Abstract
Eczema is a classical characteristic not only in atopic dermatitis but also in various genodermatosis. Patients suffering from primary immunodeficiency diseases such as hyper-immunoglobulin E syndromes, Wiskott-Aldrich syndrome, immune dysregulation, polyendocrinopathy, enteropathy, X-linked syndrome, STAT5B deficiency, Omenn syndrome, atypical complete DiGeorge syndrome; metabolic disorders such as acrodermatitis enteropathy, multiple carboxylase deficiency, prolidase deficiency; and other rare syndromes like severe dermatitis, multiple allergies and metabolic wasting syndrome, Netherton syndrome, and peeling skin syndrome frequently perform with eczema-like lesions. These genodermatosis may be misguided in the context of eczematous phenotype. Misdiagnosis of severe disorders unavoidably affects appropriate treatment and leads to irreversible outcomes for patients, which underlines the importance of molecular diagnosis and genetic analysis. Here we conclude clinical manifestations, molecular mechanism, diagnosis and management of several eczema-related genodermatosis and provide accessible advice to physicians.
1. Introduction
Atopic dermatitis is one of the most prevalent inflammatory skin diseases, affecting 15% to 20% among children and up to 10% among adults [1]. The pathogenesis of atopic dermatitis is complex, involving interactions among genetic and environmental factors, epidermal barrier dysfunction, immune dysregulation, and microbial imbalance [2]. Atopic dermatitis is characterized by chronic, recurrent, and pruritic eczema, often with seasonal fluctuations. The skin lesions can manifest as erythema, papules, oedema, crusting, scaling, and hyperpigmentation and/or hypopigmentation. Associated clinical signs show dry skin, ichthyosis, lichenification and hyperlinear palms. Other manifestations of allergies such as food allergy, asthma and allergic rhinoconjunctivitis may occur. Atopic dermatitis is a clinical diagnosis, and its renewed diagnosis criteria formulated by the American Academy of Dermatology consist of three sections: (1) essential features that must be present for diagnosis including chronic or relapsing history, eczema (acute, subacute, chronic), pruritus, and typical morphology and age-specific patterns of skin lesions; (2) important features that support the diagnosis, such as atopy (personal or family history), early-onset age, IgE reactivity, and xerosis; and (3) associated features that is suggestive for the diagnosis but nonspecific, such as hyperlinear palms, lichenification. Indeed, atopic dermatitis-like lesions are frequently present in several genoderamtosis associated with immunological, metabolic or keratinization dysfunctions. In this review, the genetic disorders with eczematous phenotype are divided into three categories, including immunodeficiency related diseases, inherited metabolic diseases, and rare syndromes. We discuss the genetic molecular mechanisms and clinical manifestations of these diseases, and further provide feasible advice for their differential diagnosis and managements.
2. Immunodeficiency Related Diseases
Primary immunodeficiency diseases (PIDDs) is a group of mostly monogenic disorders with immune system dysfunction, characteristically presenting with recurrent infections [3]. As for skin manifestations, atopic dermatitis-like lesion is a common finding among several PIDDs, being reported in 13% to 57% of patients with PIDDs in previous studies [4,5,6,7]. In particular, the skin lesion might be the early or heralding manifestation of some immunodeficiency diseases, emphasizing the importance to distinguish it from atopic dermatitis and detect the underlying life-threatening immunologic defects [4]. Here, we focus on the PIDDs commonly presented with atopic dermatitis-like skin lesions, including hyper-immunoglobulin E (IgE) syndromes (HIES), Wiskott-Aldrich syndrome (WAS), immune dysregulation, polyendocrinopathy, enteropathy, X-linked (IPEX) syndrome, STAT5B deficiency, Omenn syndrome (OS), atypical complete DiGeorge syndrome, and X-linked agammaglobulinemia (XLA).
2.1. Hyper-IgE Syndromes
Characterized by a triad of eczema, recurrent infections along with elevated IgE levels, hyper-IgE syndromes refer to a heterogeneous group of monogenic inborn immune disorders with either autosomal dominant (AD) or autosomal recessive (AR) inheritance, namely AD-HIES or AR-HIES, respectively [8,9]. Great progress in the identification of pathogenic genes has been made over the past decade. Heterozygous mutations in the signal transducer and activator of transcription-3 (STAT3) gene with dominant negative (DN) effect are recognized as the classical AD-HIES [10,11], also known as STAT3-HIES or Job syndrome. Biallelic mutations in the phosphoglucomutase 3 (PGM3) [12] and the dedicator of cytokinesis-8 (DOCK8) [13,14] are common causes of AR-HIES. In the 2019 update of the International Union of Immunological Societies (IUIS) genetic and phenotypical classification [15], five newly identified monogenic disorders with mutations associated with the STAT3 pathway, including ZNF431 [16], IL-6R [17], IL6ST [18], ERBIN [19], and TGFBR [20] gene defects, as well as CARD11 [21] deficiencies were categorized as HIES, expanding the genetic profile of HIES. In addition, the DOCK8 deficiency [13], previously recognized as AR-HIES, is now considered as a combined immunodeficiency (CID) disease [15]. Patients with HIES exhibit eczema resembling atopic dermatitis features, but in more severe phenotype. Differentiation of HIES from severe atopic dermatitis (SAD) can facilitate early diagnosis and treatment.
2.1.1. The Genetic and Clinical Spectrums of HIES
STAT3-HIES (OMIM147060)
AD-HIES (or STAT3-HIES), previously described as Job syndrome, was found to be caused by heterozygous mutations with DN effect in STAT3 gene by Mingegishi et al. in 2007 [11]. STAT3 is a member of the STAT protein family and plays a key role in the signal transduction of many cytokines, such as interleukin (IL)-6, IL-10, IL-11 and IL-21 [22]. IL-6 is crucial for the differentiation of T-helper 17 (Th17) cells, which can contribute to the defense of extracellular bacterial and fungal pathogens by further secreting cytokines IL-17 and IL-22 [22]. STAT3 is also involved in B-cell differentiation and IgE production via IL-21 signaling [23,24]. IL-11 signaling, essential for the normal development of craniofacial bones and teeth, can restrict suture fusion and tooth number [25].
Patients with STAT3 mutations can present with both immunological and non-immunological manifestations [11]. The clinical features of STAT3-HIES include early-onset eczema, recurrent skin infections and skin abscesses, recurrent pneumonia and resultant pneumatoceles, chronic mucocutaneous infection, along with skeletal, dental, and connective tissue abnormalities, such as characteristic facial features (a prominent forehead, deep-set eyes, broadened nasal bridge, and high-arched palate), retained primary teeth, bone fragility and scoliosis [11,26,27,28,29]. Early-onset (within the first week of life) papulopustular rash on the face and scalp is often the first present symptom and can develop to eczematous dermatitis within the first year. This chronic dermatitis is strongly associated with the colonization of Staphylococcal aureus [27,30]. Newborn rash, skin abscesses, chronic mucocutaneous candidiasis and pneumonia are highly specific of STAT3-HIES [27]. Arterial abnormalities predominantly with coronary artery ectasia or aneurysm [31], gastrointestinal manifestations like infection-related intestinal perforation [32], and malignancy hematopoietic in origin [26] have also been reported.
STAT3-Related Deficiencies
STAT3-related deficiencies share clinical phenotypes with STAT3-HIES due to the relevance of STAT3-dependent pathway. Here we classified ZNF431 deficiency (OMIM618282), IL-6R deficiency (OMIM618944), IL6ST deficiency (OMIM618523, 619752), TGFBR deficiency (OMIM609192, 610168), and ERBIN deficiency (OMIM606944) as STAT3-related deficiencies.
Homozygous loss-of-function (LOF) mutations of ZNF341 gene in patients with HIES were first identified by Frey-Jakobs et al. in 2018 [33]. Zinc finger protein 431 (ZNF431) is a transcription factor that regulates the transcription of STAT3 [16,33]. Patients with biallelic ZNF431 mutations have low constitutive levels of STAT3 mRNA and present with atopy, elevation of IgE, and susceptibility to Candida and Staphylococcal aureus infections, but they show mild skeletal or connective tissue abnormalities. Cutaneous manifestations involve dermatitis, pruritis, excoriated skin lesions, eczema, and skin infections particularly with Staphylococcal aureus [16]. Similar clinical features were also detected in patients with homozygous mutations in IL-6 receptor (IL-6R) gene by Spencer et al. in 2019 [17], manifesting as recurrent infections, abnormal acute-phase responses, elevated IgE, eczema, and eosinophilia. The binding of IL-6 and IL-6R further ligates glycoprotein 130 (GP130), leading to the phosphorylation of STAT3 and Janus kinase (JAK), as well as the nucleus transfer of STAT3 dimer [17].
The homozygous mutation of IL6ST was firstly identified in a patient as a novel immunodeficiency with phenotypic similarities to STAT3-HIES by Schwerd et al. in 2017 [18]. Then in 2020, DN IL6ST variants were described as another form of AD-HIES by Beziat et al. [34]. The patients have clinical features with greater similarity to that of STAT3-HIES, presenting with recurrent infections, bronchiectasis, eczema, high IgE, eosinophila, impaired acute-phase response, as well as skeletal and connective abnormalities (craniosynostosis and scoliosis) [34]. IL6ST encodes GP130, the co-receptor subunit for IL-6 family cytokines [18]. GP130 signaling is mediated via JAK/STAT3 pathway, accounting for the phenotypic overlap of DN IL6ST and STAT3 mutations [18].
TGFBR deficiency, known as Loeys-Dietz syndrome, is caused by monoallelic mutations in TGFBR1 and TGFBR2 genes that encode for subunits of the transforming growth factor β (TGF-β) receptors. TGF-β signaling plays a crucial role in the immune system by regulating T-cell differentiation and regulatory T-cells (Tregs) function [35]. Patients with TGFBR1 or 2 mutations showed defects of the SMAD2-dependent pathway and enhanced TGF-β signaling [36]. K. Felgentreff et al. reported in 2014 that patients with heterozygous mutations in TGFBR1 and TGFBR2 presented with immune dysregulation resembling STAT3-HIES, including highly elevated IgE, severe eczema, recurrent respiratory infections, skeletal and connective tissue abnormalities [20]. ERBIN deficiency caused by LOF mutations in ERBB2-interacting protein (ERBB2IP) was first described by J. Lyons et al. in 2017 [19]. It is an AD disorder that share similar clinical features with TGFBR deficiency. STAT3 can negatively regulate TGF-β signaling via the formation of a STAT3-ERBIN-SMAD2/3 complex. Therefore, the deficiency of ERBIN leads to the activation of TGF-β signaling [19]. These interactions may be the reason for the overlaps of their clinical manifestations.
DOCK8 Deficiency (OMIM243700)
Homozygosity or compound heterozygosity for deletions or mutations in DOCK8 gene resulting in LOF protein were first identified by Zhang et al. in 2009 [14]. DOCK8 protein, a member of the DOCK180 superfamily of guanine nucleotide exchange factors, plays an important role in actin-cytoskeletal rearrangement, cellular migration and the formation of immune synapses [37]. Dendritic cells fail to migrate to local lymph nodes for T-cell priming in Dock8 knockout murine model [38]. What is more, the DOCK8 deficiency leads to early death of T cells [39] and natural killer (NK) cells [40], which further prevents the generation of memory CD8+ T cells [41]. DOCK8 also regulates the activation of B cells and the generation of memory B cells, thus B cells from Dock8-deficient mice have difficulty in affinity maturation and antibody response [37]. Taken together DOCK8 deficiency have a broad effect on the immune system, characterized as a CID syndrome [14].
Apart from the classical manifestations of HIES like early-onset eczema, skin infections, pneumonia and elevated IgE, allergic manifestations including food allergies and asthma are more prevalent in DOCK8 deficiency than in STAT3 deficiency [42,43]. In addition, there is a susceptibility to cutaneous viral infections such as human papillomavirus (HPV), human simplex viruses (HSV), and molluscum contagiosum [43,44]. The poor control of virus infections further contributes to the higher risk of specific malignancy [44]. Progressive T-cell lymphoma and other cancers unrelated with viral infections have also been reported in DOCK8-deficienct patients [14]. Other reported manifestations include recurrent sinopulmonary infections, aortic and abdominal arterial vasculitis [45].
PGM3 Deficiency (OMIM172100)
PGM3 deficiency is caused by AR mutations of PGM3 gene and described in affected patients with atopy, immunodeficiency, and neurological impairment by Zhang et al. and Stray-Pedersen et al. in 2014 [12,46]. PGM3 is a crucial enzyme in the synthesis of uridine diphosphate N-acetylglucosamine (UDP-GlcNAc), which is an important precursor for protein glycosylation. The pathogenesis of PGM3 deficiency resulting in HIES remains to be elucidated, but the inaccurate glycosylation of protein can lead to multisystem abnormalities, such as the impaired immune function and abnormal neuronal development [12,46]. Complete loss of Pgm3 causes embryonic lethality in mice [47], whereas Pgm3 gene-trap mice can survive along with body weight loss, B-cell defects, and neurologic abnormalities [48].
PGM3 deficiency has variable clinical features ranging from the phenotype of STAT3 deficiency to severe combined immune deficiency (SCID) [46,49]. The patients present with atopic disease (eczema, allergy and asthma), recurrent skin and pulmonary infections, elevated IgE and eosinophilia, characteristic facial features (wide nostrils and prominent lips), skeletal abnormalities (scoliosis) [46,48]. Neonatal-onset SCID and neutropenia were reported in patients with deleterious PGM3 mutations [46,50]. Notably, neurocognitive impairments occurred in early life can be detected in almost all patients, including development delay, low intelligence quotient, hypotonia, ataxia, dysarthria, sensorineural hearing loss, and myoclonus [12].
DN Mutations in CARD11 (OMIM 617638)
Caspase recruitment domain family member 11 (CARD11) is a scaffold protein and involved in lymphocyte receptor signaling [51]. Heterozygous mutations in CARD11 with DN effect were first reported to cause CID and atopy features by Ma et al. in 2017, presenting with cutaneous viral infections, respiratory tract infections, early-onset moderate to severe atopic dermatitis, early-onset asthma and food allergy, elevated IgE and eosinophilia. Skeletal, dental and connective tissue abnormalities are absent [21,51].
2.1.2. The Diagnosis of HIES
The clinical heterogeneity of HIES throws challenges for the differential diagnosis. The characteristic clinical manifestation and laboratory findings of each condition can help clinician to predict the underlying gene defect in HIES, and genetic analysis is essential for definitive diagnosis [28,52].
Clinical Manifestations
The atopic dermatitis-like skin lesions of HIES can be confused with the features of severe atopic dermatitis (SAD). Atopic dermatitis is prevalent with a fifth of affected population in developed countries [53]. The disease occurs during the first year of life (often after the 2 month of age) with a long clinical course of a relapsing-remitting nature [53]. Patients with SAD can also exhibit increased IgE and eosinophilia [54]. Cutaneous infections can be the secondary lesion of atopic dermatitis. The colonization of Staphylococcal aureus is the leading cause of skin and soft-tissue infections. Eczema herpeticum with HSV infection tends to occurs in SAD patients. Children are more susceptible to molluscum contagiosum virus infection [54].
There are some red flags for the hint of HIES, including early-onset eczematous skin lesion (prior to 2 months of life), recurrent viral, bacterial, or fungal infections, present of other affected family members, and other characteristic immunological manifestations [52,55]. Chronic mucocutaneous candidiasis (CMC) was significantly more prevalent in HIES compared to SAD, particularly in DOCK8-deficient patients [56]. STAT3-HIES is featured by skeletal and connective tissue abnormalities along with characteristic facial appearance [55]. Severe allergic symptoms, skin viral infections, and malignancies are more specific for DOCK8-deficient patients. They are more susceptible to common virus infections, including HSV and HPV. As for PGM3-deficient patients with variable manifestations, neurocognitive impairments can be peculiarly detected.
The National Institutes of Health (NIH) clinical scoring system [57] based on the presence or severity of 21 clinical and laboratory findings was established and developed specially for the identification of STAT3-HIES, suggesting with scores of at least 40 points. Woellner, C. et al. [10] proposed diagnostic guidelines for STAT3-deficient HIES in 2010, considering IgE > 1000 IU/mL, clinical features (recurrent pneumonia, newborn rash, pathologic bone fractures, characteristic face, and high palate), and lack of Th17 cells or a family history for definitive HIES, along with a DN heterozygous mutation in STAT3 as the definitive diagnosis of STAT3-HIES.
Laboratory Findings and Genetic Analysis
Laboratory examinations including immunoglobulin levels, differential blood counts, as well as a lymphocyte subset analysis can be helpful during the process of diagnosis. Elevated serum IgE (often higher than 2000 IU/mL) and eosinophilia are the most typical laboratory findings [58]. For STAT3-HIES, immunoglobulin levels are usually normal, but a decreased percentage of Th17 cells (<0.3% of CD4+ T cells) in lymphocyte subset analysis can be a strong indication, especially for the differential diagnosis from atopic dermatitis [10,59]. In STAT3-associated disorders, decreased Th17 cell counts can also be detected. Lymphopenia, especially T-cell lymphopenia and less frequently B-cell lymphopenia, and low IgM serum levels are a diagnostic hallmark of DOCK8 deficiency [42]. Flow cytometry-based assays to detect the DOCK8 protein is a reliable method for its diagnosis [60]. PGM3 deficiency can present with lymphopenia and neutropenia. The T-cell lymphopenia affecting predominantly the CD4+ subset can result in reversed CD4:CD8 ratio [48].
Genetic analysis is necessary for the definitive diagnosis of HIES. The identification of the underlying gene mutations can be beneficial to disease managements and genetic counseling [58]. For those patients with suspected HIES, but lacking characteristic clinical and laboratory findings, genetic testing is essential to early diagnosis and appropriate treatment before the onset of complications.
2.1.3. The Managements of HIES
Therapeutic options for HIES should be tailored to the type of mutation and the clinical manifestations of individuals. The prevention and treatment of infections of HIES patients is pivotal to the disease management [26,49]. Early diagnosis of HIES and timely implementation of antimicrobials enable vigilance of skin or pulmonary infections, further preventing the occurrence of complications, especially for STAT3-HIES patients who lack the classical signs of infection [8]. In addition, the management of atopic dermatitis can help to protect skin barriers and prevent viral or bacterial super-infection [61]. Emollients, topical steroids and topical tacrolimus can be applied, or oral steroids can be considered in severe cases.
Oral anti-staphylococcal antibiotics combined with conventional topical agents are recommended in STAT3-HIES patients because the recurrent skin infection of Staphylococcal aureus can exacerbate eczematous dermatitis. Antifungal therapy is also needed due to the susceptibility of CMC [8,26]. For the treatment of chronic lung diseases, antibiotic and antifungal, along with proper airway management are beneficial [52]. Immunoglobulin replacement can be strongly considered in patients with recurrent pulmonary infections. The employ of hematopoietic stem cell transplantation (HSCT) in STAT3 deficiency patients remains to be controversial [8,52].
