; Gouriet, F.; La Scola, B.; Fournier, P.-E.; Rolain, J.M.; Raoult, D. Ongoing revolution in bacteriology: Routine identification of bacteria by matrix-assisted laser desorption ionization time-of-flight mass spectrometry. Clin. Infect. Dis. 2009, 49, 543–551. [Google Scholar] [CrossRef]
- AbdulWahab, A.; Taj-Aldeen, S.J.; Ibrahim, E.B.; Talaq, E.; Abu-Madi, M.; Fotedar, R. Discrepancy in MALDI-TOF MS identification of uncommon Gram-negative bacteria from lower respiratory secretions in patients with cystic fibrosis. Infect. Drug Resist. 2015, 8, 83–88. [Google Scholar] [CrossRef]
- Petti, C.A.; Polage, C.R.; Schreckenberger, P. The role of 16S rRNA gene sequencing in identification of microorganisms misidentified by conventional methods. J. Clin. Microbiol. 2005, 43, 6123–6125. [Google Scholar] [CrossRef]
- Han, X.Y.; Falsen, E. Characterization of oral strains of Cardiobacterium valvarum and emended description of the organism. J. Clin. Microbiol. 2005, 43, 2370–2374. [Google Scholar] [CrossRef]
- Woo, P.C.; Ng, K.H.; Lau, S.K.; Yip, K.T.; Fung, A.M.; Leung, K.W.; Tam, D.M.; Que, T.L.; Yuen, K.Y. Usefulness of the MicroSeq 500 16S ribosomal DNA-based bacterial identification system for identification of clinically significant bacterial isolates with ambiguous biochemical profiles. J. Clin. Microbiol. 2003, 41, 1996–2001. [Google Scholar] [CrossRef]
- Bosshard, P.P.; Abels, S.; Altwegg, M.; Böttger, E.C.; Zbinden, R. Comparison of conventional and molecular methods for identification of aerobic catalase-negative gram-positive cocci in the clinical laboratory. J. Clin. Microbiol. 2004, 42, 2065–2073. [Google Scholar] [CrossRef][Green Version]
- Wood, D.E.; Salzberg, S.L. Kraken: Ultrafast metagenomic sequence classification using exact alignments. Genome Biol. 2014, 15, R46. [Google Scholar] [CrossRef] [PubMed]
- Wood, D.E.; Lu, J.; Langmead, B. Improved metagenomic analysis with Kraken 2. Genome Biol. 2019, 20, 257. [Google Scholar] [CrossRef] [PubMed]
- Lu, J.; Rincon, N.; Wood, D.E.; Breitwieser, F.P.; Pockrandt, C.; Langmead, B.; Salzberg, S.L.; Steinegger, M. Metagenome analysis using the Kraken software suite. Nat. Protoc. 2022, 17, 2815–2839. [Google Scholar] [CrossRef]
- Bradford, L.M.; Carrillo, C.; Wong, A. Managing false positives during detection of pathogen sequences in shotgun metagenomics datasets. BMC Bioinform. 2024, 25, 372. [Google Scholar] [CrossRef] [PubMed]
- Marini, S.; Barquero, A.; Wadhwani, A.A.; Bian, J.; Ruiz, J.; Boucher, C.; Prosperi, M. OCTOPUS: Disk-based, Multiplatform, Mobile-friendly Metagenomics Classifier. bioRxiv 2024. [Google Scholar] [CrossRef]
- Breitwieser, F.P.; Baker, D.N.; Salzberg, S.L. KrakenUniq: Confident and fast metagenomics classification using unique k-mer counts. Genome Biol. 2018, 19, 198. [Google Scholar] [CrossRef]
- Flajolet, P.; Fusy, É.; Gandouet, O.; Meunier, F. HyperLogLog: The analysis of a near-optimal cardinality estimation algorithm. Discret. Math. Theor. Comput. Sci. 2007, 137–156. [Google Scholar] [CrossRef]
