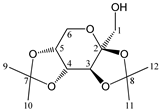
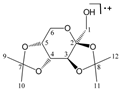
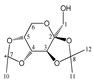
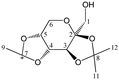
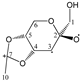
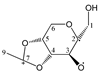
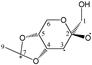
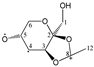
Abstract
Our study found that quantum calculations can differentiate fragmentation energies into isomeric structures with asymmetric carbon atoms, such as those of acetalized monosaccharides. It was justified by the good results that have been published in recent years on the discrimination of structural isomers and diastereomers by correlating the calculated mass energy fragmentation profiles with their mass spectra. Based on the quantitative structure–fragmentation relationship (QSFR), this technique compares the intensities of primary ions from the experimental spectrum using the mass energy profiles calculated for the candidate structures. Maximum fit is obtained for the true structure. For a preliminary assessment of the accuracy of the identification of some di-O-isopropylidene monosaccharide diastereomers, we used fragmentation enthalpies (ΔfH) and Gibbs energies (ΔfG) as the energetic descriptors of fragmentation. Four quantum chemical methods were used: RM1, PM7, DFT ΔfH and DFT ΔfG. The mass energy database shows that the differences between the profiles of the isomeric candidate structures could be large enough to be distinguished from each other. This database allows the optimization of energy descriptors and quantum computing methods that can ensure the correct identification of these isomers.
1. Introduction
The chemical structure identification of small molecules fulfills an important role in modern life sciences and technologies. A special function is the establishment of the structure of natural compounds in which molecular geometry and biological activity are drastically influenced by the configuration of asymmetric carbon atoms that are present in the molecule, e.g., in silico investigations of the biological interactions of phytochemicals for the rapid discovery of potential drugs, their exact stereochemical structure must be known [1]. Despite advances in instrumentation, the structure elucidation of unknown small molecules using mass spectrometry (MS) is often ambiguous due to the large number of possible isomers. The use of additional orthogonal filters can decrease the number of candidate structures [2,3]. Thus, fragmentation tree alignments have been developed for automated structural comparison [4]. The latest cheminformatics trends in the field of spectrum annotation, prediction and metabolite identification use quantum chemical calculations, thereby allowing for the study of the thermodynamics and kinetics of the observed processes [5,6]. Significant progress has been reported in the identification of the most likely fragmentation pathways and their final intensities in electron ionization (EI) mass spectra using calculated reactions or activation energies [7,8,9]. Unfortunately, the alignments of fragmentation trees and the search of structures in the MS databases of chemical standards or in computer-generated libraries present multiple solutions and the resulting candidate structures need to be ranked. Additionally, the technique for the comparison of experimental and calculated breakdown graphs (ionic intensities vs. impact energies) has not been developed as an automatic analytical method, although it was elaborated a long time ago [10]. Therefore, new methods may prove useful for the discrimination of isomers with similar fragmentations [11]. The coupling of chromatography with mass spectrometry has offered the possibility of analyzing complex mixtures of analytes. Although originally designed as a separation method, chromatography was later perfected to become a formidable analytical method using chemical standards. Recent studies have shown that the qualitative analysis of some symmetric and asymmetric isomers can also be performed by establishing the elution order, based on the thermodynamic characteristics of adsorption, using molecular statistical calculations [12].
Some techniques that use the quantitative structure–fragmentation relationship (QSFR) can filter the candidate structures using the reverse ordering of the calculated formation enthalpies (ΔfH) of their primary ions, together with the corresponding experimental ionic currents obtained by mass spectrometry. These are, however, limited to groups of isomers with a high similarity, where n possible structures are attributed to n unknown isomers (n = 2 to 6) or they require at least two isomeric standards of the analyte [13,14,15,16,17].
The extension of the study of these QSFR techniques to many classes of substances is necessary if their generalization is to be pursued. At the same time, a new variant that uses precalculated databases of primary mass energy fingerprints is subject to attention. Our project comprised three stages: (i) the elaboration of calculated mass energy profiles; (ii) the optimization and validation of the method using the mass spectra of some isomeric chemical standards with different molecular symmetries; (iii) the individual identification of analytes through the filtering of candidate structures. The current article presents the creation of a database with fragmentation mass energy profiles for ten candidate structures of di-O-isopropylidene monosaccharides. The enthalpies (ΔfH) and Gibbs energies (ΔfG) obtained using four variants of quantum chemical methods were used to calculate the theoretical descriptors. Graphical and statistical interpretations allowed for assessments of accuracy for the future analysis of these isomers by mass spectrometry.
2. Materials and Methods
2.1. Candidate Structures
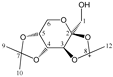
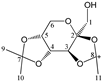
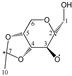
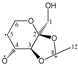
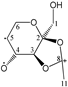
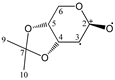
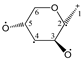
Enthalpies (ΔfH) and Gibbs energies (ΔfG) were calculated for ten structural isomers and diastereomers (1–10). These isomers had four or five asymmetric carbon atoms. They were chosen to present small and very small structural differences, so as to be useful in assessing the level of accuracy that the studied technique can offer. The candidate structures (1–5), presented in Figure 1, were:
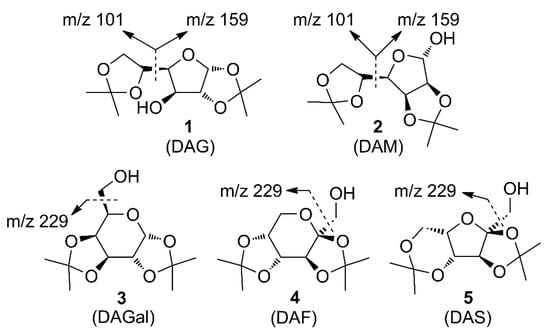
Figure 1.