The management of DOCK8 deficiency includes screening for and treatment of complications, immunoglobulin replacement, and definitive therapy with HSCT [42,62]. The potential infections should be evaluated and actively treated with prophylactic antimicrobials. Immunoglobulin prophylaxis is recommended based on impaired long-lived antibody responses in DOCK8-deficient patients [43]. HSCT has been the only curative treatment for DOCK8 deficiency, supported by multiple successful treatment cases [63,64]. HSCT is recommended at early stages of the disease, and the complications of infection or malignancy should be aggressively managed as well [8].
2.2. Wiskott-Aldrich Syndrome (WAS, OMIM301000)
2.2.1. Inherited Pattern
WAS is a rare X-linked primary immunodeficiency disorder with an estimated incidence of 1:250,000 male birth [65]. WAS, as its pathogenic gene, was identified in 1994 by Derry et al. and located on the short arm of the X-chromosome (Xp11.23), encoding the Wiskott-Aldrich syndrome protein (WASp) [66]. In the Human Gene Mutation Database (HGMD) professional 2021.4, over 450 mutations have been detected, with missense mutations ranking as the first, followed by deletions mutations, splicing mutations, insertions, indels, and complex mutations [67].
2.2.2. Molecular Mechanism
WASp is a member of the actin nucleation-promoting factors family with multiple functional domain, and predominantly expressed in hematopoietic cells [68]. WASp mainly remains in the resting state by autoinhibition in the cytoplasm. Upon activation, WASp can interact with Arp2/3 complex through its C-terminal VCA (verprolin homology domain, central hydrophobic region, acidic region) domain, thus mediating actin polymerization and cytoskeletal remodeling [68,69,70]. What is more, WASp also involves the transcription of TBX21 gene, which encodes the key regulator of Th1 immune response [71].
Actin cytoskeleton is required for several cellular processes, including immunological synapse formation, cell motility and migration, autophagy and inflammasome regulation. Resultantly, loss of WASp activity leads to defect in hematopoietic and immune cell functions [68,72]. These deficiencies explain the abnormalities of both B- and T-cell functions, impaired monocyte chemotaxis, abnormal morphology of stimulated dendritic cells, as well as lymphopenia in patients of WAS [73]. The underlying mechanism of microthrombocytopenia is not fully understood. It is probably associated with the megakaryocyte dysfunction which leads to abnormal formed platelets [74]. Recent research proposed its correlation with friend leukemia integration 1 (FLI1), which can be induced by WASp depletion and play a crucial role in megakaryocyte differentiation [75].
2.2.3. Clinical Manifestations
WAS has a large clinical spectrum characteristically associated with immunodeficiency, eczema, thrombocytopenia, a high incidence of autoimmunity and malignancies. The clinical presentation usually occurs after birth, presenting with bruises and spontaneous or prolonged bleeding due to low numbers of circulating platelets [70]. Eczema occurs in most affected patients during their first year of life. Bacterial infections (frequently Streptococcus pneumoniae, Neisseria meningitidis, and Haemophilus influenzae) manifesting as otitis media or pneumonia are common in WAS patients. They also have higher risks of viral infections and opportunistic infections [70,72,76]. Autoimmune manifestations including hemolytic anemia, vasculitis, IgA nephropathy, and inflammatory bowel disease have been reported [72,76]. Patients with WAS also have higher risks of malignancies with lymphoma and leukemia [72].
X-linked thrombocytopenia (XLT) is a relatively milder disorder caused by WAS gene mutations, with less problems in eczema and infections [69]. A 5-point severity score of WAS has been developed to differentiate XLT (score 0.5–3) from the classical WAS (score 3–5), on the basis of severity of thrombocytopenia, eczema, immunodeficiency and infections, and presence of autoimmunity or malignancy [69]. Genetic analysis and protein levels can help to differentiate XLT and WAS. According to the strong phenotype-genotype correlation, patients with mutations resulting in the absence or truncation of WASp are more likely to have the classical WAS phenotype, while patients with mutations that allowed the expression of normal-sized mutated WASp, often in reduced quantities, developed the XLT phenotype [73,77,78].
2.2.4. The Diagnosis of WAS
The diagnosis of WAS should be considered in any boy with petechiae, easy bruising, spontaneous or prolonged bleeding, with or without eczema and/or recurrent infections, autoimmunity diseases, and/or malignancies [69]. The Laboratory examinations and genetic analysis can help to diagnose definitively.
Complete blood cell count and peripheral blood smear are necessary [76]. The reduced platelet count and platelet volume show favorable diagnostic value. However, the degree of thrombocytopenia is variable and sometimes lies in normal level, which can disturb the diagnosis [72]. Reduced platelet volume is pathognomonic for WAS/XLT, but mean platelet volume is difficult to measure in moderate to severe thrombocytopenia [76]. Peripheral blood smear demonstrating small platelets is more reliable in this condition [69].
Flow cytometry can investigate WASp expression in peripheral blood leukocytes, which predict the severity of clinical phenotype and can be a quick diagnostic indicator of WAS if the result shows a complete absence of WASp [79]. Genetic analysis is the gold standard for definitive diagnosis of WAS [69,76].
2.2.5. The Managements of WAS
The management of WAS including supportive therapy and definitive therapy should be tailored to patients’ clinical manifestations and disease severity [69,72].
Once a child is diagnosed with WAS, definitive therapy should be considered, either HSCT or stem cell gene therapy (GT) [72]. Early definitive treatment is recommended for patients with absent WASp and WAS mutation consistent with classical WAS, regardless of their initial clinical presentation [72]. The HSCT for WAS patients has achieved excellent outcomes, with survival rates over 97% [80,81,82]. The post-transplant complications include graft-versus-host disease (GvHD), infections in the context of prior splenectomy, autoimmunity and infertility [72,80,81,82]. Gene therapy is a valuable therapeutic alternative of HSCT with lower risks of rejection reaction, GvHD, and infections [79]. Available data from recent GT clinical trials using lentiviral vectors supported the safety and efficacy of this therapeutic approach in the short- and medium-term, but information on the very long-term safety of GT (>10 years) is still limited [83,84,85].
Supportive therapy consists of the management of infection, thrombocytopenia, and autoimmune diseases before definitive treatment [72,76]. The employment of prophylactic antimicrobials and the strategies of anti-viral or anti-fungal medications should be decided based on the conditions of individuals [69]. It is recommended that all patients with classical WAS commence immunoglobulin replacement treatment after diagnosis [72]. Before definitive treatment, platelet transfusion is not supported in the absence of active bleeding due to the development of anti-platelet and anti-human leucocyte antigen antibodies after platelet transfusion [72]. The occurrence of autoimmunity can be managed with supportive care and immunosuppression [76]. For mild to moderate eczema, emollients, topical steroids and topical tacrolimus are recommended for the management. Oral steroids can be considered in severe cases [69,76].
2.3. Immune Dysregulation, Polyendocrinopathy, Enteropathy, X-Linked Syndrome (IPEX Syndrome, OMIM304790)
2.3.1. Inherited Pattern
IPEX syndrome is a rare monogenic inborn error of immunity with the incidence below 1:1,000,000 [86]. It is a hemizygous disorder caused by the forkhead box P3 (FOXP3) mutations firstly identified in 2001 and inherited predominantly in boys in an X-linked recessive manner. FOXP3 gene is located in the centromeric region of chromosome (Xp11.23) and consists of 12 exons [86,87,88,89]. Over 90 mutations have been reported in HGMD, including missense/nonsense mutations, splicing mutations, small deletions and insertions [90].
2.3.2. Molecular Mechanism
As a member of the forkhead/winged-helix family of transcriptional regulators, FOXP3 is encoded by FOXP3 gene and acts as a DNA-binding protein during transcription that controls the development and function of Treg cells, thereby contributing to peripheral immune tolerance [91]. Patients with defect of FOXP3 show low-level Treg cells and elevated IgE levels, as well as higher rates of autoantibodies [89,92]. Impaired Treg cell function is associated with the development of autoimmunity, severe food allergy, and an unbalanced Th2 response [92,93]. The mice with mutations in Foxp3 gene demonstrated similar phenotype with IPEX syndrome and multi-organ lymphocytic infiltration [94].
2.3.3. Clinical Manifestations
The clinical features of IPEX syndrome are dominated by autoimmunity and characterized by a triad of enteropathy, polyendocrinopathy, and dermatitis [95]. Enteropathy associated with watery diarrhea often occurs within the first year of life, with the mean onset age of 7.8 months [96]. Insulin-dependent diabetes mellitus (IDDM) and thyroiditis are most commonly reported endocrinopathy and can be concurrent [89]. Other autoimmune disorders, including autoimmune hemolytic anemia, neutropenia, autoimmune gastrointestinal disorders and cutaneous disorders have also been reported [89,95]. Skin disorders are widespread, manifesting as eczema, exfoliative dermatitis, psoriasiform dermatitis, pemphigoid nodularis, cheilitis, onychodystrophy, or alopecia [88]. Life-threatening infections including sepsis, meningitis, peritonitis, and pneumonia occurred in over a quarter of IPEX syndrome patients, mostly infected by Staphylococcus, Cytomegalovirus, and Candida [88,89].
2.3.4. The Diagnosis of IPEX
IPEX syndrome should be considered in male patients with enteropathy, dermatitis, autoimmunity, endocrinopathy. The combination of laboratory findings and genetic analysis are pivotal for diagnosis and differential diagnosis [86].
Elevated serum IgE level and eosinophilia are typical laboratory findings [88]. Flow cytometry can detect decreased Treg lymphocyte subset, while absolute lymphocyte counts and CD4+ and CD8+ lymphocyte subsets are usually normal [86]. A higher rate of autoantibodies mostly against enterocyte can be detected in patients with IPEX syndrome [89].
2.3.5. The Managements of IPEX
The management of patients with IPEX syndrome can be divided into two strategies. The primary therapeutic approach is allogeneic HSCT. For those patients without HSCT, immunosuppressive therapy and supportive therapy are recommended [86,88].
HSCT is the only potentially curative therapy for IPEX syndrome [87]. Early HSCT before the development of organ damages seems to lead to better outcomes in patients. Two long-term follow-ups respectively reported a 10-year survival of 72.8% [88] and a 15-year survival of 73.2% [95] in patients who underwent HSCT. Complications of post-HSCT include acute or chronic GvHD, transplant-related toxicity, and multiple infections [95].
A broad spectrum of supportive therapies should be carried out at once, comprising the prevention and treatment of infections (e.g., prophylactic antibiotics), as well as the measurements for endocrine disorders (e.g., insulin and/or thyroid hormones) and autoimmune disorder (e.g., intravenous immunoglobulins) [87]. Immunosuppressive therapy, often a combination of systematic corticosteroids and immunosuppressive drugs including calcineurin inhibitors, rapamycin and some monoclonal antibodies, can lead to remission of autoimmunity and plays a crucial role for the management of initial acute manifestation. Nutritional support, either enteral or parenteral is necessary for patients receiving IS [95].
2.4. STAT5B Deficiency (OMIM618985, 245590)
2.4.1. Inherited Pattern
STAT5B deficiency is considered as one of IPEX-like syndromes with similar phenotype [88,89]. The signal transducer and activator of transcription 5B (STAT5B) gene mutations have been described with both AD and AR inheritance by Kofoed et al. in 2003 [97] and Klammt et al. in 2018 [98], respectively. In HGMD, 19 mutations have been detected, including missense/nonsense mutations, deletions, insertions, and regulatory mutations [99].
2.4.2. Molecular Mechanism
The protein STAT5B encoded by STAT5B is involved in homeostasis and maintenance of Treg cells [100,101], as well as terminal NK cell maturation and NK cell lytic synapse formation [102]. Therefore, STAT5B deficiency can impair T-cell homeostasis and natural NK-cell function [102]. What is more, JAK2-STAT5B pathway involves the signal transduction of circulating growth hormone (GH), which is important for normal human growth [103].
2.4.3. Clinical Manifestations
The clinical presentation of STAT5B deficiency is featured by severe postnatal growth failure with growth hormone insensitivity (GHI) syndrome and insulin-like growth factor (IGF)-I deficiency [104]. Other manifestations include severe eczema, immune deficiency (e.g., recurrent infections), and autoimmune disease (e.g., juvenile idiopathic arthritis, autoimmune thyroiditis, idiopathic thrombocytic purpura), which are thought to be associated with Treg-cell dysfunction [100]. STAT5B deficiency with heterozygous mutation shows a milder clinical GHI without severe immune and pulmonary problems [98].
2.4.4. The Diagnosis of STAT5B Deficiency
Patients should be suspected with STAT5B deficiency if they present with both growth disorders and immunological abnormalities.
Patients usually have normal levels of GH at baseline, and show elevated GH concentration after stimulation [100]. In contrast, the concentration of serum IGF-I remains to be low even upon administration of GH [100]. Elevated prolactin levels can be detected [104]. Immunological related examinations can show moderate lymphopenia with low numbers of NK and T cells, as well as elevated B cell populations and IgG levels [101].
2.4.5. The Managements of STAT5B Deficiency
The managements of STAT5B deficiency have two sections according to its clinical presentations. One is therapies for postnatal growth failure, GH therapy is not ideal due to the GHI [97], while IGF-I therapy is presumed to be effective [100]. The management of immunological disorders is another crucial section, including treatments for infections and immunosuppressive therapies for autoimmune conditions.
2.5. The Atypical Complete Digeorge Syndrome
2.5.1. Inherited Pattern
DiGeorge syndrome (DGS) is a congenital disorder mostly caused by a hemizygous deletion of chromosome 22q11.2, a frequent chromosomal 1.5–3 Mb deletion with an incidence of 1 in 4000–6000 livebirths [105,106]. Sulagna C, et al. proposed that the deletion was attributable to aberrant meiotic exchange event [107]. More than 35 genes are present within the deleted region, among which TBXI was identified to be a major genetic determinant [108].
2.5.2. Molecular Mechanism
The mechanism of atypical complete DGS still remains unclear. It has been proposed that patients with atypical complete DGS have extrathymic differentiation and oligoclonal expansion of a small number of T cells [109,110]. Other hypothesis suggested that autoreactive T cells might result in impaired immune tolerance and defective immune regulation [111].
2.5.3. Clinical Manifestations
DGS is a congenital malformation characterized by cardiac anomalies, hypoparathyroidism, and thymus hypoplasia or aplasia [112,113]. DGS can be divided into complete and incomplete DGS according to the phenotype of thymus abnormalities [114]. Patients with complete DGS are athymic and account less than 1% of all DGS patients [114]. The atypical complete DGS is a subset of complete DGS. The patients manifest with eczematous dermatitis, oligoclonal T-cell expansion, and lymphadenopathy [111,112,113,114,115]. The cutaneous manifestations are diverse, varying from diffuse erythroderma with scale formation to a greasy seborrheic dermatitis appearance [115].
2.5.4. The Diagnosis of Atypical Complete DGS
Cutaneous manifestations, lymphadenopathy, along with laboratory findings of an oligoclonal T-cell population in the peripheral blood can help for early diagnosis [115]. Unique histopathological findings contribute to differential diagnosis, with characteristics of satellite cell necrosis and dyskeratosis, as well as brisk peri- and intra-eccrine inflammation [115]. Analysis of the immunophenotype and T cell receptor Vβ repertoire of T lymphocytes help to determine whether the patient is athymic [116,117], further discriminate atypical complete DGS from other conditions such as partial DGS or Omenn syndrome.
2.5.5. The Managements of Atypical Complete DGS
Immunosuppressive therapy is required for atypical complete DGS [110,111]. Thymic transplantation can correct severe immunodeficiency. M. Louise, et al. reported the 1-year survival rate of 73% in 50 infants with complete DGS after transplantation of thymus tissue. Most deaths occurred prior to development of T cells due to severe infection [118]. They also reported the efficiency of thymus transplantation with immunosuppression in six infant patients. Among them, four patients with atypical complete DGS received immune reconstitution as polyclonal T-cell populations developed in 1 year after transplantation [119]. The reported adverse events after transplantation include hashimoto thyroiditis, autoimmune hepatitis, cytopenias, enteritis/colitis, and autoimmune hepatitis [119,120].
2.6. Omennsyndrome. (OS, OMIM603554)
2.6.1. Inherited Pattern
OS is in a genetically heterogeneous condition with a frequency of 1:5,000,000 [121]. It was first confirmed to be related with LOF mutations in RAG1 and RAG2 genes [122,123]. Up to date, DCLRE1C [124], RMRP [125], IL7RA [126] and IL2RG genes are linked to OS.
2.6.2. Molecular Mechanism
All pathogenic genes associated with OS impede the formation and maturation of immunology system. Take RAGs as an example. A process termed V(D)J site-specific recombination takes responsibility of diversity of the immune repertoire which is generated by somatic assembly of the antigen receptor variable gene segments. The specific binding is essential to this conserved recombination. RAG1/RAG2 dysfunction impedes the initiation of V(D)J recombination and leads to complete absence of mature B and T cells [127,128].
2.6.3. Clinical Manifestations
OS is in SCID condition characterized by neonatal diffuse erythematous/eczematous rash, exfoliative dermatitis with dry and flaky skin, hypotrichosis, lymphadenopathy, hepatosplenomegaly, repeated infections, eosinophilia and elevated IgE [129,130]. Patients would frequently present with chronic diarrhea and pneumonitis in the first year of life and then failure to thrive [131,132]. Without proper treatment, OS shows a high mortality.
2.6.4. The Diagnosis of OS
Cutaneous biopsy shows that early skin biopsy is beneficial to diagnosing OS. Key histopathological pathological features include epidermal hyperplasia, necrotic keratinocytes, acanthosis and parakeratosis and dermal lymphocytic infiltrate (CD3+) [133]. Abnormal expansion of T-cell clones in peripheral blood and tissues, along with a low number of CD19+ B cells are also the characteristic markers. Genetic analysis gains the concise diagnosis of OS.
2.6.5. The Managements of OS
Outcomes of OS are greatly improved by early diagnosis and treatment. As the first-line therapy, HSCT shows fine improvement in survival rates [134]. Additionally, immunosuppressive drugs including prednisone and cyclosporin A are introduced to treat OS patients [131].
2.7. X-Linked Agammaglobulinemia (XLA, OMIM300755)
2.7.1. Inherited Pattern
XLA is an immunodeficiency characterized by maturational arrest of B cells development with the incidence of 1:100,000–200,000. XLA, the most common genetic agammaglobulinemia, comprises 85% of subjects of agammaglobulinemia [135]. Bruton tyrosine kinase (BTK), the affected gene in XLA, was firstly discovered in 1993 by S Tsukada [136] and D Vetrie [137]. It is reported that missense mutations accounting for the most common mutation type in BTK, followed with insertion or deletions, nonsense mutations, splice site mutations, and large deletions [138,139]. Agammaglobulinemia with other inherited pattern were also concluded: AR agammaglobulinemia was associated with mutations in several other genes such as IGHM, IGLL1, CD79A, CD79B, BLNK, PIK3R1, and TCF3 genes; AD-related agammaglobulinemia was also found with LRRC8A and TCF3 gene mutations [135].
