Identification of Hotspot Mutations in the N Gene of SARS-CoV-2 in Russian Clinical Samples That May Affect the Detection by Reverse Transcription-PCR
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sample Collection and Handling
2.2. Nucleic Acid Extraction
2.3. Real-Time Quantitative RT-PCR
2.4. Sanger Sequencing
3. Results
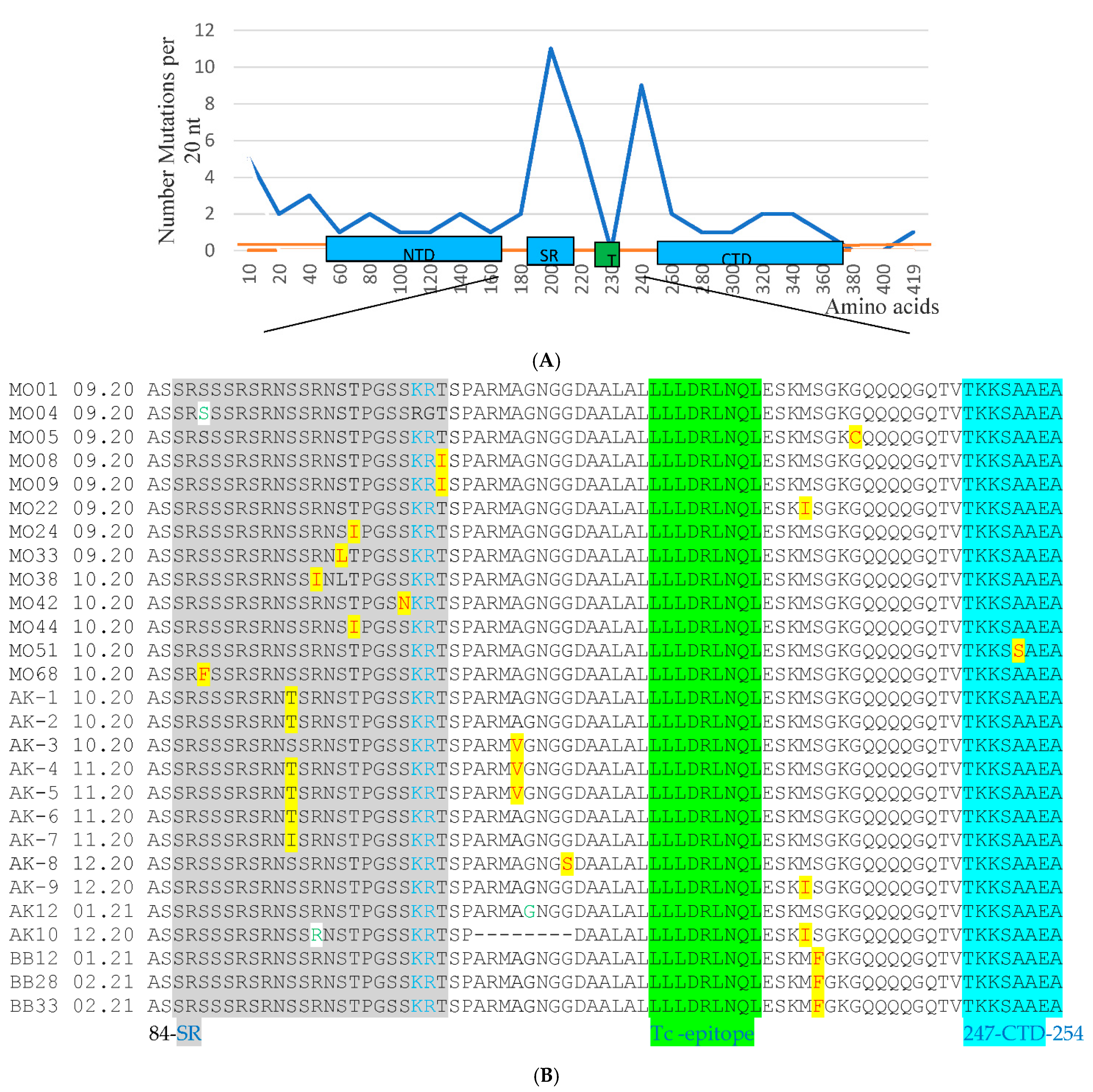
3.1. PCR and Sequencing Analysis of 12Russian Isolates with the Failure of Detection in the N Gene
3.2. Sequencing the Entire N and E Genes in 195 Clinical Samples
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Van Kasteren, P.B.; van der Veer, B.; van den Brink, S.; Wijsman, L.; de Jonge, J.; van den Brandt, A.; Molenkamp, R.; Reusken, C.B.E.M.; Meijer, A. Comparison of seven commercial RT-PCR diagnostic kits for COVID-19. J. Clin. Virol. 2020, 128, 104412. [Google Scholar] [CrossRef] [PubMed]
- Peng, Y.; Du, N.; Lei, Y.; Dorje, S.; Qi, J.; Luo, T.; Gao, G.F.; Song, H. Structures of the SARS-CoV-2 nucleocapsid and their perspectives for drug design. EMBO J. 2020, 39, e105938. [Google Scholar] [CrossRef]
- Dutta, N.K.; Mazumdar, K.; Gordy, J.T. The nucleocapsid protein of SARS-CoV-2: A target for vaccine development. J. Virol. 2020, 94, e00647-20. [Google Scholar] [CrossRef] [PubMed]
- Zeng, W.; Liu, G.; Ma, H.; Zhao, D.; Yang, Y.; Liu, M.; Mohammed, A.; Zhao, C.; Yang, Y.; Xie, J.; et al. Biochemical characterization of SARS-CoV-2 nucleocapsid protein. Biochem. Biophys. Res. Commun. 2020, 527, 618–623. [Google Scholar] [CrossRef]
- Lu, R.J.; Zhao, L.; Huang, B.Y.; Ye, F.; Wang, W.L.; Tan, W.J. Real-time reverse transcription-polymerase chain reaction assay panel for the detection of severe acute respiratory syndrome coronavirus 2 and its variants. Chin. Med. J. Engl. 2021, 134, 2048–2053. [Google Scholar] [CrossRef] [PubMed]
- Hoque, M.N.; Chaudhury, A.; Akanda, M.A.M.; Hossain, M.A.; Islam, M.T. Genomic diversity and evolution, diagnosis, prevention, and therapeutics of the pandemic COVID-19 disease. PeerJ 2020, 8, e9689. [Google Scholar] [CrossRef] [PubMed]
- Venkatagopalan, P.; Daskalova, S.M.; Lopez, L.A.; Dolezal, K.A.; Hogue, B.G. Coronavirus envelope (E) protein remains at the site of assembly. Virology 2015, 478, 75–85. [Google Scholar] [CrossRef]
- Rahman, M.S.; Hoque, M.N.; Islam, M.R.; Islam, I.; Mishu, I.D.; Rahaman, M.M.; Sultana, M.; Hossain, M.A. Mutational insights into the envelope protein of SARS-CoV-2. Gene Rep. 2021, 22, 100997. [Google Scholar] [CrossRef]
- Toto, A.; Ma, S.; Malagrin, F.; Visconti, L.; Pagano, L.; Stromgaard, K.; Gianni, S. Comparing the binding properties of peptides mimicking the envelope protein of SARS-CoV and SARS-CoV-2 to the PDZ domain of the tight junction-associated PALS1 protein. Protein Sci. 2020, 29, 2038–2042. [Google Scholar] [CrossRef] [PubMed]