- Heule, S.; Nunkesser, M.; Hall, A. HyperLogLog in practice: Algorithmic engineering of a state of the art cardinality estimation algorithm. In Proceedings of the 16th International Conference on Extending Database Technology, Genoa, Italy, 18–22 March 2013; pp. 683–692. [Google Scholar]
- Pinheiro, E.T.; Karygianni, L.; Candeiro, G.T.M.; Vilela, B.G.; Dantas, L.O.; Pereira, A.C.C.; Gomes, B.P.F.A.; Attin, T.; Thurnheer, T.; Russo, G. Metatranscriptome and Resistome of the Endodontic Microbiome. J. Endod. 2024, 50, 1059–1072.e4. [Google Scholar] [CrossRef]
- Shahzad, M.; Saeedullah, A.; Shabbir Khan, M.; Ali Ahmad, H.; Iddrissu, I.; Andrews, S.C. 16S rRNA gene amplicon sequencing data from the gut microbiota of adolescent Afghan refugees. Data Brief. 2024, 55, 110636. [Google Scholar] [CrossRef]
- Govender, K.N.; Eyre, D.W. Benchmarking taxonomic classifiers with Illumina and Nanopore sequence data for clinical metagenomic diagnostic applications. Microb. Genom. 2022, 8, mgen000886. [Google Scholar] [CrossRef]
- QCMD—Quality Control for Molecular Diagnostic. Available online: https://www.qcmd.org/fr/qcmd_eqa_schemes-fr/ (accessed on 29 June 2025).
- Moumile, K.; Merckx, J.; Glorion, C.; Berche, P.; Ferroni, A. Osteoarticular infections caused by Kingella kingae in children: Contribution of polymerase chain reaction to the microbiologic diagnosis. Pediatr. Infect. Dis. J. 2003, 22, 837–839. [Google Scholar] [CrossRef]
- Illumina. 16S Metagenomic Sequencing Library Preparation. Available online: http://support.illumina.com/content/dam/illumina-support/documents/documentation/chemistry_documentation/16s/16s-metagenomic-library-prep-guide-15044223-b.pdf (accessed on 1 May 2025).
- Simmon, K.E.; Croft, A.C.; Petti, C.A. Application of SmartGene IDNS software to partial 16S rRNA gene sequences for a di-verse group of bacteria in a clinical laboratory. J. Clin. Microbiol. 2006, 44, 4400–4406. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Woo, P.C.; Lau, S.K.; Teng, J.L.; Tse, H.; Yuen, K.Y. Then and now: Use of 16S rDNA gene sequencing for bacterial identification and discovery of novel bacteria in clinical microbiology laboratories. Clin. Microbiol. Infect. 2008, 14, 908–934. [Google Scholar] [CrossRef]
- Mignard, S.; Flandrois, J.P. 16S rRNA sequencing in routine bacterial identification: A 30-month experiment. J. Microbiol. Methods 2006, 67, 574–581. [Google Scholar] [CrossRef] [PubMed]
- Becker, K.; Harmsen, D.; Mellmann, A.; Meier, C.; Schumann, P.; Peters, G.; von Eiff, C. Development and evaluation of a quality-controlled ribosomal sequence database for 16S ribosomal DNA-based identification of Staphylococcus species. J. Clin. Microbiol. 2004, 42, 4988–4995. [Google Scholar] [CrossRef]
- Cloud, J.L.; Conville, P.S.; Croft, A.; Harmsen, D.; Witebsky, F.G.; Carroll, K.C. Evaluation of partial 16S ribosomal DNA sequencing for identification of Nocardia species by using the MicroSeq 500 system with an expanded database. J. Clin. Microbiol. 2004, 42, 578–584. [Google Scholar] [CrossRef]