2.7.2. Molecular Mechanism
BTK, expressed mainly in hematopoietic cells, is a non-receptor kinase that plays a crucial role in both B cell development and function of mature B cells [140]. Through the interaction of its pleckstrin homology domain with phosphatidylinositol-3,4,5-triphosphate (PIP3), BTK is transiently recruited to the membrane. After phosphorylated by SYK or SRC family at Y551 site, BTK autophosphorylates at Y223, which stabilizes the active conformation and fully activates BTK kinase activity. BTK is essential to B cell receptor signaling, B cell development in bone marrow and B cell malignancies. Alterations in the BTK can lead to primary B cell immunodeficiency and humoral immunodeficiency with decreased or diminished immunoglobulin production [141].
2.7.3. Clinical Manifestations
Due to a severe block of B cell development in the bone marrow, decreased B cells in the circulation and almost completely absent antibodies in the serum are frequently discovered in XLA patients. Resultantly, recurrent infections including respiratory infections, otitis, sinusitis, skin infections, cervical lymphadenitis, epididymitis, osteomyelitis, urinary tract infection, gastroenteritis and chronic diarrhea, invasive infections (sepsis, meningitis, encephalitis), arthritis and hepatitis are described [139].
Cutaneous manifestations in XLA mainly present with severe skin infections like chronic skin ulcers, refractory cellulitis, skin abscesses, skin edema and discoloration and folliculitis [142,143]. Wheal and erythema, typical atopic dermatitis, and skin cancer were also reported in XLA patients [144,145,146].
2.7.4. The Diagnosis of XLA
A male with paucity of lymphoid tissue and clinical history such as recurrent infections should be noted. Laboratory findings include: (1) marked reduction of serum immunoglobulins including IgG, IgM and IgA; (2) markedly reduced numbers of CD19-/CD20-expresssing B lymphocytes in the peripheral circulation; (3) low antibody titers to vaccine antigens; (4) severe neutropenia. However, assessment of BTK and family history of XLA are mandatory for the diagnosis of XLA [147,148].
2.7.5. The Managements of XLA
XLA patients should be treated with replacement immunoglobulin and prophylactic antibiotics to prevent infections. Intravenous immunoglobulin (IVIG) at intervals of 2–4 weeks or subcutaneous immunoglobulin (SCIG) at intervals of 1–14 days should be used and adjusted by IgG level associated with clinical course. It is reported that increased IgG level is beneficial to decreased pneumonia incidence in patients with primary immunodeficiency [149,150].
3. Inherited Metabolic Diseases
3.1. Acrodermatitis Enteropathy (AE, OMIM201100)
3.1.1. Inherited Pattern
Being a rare AR disorder, AE was first described by Brandt et al. in 1936 [151] and confirmed to be related with LOF mutations in SLC39A4 gene, which codes the zinc (Zn) transporter ZIP4 [152,153]. In the HGMD, a total of 56 mutations have been described including: missense/nonsense, and splicing site mutations; small insertions/deletions; and few gross rearrangements [154]. Inherited AE occurs worldwide with an estimated incidence of 1 per 500,000 children and has no apparent preference for race or sex [155].
3.1.2. Molecular Mechanism
Zn is stably maintained in the weight of 2–3 g in a human body. In all organs, skin is the third most Zn-abundant tissue while skeletal muscle, bones, liver and skin contains 60%, 30%, 5% and 5% respectively [156]. Flawed function of ZIP4 leads to disability of Zn absorption within the gastrointestinal tract and causes Zn deficiency. Compromised immunity is one of the first signs of zinc deficiency and the role of zinc in the immune system is known for a firm molecular basis [157].The skin inflammation pattern of AE is essentially similar to contact dermatitis induced by multiple irritants in dietary zinc-deficient mice [158]. As the particular locations which are primarily exposed to external irritants, perioral, acral and anogenital areas seem to reasonably present typical lesions [158].
3.1.3. Clinical Manifestation
AE patients often suffer from acral and perioral dermatitis, alopecia, and diarrhea [159,160,161,162]. For the reason that breast milk usually contains adequate Zn with a high concentration (>3 mg/L) but progressively declines to <1 mg/L by 6 months [163], typical clinical manifestations frequently present at the time of weaning from breast or formula feeding. However, the complete symptoms are seen only in one-third of patients [164].
Cutaneous manifestations of AE consist of sharply demarcated eczematous or psoriasiform plaques, which symmetrically start with perioral and retro-auricular sites and spread to extremities. Vescicolo-bullous, pustular, and erosive lesions may also develop in the sites of mouth, eyes, noses, the scalp and perineum. Nail, oral and ocular disorders may occur [165]. Additional features could be loss of appetite, irritability and apathy; development retardation, testicular atrophy; neuropsychiatric features, hyposmia and hypogeusia [166]. Without proper management, extensive erosions associated with a predisposition for fungal (especially Candida albicans) or bacterial (e.g., Staphylococcus aureus, Pseudomonas aeruginosa [164]) colonization may follow.
3.1.4. The Diagnosis of AE
Skin histological findings of AE are nonspecific [167,168,169]. The most common findings are alternating orthokeratosis and parakeratosis. Decreased stratum granulosum, acanthosis and focal acantholysis could also be seen. Dilated capillaries and sparse lymphocytic infiltration in the papillary dermis are the subsequent findings. Secondary skin lesions may present the characteristic of the bullae, while chronic lesions sometimes show a psoriasiform pattern [164,169,170].
Fasting Zn levels of less than 70 μg/dL (10.71 μmol/L) or post-meal levels of less than 65 μg/dL (9.95 μmol/L) are indicative for Zn deficiency. However, low Zn levels in plasma or serum, which only accounts for 0.1% in the whole body, do not directly indicate Zn deficiency [167], which indicates the importance of SLC39A4 gene analysis. Additionally, Zn-dependent enzymes, such as alkaline phosphatase, can be measured as a hallmark [171].
3.1.5. The Managements of AE
Oral administration of zinc sulfate is the base treatment for AE. Initial Zn dose of 5–10 mg/kg/d and maintenance doses of 1–2 mg/kg/d are recommended [170]. Clinical improvement can be observed after just a few days, whereas hereditary AE requires life-long replacement therapy [172]. However, the interruption of treatment could inevitably lead to relapses and the cutaneous manifestations are the first to recur [165].
3.2. Multiple Carboxylase Deficiency
Biotin, an water-soluble vitamin, is necessary for the activation of carboxylases and crucial in glucose, amino acid and fatty acid metabolism [173]. Two metabolic syndromes with decreased biotin, caused by disorders of holocarboxylase or biotinidase synthetase, are referred to multiple carboxylase deficiency [174]. Age of onset is used to differentiate between holocarboxylase synthetase (HLCS) deficiency and biotinidase (BT) deficiency. HLCS deficiency frequently occurs hours to weeks of birth while biotinidase deficiency generally presents after 3 months [175]. however, there are exceptions for both disorders.
3.2.1. Holocarboxylase Synthetase Deficiency (HLCSD, OMIM253270)
Inherited Pattern
HLCSD, which typically presents at birth or affects children below the age of 2 months, is a rare AR inborn error of the biotin metabolism. The estimated incidence of the disease is less than 1 in 200,000 live births. Y Suzuki, et al. cloned the human HLCS cDNA and mapped HLCS gene to chromosome 21q22.1 in 1994 [176]. In the HGMD, a total of 64 mutations have been described including: missense/nonsense, and splicing site mutations; small insertions/deletions; and few gross deletions and complex rearrangements [177]. And there is a relationship between clinical biotin responsiveness and the residual activity of HLCS [178].
Molecular Mechanism
Since the early part of the last century, biotin has been recognized as an essential nutrient. Biotin requirement is fulfilled in through diet, endogenous reutilization of biotin and capture of biotin generated in the intestinal flora [179]. However, biotin deficiency is associated with protein malnutrition [180], and marginal biotin deficiency in pregnant women may be teratogenic [181].
Free-form biotin can directly enter the biotin pool and is used to convert four carboxylases from the inactive to the active form. HLCS takes the responsibility for covalently linking biotin to four carboxylases: propionyl-CoA carboxylase (PCC), 3-methylcrotonyl-CoA carboxylase (MCC), pyruvate carboxylase (PC) and acetyl-CoA carboxylase (ACC) [182,183]. Failure to approach biotin causes reduced activity of these carboxylases and leads to multiple carboxylase deficiency. Hlcs-knockout mice is embryonically lethal in both early stages and midway through pregnancy due to a depletion of biotinylated carboxylases [184].
Clinical Manifestations
The typical symptoms of HLCSD manifest as metabolic acidosis (emesis, hypotonia, lethargy, seizures, hyperammonemia, tachypnea and coma). Cutaneous lesions present with localized erythematous dermatitis [185], psoriasis-like dermatitis [186], periorificial and intertriginous dermatitis [187], keratoconjunctivitis and nonscarring diffuse alopecia with loss of hair luster. The erythema is well defined around the eyes, nose, mouth and on the perineum. Other symptoms such as feeding problems, a variety of central nervous system abnormalities and developmental delay could also be seen in HLCSD.
The Diagnosis of HLCSD
Cutaneous histopathology shows superficial perivascular lymphocytic infiltration with regular acanthosis, hyperkeratosis and parakeratosis and hypogranulosis [187], which is nonspecific.
HLCSD diagnosis is suggested by blood enzymatic determination, urine organic acids and verified by HLCS gene variants [188]. Tandem mass spectroscopy analysis of analytes such as amino acids, acylcarnitines and multiplex enzyme analysis on dried blood spots are the mainstay of testing for HLCSD. The detection of elevated hydroxypentanoylcarnitine (C5-OH) could be screened for HLCSD. Additionally, urine organic acid profile may demonstrate elevated lactic, 3-OH isovaleric, 3-OH propionic, methylcitric, and tiglylglycine consistent with LOF biotin-attached carboxylases [183]. However, biotinidase and zinc levels are normal.
The Managements of HLCSD
Without early diagnosis and proper treatment, HLCSD is associated with high morbidity and mortality [189]. However, prompt biotin supplement is highly associated with regression of the disease and the fine clinical outcomes, especially before antennal stage [190]. Oral biotin is the effective treatment and the doses ranging from 3 to 200 mg/day [191,192]. The metabolic disorders could be corrected within 2 days to 2 weeks and continued oral biotin therapy is essential to the improvement of the prognosis [183].
3.2.2. Biotinidase Deficiency (BTD, OMIM253260)
Inherited Pattern
Mutations in the BTD gene produce the AR disorder, BTD, known as the late-onset MCD whose incidence is 1 in 60,000 births [193,194]. 284 variants of BTD that alter BT activity have been reported so far. All types of variants have been observed: missense/nonsense, splicing, regulatory, small insertion/deletion/indel, gross deletion and complex rearrangement [195]. Six most severe and common pathogenic allelic variants of BTD are founded: c.98_104delinsTCC, c.511G > A and c.1330G > C, c.1612C > T, c.1368A > C, c.1330G > C [196].
Molecular Mechanism
BT is the enzyme that recycles the biotin [197]. Dietary protein-linked and carboxylase-bounded biotin must be degraded to release biocytin. Small biotinyl-peptides which could be further cleaved by BT, release the free-form biotin to the pool [198]. Patients with inherited BTD are unable to utilize the biotin in food or recycle the biotin in carboxylase turnover.
Clinical Manifestations
The clinical presentation of BTD includes hypotonia, seizures, organic aciduria, mild hyperammonemia, feeding problems, developmental delay and breathing problems such as hyperventilation, laryngeal stridor, and apnea [199]. Cutaneous manifestation exhibits the following symptoms: eczematous skin rash and palmoplantar keratosis, diffuse alopecia or fragile hair [200], and recurrent viral or fungal infections. Seborrheic dermatitis-like eruptions and scaly erythematous plaques over the flexors and perioral areas can be detected [201,202]. The secondary lesions such as crusting, lichenification, and open lesions can be seen [203]. Some patients develop neurological disorders, involving ataxia, mental retardation, hearing loss, optic atrophy, myelopathy, and Leigh syndrome [204]. Symptoms of untreated BTD frequently appear between the ages of 1 week and 10 years, with a mean age of 3.5 months [205].
The Diagnosis of BTD
Quantitative determination of BT enzyme activity in serum/plasma with BTD genetic analysis is the golden standard to diagnose the BTD. Other biochemical abnormalities such as metabolic ketolactic acidosis, hyperammonemia, organic aciduria and elevated carnitine (in plasma and/or urinary) are also useful to identifying BTD [199]. Individuals with severe BTD have lower than 10% mean normal serum enzyme activity, while partial BTD patients have 10–30% of mean normal serum BT activity [206]. However, the dermatopathology shows no clinical specificity.
The Managements of BTD
Excluding the neurological abnormality, all clinical and biochemical manifestations of BTD can be alleviated or reversed with biotin (5–20 mg/day) [197]. As the child grows, the dosage of biotin decreases. The index of urine organic acid is used to estimate whether the biotin is enough [207].
3.3. Prolidase Deficiency (PD, OMIM170100)
3.3.1. Inherited Pattern
PD is a rare AR multisystem disorder associated with PEPD mutations, which is verified by A Tanoue in 1990 [208]. The estimated occurrence of PD is 1 case in 2,000,000 births [209]. More than 90 cases are reported with 42 PEPD variants [210].
3.3.2. Molecular Mechanism
Prolidase, a cytosolic manganese-dependent peptidase, is the only enzyme which hydrolyzes the tertiary amide bond involved in imidodipeptides. So prolidase performs crucially in protein synthesis and degradation, especially of proteins rich in iminopeptides such as fibrillar collagens [211,212].
3.3.3. Clinical manifestations
Clinical symptoms of PD patients show a wide range from mild to severe. Cutaneous lesions such as refractory ulcerations, telangiectasis, impetigo-like eruptions, necrotic papules, premature graying of the hair, photosensitivity, erythematous maculopapular rash, and hypertrichosis can be observed in PD [213]. Anaphylaxis to multiple food allergens, allergic rhinitis, and asthma, shows severe atopy [214]. Other symptoms include bone disorders, mental retardation, respiratory infections and facial dysmorphisms, autoimmunity, and splenomegaly [215].
3.3.4. The Diagnosis of PD
For diagnosing PD, methods are described as followed: (1) based on the determination of enzyme activity in leukocytes, erythrocytes, and skin fibroblast culture; (2) elevated levels of imidodipeptids in uria; (3) PEPD variants with classical clinical symptoms [216]. Skin biopsy found nonspecific changes including slight hyperkeratosis, edema, and parakeratosis with neutrophil leukocyte and lymphocyte infiltration [213].
3.3.5. The Management of PD
The treatment of PD is symptomatic and has no curative regimen. Topical treatments including proline, GH, and tacrolimus, are used to replace prolidase or stop ulcerative progression [217,218,219]. Usage of systemic immunosuppressive medications or packed red blood cells transfusions have been also reported [220,221].
4. Rare Syndromes
4.1. Severe Dermatitis Multiple Allergies and Metabolic Wasting (SAM) Syndrome (SAM Syndrome, OMIM615508)
4.1.1. Inherited Pattern
SAM syndrome is a rare genodermatitis caused by recessive homozygous or compound heterozygous LOF mutations in desmoglein 1 (DSG1) gene, or dominant heterozygous mutations in desmoplakin (DSP) gene [222,223]. Since Samuelov L et al. first found the DSG1-related SAM syndrome [222], a total of 16 individuals were reported.
4.1.2. Molecular Mechanism
Desmoplakin and desmogleins form the integral part of desmosomes, which serves as an anchor for adjacent epithelial cells to link to one another. Proteins in desmosomes play a role in cell signaling and skin barrier function [224].
4.1.3. Clinical Manifestations
SAM syndrome presents with a broad spectrum of skin phenotypes and multi-system manifestations. Cutaneous symptoms include erythroderma, palmoplantar keratoderma, and ichthyosis. Besides, pruritus, hypotrichosis, woolly hair, and nail abnormalities are common in SAM syndrome. Other disorders such as food sensitization, dental abnormalities, developmental delay, recurrent infections, elevated IgE, eosinophilia, ophthalmic abnormalities cardiac abnormalities, gastrointestinal problems, brain abnormalities, can also be observed in SAM syndrome [223,225,226,227,228].
4.1.4. The Diagnosis of SAM Syndrome
Skin histopathology shows psoriasiform hyperplasia and hyperkeratosis, which presents no specificity. Elevated eosinophils, reduced 25-hydroxy-vitamin D and elevated IgE could be detected in SAM syndrome while all of these could not be used as the diagnosis standards. Genetic analysis with the typical clinical manifestation is the authoritative diagnostic criteria for SAM syndrome [228].
4.1.5. The Managements of SAM Syndrome
No proper treatment is recommended to the SAM syndrome. However, several groups made novel trials: DSP-related SAM syndrome performed a favorable response to ustekinumab [229]; oral acitretin and topical pimecrolimus released cutaneous eruption and oral gabapentin relieved pruritus [230]; successful treatment with secukinumab in SAM syndrome and SAM-like syndrome were also reported [231].
4.2. Netherton Syndrome (NS, OMIM256500)
4.2.1. Inherited Pattern
NS is a rare, multisystemic, AR genodermatosis with an incidence of one patient in 200,000 newborns, which is also thought to be the cause of up to 18% of congenital erythrodermas. It was first depicted by Comel in 1949 [232] and Netherton in 1958 [233]. So far, a total of 108 variants are found, which involve all types of mutations [234]. In addition, nine lethal variants are depicted: c.153delT, c.238insG, c.375_376delAT, c.C649T, c.997C > T, c.1111C > T, c1431–12G > A, c.715insT, and 375delAT [235].
4.2.2. Molecular Mechanism
SPINK5 (serine protease inhibitor of Kazal type 5), localized on chromosome 5q32, encodes LEKTI (lympho-epithelial Kazal type related inhibitor), a multidomain serine protease inhibitor expressed in the thymus and epithelia [236]. Deficiency in LEKTI causes the increased activity of the epidermal serine proteases kallikrein 5 (KLK5), which degrades DSG1, leading to epidermal desquamation and altered skin barrier function [237]. Hyperactivity of KLK5 could increase the expression of thymic stromal lymphopoietin (TSLP), which elevates levels of IgE [238].
4.2.3. Clinical Manifestation
The clinical manifestation of NS is characterized by a triad of trichorrhexis invaginata, icthyosis linearis circumflexa (ILC), and an atopic diathesis. ILC performs as pruritic erythematous and serpiginous plaques with double-edged desquamation showing polycyclic and serpiginous borders. The recalcitrant intensely pruritic dermatosis frequently comes with hair abnormalities, such as bamboo hairs (trichorrhexis invaginata), golf tee, and matchstick hairs. Atopic dermatitis, eczema-like eruptions, pruritus, asthma, allergic rhinitis, angioedema, elevated IgE, and/or hypereosinophilia and sensitivity to multiple allergens are included in the atopic diathesis [239]. Other symptoms present as a mental deficiency, neurologic disorders, development delay, recurrent infections, aminoaciduria, and hypergammaglobulinemia [240,241].
