Abstract
Conventional cancer treatments lack specificity and often cause severe side effects. Targeted therapeutic approaches are therefore preferred, including the use of immunotoxins (ITs) that comprise cell-binding and cell death-inducing components to allow the direct and specific delivery of pro-apoptotic agents into malignant cells. The first generation of ITs consisted of toxins derived from bacteria or plants, making them immunogenic in humans. The recent development of human cytolytic fusion proteins (hCFP) consisting of human effector enzymes offers the prospect of highly-effective targeted therapies with minimal side effects. One of the most promising candidates is granzyme B (GrB) and this enzyme has already demonstrated its potential for targeted cancer therapy. However, the clinical application of GrB may be limited because it is inactivated by the overexpression in tumors of its specific inhibitor serpin B9 (PI-9). It is also highly charged, which means it can bind non-specifically to the surface of non-target cells. Furthermore, human enzymes generally lack an endogenous translocation domain, thus the endosomal release of GrB following receptor-mediated endocytosis can be inefficient. In this review we provide a detailed overview of these challenges and introduce promising solutions to increase the cytotoxic potency of GrB for clinical applications.
1. Introduction
Typical treatment strategies for cancer include surgery, chemotherapy and radiotherapy, but success is limited by the concurrent killing of nonmalignant cells, which can result in serious and often life-threatening side effects. Many tumor cells also become resistant to drugs, which substantially reduces the response to treatment and results in a poor prognosis. Surgical resection of primary tumors leads to high relapse rates reflecting the incomplete removal of tumor cells as well as the ineffective control of metastatic disease. Therefore, there is a strong demand for more potent and selective treatment options.
Targeted immunotherapeutic approaches for the specific elimination of malignant cells have the potential to meet these criteria. Immunotherapy generally involves the induction of several mechanisms that interfere with carcinogenesis and tumor cell proliferation, although active immunotherapy is a more specialized approach in which the immune system itself is stimulated to remove malignant cells [1,2]. Targeted therapy involves either the direct activation or inactivation of (soluble) enzymes, growth factors or surface receptors in signaling pathways that are necessary for tumor maintenance and proliferation (e.g., growth factor receptors such as EGFR (Epidermal Growth Factor Receptor), Her2, BRAF and Kit) or the indirect targeted delivery of effector molecules to tumor cells, leading to growth arrest or remission [3,4]. This may involve drugs that penetrate the tumor cell plasma membrane allowing the targeting of intracellular tumor antigens, or the use of monoclonal antibodies (mAbs) that target cell surface molecules.
Monoclonal antibody therapy (passive immunotherapy) is a rapidly-expanding immunotherapeutic treatment platform for cancer, and at least 12 therapeutic mAbs for oncologic indications have already been approved by the FDA [5] while several more are in clinical development [6]. However, the activity of mAbs is limited by their relatively large size, resulting in poor tumor penetration and the need for high doses to compete with serum IgGs, leading to severe side-effects. Furthermore the unfavorable Fc receptor polymorphisms can reduce the impact of Fc-mediated effector functions. Therefore, unmodified antibodies are rarely suitable as therapeutics [7,8].
Several strategies have been developed to enhance the anti-tumor activity of mAbs, including the humanization of antibodies [9], the optimization of antibody effector functions [10] and the fusion of antibodies to a potent cytotoxic drug. In the early 1900s, Paul Ehrlich considered the idea of a “magic bullet” forming the basis of the antibody-drug conjugate (ADC) and immunotoxin (IT) concepts in which antibodies or antibody fragments specifically targeting tumor antigens deliver cytotoxic payloads and trigger their internalization into abnormal cells [11]. ADCs usually comprise a full-size antibody chemically linked to a toxin, drug or radionuclide. The therapeutic potential of ADCs has been demonstrated in numerous preclinical studies [7,12]. The FDA has approved Brentuximab vedotin, a CD30-directed ADC consisting of the anti-CD30 mAb brentuximab linked to the antimitotic agent monomethyl auristatin E for the treatment of Hodgkin’s lymphoma and anaplastic large cell lymphoma [13]. The FDA also approved gemtuzumab ozogamicin (Mylotarg), a humanized anti-CD33 mAb conjugated to calicheamicin for the treatment of acute myeloid leukemia (AML) [14], although this has now been withdrawn from the market due to systemic adverse effects.
In the case of ITs the effector domain is a naturally-occurring toxin (initially of bacterial or plant origin) that inhibits protein synthesis or induces apoptosis by modifying signal transduction pathways [15,16]. Cell death-inducing toxins and chemotherapeutics use different mechanisms of action, therefore ITs are promising for the treatment of chemoresistant tumors. The first generation of ITs, comprising full-size antibodies chemically conjugated to whole-protein toxins, have now been replaced by second and third generation ITs comprising truncated toxins (with their natural binding domain deleted to reduce adverse effects) genetically linked to shorter antibody fragments (scFv) or natural ligands which makes them easier to manufacture and also improves stability and tumor penetration [17].
First-generation ITs have shown promising results in in vitro studies and preclinical tests [18,19,20,21,22,23,24,25,26,27,28] and have also been tested in a small number of clinical trials [29,30]. The most prominent example is a recombinant IT comprising the diphtheria toxin and interleukin-2, which has been approved by the FDA for the treatment of T-cell lymphoma [31]. However, the clinical deployment of first-generation ITs is limited by their immunogenicity, which results in the generation of neutralizing antibodies that prevent the use of repetitive doses.
The complete humanization of ITs is therefore an important criterion for the development of fourth-generation molecules [29,32], named human cytolytic fusion proteins (hCFPs). This concept is supported by the use of endogenous human enzymes that induce cell death, fused to humanized or even fully human antibody fragments. Candidate human enzymes include proteases, RNases and kinases [32,33,34,35].
In this review we focus on granzyme B (GrB), a human immunoprotease involved in the granule-mediated apoptosis of virus-infected or transformed tumor cells. This enzyme has several inherent advantages as an IT component such as its high cytotoxic efficacy reflecting its status as an effector molecule in the cellular immune system, its broad portfolio of apoptosis inducing mechanisms, and its low immunogenicity. Even so, there are three major factors that have limited the clinical application of GrB: (i) it is specifically inhibited by serpin B9, which usually protects cells from misdirected GrB but is also upregulated in certain tumors; (ii) it has a high isoelectric point, resulting in a positive surface charge contributing to non-specific binding to healthy cells; and (iii) it is released slowly after uptake by the receptor-mediated endocytosis which reduces its cytotoxic impact.
Our group and others have developed a number of innovative strategies to optimize hCFPs based on GrB, including the production of engineered variants that are insensitive to PI-9, having lower surface charge to reduce off-target effects, and to promote endosomal release by the insertion of functional elements.
2. Generation of hCFPs
2.1. Humanization of the Tumor-Targeting Component
The major limitation of second and third generation ITs reflects the long-term nature of cancer treatment and the need for repeated doses, which means any component of nonhuman origin can induce neutralizing antibodies in patients with a functional immune system, thereby reducing therapeutic efficiency [36,37,38,39]. These problems can be caused either by the tumor-selective ligand or the cytotoxic component so that the complete humanization of ITs is preferable when repeated dose schedules are required, to improve safety and to ensure a positive clinical outcome.
If the tumor-selective ligand is an antibody or antibody fragment of murine origin, it can induce a human anti-mouse antibody (HAMA) response in some patients [40,41]. HAMA responses not only reduce treatment efficiency, but also have been shown to cause allergic reactions, in extreme cases leading to life-threatening complications such as renal failure.
Several strategies can be used to humanize the tumor-selective ligand including the fusion of the effector moiety to human cytokines and growth factors that target tumor receptors, e.g., IL-2, IL-4, EGF (epidermal growth factor), TGFα (tumor growth factor alpha) or the CD30 ligand [29,34,42]. Recombinant DNA technology can also be used to engineer antibodies for reduced immunogenicity [43]. The generation of chimeric antibodies comprising a murine Fab fragment and a human Fc region can reduce HAMA responses because the constant region makes the most significant contribution to immunogenicity [44,45]. Another strategy is the generation of humanized antibodies by grafting murine complementarity determining regions onto human V-region frameworks, an approach known as CDR replacement [46,47,48]. Complete humanization is also possible by selecting the V-regions from human antibody libraries or using transgenic knock-out mice or rats in which the gene loci for the light and heavy antibody chains are replaced by their human counterparts [49,50]. Single chain fragment variable (scFv) antibodies used for the production of fourth-generation ITs can be derived from humanized or human monoclonal antibodies or selected directly from a human scFv library.
2.2. Humanization of the Cell-Death Inducing Component
The conventional use of plant or bacterial toxins can induce an undesirable immune response and therefore several strategies have been investigated to reduce the immunogenicity of the cytotoxic component. These include the identification and removal (by mutation) of major immunogenic B-cell and T-cell epitopes within the toxins [51,52] as well as the derivatization of ITs using polyethylene glycol [53] or the co-application of immunosuppressive drugs [54,55].
A more favorable strategy for complete humanization is the use of endogenous human cytolytic enzymes, such as enzymes that induce cell death by a variety of mechanisms. Pro-apoptotic human proteins that are potentially useful for targeted cancer therapy include proteases and RNases such as angiogenin as well as death receptor ligands such as sTRAIL and FASL and pro-apoptotic members of the Bcl-2 family [32]. In addition to pro-apoptotic proteins, enzymes with tumor suppressive function such as DAPK2 (death-associated protein kinase 2) allow the targeted restoration of tumor suppressor activity in cancer cells [33,42].
3. Granzyme B in Targeted Cancer Therapy: Advantages and Challenges
3.1. Resistance of Tumor Cells to the Induction of Apoptosis
The efficacy of ITs/hCFPs depends on the sensitivity of tumor cells to pro-apoptotic signals. One of the major challenges in both targeted cancer therapy and standard chemotherapy approaches that induce programmed cell death is the tendency for tumor cells to develop resistance to pro-apoptotic signals allowing the invasion of host immunosurveillance [56]. Tumor cells can evolve multiple intrinsic and extrinsic escape mechanisms to overcome innate and adaptive immune responses. These include impaired tumor antigen presentation by the downregulation of MHC class I molecules, and the expression of factors that either directly kill or inhibit effector cells or recruit regulatory T-cells and thereby induce immunotolerance [57].
Tumor cells can regulate apoptosis by overexpressing different classes of anti-apoptotic proteins. For example, FLICE-inhibitory proteins (FLIPs) interfere with death receptor signal transduction pathways [58], whereas anti-apoptotic proteins of the Bcl-2 family regulate apoptosis at the mitochondrial level [59,60] and IAPs (inhibitors of apoptosis proteins) inhibit caspase activity [61]. Apoptosis-related escape mechanisms in tumor cells can reduce the efficiency of targeted therapy using human pro-apoptotic enzymes, but GrB is unusual because it utilizes multiple apoptosis induction mechanisms (see below) and is therefore more likely to overcome resistance mechanisms that evolve in tumors. The inherent pro-apoptotic activity of recombinant GrB in vitro emphasizes its potential in targeted cancer therapy (Table 1).
3.2. Cell Death Pathways Induced by Granzyme B
Granzyme B, a pro-apoptotic serine protease produced by effector cells of the innate and adaptive immune system, is involved in both non-specific and specific host rejection of tumors and cells infected with viruses [62,63]. GrB belongs to a family of serine proteases stored in the cytotoxic granules of natural killer (NK) cells and cytotoxic T-lymphocytes (CTLs) [64]. Five different human granzymes have been identified (A, B, H, K and M), each with unique expression profiles and substrate specificities [65]. The most intensively studied enzymes are GrB and GrA, both of which are considered to be major inducers of granule-mediated apoptosis [66]. Active GrB is stored as a complex on a scaffold comprising the chondroitin sulfate proteoglycan serglycin, the pore-forming protein perforin (PFN) and other members of the granzyme family [67,68]. During NK cell-mediated immunosurveillance, secretory granules are deposited into an immunological synapse that forms following the specific recognition of abnormal cells through an elaborate repertoire of cell surface receptors [69]. The precise mechanism by which GrB is taken up into cells and released from the endosome into the cytosol is still controversial. Two potential models for delivery into target cells have been reviewed in detail [70]. Following the recognition of the target cell, granules release their cytolytic contents into the immunological synapse and this directs GrB, probably with the assistance of perforin, into the cytosol of the target cell. Once released, GrB activates different signal transduction pathways to induce cell death.
The classical induction of apoptosis by GrB is comparable to the Fas mediated pathway, and involves the cleavage of several executive and downstream caspases mediated by both the caspase-pathway and the BID-pathway, the latter associated with the BAX/BAK-dependent release of cytochrome c from the mitochondria into the cytosol [71,72,73]. Direct activation of the effector caspase 3 is controlled by the inhibitor of apoptosis protein (IAP), and this must be inactivated, e.g., by HtrA2/OMI, a pro-apoptotic effector released from disrupted mitochondria (see below). Successful processing of pro-caspase-3 leads to the cleavage of several downstream death substrates including the inhibitor of caspase-activated DNase (ICAD), poly(ADP ribose) polymerase (PARP), DNA-dependent protein kinase (DNA-PK), the nuclear mitotic apparatus protein (NuMA) and the nuclear-envelope intermediate-filament protein lamin B. This results in the typical morphological changes associated with apoptosis, i.e., DNA fragmentation, chromatin condensation, membrane blebbing, cell shrinkage and the formation of apoptotic bodies [71,74,75,76,77].

Table 1.