The candidate isomeric structures (1–5).
1,2:5,6-di-O-isopropylidene-α-d-glucofuranose (1);
2,3:5,6-di-O-isopropylidene-α-d-mannofuranose (2);
1,2:3,4-di-O-isopropylidene-α-d-galactopyranose (3);
2,3:4,5-di-O-isopropylidene-β-d-fructopyranose (4); and
2,3:4,6-di-O-isopropylidene-α-l-sorbofuranose (5).
For the ease of reference, they were assigned the following abbreviations: DAG (Diacetone-d-glucose) (1); DAM (Diacetone-d-mannose) (2); DAGal (Diacetone-d-galactose) (3); DAF (Diacetone-d-fructose) (4); and DAS (Diacetone-l-sorbose) (5). They were used as chemical standards in the optimization and validation of the qualitative analysis.
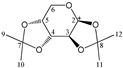
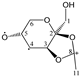
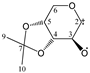
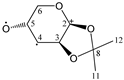
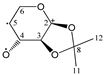
The candidate structures (6–10) that are presented in Figure 2 were the isomers of Structures 1–5. Similarly, for the ease of reference, they were also assigned the following abbreviations: DAAlo (Diacetone-d-allose) (6); DAF_spiran (Diacetone-d-frutose_spirane) (7); DAGal_furan (Diacetone-d-galactose_furan) (8); DAM_beta (Diacetone-d-mannose_beta) (9); and DAS_spiran (Diacetone-l-sorbose_spirane) (10). These abbreviations corresponded to:
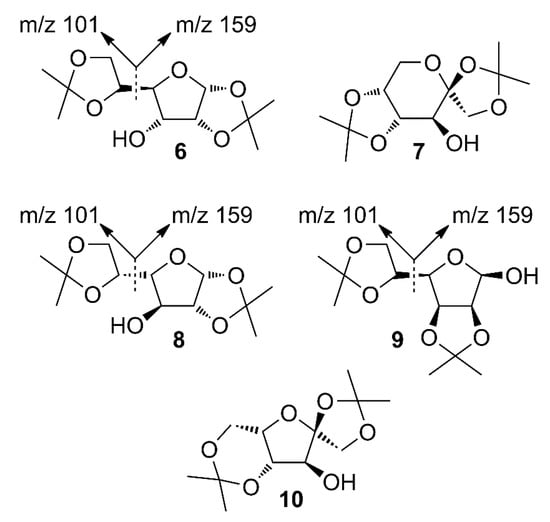
Figure 2.
The candidate isomeric structures (6–10).
1,2:5,6-di-O-isopropylidene-α-d-allofuranose (6);
1,2:4,5-di-O-isopropylidene-β-d-fructopyranose (7);
1,2:5,6-di-O-isopropylidene-α-d-galactofuranose (8);
2,3:5,6-di-O-isopropylidene-β-d-mannofuranose (9); and
1,2:4,6-di-O-isopropylidene-α-l-sorbofuranose (10).
Structures 1, 6 and 8 were diastereomers, as were Structures 2 and 9.
Thus, each experimental mass energy profile of the chemical standard was compared to the ten calculated profiles. Maximum fit was obtained for the true structure when the calculated energy descriptors correctly described the fragmentation.
Calculations were made only for the primary ions because their structure is obvious and the fragmentation energy is easier to obtain. The formation of these ions occurs with minimum energy consumption because they result from breaking a single chemical bond and, eventually, the elimination of small molecules. Another advantage is that the signals of the primary ions also appear in the mass spectra that are measured at low impact energies. Seven such ions were used for di-O-isopropylidene monosaccharides (Table 1).

Table 1.
The seven considered ions of isomers 1–10.
2.2. Activation Energy Descriptor
The fragmentation enthalpies and Gibbs energies can reasonably approximate the activation energy and the quantum chemical calculations (QCC) can provide reliable values for them when no reverse activation energy of fragmentation is present. After vertical ionization, molecular ions explore many pathways up to their respective transition states and prefer those that are thermodynamically and kinetically favorable [18,19]. Thus, the minimum fragmentation enthalpies or Gibbs energies should best describe the activation energy profile used via this technique. For the fragmentation of the molecule M into the ion Ii+ and fragments Fi (radicals and/or molecules), the enthalpies (ΔfHfrag) and Gibbs energies (ΔfGfrag) were calculated with Equations (1) and (2), respectively:
where ΣΔfH (Fi) is the sum of the formation enthalpies of the accompanying fragments, ΔfH (Ii+) is the formation enthalpy of the resulting fragmentation ion and ΔfH (M) is the molecular enthalpy of the candidate structure. The meanings are similar for ΔfG.