- De Maio, F.; Lo Cascio, E.; Babini, G.; Sali, M.; Della Longa, S.; Tilocca, B.; Roncada, P.; Arcovito, A.; Sanguinetti, M.; Scambia, G.; et al. Improved binding of SARS-CoV-2 Envelope protein to tight junction-associated PALS1 could play a key role in COVID-19 pathogenesis. Microbes Infect. 2020, 22, 592–597. [Google Scholar] [CrossRef]
- Majumdar, P.; Niyogi, S. SARS-CoV-2 mutations: The biological trackway towards viral fitness. Epidemiol. Infect. 2021, 149, e110. [Google Scholar] [CrossRef]
- Rahman, S.; Islam, R.; Alam, R.; Islam, I.; Hoque, N.; Akter, S.; Rahaman, M.M.; Sultana, M.; Hossain, M.A. Evolutionary dynamics of SARS-CoV-2 nucleocapsid protein and its consequences. J. Med. Virol. 2021, 93, 2177–2195. [Google Scholar] [CrossRef]
- Ziegler, K.; Steininger, P.; Ziegler, R.; Steinmann, J.; Korn, K.; Ensser, A. SARS-CoV-2 samples may escape detection because of a single point mutation in the N gene. Eurosurveillance 2020, 25, 2001650. [Google Scholar] [CrossRef]
- Corman, V.M.; Landt, O.; Kaiser, M.; Molenkamp, R.; Meijer, A.; Chu, D.K.; Bleicker, T.; Brünink, S.; Schneider, J.; Schmidt, M.L.; et al. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Eurosurveillance 2020, 25, 2000045. [Google Scholar] [CrossRef] [Green Version]
- Mathuria, J.P.; Yadav, R.; Rajkumar. Laboratory diagnosis of SARS-CoV-2—A review of current methods. J. Infect. Public Health 2020, 13, 901–905. [Google Scholar] [CrossRef] [PubMed]
- Cun, Y.; Li, C.; Shi, L.; Sun, M.; Dai, S.; Sun, L.; Shi, L.; Yao, Y. COVID-19 coronavirus vaccine T cell epitope prediction analysis based on distributions of HLA class I loci (HLA-A, -B, -C) across global populations. Hum. Vaccin. Immunother. 2021, 17, 1097–1108. [Google Scholar] [CrossRef] [PubMed]
- Bouhaddou, M.; Memon, D.; Meyer, B.; White, K.M.; Rezelj, V.V.; Correa Marrero, M. The global phosphorylation landscape of SARS-CoV-2 infection. Cell 2020, 182, 685–712.e19. [Google Scholar] [CrossRef] [PubMed]
- Gand, M.; Vanneste, K.; Thomas, I.; Van Gucht, S.; Capron, A.; Herman, P.; Roosens, N.H.C.; De Keersmaecker, S.C.J. Deepening of In Silico Evaluation of SARS-CoV-2 Detection RT-qPCR Assays in the Context of New Variants. Genes 2021, 12, 565. [Google Scholar] [CrossRef]
- Artesi, M.; Bontems, S.; Göbbels, P.; Franckh, M.; Maes, P.; Boreux, R.; Meex, C.; Melin, P.; Hayette, M.P.; Bours, V.; et al. A Recurrent Mutation at Position 26340 of SARS-CoV-2 Is Associated with Failure of the E Gene Quantitative Reverse Transcription-PCR Utilized in a Commercial Dual-Target Diagnostic Assay. J. Clin. Microbiol. 2020, 58, e01598-20. [Google Scholar] [CrossRef]