Overview of different granzyme B-based cytolytic fusion proteins (CFPs) published so far.
| CFP (expression system) | Indication | Target receptor / antigen | Target cells | IC50 values | PI-9 expression reported from others | Structural hallmarks | Reference | |
|---|---|---|---|---|---|---|---|---|
| Gb-H22 (scFv) (HEK293T) | AML, CD64+ malignancies | CD64 | U937 | 1.7–17 nM | no * | In vitro activation via EK | [78,79] | |
| AML cells (ex vivo) | -- | |||||||
| HL60 | ~ 4–7 nM | no * | ||||||
| GrB-scFvMEL (E.coli) | Melanoma | gp240 | A375-M | ~ 20 nM | n.d. | In vitro activation via EK | [80] | |
| GrB-scFvMEL (E.coli) | Melanoma | gp240 | A375-M (in vivo) | ~30 nM | n.d. | In vitro activation via EK | [81] | |
| MEL-526 | ~50 nM | n.d. | ||||||
| TXM-18L | ~150 nM | n.d. | ||||||
| GrB-TGFα | Breast carcinoma | EGFR, | MDA-MB468 | 0.25 nM | n.d. | In vivo activation by Kex2 | [82] | |
| GrB-scFv (FRP5) | ErbB2 (Her2) | 0.29 nM | ||||||
| (P.pastoris) | (+chloroquin for both) | |||||||
| B3-GzmB*, GzmB-CD8* | Breast carcinoma | Lewis Y antigen | SK-BR3 | 98 nM, 1595 nM | Yes [83] | Polyionic adapters between GrB and dsFv-B3 | [85] | |
| (E.coli) | Yes, after induction by estrogens [84] | |||||||
| MCF-7 (MCF-7casp3) | 140 nM (35 nM), 198 nM (394 nM) | |||||||
| GrB-VEGF121 | Tumor vascular endothelial cells | Vascular endothelial growth factor | PAE/FLK-1 | ~10 nM | n.d. | In vitro activation via EK | [86] | |
| (E.coli) | PAE/FLT-1 | -- | n.d. | |||||
| ImmunoGrB-PEAII | Breast and ovarian carcinoma | Her2 | SK-BR3 (in vivo) | Not applicable since supernatant was used | Yes [83] | PEAII translocation motif to allow endogenous furin activation: e23sFv-PEAII-GrB | [87] | |
| (HeLa, Jurkat) | n.d. | |||||||
| SKOV-3 | ||||||||
| ImmunoGrB- Fpe/Fdt/R9 # | Breast, gastric and hepatocellular carcinoma | Her2 | SK-BR3 | Not applicable since supernatant was used | Yes [83] | Introduction of novel furin cleavable sites between e23Fv and GrB (compare above) | [88] | |
| n.d. | ||||||||
| (HeLa, Jurkat) | SGC-7901 (in vivo) | n.d. | ||||||
| AGS Hep G2 | n.d. | |||||||
| GrB-YCG | Ovarian, breast, endometrial and prostate carcinoma | hLHR | MA-10 (murine) | 0.16 µM | No PI-9 expected in murine cell line | In vitro activation via EK prior to use | [89] | |
| (insect Sf9 cells) | MCF-7 | -- | ||||||
| PC-3 | -- | |||||||
Abbreviations: hLHR: human luteinizing hormone receptor; Her2: human epidermal growth factor receptor 2; Kex2: killer expression defective 2; EGFR: epidermal growth factor receptor; dsFv: disulfide stabilized Fv fragment; n.d.: not determined in other publications; EK: Enterokinase; PEAII: second domain of Pseudomonas Exotoxin A (residues 253-364); (in vivo): cell line was successful killed in mice; * compare Table 2; # Fpe: furin site sequence from PEA (amino acids 273-282), Fdt: furin site sequence from diphtheria toxin (amino acids 187-196), R9: poly-arginine tract.
The activation of the BID pathway (either directly by GrB or indirectly via caspase-8) leads to the cleavage of mitochondrial BID (BH3-interacting domain death agonist) involving BAX (BCL-2-associated X protein) and BAK (BCL-2 antagonist). The translocation of tBID (truncated BID) to the mitochondria results in the permeabilization of the outer mitochondrial membrane and thus mitochondrial depolarization and the subsequent release of cytochrome c (cyt c) and other pro-apoptotic molecules such as endonuclease G and HtrA2/OMI [90,91]. Cytochrome c must be present in the cytosol for the apoptosome to form, which requires the assembly of apoptotic protease activation factor APAF-1 which in turn leads to the autocatalytic activation of pro-caspase-9 and the subsequent processing of the effector caspase-3. In addition to the BID-mediated loss of mitochondrial transmembrane potential, GrB can directly induce mitochondrial depolarization without the assistance of cytosolic mediators, but the exact mechanisms remain unclear [59,92,93,94,95]. The natural apoptotic pathways induced by GrB have been comprehensively reviewed [96,97,98].
GrB cleaves downstream of aspartic or glutamic acid residues and therefore has a substrate specificity similar to that of caspases. GrB can therefore also directly target and activate the downstream caspase substrates ICAD, PARP, DNA-PK, NuMA and lamin B [99,100]. Taken together, these data show that GrB is not only a highly potent inducer of apoptosis due to its status as the major mediator of cellular immunity, but also because of its ability to induce apoptosis at multiple levels using distinct pathways.
3.3. Heterologous Expression of Active Granzyme B
Production according to good manufacturing practice (GMP) guidelines is a prerequisite before biopharmaceuticals can enter clinical studies. The production of GrB (and CFPs derived therefrom) in various heterologous expression systems is relatively straightforward, and success has been achieved in Escherichia coli, Pichia pastoris, insect cells and mammalian cells such as HEK 293T, HeLa, Jurkat and COS [78,82,85,87,88,89,101,102,103]. GrB requires a free N-terminus for enzymatic activity, therefore native GrB is expressed as an inactive zymogen containing an N-terminal pre-pro leader sequence. The signal peptide is cleaved by a signal peptidase in the endoplasmic reticulum to produce an inactive proenzyme with an N-terminal pro-peptide, protecting the cell from the induction of apoptosis, thus the pro-peptide must be removed to generate an active protease [104]. In the Golgi apparatus, a mannose-6-phosphate tag is attached which directs GrB to the cytotoxic granules [105]. The pro-peptide is removed by the dipeptidyl peptidase I (DPPI) (also known as cathepsin C) [106].
Several strategies have been developed to accomplish the correct in vitro or in vivo processing and activation of recombinant GrB, including the insertion of an enterokinase cleavage site (ECS) upstream of the sequence of the mature GrB polypeptide, to allow activation after purification by in vitro processing [78,80,81,86,89]. Secretory expression in the methylotropic yeast P. Pastoris has been achieved by the direct N-terminal fusion of the mature polypeptide to the Saccharomyces cerevisiae mating factor α prepro-leader sequence, allowing the straightforward purification of active GrB from the culture supernatant based on local Kex2-protease activity in the trans-Golgi compartment [82,107]. Secretion and functional in vivo activation by a host cell signal peptidase has been achieved in insect cells and COS cells using the native sequence of the GrB precursor protein with the propeptide deleted, or the genetic fusion of a furin recognition motif allowing in vivo processing by endogenous P. pastoris furin-like proteases [102,103,108]. In addition, in vitro and in vivo activation has been achieved by either coexpression of rat DPPI in COS cells or subsequent incubation with bovine spleen DPPI [109].
3.4. Clinical Limitations of Granzyme B
Despite encouraging data confirming the targeted cytotoxicity of GrB in the context of hCFPs, the involvement of GrB in natural cellular defense is a mixed blessing in targeted cancer therapy. On one hand, the induction of multiple apoptosis pathways allows GrB to evade typical apoptosis inhibitors and it is easy to express in heterologous systems. On the other hand however, two structural features reduce its therapeutic potential.
First, the endogenous GrB inhibitor PI-9 protects cytotoxic lymphocytes from misdirected GrB during an immune response, but the same molecule can be overexpressed by tumor cells allowing them to escape from the impact of GrB secreted by NK cells and CTLs (compare Table 2 and Table 3). Second, GrB has a significant positive surface charge which allows it to bind non-specifically to the negatively-charged cell surface and cause off-target effects. CFPs based on human enzymes have shown promising results in vitro and in preclinical evaluations, but are less efficacious than the more potent bacterial ITs. This probably reflects the lack of an endogenous translocation domain typically present in bacterial toxins, which is required for efficient delivery to the cytosol where the apoptotic machinery is located. The endosomal trapping of human pro-apoptotic enzymes is a major bottleneck in the development of highly effective humanized ITs.

Table 2.
Overview of PI-9 expression on protein level based on western blot analysis within different cell lines used in our laboratory.
| Cell line | Cell type | PI-9 expression | Reference |
|---|---|---|---|
| L3.6pl | Pancreatic carcinoma | + | [79] |
| L428 | Hodgkin lymphoma | + | [79] |
| L1236 | Hodgkin lymphoma | + | [79] |
| L540 | Hodgkin lymphoma | - | [79] |
| K562 | Chronic myeloid leukemia in blast crisis | + | [110] |
| A431 | Epidermoid carcinoma | - | [79] |
| Jurkat | T cell leukemia | - | [110] |
| PT45 | Pancreatic adenocarcinoma | - | [79] |
| Kasumi 1 | Acute myeloid leukemia | (+) | [79] |
| Karpas 299 | T cell lymphoma | - | [79] |
| HL60 | Acute myeloid leukemia | - | [79] |
| U937 | Histiocytic lymphoma | - | [79] |

Table 3.
Overview of PI-9 expression in different carcinomas / melanomas.
| Cancer type | Detection | Hallmarks | Reference |
|---|---|---|---|
| Lung cancer | In vitro (cell lines) | PI-9 highly expressed in lung cancer cell lines | [111] |
| In vivo (primary cancer cells) | PI-9 expression was increased in primary lung cancer cells and significantly correlated with cancer stage (dependent on granzyme B expression of CTLs) | ||
| Prostate cancer | In vitro (cell lines → qPCR and flow cytometry) | PI-9 expression in cell lines PC3 and DU-145 | [112] |
| In vivo (prostate tumor tissue → qPCR and immunohistochemistry) | PI-9 expression is up-regulated in pre-cancerous states, which is dysregulated in later stages whereas it remains in some tumors (pilot study) | ||
| Non-small cell lung carcinoma cells (NSCLCs) and tissues | In vitro (cell lines → RT-PCR and western blot analysis) | Strong PI-9 expression in 6 and low in 4 of 10 cell lines on mRNA level correlating with detection on protein level | [113] |
| In vivo (lung tissue samples from biopsies → RT-PCR) | PI-9 mRNA and protein expression in all of 150 patients at variable levels in NSCLC cells and tumors, the less differentiated lung adenocarcinomas showed significantly higher expression of PI-9 mRNA as compared to the well-differentiated tumors | ||
| Breast cancer | In vitro (cell line MCF-7 → western blot and QRT-PCR) | PI-9 expression is induced by estrogens and depends on an interplay between estrogens, estrogen receptor and EGF/EGFR | [84] |
| In vivo (MCF-7 xenograft mouse model) | Induction of PI-9 by the estrogen “genistein” | [114] | |
| Stage III and IV melanoma | In vitro (cell lines → western blot of cell lysates) | PI-9 expression in 6 of 14 melanoma cell lines | [115] |
| In vivo (paraffin embedded tissue of patients → immunohistochemistry) | PI-9 expression in 21 of 26 cases of primary melanoma and in 22 of 28 metastases | ||
| After categorizing in respect to percentage of PI-9 expressing tumor cells (+ if > 50% of cells expressed PI-9, −if < 50% of cells expressed PI-9) 15 of 26 primary tumors and 12 of 28 metastases were + | |||
| PI-9 expression in melanoma metastases correlates with poor clinical outcome following active specific immunotherapy (ASI) therapy of stage III and IV melanoma patients | |||
| Nasopharyngeal carcinoma | In vivo (formalin-fixed, paraffin-embedded tumor biopsies → immunohisto-chemistry) | PI-9 expression in 3 of 43 cases | [116] |
| Presence of many tumor infiltrating activate CTLs within patient biopsies is related to bad clinical outcome | |||
| Melanoma, breast, cervical and colon carcinoma | In vitro (cell lines → PCR) | PI-9 expression in a subset of determined tumor lines (e.g., MCF-7, SK-BR3) | [83] |
| In vivo (primary colon carcinoma cells → PCR) | PI-9 expression in 2 of 4 primary surgical specimens |
4. Bioengineering of Granzyme B
4.1. Granzyme B Variants Insensitive to PI-9—Motivation and Strategies
4.1.1. Expression of PI-9 in Different Cell Types
Because granzymes are an integral component of the human immunosurveillance system they must be tightly controlled at the posttranslational level. This is accomplished by the serpin superfamily, a group of structurally-related proteins that regulate proteases in the vertebrate adaptive and innate immune systems [117,118]. The best-known granzyme-regulating serpin is PI-9, the most important inhibitor of GrB. This 42 kDa protein is a member of the clade B serpin superfamily, and accumulates in the cytosol of lymphocytes to avoid self-inflicted injury caused by granule leakage and the subsequent misdirected activity of GrB. Cells at immune-privileged sites therefore express PI-9, including the lens capsula, ovaries, placenta, testes and embryonic stem cells [119,120]. The inhibition of GrB by PI-9 involves a stoichiometric interaction leading to the irreversible inactivation of both the enzyme and the inhibitor.
Many studies have focused on the expression profile of PI-9 and its induction by different stimuli. PI-9 is also expressed in inflammatory cells [121] and in human peripheral blood mesenchymal stem cells [122]. It is also present in cells infected by cytomegalovirus, Epstein-Barr virus and the BK polyomavirus [123,124,125] as well as in endothelial and mesothelial cells [126] and dendritic cells [119]. PI-9 can be induced in different cell lines by nuclear factor-κB (NF-κB), activator protein-1 (AP-1), pro-inflammatory cytokines such as TNFα, and lipopolysaccharides [121,125,127]. PI-9 is also induced by phorbol myristate acetate and interleukin (IL-2) in CD8+ T cells [128,129], by viral dsRNA sensors in human renal tubular epithelial cells [123] and by estrogens and estrogen receptor-α in breast cancer cells [84,114]. The expression of PI-9 in renal tubular cells is upregulated following renal allograft rejection, suggesting that its induction could be used to confer resistance against GrB-mediated cytotoxic effects leading to improved graft survival [130]. The widespread expression of PI-9 is to be taken into account when developing therapeutic strategies based on GrB, particularly because malignant cells can also involve multiple mechanisms to evade the pro-apoptotic signals induced by CTLs and NK cells [131].