4.2.4. The Diagnosis of NS
Skin biopsy usually exhibits hyperkeratosis, acanthosis, focal hypergranulosis, and superficial perivascular lymphocytic infiltration, which is apparently unspecific [242]. Hair shaft abnormality could be uneasy to find because only 20–50% of hairs are affected. Laboratory findings are significant in increased peripheral eosinophilia and elevated IgE levels [240]. Finding gene mutations in SPINK5 is essential to diagnosing NS.
4.2.5. The Managements of NS
NS could be confused with atopic dermatitis but shows an unsatisfactory response to topical corticosteroid treatment. However, topical tacrolimus and pimecrolimus or IVIG show good effects in NS treatment [241,243]. UVB treatment could also lead to positive improvement of lesions [244]. Oral retinoids are also a nice try to treat NS patients [245].
4.3. Peeling Skin Syndrome Type B (PSS-B, OMIM 270300)
4.3.1. Inherited Pattern
Generalized PSS (peeling skin syndrome) has been subclassified into a noninflammatory type (peeling skin syndrome type A), and an inflammatory type (PSS-B) [246]. PSS-B, a rare AR genodermatosis, results from LOF mutations in the CDSN gene, which was first found in 2010 by Vinzenz Oji et al. [247]. So far, a total of 18 individuals with 16 variants are reported, including missense/nonsense, splicing, small deletions or insertions, and gross deletions [248].
4.3.2. Molecular Mechanism
Corneodesmosin (CDSN), an extracellular component of corneodesmosomes, locates in the corneodesmosomal core and is covalently linked to the cornified envelope of corneocytes. CDSN is highly expressed in hair follicles and cornified epithelia, and plays an important role in maintaining desmosome integrity, the proper development and function of the skin barrier, and normal hair follicle formation [249,250]. Cdsn deficiency mice showed stratum corneum detachment resulting from abnormal desmosome formation [251].
4.3.3. Clinical Manifestations
Cutaneous symptoms of PSS-B present with generalized, pruritic scales and ichthyosiform erythroderma which begin at birth and evolve to mild erythematous areas worsening in summer, with recurrent skin infections and vesiculations. Along with hypotrichosis, hairs of PSS-B patients can be easily plucked. Nail abnormalities include onychodystrophy and white nail changes [247,252,253,254,255,256,257]. Other symptoms including food allergies, repeated episodes of angioedema, urticaria and/or asthma, feeding difficulty, failure to thrive, hypereosinophilia and hypoproteinemia, and microadenoma of the pituitary gland, nephrocalcinosis with hypercalciuria, dysphonia, and intellectual disability can be observed in PSS-B [258].
4.3.4. The Diagnosis of PSS-B
Histopathology reveals the extensive subcorneal cleavage in the epidermis with irregular acanthosis [247]. Elevated IgE and CDSN gene analysis are important clues to diagnosing PSS-B [259].
4.3.5. The Managements of PSS-B
Ustekinumab [258] and low-potency topical corticosteroid ointment, oral retinoids fail to alleviate the skin lesions, while topical 0. 005% calcipotriol ointment could reduce skin peeling and erythema [260]. PSS can usually be well managed with topical treatments and hygienic measures alone, and it seems to improve with age [259].
5. Conclusions
A topic dermatitis-like lesions can frequently present in patients with genetic diseases. In this review, three main categories including immunological disorders, metabolic diseases, and rare syndromes are discussed. We have presented the diagnostic clues that simplify the differential diagnosis, especially for immunological disorders, shown in Figure 1 and Table 1.
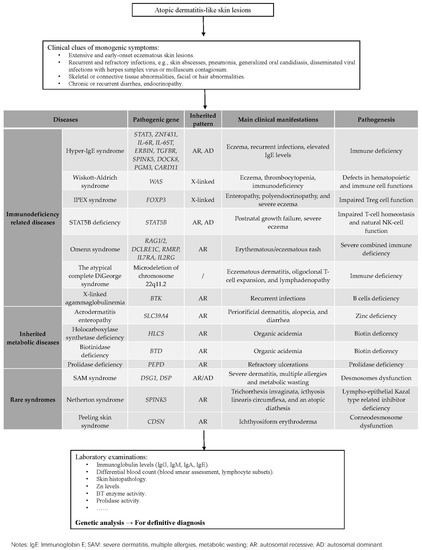
Figure 1.
How to distinguish diseases performed with atopic dermatitis-like lesions.

Table 1.
The genetic and clinical spectrums of hyper IgE syndromes.
The clinical indications of an underlying PIDD conclude severe and/or early-onset eczema, features of immunodeficiency. Notably, the phenotypes of PIDDs and atopic dermatitis partially overlap, presenting with increased serum IgE levels, eosinophilia, and eczema lesions, which indicates the shared immune pathways. Indeed, atopic dermatitis is an inflammatory skin disease caused by multiple factors, among which dysregulated type 2 immunity driven by Th2 cells plays a crucial role. Here, HIES including STAT3 deficiency and STAT3-related deficiency is also associated with activated Th2 responses and decreased Th17 generations; CARD11 deficiency, IPEX syndrome, STAT5B deficiency, and OS affect Treg diversity or function that regulates Th2 immunity. WAS and DOCK8 deficiency are associated with impaired actin assembly and immunological synapse formation, and XLA shows B cells deficiency. These discoveries provide additional insights into the pathogenesis of atopic disorders. For the diagnosis of PIDDs with atopic dermatitis-like lesions, laboratory tests that consist of blood counts, lymphocyte subsets count and serum immunoglobulin levels are recommended for suspected patients. Low lymphocyte counts can be indicative of OS, XLA, or DOCK8 deficiency, and WAS is supported if showing low mean platelet volume. Flow cytometric analysis is a valuable tool especially for identifying STAT3-HIES that low Th17 cell counts can point toward STAT3 related disorders. However, assessment of genes and family history of affected subjects are mandatory for differentiation.
As for metabolic genodermatitis, indicators in metabolism show great importance: Zn deficiency is associated with the phenotype of perioral and perianal dermatitis, diarrhea highly suggests AE resulting from SLC39A4 mutations; organic acidemia reminds us to pay attention to MCD related to BT and HLCS variants; reduced prolidase enzyme activity and elevated levels of imidodipeptids hint us with PEPD-associated PD.
Apart from all the immunologic and metabolic diseases, rare syndromes should always be taken into consideration in diagnosing eczema-like dermatitis. However, it is so hard to distinguish rare syndromes due to their complex clinical manifestations. Genetic analysis provides us with clear diagnosis clues: SAM syndromes are linked with DSG1 and DSP mutations; NS is due to LOF mutations in the SPINK5 gene; CDSN variants are always detected in PSS-B patients.
The recognition of these pathogeneses shed new light on clinical and mechanistic associations among allergy, metabolism, and desquamation. Comprehensive knowledge of the regulation of immunological, metabolic, and keratinization functions and the potential therapeutic agents are depicted. Emphatically, genetic analysis is still crucial and essential for definitive diagnosis. Early diagnosis allows for prompt and specific treatments prior to the onset of complications. Aside from the direct influence on the affected subject, research on the atopic dermatitis-like syndrome not only broadens our understanding of human biology but also promotes development in the management of common conditions.
Author Contributions
Writing: original draft preparation, review, and editing: C.P. and A.Z.; writing: review and editing: M.L.; Table 1 preparation: A.Z. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
Abbreviation
| PIDDs | Primary immunodeficiency diseases |
| HIES | Hyper- IgE syndromes |
| IgE | Immunoglobulin E |
| WAS | Wiskott-Aldrich syndrome |
| IPEX | Immune dysregulation, polyendocrinopathy, enteropathy, X-linked |
| OS | Omenn syndrome |
| XLA | X-linked agammaglobulinemia |
| AD | Autosomal dominant |
| AR | Autosomal recessive |
| STAT | Signal transducer and activator of transcription-3 |
| DN | Dominant negative |
| PGM3 | Phosphoglucomutase 3 |
| DOCK8 | Dedicator of cytokinesis-8 |
| IUIS | International Union of Immunological Societies |
| SAD | Severe atopic dermatitis |
| LOF | Homozygous loss-of-function |
| ZNF | Zinc finger protein |
| TGF-β | Transforming growth factor β |
| Tregs | Regulatory T-cells |
| ERBB2IP | ERBB2-interacting protei |
| CID | Combined immunodeficiency |
| HPV | Human papillomavirus |
| HSV | Human simplex viruses |
| CARD11 | Caspase recruitment domain family member 11 |
| CMC | Chronic mucocutaneous candidiasis |
| HSCT | Hematopoietic stem cell transplantation |
| WASp | Wiskott-Aldrich syndrome protein |
| HGMD | Human Gene Mutation Database |
| XLT | X-linked thrombocytopenia |
| GT | Gene therapy |
| GvHD | Graft-versus-host disease |
| FOXP3 | Forkhead box P3 |
| GH | Growth hormone |
| GHI | Growth hormone insensitivity |
| DGS | DiGeorge syndrome |
| BTK | Bruton tyrosine kinase |
| IVIG | Intravenous immunoglobulin |
| SCIG | subcutaneous immunoglobulin |
| AE | Acrodermatitis enteropathy |
| HLCSD | Holocarboxylase synthetase deficiency |
| BTD | Biotinidase deficiency |
| PD | Prolidase deficiency |
| SAM syndrome | Severe dermatitis multiple allergies and metabolic wasting syndrome |
| NS | Netherton syndrome |
| SPINK5 | Serine protease inhibitor of Kazal type 5 |
| TSLP | Thymic stromal lymphopoietin |
| ILC | Icthyosis linearis circumflexa |
| PSS-B | Peeling skin syndrome type B |
| CDSN | Corneodesmosin |
References
- Ständer, S. Atopic Dermatitis. N. Engl. J. Med. 2021, 384, 1136–1143. [Google Scholar] [CrossRef] [PubMed]
- Nutten, S. Atopic dermatitis: Global epidemiology and risk factors. Ann. Nutr. Metab. 2015, 66, 8–16. [Google Scholar] [CrossRef] [PubMed]
- Lehman, H. Skin manifestations of primary immune deficiency. Clin. Rev. Allergy Immunol. 2014, 46, 112–119. [Google Scholar] [CrossRef] [PubMed]
- Al-Herz, W.; Zainal, M.; Nanda, A. A Prospective Survey of Skin Manifestations in Children With Inborn Errors of Immunity From a National Registry Over 17 Years. Front. Immunol. 2021, 12, 751469. [Google Scholar] [CrossRef]
- Berron-Ruiz, A.; Berron-Perez, R.; Ruiz-Maldonado, R. Cutaneous markers of primary immunodeficiency diseases in children. Pediatr. Dermatol. 2000, 17, 91–96. [Google Scholar] [CrossRef]
- Dhouib, N.G.; Ben Khaled, M.; Ouederni, M.; Ben-Mustapha, I.; Kouki, R.; Besbes, H.; Barbouche, M.R.; Mellouli, F.; Bejaoui, M. Cutaneous Manifestations of Primary Immunodeficiency Diseases in Tunisian Children. Mediterr. J. Hematol. Infect Dis. 2018, 10, e2018065. [Google Scholar] [CrossRef]
- López-Quintero, W.; Cleves, D.; Gomez-Vasco, J.D.; Pérez, P.; Patiño, J.; Medina-Valencia, D.; Pachajoa, H.; Torres-Canchala, L.; Vidal, A.; Olaya, M. Skin manifestations in pediatric patients with primary immunodeficiency diseases (PIDs) in a tertiary care hospital in Colombia. World Allergy Organ. J. 2021, 14, 100527. [Google Scholar] [CrossRef]
- Bergerson, J.R.E.; Freeman, A.F. An Update on Syndromes with a Hyper-IgE Phenotype. Immunol. Allergy Clin. N. Am. 2019, 39, 49–61. [Google Scholar] [CrossRef]
- Al-Shaikhly, T.; Ochs, H.D. Hyper IgE syndromes: Clinical and molecular characteristics. Immunol. Cell Biol. 2019, 97, 368–379. [Google Scholar] [CrossRef]
- Woellner, C.; Gertz, E.M.; Schäffer, A.A.; Lagos, M.; Perro, M.; Glocker, E.O.; Pietrogrande, M.C.; Cossu, F.; Franco, J.L.; Matamoros, N.; et al. Mutations in STAT3 and diagnostic guidelines for hyper-IgE syndrome. J. Allergy Clin. Immunol. 2010, 125, 424–432.e428. [Google Scholar] [CrossRef]
- Minegishi, Y.; Saito, M.; Tsuchiya, S.; Tsuge, I.; Takada, H.; Hara, T.; Kawamura, N.; Ariga, T.; Pasic, S.; Stojkovic, O.; et al. Dominant-negative mutations in the DNA-binding domain of STAT3 cause hyper-IgE syndrome. Nature 2007, 448, 1058–1062. [Google Scholar] [CrossRef]
- Zhang, Y.; Yu, X.; Ichikawa, M.; Lyons, J.J.; Datta, S.; Lamborn, I.T.; Jing, H.; Kim, E.S.; Biancalana, M.; Wolfe, L.A.; et al. Autosomal recessive phosphoglucomutase 3 (PGM3) mutations link glycosylation defects to atopy, immune deficiency, autoimmunity, and neurocognitive impairment. J. Allergy Clin. Immunol. 2014, 133. 1400–1409, 1409.e1401–1409.e1405. [Google Scholar] [CrossRef]
- Engelhardt, K.R.; McGhee, S.; Winkler, S.; Sassi, A.; Woellner, C.; Lopez-Herrera, G.; Chen, A.; Kim, H.S.; Lloret, M.G.; Schulze, I.; et al. Large deletions and point mutations involving the dedicator of cytokinesis 8 (DOCK8) in the autosomal-recessive form of hyper-IgE syndrome. J. Allergy Clin. Immunol. 2009, 124. 1289–1302.e1284. [Google Scholar] [CrossRef]
- Zhang, Q.; Davis, J.C.; Lamborn, I.T.; Freeman, A.F.; Jing, H.; Favreau, A.J.; Matthews, H.F.; Davis, J.; Turner, M.L.; Uzel, G.; et al. Combined immunodeficiency associated with DOCK8 mutations. N. Engl. J. Med. 2009, 361, 2046–2055. [Google Scholar] [CrossRef]
- Tangye, S.G.; Al-Herz, W.; Bousfiha, A.; Chatila, T.; Cunningham-Rundles, C.; Etzioni, A.; Franco, J.L.; Holland, S.M.; Klein, C.; Morio, T.; et al. Human Inborn Errors of Immunity: 2019 Update on the Classification from the International Union of Immunological Societies Expert Committee. J. Clin. Immunol. 2020, 40, 24–64. [Google Scholar] [CrossRef]
- Béziat, V.; Li, J.; Lin, J.X.; Ma, C.S.; Li, P.; Bousfiha, A.; Pellier, I.; Zoghi, S.; Baris, S.; Keles, S.; et al. A recessive form of hyper-IgE syndrome by disruption of ZNF341-dependent STAT3 transcription and activity. Sci. Immunol. 2018, 3, eaat4956. [Google Scholar] [CrossRef]
- Spencer, S.; Köstel Bal, S.; Egner, W.; Lango Allen, H.; Raza, S.I.; Ma, C.A.; Gürel, M.; Zhang, Y.; Sun, G.; Sabroe, R.A.; et al. Loss of the interleukin-6 receptor causes immunodeficiency, atopy, and abnormal inflammatory responses. J. Exp. Med. 2019, 216, 1986–1998. [Google Scholar] [CrossRef]
- Schwerd, T.; Twigg, S.R.F.; Aschenbrenner, D.; Manrique, S.; Miller, K.A.; Taylor, I.B.; Capitani, M.; McGowan, S.J.; Sweeney, E.; Weber, A.; et al. A biallelic mutation in IL6ST encoding the GP130 co-receptor causes immunodeficiency and craniosynostosis. J. Exp. Med. 2017, 214, 2547–2562. [Google Scholar] [CrossRef]