- Wang, R.; Hozumi, Y.; Yin, C.; Wei, G.W. Mutations on COVID-19 diagnostic targets. Genomics 2020, 112, 5204–5213. [Google Scholar] [CrossRef]
- Nalla, A.K.; Casto, A.M.; Huang, M.W.; Perchetti, G.A.; Sampoleo, R.; Shrestha, L.; Wei, Y.; Zhu, H.; Jerome, K.R.; Greninger, A.L. Comparative Performance of SARS-CoV-2 Detection Assays Using Seven Different Primer-Probe Sets and One Assay Kit. J. Clin. Microbiol. 2020, 58, e00557-20. [Google Scholar] [CrossRef] [Green Version]
- Fox-Lewis, S.; Fox-Lewis, A.; Harrower, J.; Chen, R.; Wang, J.; de Ligt, J.; McAuliffe, G.; Taylor, S.; Smit, E. Lack of N2-gene amplification on the Cepheid Xpert Xpress SARS-CoV-2 assay and potential novel causative mutations: A case series from Auckland, New Zealand. IDCases 2021, 25, e01233. [Google Scholar] [CrossRef] [PubMed]
- Hasan, M.R.; Sundararaju, S.; Manickam, C.; Mirza, F.; Al-Hail, H.; Lorenz, S.; Tang, P. A novel point mutation in the N gene of SARS-CoV-2 may affect the detection of the virus by RT-qPCR. J. Clin. Microbiol. 2021, 59, e03278-20. [Google Scholar] [CrossRef]
- Vanaerschot, M.; Mann, S.A.; Webber, J.T.; Kamm, J.; Bell, S.M.; Bell, J.; Hong, S.N.; Nguyen, M.P.; Chan, L.Y.; Bhatt, K.D.; et al. Identification of a polymorphism in the N gene of SARS-CoV-2 that adversely impacts detection by reverse transcription-PCR. J. Clin. Microbiol. 2020, 59, e02369-20. [Google Scholar] [CrossRef] [PubMed]
- Jungreis, I.; Sealfon, R.; Kellis, M. SARS-CoV-2 gene content and COVID-19 mutation impact by comparing 44 Sarbecovirus genomes. Nat. Commun. 2021, 12, 2642. [Google Scholar] [CrossRef] [PubMed]
- Peng, T.Y.; Lee, K.R.; Tarn, W.Y. Phosphorylation of the arginine/serine dipeptide-rich motif of the severe acute respiratory syndrome coronavirus nucleocapsid protein modulates its multimerization, translation inhibitory activity and cellular localization. FEBS J. 2008, 275, 4152–4163. [Google Scholar] [CrossRef] [Green Version]
- Kozlovskaya, L.; Piniaeva, A.; Ignatyev, G.; Selivanov, A.; Shishova, A.; Kovpak, A.; Gordeychuk, I.; Ivin, Y.; Berestovskaya, A.; Prokhortchouk, E.; et al. Isolation and phylogenetic analysis of SARS-CoV-2 variants collected in Russia during the COVID-19 outbreak. Int. J. Infect. Dis. 2020, 99, 40–46. [Google Scholar] [CrossRef]
- Lee, S.; Won, D.; Kim, C.K.; Ahn, J.; Lee, Y.; Na, H.; Kim, Y.T.; Lee, M.K.; Choi, J.R.; Lim, H.S.; et al. Novel indel mutation in the N gene of SARS-CoV-2 clinical samples that were diagnosed positive in a commercial RT-PCR assay. Virus Res. 2021, 297, 198398. [Google Scholar] [CrossRef] [PubMed]