ΔfHfrag = ΔfH (Ii+) + ΣΔfH (Fi) − ΔfH (M)
ΔfGfrag = ΔfG (Ii+) + ΣΔfG (Fi) − ΔfG (M)
2.3. Computational Methods
The semi-empirical methods RM1 and PM7 were chosen because they have provided good results in previous studies and are more affordable, and DFTs were chosen because they are current and considered more efficient [13,14,15,16,17]. They also offer a variety of mass energy profiles. All structures were initially modeled using the HyperChem 8.0.10 software [20]. The starting neutral molecules, obtained after MM+ pre-optimization, were optimized using the RM1 semi-empirical method [21]. The radical cations were obtained from these structures and were finally optimized using RM1. As for “spin pairing”, RHF operators were used for neutral molecules and cations while UHF operators were employed for radicals and radical cations. The SCF “convergence limit” was set at 10−5, without using the “accelerate convergence” procedure. For geometry optimization and ΔfH calculation, the “Polak–Ribière (conjugate gradient)” algorithm was selected with an RMS gradient of 0.01 kcal/(Å·mol) because the molecules were being considered in a vacuum.
The MOPAC2012 software [22] was used for the PM7 semi-empirical method [23], with the “CHARGE = +1” option for cations and the “UHF” option for radicals. The same HyperChem starting structures were converted into “ZMT” (MOPAC Z-matrix) files and run through the MOPAC2012 interface for geometry optimization and ΔfH calculation. The line of parameters included “GNORM = 0.01”, “BONDS”, “AUX”, “GRAPHF”, “PDBOUT” and the keyword “OPT” whenever possible, as well as the keywords “SINGLET” (for neutral molecules and cations) or “DOUBLET” (for radical cations and radicals). The “EF” algorithm was used for geometry optimization and the resulting structures were analyzed with the Jmol software [24].
The theoretical calculations were also performed using the Gaussian 09 software [25]. The equilibrium geometries of the target species were optimized using the density functional theory (DFT) method at the B3LYP/6-31G level. The hybrid B3LYP functional was used for these studies because of convention, its successful use in modeling a range of gas-phase reactions and also because it is not very computationally expensive. The set 6-31G basis was deemed more than sufficient for the C, H and O elements present in carbohydrate acetals [26,27,28]. The DFT input files were the RM1-optimized structures.
3. Results
In order to establish the main currents that could have an essential contribution to the signal of the primary ions, calculations were made for all of their possible structures. For example, Table 2 presents the ΔfH data that were calculated using the RM1 method for all isomeric structures of the primary ions resulting from DAF (Structure 4). The minimum ΔfH values are in bold. These corresponded to the fragmentation pathways with the lowest activation energy and, therefore, almost exclusively frequented. No possible isomerization of the structures before fragmentation was considered. Similar calculations were made for each candidate structure (1–10).

Table 2.
Series of ΔfH values calculated using the RM1 method for all isomeric structures of primary ions resulting from DAF (Structure 4). The minimum values are bold formatted. The ion structures presented in this table are those from before optimization.
The enthalpies and Gibbs energies that were computed using semi-empirical methods RM1, PM7 and DFT (at the B3LYP/6-31G level of theory) for each candidate structure are shown in Table 3, Table 4, Table 5 and Table 6. The minimum ΔfH and ΔfG values (kcal/mol) and Equations (1) and (2) were used, where ΔfH (M) is the molecule enthalpy, ΔfH (M+) is the molecular ion enthalpy and ΣΔfH (F) is the sum of the fragment enthalpies. The meanings are similar for ΔfG. Although easy to calculate, the database did not contain ionization energies because molecular ions do not appear in the electron ionization mass spectra of di-O-isopropylidene monosaccharides [29]. The ions at m/z 245, m/z 187 and m/z 127 had calculated energy for all structures (1–10). This is because each of them could fragment into those ions by losing methyl, hydroxyl and/or one or two molecules of acetone (Table 1, Figure 1 and Figure 2). Several cells in Table 3, Table 4, Table 5 and Table 6 are empty because the structures could not form the corresponding primary ions. Thus, the ions at m/z 229 and m/z 171 appeared only for Structures 3–5 because they explicitly contain the -CH2OH group, which the other structures (1, 2 and 6–10) do not contain (Figure 1 and Figure 2, Table 3, Table 4, Table 5 and Table 6). Similarly, the ions at m/z 159 and m/z 101 appeared only for Structures 1, 2, 6, 8 and 9 as they explicitly contain the -C5H9O2 group, which the other structures (3–5, 7 and 10) do not contain (Figure 1 and Figure 2, Table 3, Table 4, Table 5 and Table 6).

Table 3.
ΔfH database (kcal/mol) calculated using RM1 and Equation (1).

Table 4.
ΔfH database (kcal/mol) calculated using PM7 and Equation (1).

Table 5.
ΔfH database (kcal/mol) calculated using DFT (B3LYP/6-31G) and Equation (1).

Table 6.
ΔfG database (kcal/mol) calculated using DFT (B3LYP/6-31G) and Equation (2).