- Lyons, J.J.; Liu, Y.; Ma, C.A.; Yu, X.; O’Connell, M.P.; Lawrence, M.G.; Zhang, Y.; Karpe, K.; Zhao, M.; Siegel, A.M.; et al. ERBIN deficiency links STAT3 and TGF-β pathway defects with atopy in humans. J. Exp. Med. 2017, 214, 669–680. [Google Scholar] [CrossRef]
- Felgentreff, K.; Siepe, M.; Kotthoff, S.; von Kodolitsch, Y.; Schachtrup, K.; Notarangelo, L.D.; Walter, J.E.; Ehl, S. Severe eczema and Hyper-IgE in Loeys-Dietz-syndrome-contribution to new findings of immune dysregulation in connective tissue disorders. Clin. Immunol. 2014, 150, 43–50. [Google Scholar] [CrossRef]
- Ma, C.A.; Stinson, J.R.; Zhang, Y.; Abbott, J.K.; Weinreich, M.A.; Hauk, P.J.; Reynolds, P.R.; Lyons, J.J.; Nelson, C.G.; Ruffo, E.; et al. Germline hypomorphic CARD11 mutations in severe atopic disease. Nat. Genet. 2017, 49, 1192–1201. [Google Scholar] [CrossRef]
- O’Shea, J.J.; Holland, S.M.; Staudt, L.M. JAKs and STATs in immunity, immunodeficiency, and cancer. N. Engl. J. Med. 2013, 368, 161–170. [Google Scholar] [CrossRef]
- Van de Veen, W.; Krätz, C.E.; McKenzie, C.I.; Aui, P.M.; Neumann, J.; van Noesel, C.J.M.; Wirz, O.F.; Hagl, B.; Kröner, C.; Spielberger, B.D.; et al. Impaired memory B-cell development and antibody maturation with a skewing toward IgE in patients with STAT3 hyper-IgE syndrome. Allergy 2019, 74, 2394–2405. [Google Scholar] [CrossRef]
- Meyer-Bahlburg, A.; Renner, E.D.; Rylaarsdam, S.; Reichenbach, J.; Schimke, L.F.; Marks, A.; Tcheurekdjian, H.; Hostoffer, R.; Brahmandam, A.; Torgerson, T.R.; et al. Heterozygous signal transducer and activator of transcription 3 mutations in hyper-IgE syndrome result in altered B-cell maturation. J. Allergy Clin. Immunol. 2012, 129, 559–562. [Google Scholar] [CrossRef]
- Keupp, K.; Li, Y.; Vargel, I.; Hoischen, A.; Richardson, R.; Neveling, K.; Alanay, Y.; Uz, E.; Elcioğlu, N.; Rachwalski, M.; et al. Mutations in the interleukin receptor IL11RA cause autosomal recessive Crouzon-like craniosynostosis. Mol. Genet. Genomic. Med. 2013, 1, 223–237. [Google Scholar] [CrossRef]
- Tsilifis, C.; Freeman, A.F.; Gennery, A.R. STAT3 Hyper-IgE Syndrome-an Update and Unanswered Questions. J. Clin. Immunol. 2021, 41, 864–880. [Google Scholar] [CrossRef]
- Lorenzini, T.; Giacomelli, M.; Scomodon, O.; Cortesi, M.; Rivellini, V.; Dotta, L.; Soresina, A.; Dellepiane, R.M.; Carrabba, M.; Cossu, F.; et al. Autosomal-dominant hyper-IgE syndrome is associated with appearance of infections early in life and/or neonatal rash: Evidence from the Italian cohort of 61 patients with elevated IgE. J. Allergy Clin. Immunol. Pract. 2019, 7. 2072–2075.e2074. [Google Scholar] [CrossRef]
- Vaseghi-Shanjani, M.; Smith, K.L.; Sara, R.J.; Modi, B.P.; Branch, A.; Sharma, M.; Lu, H.Y.; James, E.L.; Hildebrand, K.J.; Biggs, C.M.; et al. Inborn errors of immunity manifesting as atopic disorders. J. Allergy Clin. Immunol. 2021, 148, 1130–1139. [Google Scholar] [CrossRef]
- Meixner, I.; Hagl, B.; Kröner, C.I.; Spielberger, B.D.; Paschos, E.; Dückers, G.; Niehues, T.; Hesse, R.; Renner, E.D. Retained primary teeth in STAT3 hyper-IgE syndrome: Early intervention in childhood is essential. Orphanet J. Rare Dis. 2020, 15, 244. [Google Scholar] [CrossRef]
- Eberting, C.L.; Davis, J.; Puck, J.M.; Holland, S.M.; Turner, M.L. Dermatitis and the newborn rash of hyper-IgE syndrome. Arch. Dermatol. 2004, 140, 1119–1125. [Google Scholar] [CrossRef]
- Chandesris, M.O.; Azarine, A.; Ong, K.T.; Taleb, S.; Boutouyrie, P.; Mousseaux, E.; Romain, M.; Bozec, E.; Laurent, S.; Boddaert, N.; et al. Frequent and widespread vascular abnormalities in human signal transducer and activator of transcription 3 deficiency. Circ. Cardiovasc. Genet. 2012, 5, 25–34. [Google Scholar] [CrossRef] [PubMed]
- Arora, M.; Bagi, P.; Strongin, A.; Heimall, J.; Zhao, X.; Lawrence, M.G.; Trivedi, A.; Henderson, C.; Hsu, A.; Quezado, M.; et al. Gastrointestinal Manifestations of STAT3-Deficient Hyper-IgE Syndrome. J. Clin. Immunol. 2017, 37, 695–700. [Google Scholar] [CrossRef]
- Frey-Jakobs, S.; Hartberger, J.M.; Fliegauf, M.; Bossen, C.; Wehmeyer, M.L.; Neubauer, J.C.; Bulashevska, A.; Proietti, M.; Fröbel, P.; Nöltner, C.; et al. ZNF341 controls STAT3 expression and thereby immunocompetence. Sci. Immunol. 2018, 3, eaat4941. [Google Scholar] [CrossRef] [PubMed]
- Béziat, V.; Tavernier, S.J.; Chen, Y.H.; Ma, C.S.; Materna, M.; Laurence, A.; Staal, J.; Aschenbrenner, D.; Roels, L.; Worley, L.; et al. Dominant-negative mutations in human IL6ST underlie hyper-IgE syndrome. J. Exp. Med. 2020, 217, e20191804. [Google Scholar] [CrossRef] [PubMed]
- Oh, S.A.; Li, M.O. TGF-β: Guardian of T cell function. J. Immunol. 2013, 191, 3973–3979. [Google Scholar] [CrossRef] [PubMed]
- Schepers, D.; Tortora, G.; Morisaki, H.; MacCarrick, G.; Lindsay, M.; Liang, D.; Mehta, S.G.; Hague, J.; Verhagen, J.; van de Laar, I.; et al. A mutation update on the LDS-associated genes TGFB2/3 and SMAD2/3. Hum. Mutat. 2018, 39, 621–634. [Google Scholar] [CrossRef] [PubMed]
- Randall, K.L.; Lambe, T.; Johnson, A.L.; Treanor, B.; Kucharska, E.; Domaschenz, H.; Whittle, B.; Tze, L.E.; Enders, A.; Crockford, T.L.; et al. Dock8 mutations cripple B cell immunological synapses, germinal centers and long-lived antibody production. Nat. Immunol. 2009, 10, 1283–1291. [Google Scholar] [CrossRef]
- Harada, Y.; Tanaka, Y.; Terasawa, M.; Pieczyk, M.; Habiro, K.; Katakai, T.; Hanawa-Suetsugu, K.; Kukimoto-Niino, M.; Nishizaki, T.; Shirouzu, M.; et al. DOCK8 is a Cdc42 activator critical for interstitial dendritic cell migration during immune responses. Blood 2012, 119, 4451–4461. [Google Scholar] [CrossRef]
- Singh, A.K.; Eken, A.; Hagin, D.; Komal, K.; Bhise, G.; Shaji, A.; Arkatkar, T.; Jackson, S.W.; Bettelli, E.; Torgerson, T.R.; et al. DOCK8 regulates fitness and function of regulatory T cells through modulation of IL-2 signaling. JCI Insight. 2017, 2, e94275. [Google Scholar] [CrossRef]
- Kearney, C.J.; Vervoort, S.J.; Ramsbottom, K.M.; Freeman, A.J.; Michie, J.; Peake, J.; Casanova, J.L.; Picard, C.; Tangye, S.G.; Ma, C.S.; et al. DOCK8 Drives Src-Dependent NK Cell Effector Function. J. Immunol. 2017, 199, 2118–2127. [Google Scholar] [CrossRef]
- Randall, K.L.; Chan, S.S.; Ma, C.S.; Fung, I.; Mei, Y.; Yabas, M.; Tan, A.; Arkwright, P.D.; Al Suwairi, W.; Lugo Reyes, S.O.; et al. DOCK8 deficiency impairs CD8 T cell survival and function in humans and mice. J. Exp. Med. 2011, 208, 2305–2320. [Google Scholar] [CrossRef]
- Aydin, S.E.; Kilic, S.S.; Aytekin, C.; Kumar, A.; Porras, O.; Kainulainen, L.; Kostyuchenko, L.; Genel, F.; Kütükcüler, N.; Karaca, N.; et al. DOCK8 deficiency: Clinical and immunological phenotype and treatment options-a review of 136 patients. J. Clin. Immunol. 2015, 35, 189–198. [Google Scholar] [CrossRef]
- Engelhardt, K.R.; Gertz, M.E.; Keles, S.; Schäffer, A.A.; Sigmund, E.C.; Glocker, C.; Saghafi, S.; Pourpak, Z.; Ceja, R.; Sassi, A.; et al. The extended clinical phenotype of 64 patients with dedicator of cytokinesis 8 deficiency. J. Allergy Clin. Immunol. 2015, 136, 402–412. [Google Scholar] [CrossRef]
- Dimitrova, D.; Freeman, A.F. Current Status of Dedicator of Cytokinesis-Associated Immunodeficiency: DOCK8 and DOCK2. Dermatol. Clin. 2017, 35, 11–19. [Google Scholar] [CrossRef]
- Al-Herz, W.; Ragupathy, R.; Massaad, M.J.; Al-Attiyah, R.; Nanda, A.; Engelhardt, K.R.; Grimbacher, B.; Notarangelo, L.; Chatila, T.; Geha, R.S. Clinical, immunologic and genetic profiles of DOCK8-deficient patients in Kuwait. Clin. Immunol. 2012, 143, 266–272. [Google Scholar] [CrossRef]
- Stray-Pedersen, A.; Backe, P.H.; Sorte, H.S.; Mørkrid, L.; Chokshi, N.Y.; Erichsen, H.C.; Gambin, T.; Elgstøen, K.B.; Bjørås, M.; Wlodarski, M.W.; et al. PGM3 mutations cause a congenital disorder of glycosylation with severe immunodeficiency and skeletal dysplasia. Am. J. Hum. Genet. 2014, 95, 96–107. [Google Scholar] [CrossRef]
- Greig, K.T.; Antonchuk, J.; Metcalf, D.; Morgan, P.O.; Krebs, D.L.; Zhang, J.G.; Hacking, D.F.; Bode, L.; Robb, L.; Kranz, C.; et al. Agm1/Pgm3-mediated sugar nucleotide synthesis is essential for hematopoiesis and development. Mol. Cell. Biol. 2007, 27, 5849–5859. [Google Scholar] [CrossRef]
- Sassi, A.; Lazaroski, S.; Wu, G.; Haslam, S.M.; Fliegauf, M.; Mellouli, F.; Patiroglu, T.; Unal, E.; Ozdemir, M.A.; Jouhadi, Z.; et al. Hypomorphic homozygous mutations in phosphoglucomutase 3 (PGM3) impair immunity and increase serum IgE levels. J. Allergy Clin. Immunol. 2014, 133. 1410–1419, 1419.e1411–1419.e1413. [Google Scholar] [CrossRef]
- Fadil, I.; Ben-Ali, M.; Jeddane, L.; Barbouche, M.R.; Bousfiha, A.A. The Seven STAT3-Related Hyper-IgE Syndromes. J. Clin. Immunol. 2021, 41, 1384–1389. [Google Scholar] [CrossRef]
- Bernth-Jensen, J.M.; Holm, M.; Christiansen, M. Neonatal-onset T(-)B(-)NK(+) severe combined immunodeficiency and neutropenia caused by mutated phosphoglucomutase 3. J. Allergy Clin. Immunol. 2016, 137, 321–324. [Google Scholar] [CrossRef]
- Dadi, H.; Jones, T.A.; Merico, D.; Sharfe, N.; Ovadia, A.; Schejter, Y.; Reid, B.; Sun, M.; Vong, L.; Atkinson, A.; et al. Combined immunodeficiency and atopy caused by a dominant negative mutation in caspase activation and recruitment domain family member 11 (CARD11). J. Allergy Clin. Immunol. 2018, 141. 1818–1830.e1812. [Google Scholar] [CrossRef]
- Stadler, P.C.; Renner, E.D.; Milner, J.; Wollenberg, A. Inborn Error of Immunity or Atopic Dermatitis: When to be Concerned and How to Investigate. J. Allergy Clin. Immunol. Pract. 2021, 9, 1501–1507. [Google Scholar] [CrossRef]
- Weidinger, S.; Novak, N. Atopic dermatitis. Lancet 2016, 387, 1109–1122. [Google Scholar] [CrossRef]
- Thijs, J.L.; Strickland, I.; Bruijnzeel-Koomen, C.; Nierkens, S.; Giovannone, B.; Csomor, E.; Sellman, B.R.; Mustelin, T.; Sleeman, M.A.; de Bruin-Weller, M.S.; et al. Moving toward endotypes in atopic dermatitis: Identification of patient clusters based on serum biomarker analysis. J. Allergy Clin. Immunol. 2017, 140, 730–737. [Google Scholar] [CrossRef]
- Schimke, L.F.; Sawalle-Belohradsky, J.; Roesler, J.; Wollenberg, A.; Rack, A.; Borte, M.; Rieber, N.; Cremer, R.; Maass, E.; Dopfer, R.; et al. Diagnostic approach to the hyper-IgE syndromes: Immunologic and clinical key findings to differentiate hyper-IgE syndromes from atopic dermatitis. J. Allergy Clin. Immunol. 2010, 126. 611–617.e611. [Google Scholar] [CrossRef]
- Kasap, N.; Celik, V.; Isik, S.; Cennetoglu, P.; Kiykim, A.; Eltan, S.B.; Nain, E.; Ogulur, I.; Baser, D.; Akkelle, E.; et al. A set of clinical and laboratory markers differentiates hyper-IgE syndrome from severe atopic dermatitis. Clin. Immunol. 2021, 223, 108645. [Google Scholar] [CrossRef]
- Grimbacher, B.; Holland, S.M.; Gallin, J.I.; Greenberg, F.; Hill, S.C.; Malech, H.L.; Miller, J.A.; O’Connell, A.C.; Puck, J.M. Hyper-IgE syndrome with recurrent infections--an autosomal dominant multisystem disorder. N. Engl. J. Med. 1999, 340, 692–702. [Google Scholar] [CrossRef] [PubMed]
- Castagnoli, R.; Lougaris, V.; Giardino, G.; Volpi, S.; Leonardi, L.; La Torre, F.; Federici, S.; Corrente, S.; Cinicola, B.L.; Soresina, A.; et al. Inborn errors of immunity with atopic phenotypes: A practical guide for allergists. World Allergy Organ. J. 2021, 14, 100513. [Google Scholar] [CrossRef] [PubMed]
- Ma, C.S.; Chew, G.Y.; Simpson, N.; Priyadarshi, A.; Wong, M.; Grimbacher, B.; Fulcher, D.A.; Tangye, S.G.; Cook, M.C. Deficiency of Th17 cells in hyper IgE syndrome due to mutations in STAT3. J. Exp. Med. 2008, 205, 1551–1557. [Google Scholar] [CrossRef] [PubMed]
- Pai, S.Y.; de Boer, H.; Massaad, M.J.; Chatila, T.A.; Keles, S.; Jabara, H.H.; Janssen, E.; Lehmann, L.E.; Hanna-Wakim, R.; Dbaibo, G.; et al. Flow cytometry diagnosis of dedicator of cytokinesis 8 (DOCK8) deficiency. J. Allergy Clin. Immunol. 2014, 134, 221–223. [Google Scholar] [CrossRef]
- Lehman, H.; Gordon, C. The Skin as a Window into Primary Immune Deficiency Diseases: Atopic Dermatitis and Chronic Mucocutaneous Candidiasis. J. Allergy Clin. Immunol. Pract. 2019, 7, 788–798. [Google Scholar] [CrossRef]
- Biggs, C.M.; Keles, S.; Chatila, T.A. DOCK8 deficiency: Insights into pathophysiology, clinical features and management. Clin. Immunol. 2017, 181, 75–82. [Google Scholar] [CrossRef]
- McDonald, D.R.; Massaad, M.J.; Johnston, A.; Keles, S.; Chatila, T.; Geha, R.S.; Pai, S.Y. Successful engraftment of donor marrow after allogeneic hematopoietic cell transplantation in autosomal-recessive hyper-IgE syndrome caused by dedicator of cytokinesis 8 deficiency. J. Allergy Clin. Immunol. 2010, 126. 1304–1305.e1303. [Google Scholar] [CrossRef]
- Kuşkonmaz, B.; Ayvaz, D.; Tezcan, İ.; Yüce, A.; Sanal, Ö.; Çetinkaya, D.U. Successful hematopoietic stem cell transplantation after myeloablative conditioning in three patients with dedicator of cytokinesis 8 deficiency (DOCK8) related Hyper IgE syndrome. Bone Marrow Transplant. 2018, 53, 339–343. [Google Scholar] [CrossRef]
- Perry, G.S., 3rd; Spector, B.D.; Schuman, L.M.; Mandel, J.S.; Anderson, V.E.; McHugh, R.B.; Hanson, M.R.; Fahlstrom, S.M.; Krivit, W.; Kersey, J.H. The Wiskott-Aldrich syndrome in the United States and Canada (1892–1979). J. Pediatr. 1980, 97, 72–78. [Google Scholar] [CrossRef]
- Derry, J.M.; Ochs, H.D.; Francke, U. Isolation of a novel gene mutated in Wiskott-Aldrich syndrome. Cell 1994, 78, 635–644. [Google Scholar] [CrossRef]
- The Human Gene Mutation Database. Available online: http://www.hgmd.cf.ac.uk/ac/gene.php?gene=WAS (accessed on 1 July 2022).