The impact of resistance on GrB-based immunotherapeutics is controversial, with some reports claiming that resistance to perforin-dependent pathways is not a relevant escape mechanism in lymphoma [110] and that the induction of apoptosis by GrB can be achieved in cells expressing or upregulating PI-9. For example a CFP comprising GrB and dsFv-B3, targeting the antigen Lewis Y, has been shown to kill the human breast cancer lines SK-BR3 and MCF-7 in an antigen-dependent manner [85]. The reported IC50 values are rather low compared to the PE-based reference, which might reflect the expression of PI-9 in SK-BR3 cells and the ability of MCF-7 cells to upregulate PI-9 in the presence of estrogen [84]. This group also showed that the same CFP was able to kill chronic myeloid leukemia (CML) cell line K562, which expresses PI-9 at a low level [110] independent of the antigen, via the pore-forming reagent streptolysin O (SLO). However, the exaggerated delivery of GrB to the cells by SLO might allow PI-9 inhibition to be overcome by a natural mechanism, which may also explain the ability of GrB to kill SK-BR3 cells when fused to cell-specific ligands [80,87,88,101].
However, it is difficult to confirm retrospectively whether these cells actually expressed PI-9, because cell lines can diverge depending on their source and the cultivation conditions and timing [132]. Indeed, several studies using xenograft mouse models have revealed a direct correlation between PI-9 expression and the loss of GrB pro-apoptotic activity in vitro and in vivo [83,111,112,117,133]. We accordingly observed a reduction in the apoptotic efficacy of GrB based hCFPs, in the absence of SLO, in some target cell lines despite effective internalization and the presence of adequate amounts of the corresponding receptor [134]. This led us to investigate the potential influence of PI-9 on the cytotoxic potency of GrB. Therefore we studied PI-9 protein expression profiles in the target cell lines typically used in our laboratory by carrying out western blots (Table 2) and compared the results to PI-9 levels detected in primary cancer cells by other groups (Table 3 and Table 4). These studies revealed that PI-9 is expressed in a heterogeneous manner within the tumor cell population, leading to the survival of a small number of malignant cells surrounded by innate immune cells. PI-9 was expressed primarily in tumors containing many GrB-positive infiltrating CTLs and the high incidence of PI-9 positive tumor cells often predicted an unfavorable clinical outcome [135]. These data suggest that PI-9-positive tumor cells are positively selected because they evade immunosurveillance, which is clinically relevant for the treatment of hematological disorders or solid tumors in which PI-9 expression is upregulated during immunosurveillance or tumor therapy. Therefore the development of drugs that are effective against such resistant cells is highly advantageous.

Table 4.
Overview of PI-9 expression in different lymphomas.
| Cancer type | Detection | Hallmarks | Reference |
|---|---|---|---|
| Classical Hodgkin lymphoma | In vivo (lymph node sections → gene expression via microarrays) | PI-9 expression determined in 6 EBV+ and 10 EBV- cases: EBV+ are PI-9+, EBV- PI-9- | [136] |
| Leukemia | In vitro (cell lines → western blot analysis) | PI-9 expression in 4 of 6 cell lines | [137] |
| Ex vivo (monocytes from peripheral blood) | PI-9 expression in 0 of 2 ALL cases, 3 of 4 AML cases, 2 of 3 CLL cases | ||
| NK/ T cell lymphoma | In vivo (deparaffinized tissue sections of patient biopsies → immunohisto-chemistry) | PI-9 expression in 26 of 39 cases | [138] |
| PI-9 expression level was heterogeneous from case to case with clusters of negative cells suggesting the emergence of PI-9 down-regulated subclones associated with aggressiveness and invasive potential | |||
| High levels of PI-9 associated with favorable clinical outcome | |||
| Lymphomas | In vitro (cell lines → flow cytometry) | PI-9 expression in 10 of 18 lymphoma cell lines (e.g., K562) | [110] |
| Ex vivo (isolation of monocytes from peripheral blood → flow cytometry) | PI-9 expression in 9 of 14 primary lymphomas | ||
| Using highly activated CL in vitro no inhibition of the perforin-dependent cytotoxic pathway has been observed. | |||
| ALCL (anaplastic large cell lymphoma) | In vivo (biopsies of systemic ALCL patients → mmunohistochemistry) | PI-9 expression in 6 of 45 cases (percentages of PI-9+ cells ranged from 5% to 75%), primarily found in tumors harboring many Gb+ infiltrating CTLs | [135] |
| High numbers of PI-9+ tumor cells predict resistance to chemotherapy-induced apoptosis and unfavorable outcome | |||
| Different non-Hodgkin and Hodgkin lymphomas | In vivo (formalin-fixed, paraffin-embedded tumor biopsies → immunohisto-chemistry) | Sub-division into 5 categories depending on percentage of PI-9 positive cells | [139] |
| PI-9 expression in neoplastic cells in 36 of 92 cases of T-cell lymphoma, 20 of 75 of B-cell lymphoma and 6 of 57 of Hodgkin lymphoma (expression varied between 5 and 100 %) | |||
| For further differentiation of PI-9 expression and lymphoma type see reference. |
4.1.2. Downregulation of PI-9 Expression and Activity
Strategies to optimize the efficacy of GrB-based CFPs on primarily PI-9-positive cells include either the downregulation of PI-9 expression/activity in target cells or the generation of GrB variants that are insensitive to PI-9. The former may be achieved by RNA interference (RNAi) or antisense technology to knock down PI-9 gene expression. The successful use of siRNA to inhibit the expression of PI-9 has been described in stem cells [122] and in breast cancer [84]. However, the development of siRNA-based therapies has fallen out of favor following the Phase III failure of Bevasiranib, which targeted the expression of vascular endothelial growth factor (VEGF) for the treatment of age-related wet macular degeneration. In alternative approaches, the NFκB inhibitor pyrrolidin dithiocarbamate (PTDC) has been used to suppress PI-9 gene expression in PBMCs cultivated in vitro [125], whereas the serine protease granzyme M (found in the cytotoxic granules of NK cells) can inhibit PI-9 at the protein level [140]. We propose that the genetic fusion of granzyme M to a specific binding domain should facilitate cellular uptake, leading to the inactivation of endogenous PI-9. Therefore, the co-administration of granzymes M and B could potentially clear the way for GrB-based therapy.
4.1.3. Generation of Granzyme B Variants Insensitive to PI-9
The strategies described above are indirect approaches which require the administration of GrB and a separate therapeutic modality. In contrast, GrB can be modified in such a way that it retains its enzymatic activity but becomes insensitive to PI-9. In an analogous approach, a human tissue-type plasminogen activator was mutated at a few critical contact residues to achieve resistance against a complex mixture of endogenous serpins [70,141]. The activity of serpins has been elucidated and reviewed already [118,142,143,144]. The three-dimensional structure of serpins comprises nine α-helices, three β-helices and a variable reactive center loop (RCL), which is the primary site of interaction with target proteases and is therefore critical for serpin specificity. The amino acid residues in the RCL of PI-9 are necessary for its activity [145] and the residue at the P1 position is a glutamate, explaining its specificity for GrB, favoring acidic P1 amino acids such as glutamate or asparate [99,100]. The RCL of PI-9 acts as a pseudo-substrate, which is recognized by the protease so that the two molecules dock and form a reversible Michaelis complex. Rapidly, the protease cleaves at the favored P1 position resulting in a covalent stoichiometric complex, in which both proteins remain inactive [143,146]. Thus, PI-9 acts as a suicide substrate. The interaction between GrB and PI-9 has a Kass value of 1.7 × 106 M−1s−1; SI ~ 1 [99].
We generated GrB mutants insensitive to PI-9 by computationally identifying the most important amino acids responsible for the interaction between the serine protease and the RCL of its specific inhibitor leading to the irreversible inactivation of the enzyme by the formation of a covalent complex. In order to investigate complex formation in detail and thereby predict the structure of the Michaelis complex, we chose the published crystal structure of GrB and a crystallized complex of rat trypsin and Maduca sexta serpin [147], which are structurally similar to GrB and PI-9, respectively. We used homology modeling to adapt the backbone of human GrB to rat trypsin, choosing the X-ray structure of GrB which is bound to a five-residue peptide solved at 2 Å resolution (PDB ID 1IAU) [148]. The structure of PI-9 has not been determined so we built the structure by homology modeling based on the X-ray structure of the rat trypsin-Maduca sexta serpin B1 complex. At first we screened for key interactions at the interface of the two proteins using the ROBETTA server [149]. Residues R28 and R201 were found to be the most relevant, using the numbering of residues in GrB according to a previous publication [150], but they were not located in the active site so they were not expected to affect enzymatic activity. We substituted arginine with alanine, glutamate and lysine (neutral, opposite and same charge as the original amino acid, respectively) by site-directed mutagenesis. In total, seven putative PI-9 resistant variants were generated: R28A, R28K, R28E, R201A, R201K, R201E and the double mutant R28A-R201A. Molecular dynamic simulations of PI-9 complexes with wild-type and mutated GrB were then carried out in aqueous solution, showing that R28K, R201A and R201K were the most promising variants, fulfilling the requirements for preventing GrB inhibition by PI-9. These in silico findings were confirmed by in vitro studies, in which the proteolytic activity of wild-type and mutant GrB was analyzed before and after challenge with recombinant PI-9 in a comparative in vitro enzymatic assay. In the presence of PI-9, GrBR28K retained 76% of its original activity, and GrBR201A retained 46%. The enzymatic activity of GrBR201K was almost the same in the presence and absence of PI-9 (6% reduction after PI-9 challenge) so this variant was the most promising. In contrast to the computational findings, GrBR28A retained 54% of its activity. With the combination of these techniques we were able to identify novel GrB mutants which retained their enzymatic activity even after incubation with recombinant PI-9 [150]. The PI-9 insensitivity of the recently described GrB variant K27A by Sun et al. [145] could not be reproduced during our in silico and in vitro experiments. Cell-based studies using the three most promising mutants from the in silico and in vitro experiments as fusion proteins with tumor antigen specific scFvs are now in progress and are showing promising results [134]. The corresponding schematic structure of a GrB-based CFP including R201K is shown in Figure 1. A GrB variant that can kill both PI-9 positive and PI-9 negative tumor cells will provide a significant advantage, particularly for the treatment of relapsing tumors.
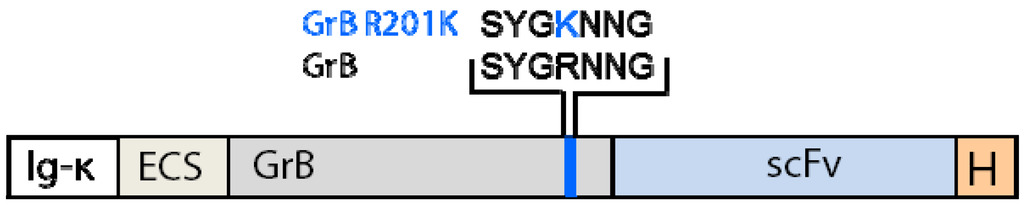
Figure 1.
Schematic structure of granzyme B (GrB) -based cytolytic fusion protein (CFP) containing the R201K modification.
4.2. Reduction of Off-Target Effects
Recombinant GrB-based CFPs have been shown to bind non-specifically to non-target cells which has the potential to cause adverse effects [151]. Different receptors have been shown to facilitate the uptake of GrB after its release from cytotoxic granules, which could lead to the non-specific delivery of recombinant GrB after systemic administration. GrB may bind non-specifically to the mannose-6-phosphate receptor [152,153,154,155] but this can be prevented by deglycosylation following secretory expression in mammalian cells, or by manufacturing the protein in Escherichia coli, which does not glycosylate proteins, or the yeast Pichia pastoris, which does not support mannose-6-phosphate modification [156]. Glycosylation has no impact on the cytotoxic activity of GrB [80,85,86] and might improve the clinical potential of GrB-based ITs. Other structures that may interact with GrB include the CD44 receptor [157] and cell surface-bound heat shock protein 70 (Hsp70) [158], which should be considered when designing GrB-based ITs to prevent off-target effects.
The high isoelectric point (pI) of GrB is a more challenging issue because the net positive charge on the protein surface facilitates electrostatic interactions with the negatively-charged heparan sulfate proteoglycans found on the surface of almost all cells [154,159,160]. The non-specific binding of GrB to the cell-surface does not generally increase off-target cytotoxic activity because the enzyme is not taken up into the cytosol so that it does not interact with its intracellular substrates, but this nevertheless reduces the therapeutic efficacy of the enzyme because it becomes trapped on non-target tissues [151]. Endogenous GrB is stored as a complex with perforin with the negatively-charged chondroitin sulfate proteoglycan serglycin, helping to shield the positive charges [64,161]. The pI of recombinant GrB can be reduced in the context of CFPs by engineering the antibody component with a low pI so that the overall charge is lower but the cytotoxic efficacy is unaffected [162]. Additionally, extracellular proteolytic GrB activity is important for tissue remodeling during an immune response [63], during which the most prominent proteins cleaved by GrB are vitronectin, fibronectin and laminin [163]. Excessive extracellular GrB activity can cause pathophysiological tissue destruction, resulting in disorders such as rheumatoid arthritis [164,165,166], atherosclerosis or abdominal aortic aneurysm [166,167,168].
The amino acids that confer a positive surface charge and thereby facilitate the interaction with glucosaminoglycans on the cell surface are mainly found within the two major heparin-binding motifs RKAKRTRA (residues 96–103 of mature GrB) and KKTMKR (residues 221–226 of mature GrB). The basic amino acid residues involved in heparan sulfate binding were substituted for alanine to reduce cell binding and endocytosis [159]. The substitution of cationic sequences has no impact on the enzymatic and pro-apoptotic activity of GrB [159,169] so that a recombinant version of GrB modified in this matter is evaluated in the context of a hCFP targeting EFGR-positive cancer cells, by exchanging the following amino acids for alanine by site-directed mutagenesis: R96, R100, R102, K221, K222, K225 and R226 [151]. The mutant version of GrB was then fused to the EGFR ligand TGFα, expressed in P. Pastoris and purified from the culture supernatant [82].