4. Discussion
The mass spectrum does not indicate the absolute value of ionic currents but does represent their relativity in arbitrary units. Thus, it was expected that the most useful mass energy profiles for structure searching would be those whose relativities ensured the best inverse correlation with the profiles of the corresponding ionic currents [17]. Obviously, this was established only when validating with the mass spectra of the chemical standards.
However, preliminary assessments of databases are very important. Thus, the comparison of the profiles resulting from the four methods for the same structure showed their similarity, according to the correlation index R (Table A1) and their average, which was higher than 0.90 (Table 7). Here, we showed that some semi-empirical methods, which were not designed for cation radicals (although the settings do allow this) and do not work well for species with many lone-pair electrons, can provide a series of fragmentation energies with appropriate relativities. Therefore, these calculations could satisfactorily describe the fragmentation of the candidate structures and the small differences between the calculated energy series could provide optimization methods for structural searches [16]. The Pearson linear correlation function normalizes covariance. It is suitable for comparing mass energy profiles because it eliminates the systematic differences that appear between semi-empirical or quantum methods. This is possible because the function does not change its value when the terms of one of the compared strings are added (subtracted) or multiplied by the same number. This function is part of a relationship that can accurately highlight the relative differences between calculated profiles and can order them according to the matched experimental energy profiles [17].

Table 7.
The means of the R correlation indices for the fragmentation energy profiles of Structures 1–10 using the four methods for quantum calculations. Linear correlations were made between pairs of strings with energies calculated using two different methods, with the order of values being that in the relevant columns of Table 3, Table 4, Table 5 and Table 6.
However, the most important analytical aspect was to assess whether there were mass energy differences between these ten isomers that were large enough to distinguish their chemical structures. Radar charts (in MS Excel) of the fragmentation energies as a function of the mass of the primary ions resemble fingerprints. They can be a means for the quick and accurate visual comparison of profiles (Figure 3). In these representations, only the points that correspond to the successive ionic masses in the graph are joined by a line. The points and lines, which were placed differently from the gridlines of energy, highlighted three groups of profiles for the structures: (1, 2, 6, 8, 9), (3–5) and (7, 10). In each group, the candidate structures showed only small stereochemical differences, with small exceptions. Even so, there were differences in the mass energy profiles that could be decisive in their correct identification.
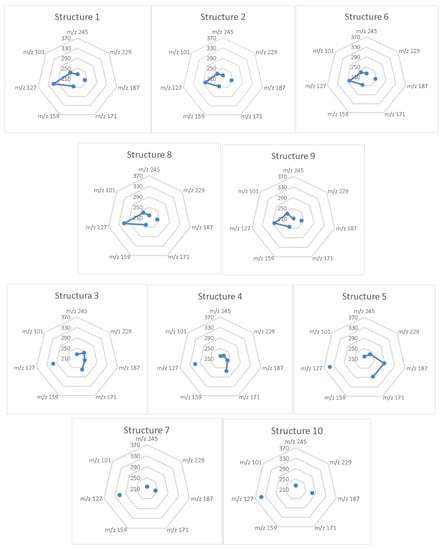
Figure 3.
The radar charts (MS Excel) of fragmentation energies (kcal/mol) corresponding to the primary ions, calculated using the DFT ΔG method. Similar profiles (fingerprints) are grouped: (1, 2, 6, 8, 9), (3–5) and (7, 10). In groups (1, 6, 8) and (2, 9), the structures were diastereomeric.
An ANOVA of the energy rows (calculated using RM1) of Structures 1, 2, 6, 8 and 9 showed that, for the fragmentation M -> m/z 127, the variance was much higher than that of the other rows (Table 8). This string contained aberrant values and had to be removed from the profile as it reduced the selectivity of the analysis. Table 9 contains the R indices from before removing the aberrant string (yellow background in Table 8). These indices had very high values of over 0.99 and showed that the five profiles were suitable for all five diastereomers and could not differentiate them.

Table 8.
ΔfH (M frag) calculated using RM1 (selection from Table 3).

Table 9.
The linear correlation indices R of the energy columns in Table 8 before (left) and after (right) the elimination of the corresponding aberrant string M -> m/z 127.
Following the elimination of the aberrant string, the selectivity increased as all of the R indices decreased in value. Only the pairs Structures 1 and 6 and Structures 8 and 9 still had indices around 0.99, but they were still lower than initially (Table 9, right-hand side, yellow background). The situation was similar for the PM7 and DFTs profiles (Table A2, Table A3 and Table A4).
As shown in Results (Section 3), starting with ten candidate structures of di-O-isopropylidene monosaccharides, a database was built using various types of quantum chemical calculations. The calculated theoretical descriptors, i.e., the enthalpies (ΔfH) and Gibbs energies (ΔfG), could thus be used in the analysis and statistical interpretation of isomer discrimination.
This database of mass energy fragmentation descriptors is ready for validation. It is far from complete for the composition of C12H20O6, but its 40 mass energy profiles should be sufficient to identify the isomers of di-O-isopropylidene monosaccharides with this formula. All four quantum calculation methods were able to distinguish between Isomers 1–10 (Figure 1 and Figure 2) using different values of fragmentation energies that led to ions with the same mass. Even for diastereomers, only the small structural differences that were generated by the configurations of asymmetric carbon atoms could be observed. The pairs of enantiomers could not be discriminated using this technique because they have identical profiles. Given these aspects and the good results that have been presented in similar studies [11,12,13,14,15,16,17], it is expected that the correlation of the profiles with the mass spectra of the chemical standards is only maximum for the truly positive results. Optimizing the experimental impact energy and the correlation relationship could also improve the analysis.