- Maurer-Stroh, S.; Eisenhaber, B.; Eisenhaber, F. N-terminal N-myristoylation of proteins: Refinement of the sequence motif and its taxon-specific differences. J. Mol. Biol. 2002, 317, 523–540. [Google Scholar] [CrossRef]
| Sample | Test Data | RT-PCR, Cp | Detected Mutations * | ||||
|---|---|---|---|---|---|---|---|
| Gene E | Gene N | Gene E | Gene N | ||||
| Nucleotide | Amino Acid | Nucleotide | Amino Acid | ||||
| AK-1 | 06.10.20 | 27.0 | 31.7 | C26261T | S6L | G28851C C28909T | S193T G212G |
| AK-2 | 06.10.20 | 25.8 | 29.6 | G26257A | V5I | G28851C | S193T |
| AK-3 | 08.10.20 | 26.1 | 29.9 | C26261T | S6L | C28905T | A211V |
| AK-4 | 06.11.20 | 25.6 | n/d ** | n/d | n/d | G28851C C28905T | S193T A211V |
| AK-5 | 06.11.20 | 24.9 | n/d | n/d | n/d | G28376A G28851C C28905T | A35T S193T A211V |
| AK-6 | 06.11.20 | 28.2 | 33.8 | C26261T | S6L | G28851C C28909T | S193T G212G |
| AK-7 | 06.11.20 | 27.3 | 31.5 | n/d | n/d | G28851T | S193I |
| AK-8 | 12.01.21 | 25.7 | 29.6 | n/d | n/d | G28916A | G215S |
| AK-16 | 10.11.20 | 26.4 | 30.5 | n/d | n/d | C28905T | A211V |
| AK-10 | 24.12.20 | 24.0 | n/d | C26261T | S6L | del 28896-28919 G28690T A28858G G28975T | A208-G215 L139F R195 M234I |
| AK-11 | 12.01.21 | 25.3 | 29.6 | n/d | n/d | G28916A | G215S |
| AK-12 | 15.02.21 | 27.8 | 31.7 | n/d | n/d | C28909T | G212G |
| Sample | Test Data | Detected Mutations | Gene N Region * | |||
|---|---|---|---|---|---|---|
| Gene E | Gene N | |||||
| Nucleotide | Amino Acid | Nucleotide | Amino Acid | |||
| M002 | 06.09.20 | n/d ** | n/d | C28866T | T198I | SR |
| M005 | 07.09.20 | n/d | n/d | G28985T | G238C | LKR |
| M008 | 12.09.20 | n/d | n/d | C28887T | T205I | LKR |
| M009 | 12.09.20 | n/d | n/d | C28887T | T205I | LKR |
| M022 | 15.09.20 | n/d | n/d | G28975T | M234I | LKR |
| M024 | 18.09.20 | n/d | n/d | C28866T | T198I | SR |
| M033 | 22.09.20 | C26261T | S6L | C28863T | S197L | SR |
| M038 | 03.10.20 | n/d | n/d | G28857T | R195I | SR |
| M042 | 09.10.20 | n/d | n/d | G28878A | S202N | SR |
| M044 | 09.10.20 | n/d | n/d | C28866T | T198I | SR |
| M047 | 09.10.20 | n/d | n/d | G28655T | D128Y | NTD |
| M051 | 11.10.20 | n/d | n/d | G29024T | A251S | CTD |
| M068 | 15.10.20 | n/d | n/d | C28830T | S186F | SR |
| M071 | 15.10.20 | n/d | n/d | G28655T | D128Y | NTD |
| AK-9 | 09.12.20 | n/d | n/d | G28975T | M234I | LKR |
| AK-18 | 11.12.20 | C26261T | S6L | G28376A | A35T | 5′DLR |
| AK-23 | 11.12.20 | n/d | n/d | G28975T C28651T | M234I N126 | LKR NTD |
| AK-26 | 11.12.20 | G26257A | V5I | C29095T C29218T | F274 F315 | CTD CTD |
| AK-31 | 14.12.20 | n/d | n/d | G28975T | M234I | LKR |
| AK-32 | 14.12.20 | n/d | n/d | G28655T | D128Y | NTD |
| AK-37 | 14.12.20 | n/d | n/d | G28975T | M234I | LKR |
| AK-39 | 14.12.20 | n/d | n/d | C28697T | P142S | NTD |