Comparative studies with wild-type GrB involving flow cytometric binding analysis on breast cancer cell lines revealed that the modified GrB (GrBcs) showed much weaker binding to EGFR-negative MDA-MB453 cells. Furthermore, cytotoxicity assays using mixed EGFR-positive and EGFR-negative cells showed no reduction in GrBcs cytotoxicity due to trapping by non-target cells, which has been demonstrated for wild-type GrB. The mutations did not interfere with ligand binding or enzyme activity and reduced the negative impact of extracellular matrix effects. Therefore, the specific cytotoxicity of GrB against the EGFR-positive MDA-MB468 cell line could be increased by using the GrBcs variant [151].
We therefore suggest using the PI-9 insensitive and surface-charged modified GrB variant GrBR201Kcs for immunotherapeutic applications. A GrB-based CFP including both modifications is shown in Figure 2.
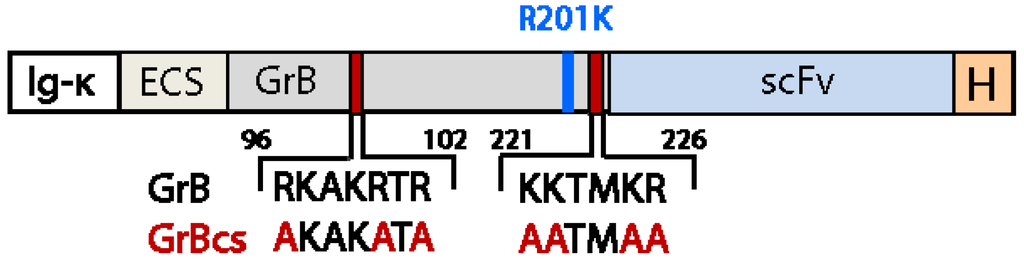
Figure 2.
Schematic structure of GrB-based CFP containing the R201K and surface charge (cs) modification.
5. Effective Endosomal Release is Facilitated by Synthetic, Multifunctional Adapters
The efficacy of receptor-targeted toxins is dependent on three major steps that deliver the therapeutic cargo to its specific site of action: (1) specific binding to cell surface receptors; (2) internalization by receptor-mediated endocytosis; and (3) directed release into the cytosol. One of the major bottlenecks limiting the development of highly efficient hCFPs is the transfer of the therapeutic cargo from the endosome into the cytosol.
Bacterial toxins usually comprise three domains, one targeting the cells, a second promoting transfer from the endosomes to the cytosol and a third catalytic domain, which induces cell death. In second and third generation ITs, the binding domain is replaced by a tumor cell-targeting component, but the toxins still possess their endogenous translocation domains facilitating the translocation of the enzymatically active domain into the cytosol. Pseudomonas exotoxin A (ETA) contains such a translocation domain downstream of a furin cleavage site, and when ETA is taken up by receptor-mediated endocytosis, local furin activity in the endosomes cleaves the toxin and thus exposes the translocation domain. Furin is a type I transmembrane endoprotease involved in the proteolytic maturation of a panel of pro-protein substrates including foreign substances such as bacterial toxins and viral coat proteins [170,171]. After endosomal processing, ETA undergoes retrograde transport through the endoplasmic reticulum to the cytosol, where it ADP-ribosylates the eukaryotic elongation factor 2 (eEF-2) and inhibits protein synthesis, which ultimately induces cell death [172,173,174]. A similar mechanism is responsible for the cellular uptake and cytosolic release of diphtheria toxin. After binding to specific receptors and following internalization, the acidification of endocytotic vesicles and processing of the toxin by local furin activity triggers the translocation of the toxic A chain into the cytosol. The catalytic domain then inhibits protein synthesis by the ADP-ribosylation of EF-2 [175,176].
Protein transduction domains (PTDs) as found on ETA and diphtheria toxin are also known as Trojan peptides, cell penetrating peptides or cell permeable peptides (CPPs), membrane transfer peptides (MTPs) or membrane transfer sequences (MTSs). They are not solely restricted to bacterial toxins, e.g., there are also viral transduction domains like the Human immunodeficiency virus (HIV) transactivation factor (TAT protein), the Influenzavirus HA2 hemagglutinin subunit, and the Herpes simplex virus VP22 protein [177,178,179,180,181]. Most PTDs are small cationic peptides that bind by electrostatic interactions to negatively-charged heparan sulfate proteoglycans found in membranes. They mediate the direct cytosolic uptake of their (chemically or genetically linked) protein cargoes, without depending on receptor-mediated endocytosis [182,183,184]. In addition to natural PTDs, synthetic peptides derived from naturally occurring sequences or selected from random peptide libraries have been screened as vehicles for the efficient delivery of therapeutic cargos [185,186]. PTDs for the delivery of therapeutic macromolecules have been comprehensively reviewed [187,188,189]. With only a few exceptions [190,191,192,193,194], PTDs contradict the concept of targeted therapy, because they lack (tumor) specificity potentially resulting in undesirable off-target effects and preventing systemic administration. Nevertheless, specificity can be engineered by restricting the activity of the cargo to target cells (e.g., the replacement of a tumor suppressor), conjugating specific-cell targeting molecules (e.g., ITs) or modifying the PTD to restrict entry solely to target cells (e.g., PTD activation by tumor-associated proteases) [189].
Human enzymes like granzyme B lack an endogenous translocation domain. Therefore endosomal release is the least efficient step in the therapeutic pathway of hCFPs because the active component can be trapped within the endosomes. To optimize the cytosolic release of human enzymes, recombinant PTDs have been inserted between the cell-targeting and cytotoxic components of the latest generation of ITs. Several proof-of-concept studies have confirmed the ability of PTDs to achieve endosomal escape. For example, two anticancer peptides have been delivered into the cytosol of cancer cells by joining them to the HIV TAT protein [195] and the translocation domain of ETA (PEAII) has been inserted into a GrB based hCFP and shown to be superior to furin-cleavable peptides from ETA and diphtheria toxin as well as a synthetic peptide [87,88]. This suggests that engineering of PTDs still has the potential to improve the translocation of effector molecules from endosomes to the cytosol, thus increasing the efficacy of hCFPs.
Previous studies have shown that PTDs used for targeted therapy can be optimized by the integration of functional, cleavable elements. For example, decoupling the toxin and cell targeting domain by inserting a synthetic, multifunctional cleavable adapter composed of a membrane transfer peptide (MTP) flanked by a cytosolic cleavable peptide (CCP) and an endosomal cleavable peptide (ECP) irreversibly targeted the toxin into the cytosol [196]. Engineering PTDs by the insertion of cleavable elements guarantees both the efficient transport of the toxic cargo into the cytosol and the prevention of its release from the cytosol to affect non-target cells. The exact mechanism of adapter-mediated toxin uptake can be summarized as follows:
- The ECP is recognized by an endogenous endosomal enzyme, which processes the CFP following receptor-mediated endocytosis, thus exposing the MTP.
- Only following the endosomal cleavage of the ECP, the MTP promotes translocation of the effector molecule across the membrane. Since the activation of the MTP is restricted to the endosomes, non-specific uptake based on the interaction between the MTP and the membranes of bystander cells is significantly reduced.
- Once in the cytosol, cleavage of the CCP releases the active enzyme (potentially facilitating its activity by avoiding steric hindrance or conformational changes).In addition, removing the MTP prevents further undesirable membrane translocation and therefore the potential non-specific impact of the effector molecule on surrounding, healthy cells.
A closer insight into the specific composition of a molecular adapter was provided by Heisler et al. combining the PreS2 domain translocation motif of the Hepatitis B virus surface antigen (TLM) with (i) cleavable peptides processed by furin in the endosomes; and (ii) cytosolic caspases. These constructs were developed to improve the cytosolic delivery of a plant-based IT targeting the EGF receptor (Saporin-EGF) [197]. Enzymatic digestion of the endosomal cleavable peptide (ECP) was based on two recognition sites for furin, derived from ETA and diphtheria toxin (see above). Enzymatic release of the toxic payload inside the cytosol was accomplished by two cleavage sites for caspases (caspase-1 and caspase-3) of the N-terminal CCP. Caspases are key mediators of apoptosis and are predominantly localized in the cytosol.
We previously described the successful use of molecular adapters as a tool to enhance the cytotoxic potency of hCFPs [198]. We could also demonstrate the efficacy of the functional domains derived from such adapters for the optimization of a hCFP. The construct comprised the human RNase angiogenin and the humanized scFv(H22) targeting CD64 (FcγR1), which is upregulated in leukemic and inflammatory diseases. The insertion of the adapter resulted in a 20-fold increase in cytotoxic activity, compared to the version lacking the adapter [199]. The engineering of GrB-based hCFPs by the insertion of functional adapter elements therefore promises to improve their therapeutic potential in the near future.
6. Conclusions
Targeted therapies provide strong and versatile weapons in the continuous fight against cancer. Using sophisticated methods, batteries of different treatment options can be developed that can even counteract tumor cell escape mechanisms and selectively eliminate them. The first and second generations of ITs are still the gold standard in terms of cytotoxic efficacy, but they have significant disadvantages in terms of immunogenicity, which remain to be overcome. Although those ITs comprised tumor-targeting components of murine origin, we now use only humanized or completely human antibody factors for this purpose. The concept of recombinant human granzyme B as effector domains instead of toxins derived from bacteria and plants was developed simultaneously in our laboratory and by the group of Michael G. Rosenblum (M.D. Anderson Cancer Center, Houston, Texas).
We have discussed the recent optimization of humanized ITs and the road to future hCFPs based on the human serine protease granzyme B (GrB). Several proof of concept studies have clearly demonstrated that engineered GrB variants can overcome drawbacks of the wild-type enzyme, such as the inactivation of GrB by the endogenous inhibitor PI-9, charge-mediated cell binding (causing drug sequestration by non-target cells) and undesirable extracellular matrix effects. Modified hCFPs containing functional elements resembling the natural translocation domain of bacterial toxins promote endosomal release and can significantly increase the cytotoxic potency of human effector domains. By combining these strategies, we have the opportunity to generate novel GrB-based hCFPs with improved qualities that compete with established IT platforms promising a new wave of hCFP-based therapies in the future.
Acknowledgments
This work was funded in part by a grant from the German province NRW from EFRE “European Fund for Regional Development” under the theme “Europe - Investment in our Future”. We thank Richard M. Twyman for critical reading of the manuscript.