5. Conclusions
Quantum calculations can highlight changes in the mass energy profiles of fragmentation by changing the configurations of the asymmetric carbon atoms in chemical structures. The quantification of energy distribution in primary molecular fragmentation by mass spectrometry, in the form of mass energy fingerprints, opens broad perspectives for full de novo structural searches using correct stereochemical assignments. This technique could be integrated into expert digital systems for automatic mass spectral interpretation.
Author Contributions
Conceptualization, M.-C.P., N.D., E.S. and M.G.; methodology, M.-C.P., N.D. and E.S.; software, C.C. and R.B.; validation, M.-C.P., N.D., E.S., A.S. and M.G.; formal analysis, M.-C.P., N.D., C.C., A.S. and E.S.; investigation, M.-C.P., N.D., C.C., E.S., A.S., R.B. and M.G.; resources, C.C., R.B. and M.G.; data curation, M.-C.P., N.D., C.C., E.S. and A.S.; writing—original draft preparation, M.-C.P., N.D., C.C., A.S. and M.G.; writing—review and editing, M.-C.P., N.D., C.C., A.S. and M.G.; visualization, M.-C.P., N.D., E.S., A.S. and M.G. supervision, M.-C.P., N.D., E.S. and M.G.; project administration, M.-C.P., N.D. and E.S. All authors have read and agreed to the published version of the manuscript.
Funding
Part of this research was conducted at the Center of Genomic Medicine at the “Victor Babes” University of Medicine and Pharmacy of Timisoara (POSCCE 185/48749; contract No. 677).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
Appendix A

Table A1.
The R correlation indices for the mass fragmentation energy profiles of Structures 1–10 using the four methods for semi-empirical and quantum calculations.
Table A1.
The R correlation indices for the mass fragmentation energy profiles of Structures 1–10 using the four methods for semi-empirical and quantum calculations.
| Structure 1 | RM1 | PM7 | DFT DH | DFT DG | Structure 6 | RM1 | PM7 | DFT DH | DFT DG |
|---|---|---|---|---|---|---|---|---|---|
| RM1 | 1 | 0.999 | 0.93 | 0.989 | RM1 | 1 | 0.991 | 0.783 | 0.949 |
| PM7 | 0.999 | 1 | 0.921 | 0.986 | PM7 | 0.991 | 1 | 0.756 | 0.933 |
| DFT DH | 0.93 | 0.921 | 1 | 0.956 | DFT DH | 0.783 | 0.756 | 1 | 0.857 |
| DFT DG | 0.989 | 0.986 | 0.956 | 1 | DFT DG | 0.949 | 0.933 | 0.857 | 1 |
| Structure 2 | RM1 | PM7 | DFT DH | DFT DG | Structure 7 | RM1 | PM7 | DFT DH | DFT DG |
| RM1 | 1 | 0.988 | 0.795 | 0.955 | RM1 | 1 | 0.956 | 1 | 0.992 |
| PM7 | 0.988 | 1 | 0.841 | 0.961 | PM7 | 0.956 | 1 | 0.955 | 0.985 |
| DFT DH | 0.795 | 0.841 | 1 | 0.89 | DFT DH | 1 | 0.955 | 1 | 0.992 |
| DFT DG | 0.955 | 0.961 | 0.89 | 1 | DFT DG | 0.992 | 0.985 | 0.992 | 1 |
| Structure 3 | RM1 | PM7 | DFT DH | DFT DG | Structure 8 | RM1 | PM7 | DFT DH | DFT DG |
| RM1 | 1 | 0.944 | 0.921 | 0.96 | RM1 | 1 | 0.922 | 0.981 | 0.986 |
| PM7 | 0.944 | 1 | 0.934 | 0.981 | PM7 | 0.922 | 1 | 0.87 | 0.957 |
| DFT DH | 0.921 | 0.934 | 1 | 0.954 | DFT DH | 0.981 | 0.87 | 1 | 0.974 |
| DFT DG | 0.96 | 0.981 | 0.954 | 1 | DFT DG | 0.986 | 0.957 | 0.974 | 1 |
| Structure 4 | RM1 | PM7 | DFT DH | DFT DG | Structure 9 | RM1 | PM7 | DFT DH | DFT DG |
| RM1 | 1 | 0.946 | 0.993 | 0.996 | RM1 | 1 | 0.984 | 0.814 | 0.916 |
| PM7 | 0.946 | 1 | 0.939 | 0.929 | PM7 | 0.984 | 1 | 0.831 | 0.948 |
| DFT DH | 0.993 | 0.939 | 1 | 0.984 | DFT DH | 0.814 | 0.831 | 1 | 0.931 |
| DFT DG | 0.996 | 0.929 | 0.984 | 1 | DFT DG | 0.916 | 0.948 | 0.931 | 1 |
| Structure 5 | RM1 | PM7 | DFT DH | DFT DG | Structure 10 | RM1 | PM7 | DFT DH | DFT DG |
| RM1 | 1 | 0.984 | 0.987 | 0.973 | RM1 | 1 | 0.992 | 0.999 | 0.996 |
| PM7 | 0.984 | 1 | 0.948 | 0.927 | PM7 | 0.992 | 1 | 0.987 | 0.976 |
| DFT DH | 0.987 | 0.948 | 1 | 0.997 | DFT DH | 0.999 | 0.987 | 1 | 0.998 |
| DFT DG | 0.973 | 0.927 | 0.997 | 1 | DFT DG | 0.996 | 0.976 | 0.998 | 1 |

Table A2.