- Thrasher, A.J.; Burns, S.O. WASP: A key immunological multitasker. Nat. Rev. Immunol. 2010, 10, 182–192. [Google Scholar] [CrossRef]
- Ochs, H.D.; Filipovich, A.H.; Veys, P.; Cowan, M.J.; Kapoor, N. Wiskott-Aldrich syndrome: Diagnosis, clinical and laboratory manifestations, and treatment. Biol. Blood Marrow Transplant. 2009, 15, 84–90. [Google Scholar] [CrossRef]
- Candotti, F. Clinical Manifestations and Pathophysiological Mechanisms of the Wiskott-Aldrich Syndrome. J. Clin. Immunol. 2018, 38, 13–27. [Google Scholar] [CrossRef]
- Taylor, M.D.; Sadhukhan, S.; Kottangada, P.; Ramgopal, A.; Sarkar, K.; D’Silva, S.; Selvakumar, A.; Candotti, F.; Vyas, Y.M. Nuclear role of WASp in the pathogenesis of dysregulated TH1 immunity in human Wiskott-Aldrich syndrome. Sci. Transl. Med. 2010, 2, 37ra44. [Google Scholar] [CrossRef]
- Rivers, E.; Worth, A.; Thrasher, A.J.; Burns, S.O. How I manage patients with Wiskott Aldrich syndrome. Br. J. Haematol. 2019, 185, 647–655. [Google Scholar] [CrossRef]
- Jin, Y.; Mazza, C.; Christie, J.R.; Giliani, S.; Fiorini, M.; Mella, P.; Gandellini, F.; Stewart, D.M.; Zhu, Q.; Nelson, D.L.; et al. Mutations of the Wiskott-Aldrich Syndrome Protein (WASP): Hotspots, effect on transcription, and translation and phenotype/genotype correlation. Blood 2004, 104, 4010–4019. [Google Scholar] [CrossRef]
- Ingrungruanglert, P.; Amarinthnukrowh, P.; Rungsiwiwut, R.; Maneesri-le Grand, S.; Sosothikul, D.; Suphapeetiporn, K.; Israsena, N.; Shotelersuk, V. Wiskott-Aldrich syndrome iPS cells produce megakaryocytes with defects in cytoskeletal rearrangement and proplatelet formation. Thromb. Haemost. 2015, 113, 792–805. [Google Scholar] [CrossRef]
- Wang, C.; Sample, K.M.; Gajendran, B.; Kapranov, P.; Liu, W.; Hu, A.; Zacksenhaus, E.; Li, Y.; Hao, X.; Ben-David, Y. FLI1 Induces Megakaryopoiesis Gene Expression Through WAS/WIP-Dependent and Independent Mechanisms; Implications for Wiskott-Aldrich Syndrome. Front. Immunol. 2021, 12, 607836. [Google Scholar] [CrossRef]
- Cavannaugh, C.; Ochs, H.D.; Buchbinder, D. Diagnosis and clinical management of Wiskott-Aldrich syndrome: Current and emerging techniques. Exp. Rev. Clin. Immunol. 2022. [Google Scholar] [CrossRef]
- Liu, D.W.; Zhang, Z.Y.; Zhao, Q.; Jiang, L.P.; Liu, W.; Tu, W.W.; Song, W.X.; Zhao, X.D. Wiskott-Aldrich syndrome/X-linked thrombocytopenia in China: Clinical characteristic and genotype-phenotype correlation. Pediatr. Blood Cancer 2015, 62, 1601–1608. [Google Scholar] [CrossRef]
- Moratto, D.; Giliani, S.; Notarangelo, L.D.; Mazza, C.; Mazzolari, E.; Notarangelo, L.D. The Wiskott-Aldrich syndrome: From genotype-phenotype correlation to treatment. Exp. Rev. Clin. Immunol. 2007, 3, 813–824. [Google Scholar] [CrossRef]
- Ferrua, F.; Marangoni, F.; Aiuti, A.; Roncarolo, M.G. Gene therapy for Wiskott-Aldrich syndrome: History, new vectors, future directions. J. Allergy Clin. Immunol. 2020, 146, 262–265. [Google Scholar] [CrossRef] [PubMed]
- Elfeky, R.A.; Furtado-Silva, J.M.; Chiesa, R.; Rao, K.; Amrolia, P.; Lucchini, G.; Gilmour, K.; Adams, S.; Bibi, S.; Worth, A.; et al. One hundred percent survival after transplantation of 34 patients with Wiskott-Aldrich syndrome over 20 years. J. Allergy Clin. Immunol. 2018, 142. 1654–1656.e1657. [Google Scholar] [CrossRef] [PubMed]
- Slatter, M.A.; Rao, K.; Abd Hamid, I.J.; Nademi, Z.; Chiesa, R.; Elfeky, R.; Pearce, M.S.; Amrolia, P.; Worth, A.; Flood, T.; et al. Treosulfan and Fludarabine Conditioning for Hematopoietic Stem Cell Transplantation in Children with Primary Immunodeficiency: UK Experience. Biol. Blood Marrow Transplant. 2018, 24, 529–536. [Google Scholar] [CrossRef] [PubMed]
- Ozsahin, H.; Cavazzana-Calvo, M.; Notarangelo, L.D.; Schulz, A.; Thrasher, A.J.; Mazzolari, E.; Slatter, M.A.; Le Deist, F.; Blanche, S.; Veys, P.; et al. Long-term outcome following hematopoietic stem-cell transplantation in Wiskott-Aldrich syndrome: Collaborative study of the European Society for Immunodeficiencies and European Group for Blood and Marrow Transplantation. Blood 2008, 111, 439–445. [Google Scholar] [CrossRef]
- Hacein-Bey Abina, S.; Gaspar, H.B.; Blondeau, J.; Caccavelli, L.; Charrier, S.; Buckland, K.; Picard, C.; Six, E.; Himoudi, N.; Gilmour, K.; et al. Outcomes following gene therapy in patients with severe Wiskott-Aldrich syndrome. JAMA 2015, 313, 1550–1563. [Google Scholar] [CrossRef]
- Burroughs, L.M.; Petrovic, A.; Brazauskas, R.; Liu, X.; Griffith, L.M.; Ochs, H.D.; Bleesing, J.J.; Edwards, S.; Dvorak, C.C.; Chaudhury, S.; et al. Excellent outcomes following hematopoietic cell transplantation for Wiskott-Aldrich syndrome: A PIDTC report. Blood 2020, 135, 2094–2105. [Google Scholar] [CrossRef]
- Ferrua, F.; Cicalese, M.P.; Galimberti, S.; Giannelli, S.; Dionisio, F.; Barzaghi, F.; Migliavacca, M.; Bernardo, M.E.; Calbi, V.; Assanelli, A.A.; et al. Lentiviral haemopoietic stem/progenitor cell gene therapy for treatment of Wiskott-Aldrich syndrome: Interim results of a non-randomised, open-label, phase 1/2 clinical study. Lancet Haematol. 2019, 6, e239–e253. [Google Scholar] [CrossRef]
- Ben-Skowronek, I. IPEX Syndrome: Genetics and Treatment Options. Genes 2021, 12, 12030323. [Google Scholar] [CrossRef]
- Barzaghi, F.; Passerini, L.; Bacchetta, R. Immune dysregulation, polyendocrinopathy, enteropathy, x-linked syndrome: A paradigm of immunodeficiency with autoimmunity. Front. Immunol. 2012, 3, 211. [Google Scholar] [CrossRef]
- Gambineri, E.; Ciullini Mannurita, S.; Hagin, D.; Vignoli, M.; Anover-Sombke, S.; DeBoer, S.; Segundo, G.R.S.; Allenspach, E.J.; Favre, C.; Ochs, H.D.; et al. Clinical, Immunological, and Molecular Heterogeneity of 173 Patients With the Phenotype of Immune Dysregulation, Polyendocrinopathy, Enteropathy, X-Linked (IPEX) Syndrome. Front. Immunol. 2018, 9, 2411. [Google Scholar] [CrossRef]
- Jamee, M.; Zaki-Dizaji, M.; Lo, B.; Abolhassani, H.; Aghamahdi, F.; Mosavian, M.; Nademi, Z.; Mohammadi, H.; Jadidi-Niaragh, F.; Rojas, M.; et al. Clinical, Immunological, and Genetic Features in Patients with Immune Dysregulation, Polyendocrinopathy, Enteropathy, X-linked (IPEX) and IPEX-like Syndrome. J. Allergy Clin. Immunol. Pract. 2020, 8. 2747–2760.e2747. [Google Scholar] [CrossRef]
- The Human Gene Mutation Database. Available online: http://www.hgmd.cf.ac.uk/ac/gene.php?gene=FOXP3 (accessed on 1 July 2022).
- Lu, L.; Barbi, J.; Pan, F. The regulation of immune tolerance by FOXP3. Nat. Rev. Immunol. 2017, 17, 703–717. [Google Scholar] [CrossRef]
- King, C.; Ilic, A.; Koelsch, K.; Sarvetnick, N. Homeostatic expansion of T cells during immune insufficiency generates autoimmunity. Cell 2004, 117, 265–277. [Google Scholar] [CrossRef]
- Torgerson, T.R.; Linane, A.; Moes, N.; Anover, S.; Mateo, V.; Rieux-Laucat, F.; Hermine, O.; Vijay, S.; Gambineri, E.; Cerf-Bensussan, N.; et al. Severe food allergy as a variant of IPEX syndrome caused by a deletion in a noncoding region of the FOXP3 gene. Gastroenterology 2007, 132, 1705–1717. [Google Scholar] [CrossRef]
- Brunkow, M.E.; Jeffery, E.W.; Hjerrild, K.A.; Paeper, B.; Clark, L.B.; Yasayko, S.A.; Wilkinson, J.E.; Galas, D.; Ziegler, S.F.; Ramsdell, F. Disruption of a new forkhead/winged-helix protein, scurfin, results in the fatal lymphoproliferative disorder of the scurfy mouse. Nat. Genet. 2001, 27, 68–73. [Google Scholar] [CrossRef]
- Barzaghi, F.; Amaya Hernandez, L.C.; Neven, B.; Ricci, S.; Kucuk, Z.Y.; Bleesing, J.J.; Nademi, Z.; Slatter, M.A.; Ulloa, E.R.; Shcherbina, A.; et al. Long-term follow-up of IPEX syndrome patients after different therapeutic strategies: An international multicenter retrospective study. J. Allergy Clin. Immunol. 2018, 141. 1036–1049.e1035. [Google Scholar] [CrossRef]
- Park, J.H.; Lee, K.H.; Jeon, B.; Ochs, H.D.; Lee, J.S.; Gee, H.Y.; Seo, S.; Geum, D.; Piccirillo, C.A.; Eisenhut, M.; et al. Immune dysregulation, polyendocrinopathy, enteropathy, X-linked (IPEX) syndrome: A systematic review. Autoimmun. Rev. 2020, 19, 102526. [Google Scholar] [CrossRef]
- Kofoed, E.M.; Hwa, V.; Little, B.; Woods, K.A.; Buckway, C.K.; Tsubaki, J.; Pratt, K.L.; Bezrodnik, L.; Jasper, H.; Tepper, A.; et al. Growth hormone insensitivity associated with a STAT5b mutation. N. Engl. J. Med. 2003, 349, 1139–1147. [Google Scholar] [CrossRef]
- Klammt, J.; Neumann, D.; Gevers, E.F.; Andrew, S.F.; Schwartz, I.D.; Rockstroh, D.; Colombo, R.; Sanchez, M.A.; Vokurkova, D.; Kowalczyk, J.; et al. Dominant-negative STAT5B mutations cause growth hormone insensitivity with short stature and mild immune dysregulation. Nat. Commun. 2018, 9, 2105. [Google Scholar] [CrossRef]
- The Human Gene Mutation Database. Available online: http://www.hgmd.cf.ac.uk/ac/gene.php?gene=STAT5B (accessed on 1 July 2022).
- Kanai, T.; Jenks, J.; Nadeau, K.C. The STAT5b Pathway Defect and Autoimmunity. Front. Immunol. 2012, 3, 234. [Google Scholar] [CrossRef]
- Cohen, A.C.; Nadeau, K.C.; Tu, W.; Hwa, V.; Dionis, K.; Bezrodnik, L.; Teper, A.; Gaillard, M.; Heinrich, J.; Krensky, A.M.; et al. Cutting Edge: Decreased Accumulation and Regulatory Function of CD4+ CD25(high) T Cells in Human STAT5b Deficiency. J. Immunol. 2006, 177, 2770. [Google Scholar] [CrossRef]
- Vargas-Hernández, A.; Witalisz-Siepracka, A.; Prchal-Murphy, M.; Klein, K.; Mahapatra, S.; Al-Herz, W.; Mace, E.M.; Carisey, A.F.; Orange, J.S.; Sexl, V.; et al. Human signal transducer and activator of transcription 5b (STAT5b) mutation causes dysregulated human natural killer cell maturation and impaired lytic function. J. Allergy Clin. Immunol. 2020, 145. 345–357.e349. [Google Scholar] [CrossRef]
- Hwa, V. Human growth disorders associated with impaired GH action: Defects in STAT5B and JAK2. Mol. Cell. Endocrinol. 2021, 519, 111063. [Google Scholar] [CrossRef]
- Hwa, V.; Nadeau, K.; Wit, J.M.; Rosenfeld, R.G. STAT5b deficiency: Lessons from STAT5b gene mutations. Best Pract. Res. Clin. Endocrinol. Metab. 2011, 25, 61–75. [Google Scholar] [CrossRef] [PubMed]
- Wilson, D.I.; Burn, J.; Scambler, P.; Goodship, J. DiGeorge syndrome: Part of CATCH 22. J. Med. Genet. 1993, 30, 852–856. [Google Scholar] [CrossRef] [PubMed]
- Botto, L.D.; May, K.; Fernhoff, P.M.; Correa, A.; Coleman, K.; Rasmussen, S.A.; Merritt, R.K.; O’Leary, L.A.; Wong, L.Y.; Elixson, E.M.; et al. A population-based study of the 22q11.2 deletion: Phenotype, incidence, and contribution to major birth defects in the population. Pediatrics 2003, 112, 101–107. [Google Scholar] [CrossRef] [PubMed]
- Saitta, S.C.; Harris, S.E.; Gaeth, A.P.; Driscoll, D.A.; McDonald-McGinn, D.M.; Maisenbacher, M.K.; Yersak, J.M.; Chakraborty, P.K.; Hacker, A.M.; Zackai, E.H.; et al. Aberrant interchromosomal exchanges are the predominant cause of the 22q11.2 deletion. Hum. Mol. Genet. 2004, 13, 417–428. [Google Scholar] [CrossRef] [PubMed]
- Yagi, H.; Furutani, Y.; Hamada, H.; Sasaki, T.; Asakawa, S.; Minoshima, S.; Ichida, F.; Joo, K.; Kimura, M.; Imamura, S.-I.; et al. Role of TBX1 in human del22q11.2 syndrome. Lancet 2003, 362, 1366–1373. [Google Scholar] [CrossRef]
- Pirovano, S.; Mazzolari, E.; Pasic, S.; Albertini, A.; Notarangelo, L.D.; Imberti, L. Impaired thymic output and restricted T-cell repertoire in two infants with immunodeficiency and early-onset generalized dermatitis. Immunol. Lett. 2003, 86, 93–97. [Google Scholar] [CrossRef]
- Vu, Q.V.; Wada, T.; Toma, T.; Tajima, H.; Maeda, M.; Tanaka, R.; Oh-Ishi, T.; Yachie, A. Clinical and immunophenotypic features of atypical complete DiGeorge syndrome. Pediatr. Int. 2013, 55, 2–6. [Google Scholar] [CrossRef]
- Louise Markert, M.; Alexieff, M.J.; Li, J.; Sarzotti, M.; Ozaki, D.A.; Devlin, B.H.; Sempowski, G.D.; Rhein, M.E.; Szabolcs, P.; Hale, L.P.; et al. Complete DiGeorge syndrome: Development of rash, lymphadenopathy, and oligoclonal T cells in 5 cases. J. Allergy Clin. Immunol. 2004, 113, 734–741. [Google Scholar] [CrossRef]
- Conley, M.E.; Beckwith, J.B.; Mancer, J.F.; Tenckhoff, L. The spectrum of the DiGeorge syndrome. J. Pediatr. 1979, 94, 883–890. [Google Scholar] [CrossRef]
- Hong, R. The DiGeorge anomaly. Clin. Rev. Allergy Immunol. 2001, 20, 43–60. [Google Scholar] [CrossRef]
- Markert, M.L.; Hummell, D.S.; Rosenblatt, H.M.; Schiff, S.E.; Harville, T.O.; Williams, L.W.; Schiff, R.I.; Buckley, R.H. Complete DiGeorge syndrome: Persistence of profound immunodeficiency. J. Pediatr. 1998, 132, 15–21. [Google Scholar] [CrossRef]
- Selim, M.A.; Markert, M.L.; Burchette, J.L.; Herman, C.M.; Turner, J.W. The cutaneous manifestations of atypical complete DiGeorge syndrome: A histopathologic and immunohistochemical study. J. Cutan. Pathol. 2008, 35, 380–385. [Google Scholar] [CrossRef]
- McLean-Tooke, A.; Barge, D.; Spickett, G.P.; Gennery, A.R. Flow cytometric analysis of TCR Vβ repertoire in patients with 22q11.2 deletion syndrome. Scand. J. Immunol. 2011, 73, 577–585. [Google Scholar] [CrossRef]
- Cancrini, C.; Romiti, M.L.; Finocchi, A.; Di Cesare, S.; Ciaffi, P.; Capponi, C.; Pahwa, S.; Rossi, P. Post-natal ontogenesis of the T-cell receptor CD4 and CD8 Vbeta repertoire and immune function in children with DiGeorge syndrome. J. Clin. Immunol. 2005, 25, 265–274. [Google Scholar] [CrossRef]
- Markert, M.L.; Devlin, B.H.; Chinn, I.K.; McCarthy, E.A. Thymus transplantation in complete DiGeorge anomaly. Immunol. Res. 2009, 44, 61–70. [Google Scholar] [CrossRef]
- Markert, M.L.; Alexieff, M.J.; Li, J.; Sarzotti, M.; Ozaki, D.A.; Devlin, B.H.; Sedlak, D.A.; Sempowski, G.D.; Hale, L.P.; Rice, H.E.; et al. Postnatal thymus transplantation with immunosuppression as treatment for DiGeorge syndrome. Blood 2004, 104, 2574–2581. [Google Scholar] [CrossRef]
- Davies, E.G.; Cheung, M.; Gilmour, K.; Maimaris, J.; Curry, J.; Furmanski, A.; Sebire, N.; Halliday, N.; Mengrelis, K.; Adams, S.; et al. Thymus transplantation for complete DiGeorge syndrome: European experience. J. Allergy Clin. Immunol. 2017, 140. 1660–1670.e1616. [Google Scholar] [CrossRef]
- Biggs, C.M.; Haddad, E.; Issekutz, T.B.; Roifman, C.M.; Turvey, S.E. Newborn screening for severe combined immunodeficiency: A primer for clinicians. CMAJ 2017, 189, e1551–e1557. [Google Scholar] [CrossRef]
- Villa, A.; Santagata, S.; Bozzi, F.; Giliani, S.; Frattini, A.; Imberti, L.; Gatta, L.B.; Ochs, H.D.; Schwarz, K.; Notarangelo, L.D.; et al. Partial V(D)J recombination activity leads to Omenn syndrome. Cell 1998, 93, 885–896. [Google Scholar] [CrossRef]
- Corneo, B.; Moshous, D.; Güngör, T.; Wulffraat, N.; Philippet, P.; Le Deist, F.L.; Fischer, A.; de Villartay, J.P. Identical mutations in RAG1 or RAG2 genes leading to defective V(D)J recombinase activity can cause either T-B-severe combined immune deficiency or Omenn syndrome. Blood 2001, 97, 2772–2776. [Google Scholar] [CrossRef]
- Ege, M.; Ma, Y.; Manfras, B.; Kalwak, K.; Lu, H.; Lieber, M.R.; Schwarz, K.; Pannicke, U. Omenn syndrome due to ARTEMIS mutations. Blood 2005, 105, 4179–4186. [Google Scholar] [CrossRef]
- Roifman, C.M.; Gu, Y.; Cohen, A. Mutations in the RNA component of RNase mitochondrial RNA processing might cause Omenn syndrome. J. Allergy Clin. Immunol. 2006, 117, 897–903. [Google Scholar] [CrossRef]