- Lane, D.J.; Pace, B.; Olsen, G.J.; Stahl, D.A.; Sogin, M.L.; Pace, N.R. Rapid determination of 16S ribosomal RNA sequences for phylogenetic analyses. Proc. Natl. Acad. Sci. USA 1985, 82, 6955–6959. [Google Scholar] [CrossRef]
- Woese, C.R.; Gutell, R.; Gupta, R.; Noller, H.F. Detailed analysis of the higher-order structure of 16S-like ribosomal ribonucleic acids. Microbiol. Rev. 1983, 47, 621–669. [Google Scholar] [CrossRef] [PubMed]
- Shine, J.; Dalgarno, L. Growth-dependent changes in terminal heterogeneity involving 3′-adenylate of bacterial 16S ribosomal RNA. Nature 1975, 256, 232–233. [Google Scholar] [CrossRef]
- Woese, C.R.; Fox, G.E.; Zablen, L.; Uchida, T.; Bonen, L.; Pechman, K.; Lewis, B.J.; Stahl, D. Conservation of primary structure in 16S ribosomal RNA. Nature 1975, 254, 83–86. [Google Scholar] [CrossRef] [PubMed]
- Kommedal, O.; Kvello, K.; Skjastad, R.; Langeland, N.; Wiker, H.G. Direct 16S rRNA gene sequencing from clinical specimens, with special focus on polybacterial samples and interpretation of mixed DNA chromatograms. J. Clin. Microbiol. 2009, 47, 3562–3568. [Google Scholar] [CrossRef] [PubMed]
- Papa Mze, N.; Fernand-Laurent, C.; Daugabel, S.; Zanzouri, O.; Marque Juillet, S. Optimization of HIV Sequencing Method Using Vela Sentosa Library on Miseq Ilumina Platform. Genes 2024, 15, 259. [Google Scholar] [CrossRef]
- Kang, S.B.; Kim, H.; Kim, S.; Kim, J.; Park, S.K.; Lee, C.W.; Kim, K.O.; Seo, G.S.; Kim, M.S.; Cha, J.M.; et al. Potential Oral Microbial Markers for Differential Diagnosis of Crohn’s Disease and Ulcerative Colitis Using Machine Learning Models. Microorganisms 2023, 11, 1665. [Google Scholar] [CrossRef] [PubMed]
- Hung, Y.M.; Lyu, W.N.; Tsai, M.L.; Liu, C.L.; Lai, L.C.; Tsai, M.H.; Chuang, E.Y. To compare the performance of prokaryotic taxonomy classifiers using curated 16S full-length rRNA sequences. Comput. Biol. Med. 2022, 145, 105416. [Google Scholar] [CrossRef]
- Lu, J.; Salzberg, S.L. Ultrafast and accurate 16S rRNA microbial community analysis using Kraken 2. Microbiome 2020, 8, 124. [Google Scholar] [CrossRef]
- Cole, J.R.; Wang, Q.; Fish, J.A.; Chai, B.; McGarrell, D.M.; Sun, Y.; Brown, C.T.; Porras-Alfaro, A.; Kuske, C.R.; Tiedje, J.M. Ribosomal database project: Data and tools for high throughput rRNA analysis. Nucleic Acids Res. 2014, 42, 633–642. [Google Scholar] [CrossRef]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2013, 41, D590–D596. [Google Scholar] [CrossRef]
- DeSantis, T.Z.; Hugenholtz, P.; Larsen, N.; Rojas, M.; Brodie, E.L.; Keller, K.; Huber, T.; Dalevi, D.; Hu, P.; Andersen, G.L. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl. Environ. Microbiol. 2006, 72, 5069–5072. [Google Scholar] [CrossRef]
- Pockrandt, C.; Zimin, A.V.; Salzberg, S.L. Metagenomic classification with KrakenUniq on low-memory computers. J. Open Source Softw. 2022, 7, 4908. [Google Scholar] [CrossRef] [PubMed]
- Crossley, B.M.; Bai, J.; Glaser, A.; Maes, R.; Porter, E.; Killian, M.L.; Clement, T.; Toohey-Kurth, K. Guidelines for Sanger sequencing and molecular assay monitoring. J. Vet. Diagn. Investig. 2020, 32, 767–775. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).