ΔfH (M frag) (kcal/mol) calculated using PM7 (selection from Table 4) and the linear correlation indices R of the energy columns before (left bottom) and after (right bottom) the elimination of the corresponding aberrant string M -> m/z 127.
Table A2.
ΔfH (M frag) (kcal/mol) calculated using PM7 (selection from Table 4) and the linear correlation indices R of the energy columns before (left bottom) and after (right bottom) the elimination of the corresponding aberrant string M -> m/z 127.
| Structures | 1 | 2 | 6 | 8 | 9 | Variance |
|---|---|---|---|---|---|---|
| M -> m/z 245 | 206.7 | 207.6 | 211.6 | 206.3 | 202 | 11.743 |
| M -> m/z 187 | 219.8 | 216 | 217.3 | 241.6 | 213.1 | 131.33 |
| M -> m/z 159 | 220.5 | 221.6 | 220.8 | 218.6 | 214.9 | 7.207 |
| M -> m/z 127 | 332.8 | 265.9 | 286.1 | 293.9 | 251.7 | 958.28 |
| M -> m/z 101 | 215.6 | 213.5 | 213.7 | 213.7 | 208.6 | 6.837 |
| Structures | 1 | 2 | 6 | 8 | 9 | |
| 1 | 1 | 0.9921 | 0.9986 | 0.9511 | 0.9879 | |
| 2 | 0.9921 | 1 | 0.9939 | 0.9440 | 0.9970 | |
| 6 | 0.9986 | 0.9939 | 1 | 0.9443 | 0.9875 | |
| 8 | 0.9511 | 0.9440 | 0.9443 | 1 | 0.9623 | |
| 9 | 0.9879 | 0.9970 | 0.9875 | 0.9623 | 1 | |
| Structures | 1 | 2 | 6 | 8 | 9 | |
| 1 | 1 | 0.9268 | 0.8833 | 0.7096 | 0.9897 | |
| 2 | 0.9268 | 1 | 0.9743 | 0.4646 | 0.9618 | |
| 6 | 0.8833 | 0.9743 | 1 | 0.5192 | 0.9410 | |
| 8 | 0.7096 | 0.4646 | 0.5192 | 1 | 0.6801 | |
| 9 | 0.9897 | 0.9618 | 0.9410 | 0.6801 | 1 |

Table A3.
ΔfH (M frag) (kcal/mol) calculated using DFT ΔH (selection from Table 5) and the linear correlation indices R of the energy columns before (left bottom) and after (right bottom) the elimination of the corresponding aberrant string M -> m/z 127.
Table A3.
ΔfH (M frag) (kcal/mol) calculated using DFT ΔH (selection from Table 5) and the linear correlation indices R of the energy columns before (left bottom) and after (right bottom) the elimination of the corresponding aberrant string M -> m/z 127.
| Structures | 1 | 2 | 6 | 8 | 9 | Variance |
|---|---|---|---|---|---|---|
| M -> m/z 245 | 211 | 207.8 | 209.4 | 206 | 199.6 | 19.47 |
| M -> m/z 187 | 210 | 216.8 | 215.5 | 211.2 | 213.7 | 8.113 |
| M -> m/z 159 | 229.4 | 230.7 | 230.6 | 224.3 | 229.3 | 6.923 |
| M -> m/z 127 | 263.5 | 239.4 | 236.6 | 265.1 | 241.6 | 192.5 |
| M -> m/z 101 | 227.8 | 223.2 | 222.2 | 222.7 | 223.1 | 5.155 |
| Structures | 1 | 2 | 6 | 8 | 9 | |
| 1 | 1 | 0.9100 | 0.9024 | 0.9900 | 0.8999 | |
| 2 | 0.9100 | 1 | 0.9956 | 0.9018 | 0.9952 | |
| 6 | 0.9024 | 0.9956 | 1 | 0.8838 | 0.9841 | |
| 8 | 0.9900 | 0.9018 | 0.8838 | 1 | 0.8949 | |
| 9 | 0.8999 | 0.9952 | 0.9841 | 0.8949 | 1 | |
| Structures | 1 | 2 | 6 | 8 | 9 | |
| 1 | 1 | 0.8730 | 0.8954 | 0.9609 | 0.8661 | |
| 2 | 0.8730 | 1 | 0.9925 | 0.9563 | 0.9906 | |
| 6 | 0.8954 | 0.9925 | 1 | 0.9499 | 0.9688 | |
| 8 | 0.9609 | 0.9563 | 0.9499 | 1 | 0.9667 | |
| 9 | 0.8661 | 0.9906 | 0.9688 | 0.9667 | 1 |

Table A4.