- Giliani, S.; Bonfim, C.; de Saint Basile, G.; Lanzi, G.; Brousse, N.; Koliski, A.; Malvezzi, M.; Fischer, A.; Notarangelo, L.D.; Le Deist, F. Omenn syndrome in an infant with IL7RA gene mutation. J. Pediatr. 2006, 148, 272–274. [Google Scholar] [CrossRef]
- Lawless, D.; Lango Allen, H.; Thaventhiran, J.; Hodel, F.; Anwar, R.; Fellay, J.; Walter, J.E.; Savic, S. Predicting the Occurrence of Variants in RAG1 and RAG2. J. Clin. Immunol. 2019, 39, 688–701. [Google Scholar] [CrossRef]
- Gan, T.; Wang, Y.; Liu, Y.; Schatz, D.G.; Hu, J. RAG2 abolishes RAG1 aggregation to facilitate V(D)J recombination. Cell Rep. 2021, 37, 109824. [Google Scholar] [CrossRef]
- Tallar, M.; Routes, J. Omenn Syndrome Identified by Newborn Screening. Clin. Perinatol. 2020, 47, 77–86. [Google Scholar] [CrossRef]
- Puck, J.M. Newborn screening for severe combined immunodeficiency and T-cell lymphopenia. Immunol. Rev. 2019, 287, 241–252. [Google Scholar] [CrossRef]
- Villa, A.; Notarangelo, L.D.; Roifman, C.M. Omenn syndrome: Inflammation in leaky severe combined immunodeficiency. J. Allergy Clin. Immunol. 2008, 122, 1082–1086. [Google Scholar] [CrossRef]
- Daley, A.; Andreae, D.A.; Horwitz, A. Not just a rash diagnosis! The allergist’s role in improving recognition of Omenn syndrome. Ann. Allergy Asthma. Immunol. 2020, 125. 244–246.e241. [Google Scholar] [CrossRef]
- Cutts, L.; Bakshi, A.; Walsh, M.; Parslew, R.; Eustace, K. Diagnosing Omenn syndrome. Pediatr. Dermatol. 2021, 38, 541–543. [Google Scholar] [CrossRef]
- Scheimberg, I.; Hoeger, P.H.; Harper, J.I.; Lake, B.; Malone, M. Omenn’s syndrome: Differential diagnosis in infants with erythroderma and immunodeficiency. Pediatr. Dev. Pathol. 2001, 4, 237–245. [Google Scholar] [CrossRef] [PubMed]
- El-Sayed, Z.A.; Abramova, I.; Aldave, J.C.; Al-Herz, W.; Bezrodnik, L.; Boukari, R.; Bousfiha, A.A.; Cancrini, C.; Condino-Neto, A.; Dbaibo, G.; et al. X-linked agammaglobulinemia (XLA):Phenotype, diagnosis, and therapeutic challenges around the world. World Allergy Organ. J. 2019, 12, 100018. [Google Scholar] [CrossRef] [PubMed]
- Tsukada, S.; Saffran, D.C.; Rawlings, D.J.; Parolini, O.; Allen, R.C.; Klisak, I.; Sparkes, R.S.; Kubagawa, H.; Mohandas, T.; Quan, S.; et al. Deficient expression of a B cell cytoplasmic tyrosine kinase in human X-linked agammaglobulinemia. Cell 1993, 72, 279–290. [Google Scholar] [CrossRef]
- Vetrie, D.; Vorechovský, I.; Sideras, P.; Holland, J.; Davies, A.; Flinter, F.; Hammarström, L.; Kinnon, C.; Levinsky, R.; Bobrow, M.; et al. The gene involved in X-linked agammaglobulinaemia is a member of the src family of protein-tyrosine kinases. Nature 1993, 361, 226–233. [Google Scholar] [CrossRef]
- Plebani, A.; Soresina, A.; Rondelli, R.; Amato, G.M.; Azzari, C.; Cardinale, F.; Cazzola, G.; Consolini, R.; De Mattia, D.; Dell’Erba, G.; et al. Clinical, immunological, and molecular analysis in a large cohort of patients with X-linked agammaglobulinemia: An Italian multicenter study. Clin. Immunol. 2002, 104, 221–230. [Google Scholar] [CrossRef]
- Lougaris, V.; Soresina, A.; Baronio, M.; Montin, D.; Martino, S.; Signa, S.; Volpi, S.; Zecca, M.; Marinoni, M.; Baselli, L.A.; et al. Long-term follow-up of 168 patients with X-linked agammaglobulinemia reveals increased morbidity and mortality. J. Allergy Clin. Immunol. 2020, 146, 429–437. [Google Scholar] [CrossRef]
- Khan, W.N.; Alt, F.W.; Gerstein, R.M.; Malynn, B.A.; Larsson, I.; Rathbun, G.; Davidson, L.; Müller, S.; Kantor, A.B.; Herzenberg, L.A.; et al. Defective B cell development and function in Btk-deficient mice. Immunity 1995, 3, 283–299. [Google Scholar] [CrossRef]
- Pal Singh, S.; Dammeijer, F.; Hendriks, R.W. Role of Bruton’s tyrosine kinase in B cells and malignancies. Mol. Cancer 2018, 17, 57. [Google Scholar] [CrossRef]
- Pituch-Noworolska, A.; Mach-Tomalska, H.; Szaflarska, A.; Adamek, D. Shulman disease (eosinophilic fasciitis) in X-linked agammaglobulinemia. Pol. J. Pathol. 2016, 67, 183–188. [Google Scholar] [CrossRef]
- Sharp, S.E. Chronic skin lesions from a patient with Bruton’s X-linked agammaglobulinemia. J. Clin. Microbiol. 2011, 49. 483, 770. [Google Scholar] [CrossRef][Green Version]
- Yamazaki, E.; Kikuchi, K.; Sasahara, Y.; Kono, M.; Akiyama, M.; Aiba, S. Atopic dermatitis without serum immunoglobulin E elevation or loss-of-function filaggrin gene mutation in a patient with X-linked agammaglobulinemia. J. Dermatol. 2020, 47, 58–60. [Google Scholar] [CrossRef]
- Sá, A.; Palha, V.; Condez, E.; Oliveira, N. Skin cancer in congenital X-linked agammaglobulinaemia. BMJ Case Rep. 2021, 14. [Google Scholar] [CrossRef]
- Peterson, R.D.; Page, A.R.; Good, R.A. Wheal and erythema allergy in patients with agammaglobulinemia. J. Allergy 1962, 33, 406–411. [Google Scholar] [CrossRef]
- Kanegane, H.; Hoshino, A.; Okano, T.; Yasumi, T.; Wada, T.; Takada, H.; Okada, S.; Yamashita, M.; Yeh, T.W.; Nishikomori, R.; et al. Flow cytometry-based diagnosis of primary immunodeficiency diseases. Allergol. Int. 2018, 67, 43–54. [Google Scholar] [CrossRef]
- Smith, C.I.E.; Berglöf, A. X-Linked Agammaglobulinemia. In GeneReviews(®); Adam, M.P., Ardinger, H.H., Pagon, R.A., Wallace, S.E., Bean, L.J.H., Gripp, K.W., Mirzaa, G.M., Amemiya, A., Eds.; University of Washington: Seattle, WA, USA, 2000. [Google Scholar]
- Orange, J.S.; Grossman, W.J.; Navickis, R.J.; Wilkes, M.M. Impact of trough IgG on pneumonia incidence in primary immunodeficiency: A meta-analysis of clinical studies. Clin. Immunol. 2010, 137, 21–30. [Google Scholar] [CrossRef]
- Albin, S.; Cunningham-Rundles, C. An update on the use of immunoglobulin for the treatment of immunodeficiency disorders. Immunotherapy 2014, 6, 1113–1126. [Google Scholar] [CrossRef]
- Brandt, T. Dermatitis in children with disturbances of the general condition and the absorption of food elements. Acta Derm. Venereol. 1936, 8. [Google Scholar]
- Küry, S.; Dréno, B.; Bézieau, S.; Giraudet, S.; Kharfi, M.; Kamoun, R.; Moisan, J.P. Identification of SLC39A4, a gene involved in acrodermatitis enteropathica. Nat. Genet. 2002, 31, 239–240. [Google Scholar] [CrossRef]
- Wang, K.; Zhou, B.; Kuo, Y.M.; Zemansky, J.; Gitschier, J. A novel member of a zinc transporter family is defective in acrodermatitis enteropathica. Am. J. Hum. Genet. 2002, 71, 66–73. [Google Scholar] [CrossRef]
- The Human Gene Mutation Database. Available online: http://www.hgmd.cf.ac.uk/ac/gene.php?gene=SLC39A4 (accessed on 1 July 2022).
- Van Wouwe, J.P. Clinical and laboratory assessment of zinc deficiency in Dutch children. A review. Biol. Trace Element Res. 1995, 49, 211–225. [Google Scholar] [CrossRef]
- Kogan, S.; Sood, A.; Garnick, M.S. Zinc and Wound Healing: A Review of Zinc Physiology and Clinical Applications. Wounds 2017, 29, 102–106. [Google Scholar]
- Haase, H.; Rink, L. Multiple impacts of zinc on immune function. Metallomics 2014, 6, 1175–1180. [Google Scholar] [CrossRef]
- Kawamura, T.; Ogawa, Y.; Nakamura, Y.; Nakamizo, S.; Ohta, Y.; Nakano, H.; Kabashima, K.; Katayama, I.; Koizumi, S.; Kodama, T.; et al. Severe dermatitis with loss of epidermal Langerhans cells in human and mouse zinc deficiency. J. Clin. Investig. 2012, 122, 722–732. [Google Scholar] [CrossRef]
- Küry, S.; Kharfi, M.; Kamoun, R.; Taïeb, A.; Mallet, E.; Baudon, J.J.; Glastre, C.; Michel, B.; Sebag, F.; Brooks, D.; et al. Mutation spectrum of human SLC39A4 in a panel of patients with acrodermatitis enteropathica. Hum. Mutat. 2003, 22, 337–338. [Google Scholar] [CrossRef]
- Nakano, A.; Nakano, H.; Nomura, K.; Toyomaki, Y.; Hanada, K. Novel SLC39A4 mutations in acrodermatitis enteropathica. J. Investig. Dermatol. 2003, 120, 963–966. [Google Scholar] [CrossRef]
- Nakano, H.; Nakamura, Y.; Kawamura, T.; Shibagaki, N.; Matsue, H.; Aizu, T.; Rokunohe, D.; Akasaka, E.; Kimura, K.; Nishizawa, A.; et al. Novel and recurrent nonsense mutation of the SLC39A4 gene in Japanese patients with acrodermatitis enteropathica. Br. J. Dermatol. 2009, 161, 184–186. [Google Scholar] [CrossRef]
- Kharfi, M.; El Fékih, N.; Aounallah-Skhiri, H.; Schmitt, S.; Fazaa, B.; Küry, S.; Kamoun, M.R. Acrodermatitis enteropathica: A review of 29 Tunisian cases. Int. J. Dermatol. 2010, 49, 1038–1044. [Google Scholar] [CrossRef]
- Krebs, N.F.; Reidinger, C.J.; Hartley, S.; Robertson, A.D.; Hambidge, K.M. Zinc supplementation during lactation: Effects on maternal status and milk zinc concentrations. Am. J. Clin. Nutr. 1995, 61, 1030–1036. [Google Scholar] [CrossRef]
- Ozkan, S.; Ozkan, H.; Fetil, E.; Corapçioglu, F.; Yilmaz, S.; Ozer, E. Acrodermatitis enteropathica with Pseudomonas aeruginosa sepsis. Pediatr. Dermatol. 1999, 16, 444–447. [Google Scholar] [CrossRef]
- Perafán-Riveros, C.; França, L.F.; Alves, A.C.; Sanches, J.A., Jr. Acrodermatitis enteropathica: Case report and review of the literature. Pediatr. Dermatol. 2002, 19, 426–431. [Google Scholar] [CrossRef]
- Livingstone, C. Zinc: Physiology, deficiency, and parenteral nutrition. Nutr. Clin. Pract. 2015, 30, 371–382. [Google Scholar] [CrossRef] [PubMed]
- Maverakis, E.; Fung, M.A.; Lynch, P.J.; Draznin, M.; Michael, D.J.; Ruben, B.; Fazel, N. Acrodermatitis enteropathica and an overview of zinc metabolism. J. Am. Acad. Dermatol. 2007, 56, 116–124. [Google Scholar] [CrossRef] [PubMed]
- Lee, M.G.; Hong, K.T.; Kim, J.J. Transient symptomatic zinc deficiency in a full-term breast-fed infant. J. Am. Acad. Dermatol. 1990, 23, 375–379. [Google Scholar] [CrossRef]
- Oztürkcan, S.; Içağasioğlu, D.; Akyol, M.; Cevit, O. A case of acrodermatitis enteropathica. J. Dermatol. 2000, 27, 475–477. [Google Scholar] [CrossRef] [PubMed]
- Weismann, K.; Hoe, S.; Knudsen, L.; Sørensen, S.S. 65Zinc absorption in patients suffering from acrodermatitis enteropathica and in normal adults assessed by whole-body counting technique. Br. J. Dermatol. 1979, 101, 573–579. [Google Scholar] [CrossRef] [PubMed]
- Corbo, M.D.; Lam, J. Zinc deficiency and its management in the pediatric population: A literature review and proposed etiologic classification. J. Am. Acad. Dermatol. 2013, 69. 616–624.e611. [Google Scholar] [CrossRef]
- Dietary reference values for food energy and nutrients for the United Kingdom. Report of the Panel on Dietary Reference Values of the Committee on Medical Aspects of Food Policy. Rep. Health Soc. Subj. 1991, 41, 1–210.
- Munnich, A.; Saudubray, J.M.; Cotisson, A.; Coudĕ, F.X.; Ogier, H.; Charpentier, C.; Marsac, C.; Carrĕ, G.; Bourgeay-Causse, M.; Frĕzal, J. Biotin dependent multiple carboxylase deficiency presenting as a congenital lactic acidosis. Eur. J. Pediatr. 1981, 137, 203–206. [Google Scholar] [CrossRef]
- Hou, J.W. Biotin responsive multiple carboxylase deficiency presenting as diabetic ketoacidosis. Chang. Gung Med. J. 2004, 27, 129–133. [Google Scholar]
- Wolf, B. Biotinidase Deficiency. In GeneReviews(®); Adam, M.P., Ardinger, H.H., Pagon, R.A., Wallace, S.E., Bean, L.J.H., Gripp, K.W., Mirzaa, G.M., Amemiya, A., Eds.; University of Washington: Seattle, WA, USA, 1993. [Google Scholar]
- Suzuki, Y.; Aoki, Y.; Ishida, Y.; Chiba, Y.; Iwamatsu, A.; Kishino, T.; Niikawa, N.; Matsubara, Y.; Narisawa, K. Isolation and characterization of mutations in the human holocarboxylase synthetase cDNA. Nat. Genet. 1994, 8, 122–128. [Google Scholar] [CrossRef]
- The Human Gene Mutation Database. Available online: http://www.hgmd.cf.ac.uk/ac/gene.php?gene=HLCS (accessed on 1 July 2022).
- Suzuki, Y.; Yang, X.; Aoki, Y.; Kure, S.; Matsubara, Y. Mutations in the holocarboxylase synthetase gene HLCS. Hum. Mutat. 2005, 26, 285–290. [Google Scholar] [CrossRef]
- Gravel, R.A.; Narang, M.A. Molecular genetics of biotin metabolism: Old vitamin, new science. J. Nutr. Biochem. 2005, 16, 428–431. [Google Scholar] [CrossRef]
- Velázquez, A. Biotin deficiency in protein-energy malnutrition: Implications for nutritional homeostasis and individuality. Nutrition 1997, 13, 991–992. [Google Scholar] [CrossRef]
- Zempleni, J.; Mock, D.M. Marginal biotin deficiency is teratogenic. Proc. Soc. Exp. Biol. Med. 2000, 223, 14–21. [Google Scholar] [CrossRef]
- Suormala, T.; Fowler, B.; Jakobs, C.; Duran, M.; Lehnert, W.; Raab, K.; Wick, H.; Baumgartner, E.R. Late-onset holocarboxylase synthetase-deficiency: Pre- and post-natal diagnosis and evaluation of effectiveness of antenatal biotin therapy. Eur. J. Pediatr. 1998, 157, 570–575. [Google Scholar] [CrossRef]
- Van Hove, J.L.; Josefsberg, S.; Freehauf, C.; Thomas, J.A.; le Thuy, P.; Barshop, B.A.; Woontner, M.; Mock, D.M.; Chiang, P.W.; Spector, E.; et al. Management of a patient with holocarboxylase synthetase deficiency. Mol. Genet. Metab. 2008, 95, 201–205. [Google Scholar] [CrossRef]
- Sadri, M.; Wang, H.; Kuroishi, T.; Li, Y.; Zempleni, J. Holocarboxylase synthetase knockout is embryonic lethal in mice. PLoS ONE 2022, 17, e0265539. [Google Scholar] [CrossRef]
- Xiong, Z.; Zhang, G.; Luo, X.; Zhang, N.; Zheng, J. Case report of holocarboxylase synthetase deficiency (late-onset) in 2 Chinese patients. Medicine 2020, 99, e19964. [Google Scholar] [CrossRef]
- Watabe, D.; Watanabe, A.; Akasaka, T.; Sakamoto, O.; Amano, H. Psoriasis-like Dermatitis in Adulthood: A Skin Manifestation of Holocarboxylase Synthetase Deficiency. Acta Derm Venereol. 2018, 98, 805–806. [Google Scholar] [CrossRef]
- Esparza, E.M.; Golden, A.S.; Hahn, S.H.; Patterson, K.; Brandling-Bennett, H.A. What syndrome is this? Infantile periorificial and intertriginous dermatitis preceding sepsis-like respiratory failure. Pediatr. Dermatol. 2011, 28, 333–334. [Google Scholar] [CrossRef]
- Cowan, T.M.; Blitzer, M.G.; Wolf, B. Technical standards and guidelines for the diagnosis of biotinidase deficiency. Genet. Med. 2010, 12, 464–470. [Google Scholar] [CrossRef]
- Thuy, L.P.; Jurecki, E.; Nemzer, L.; Nyhan, W.L. Prenatal diagnosis of holocarboxylase synthetase deficiency by assay of the enzyme in chorionic villus material followed by prenatal treatment. Clin. Chim. Acta 1999, 284, 59–68. [Google Scholar] [CrossRef]
- Packman, S.; Cowan, M.J.; Golbus, M.S.; Caswell, N.M.; Sweetman, L.; Burri, B.J.; Nyhan, W.L.; Baker, H. Prenatal treatment of biotin responsive multiple carboxylase deficiency. Lancet 1982, 1, 1435–1438. [Google Scholar] [CrossRef]
- Santer, R.; Muhle, H.; Suormala, T.; Baumgartner, E.R.; Duran, M.; Yang, X.; Aoki, Y.; Suzuki, Y.; Stephani, U. Partial response to biotin therapy in a patient with holocarboxylase synthetase deficiency: Clinical, biochemical, and molecular genetic aspects. Mol. Genet. Metab. 2003, 79, 160–166. [Google Scholar] [CrossRef]
- Gibson, K.M.; Bennett, M.J.; Nyhan, W.L.; Mize, C.E. Late-onset holocarboxylase synthetase deficiency. J. Inherit. Metab. Dis. 1996, 19, 739–742. [Google Scholar] [CrossRef]
- Pindolia, K.; Jordan, M.; Wolf, B. Analysis of mutations causing biotinidase deficiency. Hum. Mutat. 2010, 31, 983–991. [Google Scholar] [CrossRef]
- Wolf, B.; Heard, G.S. Screening for biotinidase deficiency in newborns: Worldwide experience. Pediatrics 1990, 85, 512–517. [Google Scholar] [CrossRef]
- The Human Gene Mutation Database. Available online: http://www.hgmd.cf.ac.uk/ac/gene.php?gene=BTD (accessed on 1 July 2022).