References and Notes
- Klebanoff, C.A.; Acquavella, N.; Yu, Z.Y.; Restifo, N.P. Therapeutic cancer vaccines: Are we there yet? Immunol. Rev. 2011, 239, 27–44. [Google Scholar] [CrossRef]
- Kreitman, R.J. Immunotoxins for targeted cancer therapy. AAPS J. 2006, 8, E532–E551. [Google Scholar] [CrossRef]
- Vanneman, M.; Dranoff, G. Combining immunotherapy and targeted therapies in cancer treatment. Nat. Rev. Cancer 2012, 12, 237–251. [Google Scholar] [CrossRef]
- Waldmann, T.A. Immunotherapy: Past, present and future. Nat. Med. 2003, 9, 269–277. [Google Scholar] [CrossRef]
- Scott, A.M.; Allison, J.P.; Wolchok, J.D. Monoclonal antibodies in cancer therapy. Cancer Immun. 2012, 12, 14. [Google Scholar]
- Reichert, J.M. Antibody-based therapeutics to watch in 2011. MAbs 2011, 3, 76–99. [Google Scholar] [CrossRef]
- Schrama, D.; Reisfeld, R.A.; Becker, J.C. Antibody targeted drugs as cancer therapeutics. Nat. Rev. Drug Discov. 2006, 5, 147–59. [Google Scholar] [CrossRef]
- Carter, P. Improving the efficacy of antibody-based cancer therapies. Nat. Rev. Cancer 2001, 1, 118–129. [Google Scholar] [CrossRef]
- Rybak, S.M.; Hoogenboom, H.R.; Meade, H.M.; Raus, J.C.; Schwartz, D.; Youle, R.J. Humanization of immunotoxins. Proc. Natl. Acad. Sci. USA 1992, 89, 3165–3169. [Google Scholar]
- Natsume, A.; Niwa, R.; Satoh, M. Improving effector functions of antibodies for cancer treatment: Enhancing ADCC and CDC. Drug Des. Devel. Ther. 2009, 3, 7–16. [Google Scholar]
- Carter, P.J.; Senter, P.D. Antibody-drug conjugates for cancer therapy. Cancer J. 2008, 14, 154–169. [Google Scholar] [CrossRef]
- Lambert, J.M. Drug-conjugated monoclonal antibodies for the treatment of cancer. Curr. Opin. Pharmacol. 2005, 5, 543–549. [Google Scholar] [CrossRef]
- Younes, A.; Yasothan, U.; Kirkpatrick, P. Brentuximab vedotin. Nat. Rev. Drug Discov. 2012, 11, 19–20. [Google Scholar] [CrossRef]
- Bross, P.F.; Beitz, J.; Chen, G.; Chen, X.H.; Duffy, E.; Kieffer, L.; Roy, S.; Sridhara, R.; Rahman, A.; Williams, G.; et al. Approval summary: Gemtuzumab ozogamicin in relapsed acute myeloid leukemia. Clin. Cancer Res. 2001, 7, 1490–1496. [Google Scholar]
- Kreitman, R.J. Recombinant toxins for the treatment of cancer. Curr. Opin. Mol. Ther. 2003, 5, 44–51. [Google Scholar]
- Allen, T.M. Ligand-targeted therapeutics in anticancer therapy. Nat. Rev. Cancer 2002, 2, 750–763. [Google Scholar] [CrossRef]
- Pavlinkova, G.; Beresford, G.W.; Booth, B.J.; Batra, S.K.; Colcher, D. Pharmacokinetics and biodistribution of engineered single-chain antibody constructs of MAb CC49 in colon carcinoma xenografts. J. Nucl. Med. 1999, 40, 1536–1546. [Google Scholar]
- Pirker, R.; FitzGerald, D.J.; Willingham, M.C.; Pastan, I. Enhancement of the activity of immunotoxins made with either ricin A chain or Pseudomonas exotoxin in human ovarian and epidermoid carcinoma cell lines. Cancer Res. 1988, 48, 3919–3923. [Google Scholar]
- Barth, S.; Huhn, M.; Matthey, B.; Schnell, R.; Tawadros, S.; Schinkothe, T.; Lorenzen, J.; Diehl, V.; Engert, A. Recombinant anti-CD25 immunotoxin RFT5(SCFV)-ETA' demonstrates successful elimination of disseminated human Hodgkin lymphoma in SCID mice. Int. J. Cancer 2000, 86, 718–724. [Google Scholar] [CrossRef]
- Barth, S.; Huhn, M.; Matthey, B.; Tawadros, S.; Schnell, R.; Schinkothe, T.; Diehl, V.; Engert, A. Ki-4(scFv)-ETA', a new recombinant anti-CD30 immunotoxin with highly specific cytotoxic activity against disseminated Hodgkin tumors in SCID mice. Blood 2000, 95, 3909–3914. [Google Scholar]
- Schnell, R.; Vitetta, E.; Schindler, J.; Borchmann, P.; Barth, S.; Ghetie, V.; Hell, K.; Drillich, S.; Diehl, V.; Engert, A. Treatment of refractory Hodgkin's lymphoma patients with an anti-CD25 ricin A-chain immunotoxin. Leukemia 2000, 14, 129–135. [Google Scholar] [CrossRef]
- Kreitman, R.J. Recombinant immunotoxins for the treatment of chemoresistant hematologic malignancies. Curr. Pharm. Des. 2009, 15, 2652–2664. [Google Scholar] [CrossRef]
- Kreitman, R.J.; Wilson, W.H.; Bergeron, K.; Raggio, M.; Stetler-Stevenson, M.; FitzGerald, D.J.; Pastan, I. Efficacy of the anti-CD22 recombinant immunotoxin BL22 in chemotherapy-resistant hairy-cell leukemia. N. Engl. J. Med. 2001, 345, 241–247. [Google Scholar] [CrossRef]
- Frankel, A.E.; Powell, B.L.; Hall, P.D.; Case, L.D.; Kreitman, R.J. Phase I trial of a novel diphtheria toxin/granulocyte macrophage colony-stimulating factor fusion protein (DT388GMCSF) for refractory or relapsed acute myeloid leukemia. Clin. Cancer Res. 2002, 8, 1004–1013. [Google Scholar]
- Szatrowski, T.P.; Dodge, R.K.; Reynolds, C.; Westbrook, C.A.; Frankel, S.R.; Sklar, J.; Stewart, C.C.; Hurd, D.D.; Kolitz, J.E.; Velez-Garcia, E.; et al. Lineage specific treatment of adult patients with acute lymphoblastic leukemia in first remission with anti-B4-blocked ricin or high-dose cytarabine: Cancer and Leukemia Group B Study 9311. Cancer 2003, 97, 1471–1480. [Google Scholar] [CrossRef]
- Pai, L.H.; Pastan, I. Clinical trials with Pseudomonas exotoxin immunotoxins. Curr. Top. Microbiol. Immunol. 1998, 234, 83–96. [Google Scholar]
- Pastan, I.; Hassan, R.; Fitzgerald, D.J.; Kreitman, R.J. Immunotoxin therapy of cancer. Nat. Rev. Cancer 2006, 6, 559–565. [Google Scholar] [CrossRef]
- Pastan, I.; Hassan, R.; FitzGerald, D.J.; Kreitman, R.J. Immunotoxin treatment of cancer. Annu. Rev. Med. 2007, 58, 221–237. [Google Scholar] [CrossRef]
- Madhumathi, J.; Verma, R.S. Therapeutic targets and recent advances in protein immunotoxins. Curr. Opin. Microbiol. 2012, 15, 300–309. [Google Scholar] [CrossRef]
- Becker, N.; Benhar, I. Antibody-Based Immunotoxins for the Treatment of Cancer. Antibodies 2012, 1, 39–69. [Google Scholar] [CrossRef]
- Dang, N.H.; Pro, B.; Hagemeister, F.B.; Samaniego, F.; Jones, D.; Samuels, B.I.; Rodriguez, M.A.; Goy, A.; Romaguera, J.E.; McLaughlin, P.; et al. Phase II trial of denileukin diftitox for relapsed/refractory T-cell non-Hodgkin lymphoma. Br. J. Haematol. 2007, 136, 439–447. [Google Scholar] [CrossRef]
- Mathew, M.; Verma, R.S. Humanized immunotoxins: A new generation of immunotoxins for targeted cancer therapy. Cancer Sci. 2009, 100, 1359–1365. [Google Scholar] [CrossRef]
- Tur, M.K.; Neef, I.; Jager, G.; Teubner, A.; Stocker, M.; Melmer, G.; Barth, S. Immunokinases, a novel class of immunotherapeutics for targeted cancer therapy. Curr. Pharm. Des. 2009, 15, 2693–2699. [Google Scholar] [CrossRef]
- Huhn, M.; Sasse, S.; Tur, M.K.; Matthey, B.; Schinkothe, T.; Rybak, S.M.; Barth, S.; Engert, A. Human angiogenin fused to human CD30 ligand (Ang-CD30L) exhibits specific cytotoxicity against CD30-positive lymphoma. Cancer Res. 2001, 61, 8737–8742. [Google Scholar]
- Rosenblum, M.G.; Barth, S. Development of novel, highly cytotoxic fusion constructs containing granzyme B: Unique mechanisms and functions. Curr. Pharm. Des. 2009, 15, 2676–2692. [Google Scholar] [CrossRef]
- von Minckwitz, G.; Harder, S.; Hovelmann, S.; Jager, E.; Al-Batran, S.E.; Loibl, S.; Atmaca, A.; Cimpoiasu, C.; Neumann, A.; Abera, A.; et al. Phase I clinical study of the recombinant antibody toxin scFv(FRP5)-ETA specific for the ErbB2/HER2 receptor in patients with advanced solid malignomas. Breast Cancer Res. 2005, 7, R617–R626. [Google Scholar] [CrossRef]
- Engert, A.; Diehl, V.; Schnell, R.; Radszuhn, A.; Hatwig, M.T.; Drillich, S.; Schon, G.; Bohlen, H.; Tesch, H.; Hansmann, M.L.; et al. A phase-I study of an anti-CD25 ricin A-chain immunotoxin (RFT5-SMPT-dgA) in patients with refractory Hodgkin's lymphoma. Blood 1997, 89, 403–410. [Google Scholar]
- Hall, P.D.; Virella, G.; Willoughby, T.; Atchley, D.H.; Kreitman, R.J.; Frankel, A.E. Antibody response to DT-GM, a novel fusion toxin consisting of a truncated diphtheria toxin (DT) linked to human granulocyte-macrophage colony stimulating factor (GM), during a phase I trial of patients with relapsed or refractory acute myeloid leukemia. Clin. Immunol. 2001, 100, 191–17. [Google Scholar] [CrossRef]
- Posey, J.A.; Khazaeli, M.B.; Bookman, M.A.; Nowrouzi, A.; Grizzle, W.E.; Thornton, J.; Carey, D.E.; Lorenz, J.M.; Sing, A.P.; Siegall, C.B.; et al. A phase I trial of the single-chain immunotoxin SGN-10 (BR96 sFv-PE40) in patients with advanced solid tumors. Clin. Cancer Res. 2002, 8, 3092–3099. [Google Scholar]
- Scadden, D.T.; Schenkein, D.P.; Bernstein, Z.; Luskey, B.; Doweiko, J.; Tulpule, A.; Levine, A.M. Immunotoxin combined with chemotherapy for patients with AIDS-related non-Hodgkin's lymphoma. Cancer 1998, 83, 2580–2587. [Google Scholar] [CrossRef]
- Mischak, R.P.; Foxall, C.; Rosendorf, L.L.; Knebel, K.; Scannon, P.J.; Spitler, L.E. Human antibody responses to components of the monoclonal antimelanoma antibody ricin A chain immunotoxin XomaZyme-MEL. Mol. Biother. 1990, 2, 104–109. [Google Scholar]
- Tur, M.K.; Neef, I.; Jost, E.; Galm, O.; Jager, G.; Stocker, M.; Ribbert, M.; Osieka, R.; Klinge, U.; Barth, S. Targeted restoration of down-regulated DAPK2 tumor suppressor activity induces apoptosis in Hodgkin lymphoma cells. J. Immunother. 2009, 32, 431–441. [Google Scholar] [CrossRef]
- Hwang, W.Y.; Foote, J. Immunogenicity of engineered antibodies. Methods 2005, 36, 3–10. [Google Scholar] [CrossRef]
- Boulianne, G.L.; Hozumi, N.; Shulman, M.J. Production of functional chimaeric mouse/human antibody. Nature 1984, 312, 643–646. [Google Scholar] [CrossRef]
- Morrison, S.L.; Johnson, M.J.; Herzenberg, L.A.; Oi, V.T. Chimeric human antibody molecules: Mouse antigen-binding domains with human constant region domains. Proc. Natl. Acad. Sci. USA 1984, 81, 6851–6855. [Google Scholar]
- Jones, P.T.; Dear, P.H.; Foote, J.; Neuberger, M.S.; Winter, G. Replacing the complementarity-determining regions in a human antibody with those from a mouse. Nature 1986, 321, 522–525. [Google Scholar] [CrossRef]
- Verhoeyen, M.; Milstein, C.; Winter, G. Reshaping human antibodies: Grafting an antilysozyme activity. Science 1988, 239, 1534–1536. [Google Scholar]
- Hu, W.G.; Yin, J.; Chau, D.; Negrych, L.M.; Cherwonogrodzky, J.W. Humanization and characterization of an anti-ricin neutralization monoclonal antibody. PLoS One 2012, 7, e45595. [Google Scholar]
- Lonberg, N. Fully human antibodies from transgenic mouse and phage display platforms. Curr. Opin. Immunol. 2008, 20, 450–459. [Google Scholar] [CrossRef]
- Menoret, S.; Iscache, A.L.; Tesson, L.; Remy, S.; Usal, C.; Osborn, M.J.; Cost, G.J.; Bruggemann, M.; Buelow, R.; Anegon, I. Characterization of immunoglobulin heavy chain knockout rats. Eur. J. Immunol. 2010, 40, 2932–2941. [Google Scholar] [CrossRef]
- Hansen, J.K.; Weldon, J.E.; Xiang, L.; Beers, R.; Onda, M.; Pastan, I. A recombinant immunotoxin targeting CD22 with low immunogenicity, low nonspecific toxicity, and high antitumor activity in mice. J. Immunother. 2010, 33, 297–304. [Google Scholar] [CrossRef]
- Liu, W.; Onda, M.; Lee, B.; Kreitman, R.J.; Hassan, R.; Xiang, L.; Pastan, I. Recombinant immunotoxin engineered for low immunogenicity and antigenicity by identifying and silencing human B-cell epitopes. Proc. Natl. Acad. Sci. USA 2012, 109, 11782–11787. [Google Scholar]
- Tsutsumi, Y.; Onda, M.; Nagata, S.; Lee, B.; Kreitman, R.J.; Pastan, I. Site-specific chemical modification with polyethylene glycol of recombinant immunotoxin anti-Tac(Fv)-PE38 (LMB-2) improves antitumor activity and reduces animal toxicity and immunogenicity. Proc. Natl. Acad. Sci. USA 2000, 97, 8548–8553. [Google Scholar]
- Oratz, R.; Speyer, J.L.; Wernz, J.C.; Hochster, H.; Meyers, M.; Mischak, R.; Spitler, L.E. Antimelanoma monoclonal antibody-ricin A chain immunoconjugate (XMMME-001-RTA) plus cyclophosphamide in the treatment of metastatic malignant melanoma: Results of a phase II trial. J. Biol. Response Mod. 1990, 9, 345–354. [Google Scholar]