ΔfG (M frag) (kcal/mol) calculated using DFT ΔG (selection from Table 6) and the linear correlation indices R of the energy columns before (left bottom) and after (right bottom) the elimination of the corresponding aberrant string M -> m/z 127.
Table A4.
ΔfG (M frag) (kcal/mol) calculated using DFT ΔG (selection from Table 6) and the linear correlation indices R of the energy columns before (left bottom) and after (right bottom) the elimination of the corresponding aberrant string M -> m/z 127.
| Structures | 1 | 2 | 6 | 8 | 9 | Variance |
|---|---|---|---|---|---|---|
| M -> m/z 245 | 225.9 | 222.2 | 223.8 | 218.9 | 210.9 | 34.383 |
| M -> m/z 187 | 240.2 | 245.5 | 245.5 | 240.6 | 242.2 | 6.635 |
| M -> m/z 159 | 245.2 | 245.4 | 246.3 | 240.2 | 244.4 | 5.71 |
| M -> m/z 127 | 306.9 | 281.6 | 279 | 309.6 | 283.2 | 221.59 |
| M -> m/z 101 | 244.7 | 239.6 | 238.6 | 239.7 | 239.6 | 5.873 |
| Structures | 1 | 2 | 6 | 8 | 9 | |
| 1 | 1 | 0.9694 | 0.9651 | 0.9973 | 0.9490 | |
| 2 | 0.9694 | 1 | 0.9992 | 0.9797 | 0.9922 | |
| 6 | 0.9651 | 0.9992 | 1 | 0.9756 | 0.9896 | |
| 8 | 0.9973 | 0.9797 | 0.9756 | 1 | 0.9582 | |
| 9 | 0.9490 | 0.9922 | 0.9896 | 0.9582 | 1 | |
| Structures | 1 | 2 | 6 | 8 | 9 | |
| 1 | 1 | 0.9104 | 0.8862 | 0.9611 | 0.9622 | |
| 2 | 0.9104 | 1 | 0.9959 | 0.9753 | 0.9881 | |
| 6 | 0.8862 | 0.9959 | 1 | 0.9526 | 0.9745 | |
| 8 | 0.9611 | 0.9753 | 0.9526 | 1 | 0.9941 | |
| 9 | 0.9622 | 0.9881 | 0.9745 | 0.9941 | 1 |
References
- Vicidomini, C.; Roviello, V.; Roviello, G.N. In Silico Investigation on the Interaction of Chiral Phytochemicals from Opuntia ficus-indica with SARS-CoV-2 Mpro. Symmetry 2021, 13, 1041. [Google Scholar] [CrossRef]
- Kind, T.; Fiehn, O. Advances in structure elucidation of small molecules using mass spectrometry. Bioanal. Rev. 2010, 2, 23–60. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kind, T.; Fiehn, O. Seven Golden Rules for heuristic filtering of molecular formulas obtained by accurate mass spectrometry. BMC Bioinform. 2007, 8, 105. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rasche, F.; Svatoš, A.; Maddula, R.K.; Böcker, C.; Böcker, S. Identifying the unknowns by aligning fragmentation trees. Anal. Chem. 2011, 83, 1243–1251. [Google Scholar] [CrossRef] [PubMed]
- Allen, F.; Greiner, R.; Wishart, D. Competitive fragmentation modeling of ESI-MS/MS spectra for putative metabolite identification. Metabolomics 2015, 11, 98–110. [Google Scholar] [CrossRef] [Green Version]
- Mistrík, R. Mass FrontierTM 3.0. Available online: http://www.highchem.com (accessed on 20 March 2017).
- Cautereels, J.; Claeys, M.; Geldof, D.; Blockhuys, F. Quantum chemical mass spectrometry: Ab initio prediction of electron ionization mass spectra and identification of new fragmentation pathways. J. Mass Spectrom. 2016, 51, 602–614. [Google Scholar] [CrossRef] [PubMed]
- Grimme, S. Towards first principles calculation of electron impact mass spectra of molecules. Angew. Chem. Int. Ed. Engl. 2013, 52, 6306–6312. [Google Scholar] [CrossRef] [PubMed]
- Bauer, C.; Grimme, S. How to Compute Electron Ionization Mass Spectra from First Principles. J. Phys. Chem. A 2016, 120, 3755–3766. [Google Scholar] [CrossRef] [PubMed]
- Rosenstock, H.M.; Wallenstein, M.B.; Wahrhaftig, A.L.; Eyring, H. Absolute rate theory for isolated systems and the mass spectra of polyatomic molecules. Proc. Natl. Acad. Sci. USA 1952, 38, 667–678. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vaniya, A.; Fiehn, O. Using fragmentation trees and mass spectral trees for identifying unknown compounds in metabolomics. TrAC Trends Anal. Chem. 2015, 69, 52–61. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fedorova, E.; Stavrianidi, A.; Minenkova, I.; Buryak, A. Calculations of the Thermodynamic Characteristics and Physicochemical Properties of Symmetric and Asymmetric Isomeric Compounds for Identification in Chromatography-Mass Spectrometry. Symmetry 2021, 13, 1681. [Google Scholar] [CrossRef]
- Dinca, N. Chemical structure identification by differential mass spectra. In Applications of Mass Spectrometry in Life Safety—NATO Science for Peace and Security Series A: Chemistry and Biology; Popescu, C., Zamfir, A.D., Dinca, N., Eds.; Springer: Dordrecht, The Netherlands, 2008; pp. 221–233. [Google Scholar]