- Küry, S.; Ramaekers, V.; Bézieau, S.; Wolf, B. Clinical utility gene card for: Biotinidase deficiency-update 2015. Eur. J. Hum. Genet. 2016, 24, 3–5. [Google Scholar] [CrossRef]
- Wolf, B. Biotinidase deficiency and our champagne legacy. Gene 2016, 589, 142–150. [Google Scholar] [CrossRef]
- León-Del-Río, A.; Valadez-Graham, V.; Gravel, R.A. Holocarboxylase Synthetase: A Moonlighting Transcriptional Coregulator of Gene Expression and a Cytosolic Regulator of Biotin Utilization. Annu. Rev. Nutr. 2017, 37, 207–223. [Google Scholar] [CrossRef]
- Wolf, B. Biotinidase deficiency: “if you have to have an inherited metabolic disease, this is the one to have”. Genet. Med. 2012, 14, 565–575. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Yang, J.Y.; Chen, X.J. Biotinidase deficiency characterized by skin and hair findings. Clin. Dermatol. 2020, 38, 477–483. [Google Scholar] [CrossRef] [PubMed]
- Patra, S.; Senthilnathan, G.; Bhari, N. Acrodermatitis enteropathica-like skin eruption with neonatal seizures in a child with biotinidase deficiency. Clin. Exp. Dermatol. 2020, 45, 266–267. [Google Scholar] [CrossRef] [PubMed]
- Hsu, R.H.; Chien, Y.H.; Hwu, W.L.; Chang, I.F.; Ho, H.C.; Chou, S.P.; Huang, T.M.; Lee, N.C. Genotypic and phenotypic correlations of biotinidase deficiency in the Chinese population. Orphanet J. Rare Dis. 2019, 14, 6. [Google Scholar] [CrossRef] [PubMed]
- Mock, D.M. Skin manifestations of biotin deficiency. Semin. Dermatol. 1991, 10, 296–302. [Google Scholar]
- Wolf, B. The neurology of biotinidase deficiency. Mol. Genet. Metab. 2011, 104, 27–34. [Google Scholar] [CrossRef]
- Wolf, B.; Heard, G.S.; Weissbecker, K.A.; McVoy, J.R.; Grier, R.E.; Leshner, R.T. Biotinidase deficiency: Initial clinical features and rapid diagnosis. Ann. Neurol. 1985, 18, 614–617. [Google Scholar] [CrossRef]
- Wolf, B. Clinical issues and frequent questions about biotinidase deficiency. Mol. Genet. Metab. 2010, 100, 6–13. [Google Scholar] [CrossRef]
- Canda, E.; Kalkan Uçar, S.; Çoker, M. Biotinidase Deficiency: Prevalence, Impact And Management Strategies. Pediatr. Health Med. Ther. 2020, 11, 127–133. [Google Scholar] [CrossRef]
- Tanoue, A.; Endo, F.; Matsuda, I. Structural organization of the gene for human prolidase (peptidase D) and demonstration of a partial gene deletion in a patient with prolidase deficiency. J. Biol. Chem. 1990, 265, 11306–11311. [Google Scholar] [CrossRef]
- Kokturk, A.; Kaya, T.I.; Ikizoglu, G.; Koca, A. Prolidase deficiency. Int. J. Dermatol. 2002, 41, 45–48. [Google Scholar] [CrossRef]
- The Human Gene Mutation Database. Available online: http://www.hgmd.cf.ac.uk/ac/gene.php?gene=PEPD (accessed on 1 July 2022).
- Besio, R.; Maruelli, S.; Gioia, R.; Villa, I.; Grabowski, P.; Gallagher, O.; Bishop, N.J.; Foster, S.; De Lorenzi, E.; Colombo, R.; et al. Lack of prolidase causes a bone phenotype both in human and in mouse. Bone 2015, 72, 53–64. [Google Scholar] [CrossRef]
- Wilk, P.; Uehlein, M.; Piwowarczyk, R.; Dobbek, H.; Mueller, U.; Weiss, M.S. Structural basis for prolidase deficiency disease mechanisms. FEBS J. 2018, 285, 3422–3441. [Google Scholar] [CrossRef]
- Kiratli Nalbant, E.; Karaosmanoglu, N.; Kutlu, O.; Ceylaner, S.; Eksioglu, H.M. A rare case of prolidase deficiency with situs inversus totalis, identified by a novel mutation in the PEPD gene. JAAD Case Rep. 2019, 5, 436–438. [Google Scholar] [CrossRef][Green Version]
- Alrumayyan, N.; Slauenwhite, D.; McAlpine, S.M.; Roberts, S.; Issekutz, T.B.; Huber, A.M.; Liu, Z.; Derfalvi, B. Prolidase deficiency, a rare inborn error of immunity, clinical phenotypes, immunological features, and proposed treatments in twins. Allergy Asthma Clin. Immunol. 2022, 18, 17. [Google Scholar] [CrossRef]
- Hajjar, N.; Kabbani, M.; Tannous, R.; Lebre, A.S.; Megarbane, A.; Minari, A.; El Sayed, F. Prolidase Deficiency Causing Recalcitrant Leg Ulcerations in Siblings. Adv. Skin Wound Care 2021, 34, 1–4. [Google Scholar] [CrossRef]
- Viglio, S.; Annovazzi, L.; Conti, B.; Genta, I.; Perugini, P.; Zanone, C.; Casado, B.; Cetta, G.; Iadarola, P. The role of emerging techniques in the investigation of prolidase deficiency: From diagnosis to the development of a possible therapeutical approach. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 2006, 832, 1–8. [Google Scholar] [CrossRef]
- Spodenkiewicz, M.; Spodenkiewicz, M.; Cleary, M.; Massier, M.; Fitsialos, G.; Cottin, V.; Jouret, G.; Poirsier, C.; Doco-Fenzy, M.; Lèbre, A.S. Clinical Genetics of Prolidase Deficiency: An Updated Review. Biology 2020, 9, 108. [Google Scholar] [CrossRef]
- Misiura, M.; Miltyk, W. Current Understanding of the Emerging Role of Prolidase in Cellular Metabolism. Int. J. Mol. Sci. 2020, 21, 5906. [Google Scholar] [CrossRef]
- Eni-Aganga, I.; Lanaghan, Z.M.; Balasubramaniam, M.; Dash, C.; Pandhare, J. PROLIDASE: A Review from Discovery to its Role in Health and Disease. Front. Mol. Biosci. 2021, 8, 723003. [Google Scholar] [CrossRef]
- Berardesca, E.; Fideli, D.; Bellosta, M.; Dyne, K.M.; Zanaboni, G.; Cetta, G. Blood transfusions in the therapy of a case of prolidase deficiency. Br.J. Dermatol. 1992, 126, 193–195. [Google Scholar] [CrossRef]
- Dunn, R.; Varigos, G.; Winship, I. A photographic essay of prolidase deficiency. Clin. Dysmorphol. 2011, 20, 194–199. [Google Scholar] [CrossRef]
- Samuelov, L.; Sarig, O.; Harmon, R.M.; Rapaport, D.; Ishida-Yamamoto, A.; Isakov, O.; Koetsier, J.L.; Gat, A.; Goldberg, I.; Bergman, R.; et al. Desmoglein 1 deficiency results in severe dermatitis, multiple allergies and metabolic wasting. Nat. Genet. 2013, 45, 1244–1248. [Google Scholar] [CrossRef]
- McAleer, M.A.; Pohler, E.; Smith, F.J.; Wilson, N.J.; Cole, C.; MacGowan, S.; Koetsier, J.L.; Godsel, L.M.; Harmon, R.M.; Gruber, R.; et al. Severe dermatitis, multiple allergies, and metabolic wasting syndrome caused by a novel mutation in the N-terminal plakin domain of desmoplakin. J. Allergy Clin. Immunol. 2015, 136, 1268–1276. [Google Scholar] [CrossRef]
- McGrath, J.A. Inherited disorders of desmosomes. Australas. J. Dermatol. 2005, 46, 221–229. [Google Scholar] [CrossRef]
- Has, C.; Jakob, T.; He, Y.; Kiritsi, D.; Hausser, I.; Bruckner-Tuderman, L. Loss of desmoglein 1 associated with palmoplantar keratoderma, dermatitis and multiple allergies. Br. J. Dermatol. 2015, 172, 257–261. [Google Scholar] [CrossRef] [PubMed]
- Dănescu, S.; Leppert, J.; Cosgarea, R.; Zurac, S.; Pop, S.; Baican, A.; Has, C. Compound heterozygosity for dominant and recessive DSG1 mutations in a patient with atypical SAM syndrome (severe dermatitis, multiple allergies, metabolic wasting). J. Eur. Acad. Dermatol. Venereol. 2017, 31, e144–e146. [Google Scholar] [CrossRef] [PubMed]
- Ishida-Yamamoto, A.; Igawa, S.; Kishibe, M.; Honma, M. Clinical and molecular implications of structural changes to desmosomes and corneodesmosomes. J. Dermatol. 2018, 45, 385–389. [Google Scholar] [CrossRef] [PubMed]
- Pan, C.; Cheng, R.; Li, Y.; Zhao, M.; Chen, F.; Liang, J.; Yu, X.; Wang, X.; Li, H.; Yao, Z.; et al. Deep-intronic and frameshift DSG1 variants associated with atypical severe dermatitis, multiple allergies and metabolic wasting (SAM) syndrome in a Chinese family. Eur. J. Dermatol. 2021, 31, 239–244. [Google Scholar] [CrossRef] [PubMed]
- Vakkilainen, S.; Puhakka, L.; Klemetti, P.; Heiskanen, K.; Seppänen, M.; Muona, M.; Posseme, C.; Duffy, D.; Väisänen, T.; Elomaa, O.; et al. Novel DSP Spectrin 6 Region Variant Causes Neonatal Erythroderma, Failure to Thrive, Severe Herpes Simplex Infections and Brain Lesions. Acta Derm Venereol. 2019, 99, 789–796. [Google Scholar] [CrossRef]
- Liang, J.; Li, C.; Zhang, Z.; Ni, C.; Yu, H.; Li, M.; Yao, Z. Severe dermatitis, multiple allergies and metabolic wasting (SAM) syndrome caused by de novo mutation in the DSP gene misdiagnosed as generalized pustular psoriasis and treatment of acitretin with gabapentin. J. Dermatol. 2019, 46, 622–625. [Google Scholar] [CrossRef]
- Hernández-Martín, A.; Kennedy-Batalla, R.; Cañedo, E.; Bernaldo-de-Quirós, E.; Carazo-Gallego, B.; Vera, A.; Torrelo, A.; Noguera-Morel, L.; González-Sarmiento, R.; Bolling, M.; et al. Imbalance in T-Helper 17 Cells and Targeted Therapy in an Infant with SAM-like Syndrome. N. Engl. J. Med. 2019, 381, 2176–2178. [Google Scholar] [CrossRef]
- Comel, M. Ichthyosis linearis circumflexa. Dermatologica 1949, 98, 133–136. [Google Scholar] [CrossRef]
- Netherton, E.W. A unique case of trichorrhexis nodosa; bamboo hairs. AMA Arch. Derm. 1958, 78, 483–487. [Google Scholar] [CrossRef]
- The Human Gene Mutation Database. Available online: http://www.hgmd.cf.ac.uk/ac/gene.php?gene=SPINK5 (accessed on 1 July 2022).
- Sarri, C.A.; Roussaki-Schulze, A.; Vasilopoulos, Y.; Zafiriou, E.; Patsatsi, A.; Stamatis, C.; Gidarokosta, P.; Sotiriadis, D.; Sarafidou, T.; Mamuris, Z. Netherton Syndrome: A Genotype-Phenotype Review. Mol. Diagn. Ther. 2017, 21, 137–152. [Google Scholar] [CrossRef]
- Roda, Â.; Mendonça-Sanches, M.; Travassos, A.R.; Soares-de-Almeida, L.; Metze, D. Infliximab therapy for Netherton syndrome: A case report. JAAD Case Rep. 2017, 3, 550–552. [Google Scholar] [CrossRef]
- D’Alessio, M.; Fortugno, P.; Zambruno, G.; Hovnanian, A. Netherton syndrome and its multifaceted defective protein LEKTI. G. Ital. Dermatol. Venereol. 2013, 148, 37–51. [Google Scholar]
- Small, A.M.; Cordoro, K.M. Netherton Syndrome Mimicking Pustular Psoriasis: Clinical Implications and Response to Intravenous Immunoglobulin. Pediatr.Dermatol. 2016, 33, e222–e223. [Google Scholar] [CrossRef]
- Smith, D.L.; Smith, J.G.; Wong, S.W.; deShazo, R.D. Netherton’s syndrome: A syndrome of elevated IgE and characteristic skin and hair findings. J. Allergy Clin. Immunol. 1995, 95, 116–123. [Google Scholar] [CrossRef]
- Sun, J.D.; Linden, K.G. Netherton syndrome: A case report and review of the literature. Int. J. Dermatol. 2006, 45, 693–697. [Google Scholar] [CrossRef]
- Bin Saif, G.; Al-Khenaizan, S. Netherton syndrome: Successful use of topical tacrolimus and pimecrolimus in four siblings. Int. J. Dermatol. 2007, 46, 290–294. [Google Scholar] [CrossRef]
- Hovnanian, A. Netherton syndrome: Skin inflammation and allergy by loss of protease inhibition. Cell Tissue Res. 2013, 351, 289–300. [Google Scholar] [CrossRef]
- Bonilla, F.A. Intravenous and subcutaneous immunoglobulin G replacement therapy. Allergy Asthma Proc. 2016, 37, 426–431. [Google Scholar] [CrossRef]
- Singer, R.; Çopur, M.; Yüksel, E.N.; Kocatürk, E.; Erhan, S. Ichthyosis linearis circumflexa in a child. Response to narrowband UVB therapy. J. Dermatol. Case Rep. 2015, 9, 110–112. [Google Scholar] [CrossRef]
- Leung, A.K.C.; Barankin, B.; Leong, K.F. An 8-Year-Old Child with Delayed Diagnosis of Netherton Syndrome. Case Rep. Pediatr. 2018, 2018, 9434916. [Google Scholar] [CrossRef]
- Kharfi, M.; Khaled, A.; Ammar, D.; Ezzine, N.; El Fekih, N.; Fazaa, B.; Jaafoura, H.; Kamoun, M.R. Generalized peeling skin syndrome: Case report and review of the literature. Dermatol. Online J. 2010, 16, 1. [Google Scholar] [CrossRef]
- Oji, V.; Eckl, K.M.; Aufenvenne, K.; Nätebus, M.; Tarinski, T.; Ackermann, K.; Seller, N.; Metze, D.; Nürnberg, G.; Fölster-Holst, R.; et al. Loss of corneodesmosin leads to severe skin barrier defect, pruritus, and atopy: Unraveling the peeling skin disease. Am. J. Hum.Genet. 2010, 87, 274–281. [Google Scholar] [CrossRef]
- The Human Gene Mutation Database. Available online: http://www.hgmd.cf.ac.uk/ac/gene.php?gene=CDSN (accessed on 1 July 2022).
- Jonca, N.; Leclerc, E.A.; Caubet, C.; Simon, M.; Guerrin, M.; Serre, G. Corneodesmosomes and corneodesmosin: From the stratum corneum cohesion to the pathophysiology of genodermatoses. Eur. J. Dermatol. 2011, 21 (Suppl. S2), 35–42. [Google Scholar] [CrossRef]
- Samuelov, L.; Sprecher, E. Peeling off the genetics of atopic dermatitis-like congenital disorders. J. Allergy Clin. Immunol. 2014, 134, 808–815. [Google Scholar] [CrossRef]
- Matsumoto, M.; Zhou, Y.; Matsuo, S.; Nakanishi, H.; Hirose, K.; Oura, H.; Arase, S.; Ishida-Yamamoto, A.; Bando, Y.; Izumi, K.; et al. Targeted deletion of the murine corneodesmosin gene delineates its essential role in skin and hair physiology. Proc. Natl. Acad. Sci. USA 2008, 105, 6720–6724. [Google Scholar] [CrossRef]
- Israeli, S.; Zamir, H.; Sarig, O.; Bergman, R.; Sprecher, E. Inflammatory peeling skin syndrome caused by a mutation in CDSN encoding corneodesmosin. J. Investig. Dermatol. 2011, 131, 779–781. [Google Scholar] [CrossRef] [PubMed]
- Mallet, A.; Kypriotou, M.; George, K.; Leclerc, E.; Rivero, D.; Mazereeuw-Hautier, J.; Serre, G.; Huber, M.; Jonca, N.; Hohl, D. Identification of the first nonsense CDSN mutation with expression of a truncated protein causing peeling skin syndrome type B. Br. J. Dermatol. 2013, 169, 1322–1325. [Google Scholar] [CrossRef] [PubMed]
- Telem, D.F.; Israeli, S.; Sarig, O.; Sprecher, E. Inflammatory peeling skin syndrome caused a novel mutation in CDSN. Arch. Dermatol. Res. 2012, 304, 251–255. [Google Scholar] [CrossRef] [PubMed]
- Wada, T.; Matsuda, Y.; Muraoka, M.; Toma, T.; Takehara, K.; Fujimoto, M.; Yachie, A. Alu-mediated large deletion of the CDSN gene as a cause of peeling skin disease. Clin. Genet. 2014, 86, 383–386. [Google Scholar] [CrossRef]
- Ishida-Yamamoto, A.; Furio, L.; Igawa, S.; Honma, M.; Tron, E.; Malan, V.; Murakami, M.; Hovnanian, A. Inflammatory peeling skin syndrome caused by homozygous genomic deletion in the PSORS1 region encompassing the CDSN gene. Exp. Dermatol. 2014, 23, 60–63. [Google Scholar] [CrossRef]
- Teye, K.; Hamada, T.; Krol, R.P.; Numata, S.; Ishii, N.; Matsuda, M.; Ohata, C.; Furumura, M.; Hashimoto, T. Homozygous deletion of six genes including corneodesmosin on chromosome 6p21.3 is associated with generalized peeling skin disease. J. Dermatol. Sci. 2014, 75, 36–42. [Google Scholar] [CrossRef]
- Navarro-Navarro, I.; Jiménez-Gallo, D.; de la Varga-Martínez, R.; Villegas-Romero, I.; Mora-López, F.; Linares-Barrios, M.; Youssefian, L.; Vahidnezhad, H.; Uitto, J. Novel splice mutation in CDSN gene causing type b peeling skin syndrome. J. Eur. Acad. Dermatol. Venereol. 2022, 36, e456–e460. [Google Scholar] [CrossRef]
- Mir-Bonafe, J.F.; Baselga-Torres, E.; Gonzalez-Sarmiento, R. A 2-year-old girl with skin fragility. JAMA Dermatol. 2015, 151, 225–226. [Google Scholar] [CrossRef]
- Mizuno, Y.; Suga, Y.; Hasegawa, T.; Haruna, K.; Kohroh, K.; Ogawa, H.; Ikeda, S.; Shimizu, T.; Komatsu, N. A case of peeling skin syndrome successfully treated with topical calcipotriol. J. Dermatol. 2006, 33, 430–432. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).