- Siegall, C.B.; Haggerty, H.G.; Warner, G.L.; Chace, D.; Mixan, B.; Linsley, P.S.; Davidson, T. Prevention of immunotoxin-induced immunogenicity by coadministration with CTLA4Ig enhances antitumor efficacy. J. Immunol. 1997, 159, 5168–5173. [Google Scholar]
- Igney, F.H.; Krammer, P.H. Immune escape of tumors: Apoptosis resistance and tumor counterattack. J. Leuk. Biol. 2002, 71, 907–920. [Google Scholar]
- Vesely, M.D.; Kershaw, M.H.; Schreiber, R.D.; Smyth, M.J. Natural innate and adaptive immunity to cancer. Annu. Rev. Immunol. 2011, 29, 235–271. [Google Scholar] [CrossRef]
- Griffith, T.S.; Chin, W.A.; Jackson, G.C.; Lynch, D.H.; Kubin, M.Z. Intracellular regulation of TRAIL-induced apoptosis in human melanoma cells. J. Immunol. 1998, 161, 2833–2840. [Google Scholar]
- Sutton, V.R.; Wowk, M.E.; Cancilla, M.; Trapani, J.A. Caspase activation by granzyme B is indirect, and caspase autoprocessing requires the release of proapoptotic mitochondrial factors. Immunity 2003, 18, 319–329. [Google Scholar] [CrossRef]
- Campos, L.; Rouault, J.P.; Sabido, O.; Oriol, P.; Roubi, N.; Vasselon, C.; Archimbaud, E.; Magaud, J.P.; Guyotat, D. High expression of bcl-2 protein in acute myeloid leukemia cells is associated with poor response to chemotherapy. Blood 1993, 81, 3091–3096. [Google Scholar]
- Ambrosini, G.; Adida, C.; Altieri, D.C. A novel anti-apoptosis gene, survivin, expressed in cancer and lymphoma. Nat. Med. 1997, 3, 917–921. [Google Scholar] [CrossRef]
- Russell, J.H.; Ley, T.J. Lymphocyte-mediated cytotoxicity. Annu. Rev. Immunol. 2002, 20, 323–370. [Google Scholar] [CrossRef]
- Boivin, W.A.; Cooper, D.M.; Hiebert, P.R.; Granville, D.J. Intracellular versus extracellular granzyme B in immunity and disease: Challenging the dogma. Lab. Invest. 2009, 89, 1195–1220. [Google Scholar] [CrossRef]
- Raja, S.M.; Wang, B.; Dantuluri, M.; Desai, U.R.; Demeler, B.; Spiegel, K.; Metkar, S.S.; Froelich, C.J. Cytotoxic cell granule-mediated apoptosis. Characterization of the macromolecular complex of granzyme B with serglycin. J. Biol. Chem. 2002, 277, 49523–49530. [Google Scholar]
- Grossman, W.J.; Revell, P.A.; Lu, Z.H.; Johnson, H.; Bredemeyer, A.J.; Ley, T.J. The orphan granzymes of humans and mice. Curr. Opin. Immunol. 2003, 15, 544–552. [Google Scholar] [CrossRef]
- Bots, M.; Medema, J.P. Granzymes at a glance. J. Cell Sci. 2006, 119, 5011–5014. [Google Scholar] [CrossRef]
- Galvin, J.P.; Spaeny-Dekking, L.H.; Wang, B.; Seth, P.; Hack, C.E.; Froelich, C.J. Apoptosis induced by granzyme B-glycosaminoglycan complexes: Implications for granule-mediated apoptosis in vivo. J. Immunol. 1999, 162, 5345–5350. [Google Scholar]
- Grujic, M.; Braga, T.; Lukinius, A.; Eloranta, M.L.; Knight, S.D.; Pejler, G.; Abrink, M. Serglycin-deficient cytotoxic T lymphocytes display defective secretory granule maturation and granzyme B storage. J. Biol. Chem. 2005, 280, 33411–33418. [Google Scholar]
- Lanier, L.L. NK cell recognition. Annu. Rev. Immunol. 2005, 23, 225–274. [Google Scholar] [CrossRef]
- Kurschus, F.C.; Jenne, D.E. Delivery and therapeutic potential of human granzyme B. Immunol. Rev. 2010, 235, 159–171. [Google Scholar]
- Andrade, F.; Casciola-Rosen, L.A.; Rosen, A. Granzyme B-induced cell death. Acta Haematol. 2004, 111, 28–41. [Google Scholar] [CrossRef]
- Heibein, J.A.; Goping, I.S.; Barry, M.; Pinkoski, M.J.; Shore, G.C.; Green, D.R.; Bleackley, R.C. Granzyme B-mediated cytochrome c release is regulated by the Bcl-2 family members bid and Bax. J. Exp. Med. 2000, 192, 1391–1402. [Google Scholar] [CrossRef]
- Pinkoski, M.J.; Waterhouse, N.J.; Heibein, J.A.; Wolf, B.B.; Kuwana, T.; Goldstein, J.C.; Newmeyer, D.D.; Bleackley, R.C.; Green, D.R. Granzyme B-mediated apoptosis proceeds predominantly through a Bcl-2-inhibitable mitochondrial pathway. J. Biol. Chem. 2001, 276, 12060–12067. [Google Scholar]
- Metkar, S.S.; Wang, B.; Ebbs, M.L.; Kim, J.H.; Lee, Y.J.; Raja, S.M.; Froelich, C.J. Granzyme B activates procaspase-3 which signals a mitochondrial amplification loop for maximal apoptosis. J. Cell Biol. 2003, 160, 875–885. [Google Scholar] [CrossRef]
- Talanian, R.V.; Yang, X.; Turbov, J.; Seth, P.; Ghayur, T.; Casiano, C.A.; Orth, K.; Froelich, C.J. Granule-mediated killing: Pathways for granzyme B-initiated apoptosis. J. Exp. Med. 1997, 186, 1323–1331. [Google Scholar] [CrossRef]
- Waterhouse, N.J.; Sedelies, K.A.; Trapani, J.A. Role of Bid-induced mitochondrial outer membrane permeabilization in granzyme B-induced apoptosis. Immunol. Cell Biol. 2006, 84, 72–78. [Google Scholar] [CrossRef]
- Hengartner, M.O. The biochemistry of apoptosis. Nature 2000, 407, 770–776. [Google Scholar] [CrossRef]
- Stahnke, B.; Thepen, T.; Stocker, M.; Rosinke, R.; Jost, E.; Fischer, R.; Tur, M.K.; Barth, S. Granzyme B-H22(scFv), a human immunotoxin targeting CD64 in acute myeloid leukemia of monocytic subtypes. Mol. Cancer Ther. 2008, 7, 2924–2932. [Google Scholar] [CrossRef]
- Schiffer, S. Fraunhofer Institute for Molecular Biology and Applied Ecology IME. unpublished work, Aachen: Germany, 2012. [Google Scholar]
- Liu, Y.; Cheung, L.H.; Hittelman, W.N.; Rosenblum, M.G. Targeted delivery of human pro-apoptotic enzymes to tumor cells: In vitro studies describing a novel class of recombinant highly cytotoxic agents. Mol. Cancer Ther. 2003, 2, 1341–1350. [Google Scholar]
- Liu, Y.; Zhang, W.; Niu, T.; Cheung, L.H.; Munshi, A.; Meyn, R.E., Jr.; Rosenblum, M.G. Targeted apoptosis activation with GrB/scFvMEL modulates melanoma growth, metastatic spread, chemosensitivity, and radiosensitivity. Neoplasia 2006, 8, 125–135. [Google Scholar] [CrossRef]
- Dalken, B.; Giesubel, U.; Knauer, S.K.; Wels, W.S. Targeted induction of apoptosis by chimeric granzyme B fusion proteins carrying antibody and growth factor domains for cell recognition. Cell Death Differ. 2006, 13, 576–585. [Google Scholar] [CrossRef]
- Medema, J.P.; de Jong, J.; Peltenburg, L.T.; Verdegaal, E.M.; Gorter, A.; Bres, S.A.; Franken, K.L.; Hahne, M.; Albar, J.P.; Melief, C.J.; et al. Blockade of the granzyme B/perforin pathway through overexpression of the serine protease inhibitor PI-9/SPI-6 constitutes a mechanism for immune escape by tumors. Proc. Natl. Acad. Sci. USA 2001, 98, 11515–11520. [Google Scholar]
- Jiang, X.; Ellison, S.J.; Alarid, E.T.; Shapiro, D.J. Interplay between the levels of estrogen and estrogen receptor controls the level of the granzyme inhibitor, proteinase inhibitor 9 and susceptibility to immune surveillance by natural killer cells. Oncogene 2007, 26, 4106–4114. [Google Scholar] [CrossRef]
- Kurschus, F.C.; Kleinschmidt, M.; Fellows, E.; Dornmair, K.; Rudolph, R.; Lilie, H.; Jenne, D.E. Killing of target cells by redirected granzyme B in the absence of perforin. FEBS Lett. 2004, 562, 87–92. [Google Scholar] [CrossRef]
- Liu, Y.; Cheung, L.H.; Thorpe, P.; Rosenblum, M.G. Mechanistic studies of a novel human fusion toxin composed of vascular endothelial growth factor (VEGF)121 and the serine protease granzyme B: Directed apoptotic events in vascular endothelial cells. Mol. Cancer Ther. 2003, 2, 949–959. [Google Scholar]
- Zhang, L.; Zhao, J.; Wang, T.; Yu, C.J.; Jia, L.T.; Duan, Y.Y.; Yao, L.B.; Chen, S.Y.; Yang, A.G. HER2-targeting recombinant protein with truncated pseudomonas exotoxin A translocation domain efficiently kills breast cancer cells. Cancer Biol. Ther. 2008, 7, 1226–1231. [Google Scholar]
- Wang, T.; Zhao, J.; Ren, J.L.; Zhang, L.; Wen, W.H.; Zhang, R.; Qin, W.W.; Jia, L.T.; Yao, L.B.; Zhang, Y.Q.; et al. Recombinant immunoproapoptotic proteins with furin site can translocate and kill HER2-positive cancer cells. Cancer Res. 2007, 67, 11830–11839. [Google Scholar]
- Kanatani, I.; Lin, X.; Yuan, X.; Manorek, G.; Shang, X.; Cheung, L.H.; Rosenblum, M.G.; Howell, S.B. Targeting granzyme B to tumor cells using a yoked human chorionic gonadotropin. Cancer Chemother. Pharmacol. 2011, 68, 979–990. [Google Scholar] [CrossRef]
- Hegde, R.; Srinivasula, S.M.; Zhang, Z.; Wassell, R.; Mukattash, R.; Cilenti, L.; DuBois, G.; Lazebnik, Y.; Zervos, A.S.; Fernandes-Alnemri, T.; et al. Identification of Omi/HtrA2 as a mitochondrial apoptotic serine protease that disrupts inhibitor of apoptosis protein-caspase interaction. J. Biol. Chem. 2002, 277, 432–438. [Google Scholar]
- Liu, X.; Kim, C.N.; Yang, J.; Jemmerson, R.; Wang, X. Induction of apoptotic program in cell-free extracts: Requirement for dATP and cytochrome c. Cell 1996, 86, 147–157. [Google Scholar] [CrossRef]
- Han, J.; Goldstein, L.A.; Gastman, B.R.; Froelich, C.J.; Yin, X.M.; Rabinowich, H. Degradation of Mcl-1 by granzyme B: Implications for Bim-mediated mitochondrial apoptotic events. J. Biol. Chem. 2004, 279, 22020–22029. [Google Scholar]
- Alimonti, J.B.; Shi, L.; Baijal, P.K.; Greenberg, A.H. Granzyme B induces BID-mediated cytochrome c release and mitochondrial permeability transition. J. Biol. Chem. 2001, 276, 6974–6982. [Google Scholar] [CrossRef]
- Waterhouse, N.J.; Sedelies, K.A.; Browne, K.A.; Wowk, M.E.; Newbold, A.; Sutton, V.R.; Clarke, C.J.; Oliaro, J.; Lindemann, R.K.; Bird, P.I.; et al. A central role for Bid in granzyme B-induced apoptosis. J. Biol. Chem. 2005, 280, 4476–4482. [Google Scholar]
- Cullen, S.P.; Adrain, C.; Luthi, A.U.; Duriez, P.J.; Martin, S.J. Human and murine granzyme B exhibit divergent substrate preferences. J. Cell Biol. 2007, 176, 435–444. [Google Scholar] [CrossRef]
- Chowdhury, D.; Lieberman, J. Death by a thousand cuts: Granzyme pathways of programmed cell death. Annu. Rev. Immunol. 2008, 26, 389–420. [Google Scholar] [CrossRef]
- Trapani, J.A.; Sutton, V.R. Granzyme B: Pro-apoptotic, antiviral and antitumor functions. Curr. Opin. Immunol. 2003, 15, 533–543. [Google Scholar] [CrossRef]
- Bleackley, R.C. A molecular view of cytotoxic T lymphocyte induced killing. Biochem. Cell Biol. 2005, 83, 747–51. [Google Scholar] [CrossRef]
- Sun, J.; Bird, C.H.; Sutton, V.; McDonald, L.; Coughlin, P.B.; De Jong, T.A.; Trapani, J.A.; Bird, P.I. A cytosolic granzyme B inhibitor related to the viral apoptotic regulator cytokine response modifier A is present in cytotoxic lymphocytes. J. Biol. Chem. 1996, 271, 27802–27809. [Google Scholar]
- Poe, M.; Blake, J.T.; Boulton, D.A.; Gammon, M.; Sigal, N.H.; Wu, J.K.; Zweerink, H.J. Human cytotoxic lymphocyte granzyme B. Its purification from granules and the characterization of substrate and inhibitor specificity. J. Biol. Chem. 1991, 266, 98–103. [Google Scholar]
- Zhao, J.; Zhang, L.H.; Jia, L.T.; Zhang, L.; Xu, Y.M.; Wang, Z.; Yu, C.J.; Peng, W.D.; Wen, W.H.; Wang, C.J.; et al. Secreted antibody/granzyme B fusion protein stimulates selective killing of HER2-overexpressing tumor cells. J. Biol. Chem. 2004, 279, 21343–21348. [Google Scholar]
- Caputo, A.; Garner, R.S.; Winkler, U.; Hudig, D.; Bleackley, R.C. Activation of recombinant murine cytotoxic cell proteinase-1 requires deletion of an amino-terminal dipeptide. J. Biol. Chem. 1993, 268, 17672–17675. [Google Scholar]
- Dalken, B.; Jabulowsky, R.A.; Oberoi, P.; Benhar, I.; Wels, W.S. Maltose-binding protein enhances secretion of recombinant human granzyme B accompanied by in vivo processing of a precursor MBP fusion protein. PLoS One 2010, 5, e14404. [Google Scholar]
- Kam, C.M.; Hudig, D.; Powers, J.C. Granzymes (lymphocyte serine proteases): Characterization with natural and synthetic substrates and inhibitors. Biochim. Biophys. Acta 2000, 1477, 307–323. [Google Scholar] [CrossRef]
- Griffiths, G.M.; Isaaz, S. Granzymes A and B are targeted to the lytic granules of lymphocytes by the mannose-6-phosphate receptor. J. Cell Biol. 1993, 120, 885–896. [Google Scholar] [CrossRef]