- Dinca, N.; Stanescu, M.D.; Sisu, E.; Mracec, M. Differential mass spectrometry (Diff MS) and computational chemistry. II. Diff MS and MO semi-empirical analyses of exo- and endo-5,10-methylene-10,11-dihydro-5H-dibenzo[a,d]cyclohepten-11-ols. Rev. Chim. 2004, 55, 347–350. [Google Scholar]
- Harja, F.; Bettendorf, C.; Grosu, I.; Dinca, N. Stereochemistry studies of some 1,3-dioxane derivatives by differential mass spectrometry and computational chemistry. In Applications of Mass Spectrometry in Life Safety—NATO Science for Peace and Security Series A: Chemistry and Biology; Popescu, C., Zamfir, A.D., Dinca, N., Eds.; Springer: Dordrecht, The Netherlands, 2008; pp. 185–191. [Google Scholar]
- Dinca, N.; Covaci, A. Structural identification by differential mass spectrometry as a criterion for selecting the best quantum chemical calculation of formation enthalpy for tetrachlorinated biphenyls. Rapid Commun. Mass Spectrom. 2012, 26, 2033–2040. [Google Scholar] [CrossRef] [PubMed]
- Dinca, N.; Dragan, S.; Dinca, M.; Sisu, E.; Covaci, A. New quantitative structure-fragmentation relationship strategy for chemical structure identification using the calculated enthalpy of formation as a descriptor for the fragments produced in electron ionization mass spectrometry: A case study with tetrachlorinated biphenyls. Anal. Chem. 2014, 86, 4949–4955. [Google Scholar] [PubMed]
- Gross, J. Mass Spectrometry—A Textbook, 3rd ed.; Springer: Cham, Switzerland, 2011; pp. 36–40. [Google Scholar]
- Splitter, J.S. Applications of Mass Spectrometry to Organic Stereochemistry, 1st ed.; Verlag Chemie: Weinheim, Germany, 1994; pp. 343–352. [Google Scholar]
- Hypercube, Inc. HyperChem™ Professional, Version 8.0.10 for Windows; Hypercube, Inc.: Gainesville, FL, USA. Available online: http://www.hypercubeusa.com/?tabid=360 (accessed on 20 March 2021).
- Rocha, G.B.; Freire, R.O.; Simas, A.M.; Stewart, J.J.P. RM1: A reparameterization of AM1 for H, C, N, O, P, S, F, Cl, Br, and I. J. Comput. Chem. 2006, 27, 1101–1111. [Google Scholar] [CrossRef] [PubMed]
- MOPAC2012; Stewart, J.J.P. Stewart Computational Chemistry, Colorado Springs, CO, USA. Version 15.027W. Available online: http://OpenMOPAC.net/ (accessed on 20 March 2021).
- Stewart, J.J.P. Optimization of parameters for semiempirical methods VI: More modifications to the NDDO approximations and re-optimization of parameters. J. Mol. Model. 2013, 19, 1–32. [Google Scholar] [CrossRef] [Green Version]
- Jmol: An Open-Source Java Viewer for Chemical Structures in 3D. Available online: http://www.jmol.org/ (accessed on 20 March 2021).
- Frisch, M.J.; Trucks, G.W.; Schlegel, H.B.; Scuseria, G.E.; Robb, M.A.; Cheeseman, J.R.; Scalmani, G.; Barone, V.; Mennucci, B.; Petersson, G.A.; et al. Gaussian 09. Revision B.01; Gaussian, Inc.: Wallingford, CT, USA, 2010. [Google Scholar]
- Wang, S.; Dong, C.; Yu, L.; Guo, C.; Jiang, K. Dissociation of protonated N-(3-phenyl-2H-chromen-2-ylidene)-benzenesulfonamide in the gas phase: Cyclization via sulfonyl cation transfer. Rapid Commun. Mass Spectrom. 2016, 30, 95–100. [Google Scholar] [CrossRef]
- Pinto, A.C.; Vessecchi, R.; da Silva, C.G.; Lourenço Amorim, A.C.; dos Santos Júnior, H.M.; Rezende, M.J.C.; Gates, P.J.; Rezende, C.M.; Lopes, N.P. Electrospray ionization tandem mass spectrometry analysis of isopimarane diterpenes from Velloziaceae. Rapid Commun. Mass Spectrom. 2016, 30, 61–68. [Google Scholar] [CrossRef] [Green Version]
- Osburn, S.; Plaviak, A.; Pestok, J.; Van Stipdonk, M.J. Apparent activation of H2O and elimination of H2 from gas-phase mixed-metal complexes containing silver, calcium and deprotonated glycine. Rapid Commun. Mass Spectrom. 2016, 30, 101–111. [Google Scholar] [CrossRef] [PubMed]
- Pascariu, M.C.; Șișu, E.; Ordodi, V.L.; Rusnac, L.M. Spectral Analysis of Diisopropylidenated Monosaccharides. Low Energy EI-MS Fragmentation Study. Chem. Bull. “Politeh.” Univ. 2011, 56, 6–11. [Google Scholar]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).