| AK-41 | 15.12.20 | n/d | n/d | C28887T G28195T | T205I N140K | LKR NTD |
| AK-46 | 17.12.20 | n/d | n/d | G28985T | G238C | LKR |
| AK-49 | 17.12.20 | n/d | n/d | G29024T | A251S | CTD |
| AK-56 | 18.12.20 | n/d | n/d | G28878A | S202N | SR |
| AK-57 | 18.12.20 | n/d | n/d | G28376A | A35T | 5′DLR |
| AK-72 | 14.01.21 | n/d | n/d | C28755T | L161F | NTD |
| AK-77 | 14.01.21 | n/d | n/d | C28775T | P168S | NTD |
| AK-81 | 14.01.21 | n/d | n/d | C28311T | P13L | 5′DLR |
| AK-84 | 14.01.21 | n/d | n/d | C29250T | P326L | CTD |
| AK-85 | 14.01.21 | n/d | n/d | C29153A | Q294K | CTD |
| AK-92 | 15.01.21 | n/d | n/d | G29315C | D348H | CTD |
| AK-97 | 15.01.21 | n/d | n/d | C28826T C29463T | R185C A397V | SR 3′DLR |
| AK-104 | 15.01.21 | n/d | n/d | C29067T | T265I | CTD |
| AK-113 | 16.01.21 | n/d | n/d | G28376A | A35T | 5′DLR |
| AK-118 | 16.01.21 | n/d | n/d | C29095T | F274F | CTD |
| AK-124 | 16.01.21 | n/d | n/d | G29148C | I292T | CTD |
| AK-135 | 16.01.21 | n/d | n/d | C29250T | P326L | CTD |
| AK-143 | 16.01.21 | n/d | n/d | C29149T | I292I | CTD |
| BB ***-12 | 22.01.21 | n/d | n/d | 28280GAT>CTA C28977T | D3L S235F | 5′DLR LKR |
| BB-28 | 26.02.21 | n/d | n/d | 28280GAT>CTA C28977T | D3L S235F | 5′DLR LKR |
| BB-31 | 26.02.21 | n/d | n/d | 28280GAT>CTA C28977T | D3L S235F | 5′DLR LKR |
| BB-37 | 26.02.21 | n/d | n/d | 28280GAT>CTA C28977T | D3L S235F | 5′DLR LKR |
| BB-37 | 03.03.21 | n/d | n/d | 28280GAT>CTA C28977T | D3L S235F | 5′DLR LKR |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kiryanov, S.A.; Levina, T.A.; Konopleva, M.V.; Suslov, A.P. Identification of Hotspot Mutations in the N Gene of SARS-CoV-2 in Russian Clinical Samples That May Affect the Detection by Reverse Transcription-PCR. Diagnostics 2022, 12, 147. https://doi.org/10.3390/diagnostics12010147
Kiryanov SA, Levina TA, Konopleva MV, Suslov AP. Identification of Hotspot Mutations in the N Gene of SARS-CoV-2 in Russian Clinical Samples That May Affect the Detection by Reverse Transcription-PCR. Diagnostics. 2022; 12(1):147. https://doi.org/10.3390/diagnostics12010147
Chicago/Turabian StyleKiryanov, Sergei A., Tatiana A. Levina, Maria V. Konopleva, and Anatoly P. Suslov. 2022. "Identification of Hotspot Mutations in the N Gene of SARS-CoV-2 in Russian Clinical Samples That May Affect the Detection by Reverse Transcription-PCR" Diagnostics 12, no. 1: 147. https://doi.org/10.3390/diagnostics12010147
APA StyleKiryanov, S. A., Levina, T. A., Konopleva, M. V., & Suslov, A. P. (2022). Identification of Hotspot Mutations in the N Gene of SARS-CoV-2 in Russian Clinical Samples That May Affect the Detection by Reverse Transcription-PCR. Diagnostics, 12(1), 147. https://doi.org/10.3390/diagnostics12010147