- Edwards, K.M.; Davis, J.E.; Browne, K.A.; Sutton, V.R.; Trapani, J.A. Anti-viral strategies of cytotoxic T lymphocytes are manifested through a variety of granule-bound pathways of apoptosis induction. Immunol. Cell Biol. 1999, 77, 76–89. [Google Scholar] [CrossRef]
- Giesubel, U.; Dalken, B.; Mahmud, F.; Wels, W.S. Cell binding, internalization and cytotoxic activity of human granzyme B expressed in the yeast Pichia pastoris. Biochem. J. 2006, 394, 563–573. [Google Scholar] [CrossRef]
- Xia, Z.; Kam, C.M.; Huang, C.; Powers, J.C.; Mandle, R.J.; Stevens, R.L.; Lieberman, J. Expression and purification of enzymatically active recombinant granzyme B in a baculovirus system. Biochem. Biophys. Res. Commun. 1998, 243, 384–389. [Google Scholar] [CrossRef]
- Smyth, M.J.; McGuire, M.J.; Thia, K.Y. Expression of recombinant human granzyme B. A processing and activation role for dipeptidyl peptidase I. J. Immunol. 1995, 154, 6299–6305. [Google Scholar]
- Godal, R.; Keilholz, U.; Uharek, L.; Letsch, A.; Asemissen, A.M.; Busse, A.; Na, I.K.; Thiel, E.; Scheibenbogen, C. Lymphomas are sensitive to perforin-dependent cytotoxic pathways despite expression of PI-9 and overexpression of bcl-2. Blood 2006, 107, 3205–3211. [Google Scholar] [CrossRef]
- Soriano, C.; Mukaro, V.; Hodge, G.; Ahern, J.; Holmes, M.; Jersmann, H.; Moffat, D.; Meredith, D.; Jurisevic, C.; Reynolds, P.N.; et al. Increased proteinase inhibitor-9 (PI-9) and reduced granzyme B in lung cancer: Mechanism for immune evasion? Lung Cancer 2012, 77, 38–45. [Google Scholar] [CrossRef]
- Ray, M.; Hostetter, D.R.; Loeb, C.R.; Simko, J.; Craik, C.S. Inhibition of Granzyme B by PI-9 protects prostate cancer cells from apoptosis. Prostate 2012, 72, 846–855. [Google Scholar] [CrossRef]
- Rousalova, I.; Krepela, E.; Prochazka, J.; Cermak, J.; Benkova, K. Expression of proteinase inhibitor-9/serpinB9 in non-small cell lung carcinoma cells and tissues. Int. J. Oncol. 2010, 36, 275–283. [Google Scholar]
- Jiang, X.G.; Patterson, N.M.; Ling, Y.; Xie, J.W.; Helferich, W.G.; Shapiro, D.J. Low Concentrations of the Soy Phytoestrogen Genistein Induce Proteinase Inhibitor 9 and Block Killing of Breast Cancer Cells by Immune Cells. Endocrinology 2008, 149, 5366–5373. [Google Scholar] [CrossRef]
- van Houdt, I.S.; Oudejans, J.J.; van den Eertwegh, A.J.; Baars, A.; Vos, W.; Bladergroen, B.A.; Rimoldi, D.; Muris, J.J.; Hooijberg, E.; Gundy, C.M.; et al. Expression of the apoptosis inhibitor protease inhibitor 9 predicts clinical outcome in vaccinated patients with stage III and IV melanoma. Clin. Cancer Res. 2005, 11, 6400–6407. [Google Scholar]
- Oudejans, J.J.; Harijadi, H.; Kummer, J.A.; Tan, I.B.; Bloemena, E.; Middeldorp, J.M.; Bladergroen, B.; Dukers, D.F.; Vos, W.; Meijer, C.J. High numbers of granzyme B/CD8-positive tumour-infiltrating lymphocytes in nasopharyngeal carcinoma biopsies predict rapid fatal outcome in patients treated with curative intent. J. Pathol. 2002, 198, 468–475. [Google Scholar] [CrossRef]
- Bots, M.; Liesbeth, V.A.N.B.; Rademaker, M.T.; Offringa, R.; Medema, J.P. Serpins prevent granzyme-induced death in a species-specific manner. Immunol. Cell Biol. 2006, 84, 79–86. [Google Scholar] [CrossRef]
- Huntington, J.A.; Read, R.J.; Carrell, R.W. Structure of a serpin-protease complex shows inhibition by deformation. Nature 2000, 407, 923–926. [Google Scholar] [CrossRef]
- Bladergroen, B.A.; Strik, M.C.; Bovenschen, N.; van Berkum, O.; Scheffer, G.L.; Meijer, C.J.; Hack, C.E.; Kummer, J.A. The granzyme B inhibitor, protease inhibitor 9, is mainly expressed by dendritic cells and at immune-privileged sites. J. Immunol. 2001, 166, 3218–3225. [Google Scholar]
- Buzza, M.S.; Hosking, P.; Bird, P.I. The granzyme B inhibitor, PI-9, is differentially expressed during placental development and up-regulated in hydatidiform moles. Placenta 2006, 27, 62–69. [Google Scholar] [CrossRef]
- Kannan-Thulasiraman, P.; Shapiro, D.J. Modulators of inflammation use nuclear factor-kappa B and activator protein-1 sites to induce the caspase-1 and granzyme B inhibitor, proteinase inhibitor 9. J. Biol. Chem. 2002, 277, 41230–41239. [Google Scholar] [CrossRef]
- El Haddad, N.; Moore, R.; Heathcote, D.; Mounayar, M.; Azzi, J.; Mfarrej, B.; Batal, I.; Ting, C.; Atkinson, M.; Sayegh, M.H.; et al. The novel role of SERPINB9 in cytotoxic protection of human mesenchymal stem cells. J. Immunol. 2011, 187, 2252–2260. [Google Scholar] [CrossRef]
- Heutinck, K.M.; Kassies, J.; Florquin, S.; Ten Berge, I.J.; Hamann, J.; Rowshani, A.T. SerpinB9 expression in human renal tubular epithelial cells is induced by triggering of the viral dsRNA sensors TLR3, MDA5 and RIG-I. Nephrol. Dial. Transplant. 2012, 27, 2746–2754. [Google Scholar] [CrossRef]
- Rowshani, A.T.; Strik, M.C.; Molenaar, R.; Yong, S.L.; Wolbink, A.M.; Bemelman, F.J.; Hack, C.E.; Ten Berge, I.J. The granzyme B inhibitor SERPINB9 (protease inhibitor 9) circulates in blood and increases on primary cytomegalovirus infection after renal transplantation. J. Infect. Dis. 2005, 192, 1908–1911. [Google Scholar] [CrossRef]
- Classen, C.F.; Bird, P.I.; Debatin, K.M. Modulation of the granzyme B inhibitor proteinase inhibitor 9 (PI-9) by activation of lymphocytes and monocytes in vitro and by Epstein-Barr virus and bacterial infection. Clin. Exp. Immunol. 2006, 143, 534–542. [Google Scholar] [CrossRef]
- Buzza, M.S.; Hirst, C.E.; Bird, C.H.; Hosking, P.; McKendrick, J.; Bird, P.I. The granzyme B inhibitor, PI-9, is present in endothelial and mesothelial cells, suggesting that it protects bystander cells during immune respons. Cell. Immunol. 2001, 210, 21–29. [Google Scholar] [CrossRef]
- Barrie, M.B.; Stout, H.W.; Abougergi, M.S.; Miller, B.C.; Thiele, D.L. Antiviral cytokines induce hepatic expression of the granzyme B inhibitors, proteinase inhibitor 9 and serine proteinase inhibitor 6. J. Immunol. 2004, 172, 6453–6459. [Google Scholar]
- Horie, O.; Saigo, K.; Murayama, T.; Ryo, R. Differential expression of proteinase inhibitor-9 and granzyme B mRNAs in activated immunocompetent cells. Tohoku J. Exp. Med. 2005, 205, 103–113. [Google Scholar] [CrossRef]
- Hirst, C.E.; Buzza, M.S.; Bird, C.H.; Warren, H.S.; Cameron, P.U.; Zhang, M.; Ashton-Rickardt, P.G.; Bird, P.I. The intracellular granzyme B inhibitor, proteinase inhibitor 9, is up-regulated during accessory cell maturation and effector cell degranulation, and its overexpression enhances CTL potency. J. Immunol. 2003, 170, 805–815. [Google Scholar]
- Rowshani, A.T.; Florquin, S.; Bemelman, F.; Kummer, J.A.; Hack, C.E.; Ten Berge, I.J. Hyperexpression of the granzyme B inhibitor PI-9 in human renal allografts: A potential mechanism for stable renal function in patients with subclinical rejection. Kidney Int. 2004, 66, 1417–1422. [Google Scholar] [CrossRef]
- Walker, P.R.; Saas, P.; Dietrich, P.Y. Role of Fas ligand (CD95L) in immune escape: The tumor cell strikes back. J. Immunol. 1997, 158, 4521–4524. [Google Scholar]
- Buehring, G.C.; Eby, E.A.; Eby, M.J. Cell line cross-contamination: How aware are Mammalian cell culturists of the problem and how to monitor it? In Vitro Cell. Dev. Biol. Anim. 2004, 40, 211–215. [Google Scholar] [CrossRef]
- Bird, C.H.; Sutton, V.R.; Sun, J.; Hirst, C.E.; Novak, A.; Kumar, S.; Trapani, J.A.; Bird, P.I. Selective regulation of apoptosis: The cytotoxic lymphocyte serpin proteinase inhibitor 9 protects against granzyme B-mediated apoptosis without perturbing the Fas cell death pathway. Mol. Cell. Biol. 1998, 18, 6387–6398. [Google Scholar]
- Schiffer, S. Efficacy of an adapted Granzyme B-based anti-CD30 cytolytic fusion protein against PI-9-positive classical Hodgkin lymphoma cells in a murine model. Blood Cancer J. 2012. submitted for publication. [Google Scholar]
- ten Berge, R.L.; Meijer, C.J.; Dukers, D.F.; Kummer, J.A.; Bladergroen, B.A.; Vos, W.; Hack, C.E.; Ossenkoppele, G.J.; Oudejans, J.J. Expression levels of apoptosis-related proteins predict clinical outcome in anaplastic large cell lymphoma. Blood 2002, 99, 4540–4546. [Google Scholar] [CrossRef]
- Tiacci, E.; Doring, C.; Brune, V.; van Noesel, C.J.; Klapper, W.; Mechtersheimer, G.; Falini, B.; Kuppers, R.; Hansmann, M.L. Analyzing primary Hodgkin and Reed-Sternberg cells to capture the molecular and cellular pathogenesis of classical Hodgkin lymphoma. Blood 2012, 120, 4609–4620. [Google Scholar] [CrossRef]
- Fritsch, K. Die Bedeutung des Proteinase Inhibitor 9 für die Apoptoseresistenz in leukämischen Zelllinien. Ph.D. Thesis, Albert-Ludwigs-University, Freiburg im Breisgau.
- Bossard, C.; Belhadj, K.; Reyes, F.; Martin-Garcia, N.; Berger, F.; Kummer, J.A.; Briere, J.; Baglin, A.C.; Cheze, S.; Bosq, J.; et al. Expression of the granzyme B inhibitor PI9 predicts outcome in nasal NK/T-cell lymphoma: Results of a Western series of 48 patients treated with first-line polychemotherapy within the Groupe d'Etude des Lymphomes de l'Adulte (GELA) trials. Blood 2007, 109, 2183–2189. [Google Scholar] [CrossRef]
- Bladergroen, B.A.; Meijer, C.J.; ten Berge, R.L.; Hack, C.E.; Muris, J.J.; Dukers, D.F.; Chott, A.; Kazama, Y.; Oudejans, J.J.; van Berkum, O.; et al. Expression of the granzyme B inhibitor, protease inhibitor 9, by tumor cells in patients with non-Hodgkin and Hodgkin lymphoma: A novel protective mechanism for tumor cells to circumvent the immune system? Blood 2002, 99, 232–237. [Google Scholar] [CrossRef]
- Mahrus, S.; Kisiel, W.; Craik, C.S. Granzyme M is a regulatory protease that inactivates proteinase inhibitor 9, an endogenous inhibitor of granzyme B. J. Biol. Chem. 2004, 279, 54275–54282. [Google Scholar]
- Madison, E.L.; Goldsmith, E.J.; Gerard, R.D.; Gething, M.J.; Sambrook, J.F. Serpin-resistant mutants of human tissue-type plasminogen activator. Nature 1989, 339, 721–724. [Google Scholar] [CrossRef]
- Potempa, J.; Korzus, E.; Travis, J. The serpin superfamily of proteinase inhibitors: Structure, function, and regulation. J. Biol. Chem. 1994, 269, 15957–15960. [Google Scholar]
- Marszal, E.; Shrake, A. Serpin crystal structure and serpin polymer structure. Arch. Biochem. Biophys. 2006, 453, 123–129. [Google Scholar] [CrossRef]
- Silverman, G.A.; Bird, P.I.; Carrell, R.W.; Church, F.C.; Coughlin, P.B.; Gettins, P.G.; Irving, J.A.; Lomas, D.A.; Luke, C.J.; Moyer, R.W.; et al. The serpins are an expanding superfamily of structurally similar but functionally diverse proteins. Evolution, mechanism of inhibition, novel functions, and a revised nomenclature. J. Biol. Chem. 2001, 276, 33293–33296. [Google Scholar]
- Sun, J.; Whisstock, J.C.; Harriott, P.; Walker, B.; Novak, A.; Thompson, P.E.; Smith, A.I.; Bird, P.I. Importance of the P4' residue in human granzyme B inhibitors and substrates revealed by scanning mutagenesis of the proteinase inhibitor 9 reactive center loop. J. Biol. Chem. 2001, 276, 15177–15184. [Google Scholar]
- Hedstrom, L. Serine protease mechanism and specificity. Chem. Rev. 2002, 102, 4501–4524. [Google Scholar] [CrossRef]
- Ye, S.; Cech, A.L.; Belmares, R.; Bergstrom, R.C.; Tong, Y.; Corey, D.R.; Kanost, M.R.; Goldsmith, E.J. The structure of a Michaelis serpin-protease complex. Nat. Struct. Biol. 2001, 8, 979–983. [Google Scholar]
- Rotonda, J.; Garcia-Calvo, M.; Bull, H.G.; Geissler, W.M.; McKeever, B.M.; Willoughby, C.A.; Thornberry, N.A.; Becker, J.W. The three-dimensional structure of human granzyme B compared to caspase-3, key mediators of cell death with cleavage specificity for aspartic acid in P1. Chem. Biol. 2001, 8, 357–368. [Google Scholar]
- Kortemme, T.; Baker, D. A simple physical model for binding energy hot spots in protein-protein complexes. Proc. Natl. Acad. Sci. USA 2002, 99, 14116–14121. [Google Scholar] [CrossRef]
- Losasso, V.; Schiffer, S.; Barth, S.; Carloni, P. Design of human granzyme B variants resistant to serpin B9. Proteins 2012, 80, 2514–2522. [Google Scholar]
- Jabulowsky, R.A.; Oberoi, P.; Bahr-Mahmud, H.; Dalken, B.; Wels, W.S. Surface Charge-Modification Prevents Sequestration and Enhances Tumor-Cell Specificity of a Recombinant Granzyme B-TGF alpha Fusion Protein. Bioconjug. Chem. 2012, 23, 1567–1576. [Google Scholar]
- Motyka, B.; Korbutt, G.; Pinkoski, M.J.; Heibein, J.A.; Caputo, A.; Hobman, M.; Barry, M.; Shostak, I.; Sawchuk, T.; Holmes, C.F.; et al. Mannose 6-phosphate/insulin-like growth factor II receptor is a death receptor for granzyme B during cytotoxic T cell-induced apoptosis. Cell 2000, 103, 491–500. [Google Scholar] [CrossRef]
- Veugelers, K.; Motyka, B.; Goping, I.S.; Shostak, I.; Sawchuk, T.; Bleackley, R.C. Granule-mediated killing by granzyme B and perforin requires a mannose 6-phosphate receptor and is augmented by cell surface heparan sulfate. Mol. Biol. Cell 2006, 17, 623–633. [Google Scholar]
- Kurschus, F.C.; Bruno, R.; Fellows, E.; Falk, C.S.; Jenne, D.E. Membrane receptors are not required to deliver granzyme B during killer cell attack. Blood 2005, 105, 2049–2058. [Google Scholar] [CrossRef]
- Trapani, J.A.; Sutton, V.R.; Thia, K.Y.; Li, Y.Q.; Froelich, C.J.; Jans, D.A.; Sandrin, M.S.; Browne, K.A. A clathrin/dynamin- and mannose-6-phosphate receptor-independent pathway for granzyme B-induced cell death. J. Cell Biol. 2003, 160, 223–233. [Google Scholar] [CrossRef]
- Bretthauer, R.K.; Castellino, F.J. Glycosylation of Pichia pastoris-derived proteins. Biotechnol. Appl. Biochem. 1999, 30, 193–200. [Google Scholar]
- Raja, S.M.; Metkar, S.S.; Froelich, C.J. Cytotoxic granule-mediated apoptosis: Unraveling the complex mechanism. Curr. Opin. Immunol. 2003, 15, 528–532. [Google Scholar] [CrossRef]
- Gross, C.; Koelch, W.; DeMaio, A.; Arispe, N.; Multhoff, G. Cell surface-bound heat shock protein 70 (Hsp70) mediates perforin-independent apoptosis by specific binding and uptake of granzyme B. J. Biol. Chem. 2003, 278, 41173–41181. [Google Scholar]
- Bird, C.H.; Sun, J.; Ung, K.; Karambalis, D.; Whisstock, J.C.; Trapani, J.A.; Bird, P.I. Cationic sites on granzyme B contribute to cytotoxicity by promoting its uptake into target cells. Mol. Cell. Biol. 2005, 25, 7854–7867. [Google Scholar]
- Shi, L.; Keefe, D.; Durand, E.; Feng, H.; Zhang, D.; Lieberman, J. Granzyme B binds to target cells mostly by charge and must be added at the same time as perforin to trigger apoptosis. J. Immunol. 2005, 174, 5456–5461. [Google Scholar]
- Metkar, S.S.; Wang, B.; Aguilar-Santelises, M.; Raja, S.M.; Uhlin-Hansen, L.; Podack, E.; Trapani, J.A.; Froelich, C.J. Cytotoxic cell granule-mediated apoptosis: Perforin delivers granzyme B-serglycin complexes into target cells without plasma membrane pore formation. Immunity 2002, 16, 417–428. [Google Scholar] [CrossRef]
- Onda, M.; Nagata, S.; Tsutsumi, Y.; Vincent, J.J.; Wang, Q.; Kreitman, R.J.; Lee, B.; Pastan, I. Lowering the isoelectric point of the Fv portion of recombinant immunotoxins leads to decreased nonspecific animal toxicity without affecting antitumor activity. Cancer Res. 2001, 61, 5070–5077. [Google Scholar]
- Buzza, M.S.; Zamurs, L.; Sun, J.; Bird, C.H.; Smith, A.I.; Trapani, J.A.; Froelich, C.J.; Nice, E.C.; Bird, P.I. Extracellular matrix remodeling by human granzyme B via cleavage of vitronectin, fibronectin, and laminin. J. Biol. Chem. 2005, 280, 23549–23558. [Google Scholar]
- Ronday, H.K.; van der Laan, W.H.; Tak, P.P.; de Roos, J.A.; Bank, R.A.; TeKoppele, J.M.; Froelich, C.J.; Hack, C.E.; Hogendoorn, P.C.; Breedveld, F.C.; et al. Human granzyme B mediates cartilage proteoglycan degradation and is expressed at the invasive front of the synovium in rheumatoid arthritis. Rheumatology (Oxford) 2001, 40, 55–61. [Google Scholar] [CrossRef]
- Tak, P.P.; Spaeny-Dekking, L.; Kraan, M.C.; Breedveld, F.C.; Froelich, C.J.; Hack, C.E. The levels of soluble granzyme A and B are elevated in plasma and synovial fluid of patients with rheumatoid arthritis (RA). Clin. Exp. Immunol. 1999, 116, 366–370. [Google Scholar] [CrossRef]
- Goldbach-Mansky, R.; Suson, S.; Wesley, R.; Hack, C.E.; El-Gabalawy, H.S.; Tak, P.P. Raised granzyme B levels are associated with erosions in patients with early rheumatoid factor positive rheumatoid arthritis. Ann. Rheum. Dis. 2005, 64, 715–721. [Google Scholar] [CrossRef]
- Chamberlain, C.M.; Ang, L.S.; Boivin, W.A.; Cooper, D.M.; Williams, S.J.; Zhao, H.; Hendel, A.; Folkesson, M.; Swedenborg, J.; Allard, M.F.; et al. Perforin-independent extracellular granzyme B activity contributes to abdominal aortic aneurysm. Am. J. Pathol. 2010, 176, 1038–1049. [Google Scholar] [CrossRef]
- Saito, Y.; Kondo, H.; Hojo, Y. Granzyme B as a novel factor involved in cardiovascular diseases. J. Cardiol. 2011, 57, 141–147. [Google Scholar] [CrossRef]
- Kurschus, F.C.; Fellows, E.; Stegmann, E.; Jenne, D.E. Granzyme B delivery via perforin is restricted by size, but not by heparan sulfate-dependent endocytosis. Proc. Natl. Acad. Sci. USA 2008, 105, 13799–13804. [Google Scholar] [CrossRef]
- Thomas, G. Furin at the cutting edge: From protein traffic to embryogenesis and disease. Nat. Rev. Mol. Cell Biol. 2002, 3, 753–766. [Google Scholar] [CrossRef]
- Molloy, S.S.; Anderson, E.D.; Jean, F.; Thomas, G. Bi-cycling the furin pathway: From TGN localization to pathogen activation and embryogenesis. Trends Cell Biol. 1999, 9, 28–35. [Google Scholar] [CrossRef]
- Ogata, M.; Fryling, C.M.; Pastan, I.; FitzGerald, D.J. Cell-mediated cleavage of Pseudomonas exotoxin between Arg279 and Gly280 generates the enzymatically active fragment which translocates to the cytosol. J. Biol. Chem. 1992, 267, 25396–25401. [Google Scholar]
- Theuer, C.P.; FitzGerald, D.; Pastan, I. A recombinant form of Pseudomonas exotoxin directed at the epidermal growth factor receptor that is cytotoxic without requiring proteolytic processing. J. Biol. Chem. 1992, 267, 16872–16877. [Google Scholar]
- Wolf, P.; Elsasser-Beile, U. Pseudomonas exotoxin A: From virulence factor to anti-cancer agent. Int. J. Med. Microbiol. 2009, 299, 161–176. [Google Scholar] [CrossRef]
- Collier, R.J. Understanding the mode of action of diphtheria toxin: A perspective on progress during the 20th century. Toxicon 2001, 39, 1793–1803. [Google Scholar] [CrossRef]
- Zhan, H.; Choe, S.; Huynh, P.D.; Finkelstein, A.; Eisenberg, D.; Collier, R.J. Dynamic transitions of the transmembrane domain of diphtheria toxin: Disulfide trapping and fluorescence proximity studies. Biochemistry 1994, 33, 11254–11263. [Google Scholar]
- Plank, C.; Oberhauser, B.; Mechtler, K.; Koch, C.; Wagner, E. The influence of endosome-disruptive peptides on gene transfer using synthetic virus-like gene transfer systems. J. Biol. Chem. 1994, 269, 12918–12924. [Google Scholar]
- Vives, E.; Brodin, P.; Lebleu, B. A truncated HIV-1 Tat protein basic domain rapidly translocates through the plasma membrane and accumulates in the cell nucleus. J. Biol. Chem. 1997, 272, 16010–16017. [Google Scholar] [CrossRef]
- Gius, D.R.; Ezhevsky, S.A.; Becker-Hapak, M.; Nagahara, H.; Wei, M.C.; Dowdy, S.F. Transduced p16INK4a peptides inhibit hypophosphorylation of the retinoblastoma protein and cell cycle progression prior to activation of Cdk2 complexes in late G1. Cancer Res. 1999, 59, 2577–2580. [Google Scholar]
- Vocero-Akbani, A.M.; Heyden, N.V.; Lissy, N.A.; Ratner, L.; Dowdy, S.F. Killing HIV-infected cells by transduction with an HIV protease-activated caspase-3 protein. Nat. Med. 1999, 5, 29–33. [Google Scholar] [CrossRef]
- Falnes, P.O.; Wesche, J.; Olsnes, S. Ability of the Tat basic domain and VP22 to mediate cell binding, but not membrane translocation of the diphtheria toxin A-fragment. Biochemistry 2001, 40, 4349–4358. [Google Scholar] [CrossRef]
- Ziegler, A.; Seelig, J. Interaction of the protein transduction domain of HIV-1 TAT with heparan sulfate: Binding mechanism and thermodynamic parameters. Biophys. J. 2004, 86, 254–263. [Google Scholar] [CrossRef]
- Noguchi, H.; Ueda, M.; Matsumoto, S.; Kobayashi, N.; Hayashi, S. BETA2/NeuroD protein transduction requires cell surface heparan sulfate proteoglycans. Hum. Gene Ther. 2007, 18, 10–17. [Google Scholar] [CrossRef]
- Richard, J.P.; Melikov, K.; Brooks, H.; Prevot, P.; Lebleu, B.; Chernomordik, L.V. Cellular uptake of unconjugated TAT peptide involves clathrin-dependent endocytosis and heparan sulfate receptors. J. Biol. Chem. 2005, 280, 15300–15306. [Google Scholar]
- Tilstra, J.; Rehman, K.K.; Hennon, T.; Plevy, S.E.; Clemens, P.; Robbins, P.D. Protein transduction: Identification, characterization and optimization. Biochem. Soc. Trans. 2007, 35, 811–815. [Google Scholar] [CrossRef]
- Zahid, M.; Lu, X.; Mi, Z.; Robbins, P.D. Cationic and tissue-specific protein transduction domains identification, characterization, and therapeutic application. Adv. Genet. 2010, 69, 83–95. [Google Scholar] [CrossRef]
- Fuchs, H.; Bachran, C.; Heisler, I.; Sutherland, M. A Closer Look at Protein Transduction Domains as a Tool in Drug Delivery. Curr. Nanosci. 2005, 1, 117–124. [Google Scholar] [CrossRef]
- Kabouridis, P.S. Biological applications of protein transduction technology. Trends Biotechnol. 2003, 21, 498–503. [Google Scholar] [CrossRef]
- van den Berg, A.; Dowdy, S.F. Protein transduction domain delivery of therapeutic macromolecules. Curr. Opin. Biotechnol. 2011, 22, 888–893. [Google Scholar] [CrossRef]
- Snyder, E.L.; Meade, B.R.; Dowdy, S.F. Anti-cancer protein transduction strategies: Reconstitution of p27 tumor suppressor function. J. Control Release 2003, 91, 45–51. [Google Scholar] [CrossRef]
- Plescia, J.; Salz, W.; Xia, F.; Pennati, M.; Zaffaroni, N.; Daidone, M.G.; Meli, M.; Dohi, T.; Fortugno, P.; Nefedova, Y.; et al. Rational design of shepherdin, a novel anticancer agent. Cancer Cell 2005, 7, 457–468. [Google Scholar] [CrossRef]
- Hong, F.D.; Clayman, G.L. Isolation of a peptide for targeted drug delivery into human head and neck solid tumors. Cancer Res. 2000, 60, 6551–6556. [Google Scholar]
- Wang, J.L.; Zhang, Z.J.; Choksi, S.; Shan, S.; Lu, Z.; Croce, C.M.; Alnemri, E.S.; Korngold, R.; Huang, Z. Cell permeable Bcl-2 binding peptides: A chemical approach to apoptosis induction in tumor cells. Cancer Res. 2000, 60, 1498–1502. [Google Scholar]
- Curnis, F.; Arrigoni, G.; Sacchi, A.; Fischetti, L.; Arap, W.; Pasqualini, R.; Corti, A. Differential binding of drugs containing the NGR motif to CD13 isoforms in tumor vessels, epithelia, and myeloid cells. Cancer Res. 2002, 62, 867–874. [Google Scholar]
- Snyder, E.L.; Saenz, C.C.; Denicourt, C.; Meade, B.R.; Cui, X.S.; Kaplan, I.M.; Dowdy, S.F. Enhanced targeting and killing of tumor cells expressing the CXC chemokine receptor 4 by transducible anticancer peptides. Cancer Res. 2005, 65, 10646–10650. [Google Scholar] [CrossRef]
- Keller, J.; Heisler, I.; Tauber, R.; Fuchs, H. Development of a novel molecular adapter for the optimization of immunotoxins. J. Control Release 2001, 74, 259–261. [Google Scholar] [CrossRef]
- Heisler, I.; Keller, J.; Tauber, R.; Sutherland, M.; Fuchs, H. A cleavable adapter to reduce nonspecific cytotoxicity of recombinant immunotoxins. Int. J. Cancer 2003, 103, 277–282. [Google Scholar] [CrossRef]
- Hetzel, C.; Bachran, C.; Tur, M.K.; Fuchs, H.; Stocker, M. Improved immunotoxins with novel functional elements. Curr. Pharm Des. 2009, 15, 2700–2711. [Google Scholar] [CrossRef]
- Hetzel, C.; Bachran, C.; Fischer, R.; Fuchs, H.; Barth, S.; Stocker, M. Small cleavable adapters enhance the specific cytotoxicity of a humanized immunotoxin directed against CD64-positive cells. J. Immunother. 2008, 31, 370–376. [Google Scholar] [CrossRef]
© 2013 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).