Abstract
Background/Objectives: Chemical methods for quantifying resistant starch (RS) in rice are labor-intensive, costly, and lack high repeatability, creating a bottleneck in breeding. This study aimed to develop specific, codominant molecular markers for the Wx gene to enable rapid and accurate genotype screening for RS content, thereby accelerating the development of high-RS rice varieties. Methods: Based on sequence alignment of the Wx gene in rice varieties with divergent RS content, a key single-nucleotide polymorphism was targeted. Two sets of tetra-primer amplification refractory mutation system polymerase chain reaction (ARMS-PCR) markers, T-Wx9-RS1 and T-Wx9-RS2, were designed. These markers were used to genotype diverse rice varieties and F4 segregating populations, with results validated against standard chemical assays. Results: Sequence analysis identified a critical T → C base mutation at position 202 of the ninth exon in high-RS varieties. The developed ARMS-PCR markers successfully and consistently distinguished all three possible genotypes (homozygous mutant, homozygous wild-type, and heterozygous). The genotyping results showed complete concordance with the phenotypes determined by chemical methods. Conclusions: The developed molecular markers, T-Wx9-RS1 and T-Wx9-RS2, provide a rapid, reliable, and cost-effective tool for marker-assisted selection of high resistant starch content in rice. Their implementation can significantly enhance screening efficiency and expedite the breeding pipeline for novel, nutritionally improved rice cultivars.
1. Introduction
Rice (Oryza sativa L.), a major global food crop, not only is a vital source of energy for humans but also holds significant cultural importance [1]. As health awareness has increased, the focus on rice quality has expanded from traditional yield and taste to include nutritional and health benefits. High-resistance starch (RS) rice, known for its unique physiological functions that help reduce post-meal blood sugar spikes and lower the risk of diseases such as type II diabetes and obesity, is becoming a key target in the breeding of functional rice [1,2]. Resistant starch, a component of starch that is difficult to digest in the small intestine, ferments in the gut to produce short-chain fatty acids, which are beneficial to gut health [1,2].
The starch in the rice endosperm is primarily composed of amylose and amylopectin, and its physicochemical properties affect the processing, taste, and nutritional quality of the rice and, especially, the content of RS. Granule-bound starch synthase I (GBSSI), encoded by the Wx gene, is a key enzyme that regulates the amylose content (AC) in the rice endosperm [3,4]. Numerous studies have shown that subtle changes in the expression levels and activity of the Wx gene can considerably alter the amylose content, which in turn affects the gelatinization temperature (GT), viscosity profile (RVA profile), eating and cooking quality (ECQ), and potential formation of RS [3,4,5,6]. Therefore, the Wx gene is considered a core regulatory module for improving the taste and nutritional quality of rice, particularly the content of RS [3].
Precisely regulating the expression of the Wx gene is an effective strategy for enhancing rice quality. Traditional breeding methods regulate the amylose content by selecting and combining different natural alleles (such as Wxa and Wxb). Molecular biology research has further revealed the complexity of Wx gene expression regulation, including cis-acting elements in the promoter region (such as CAAT-box and TATA-box sequences), structural changes in the 5′-UTR region [3], and the regulatory mechanisms of upstream transcription factors such as OsEBP89, OsSPL14, and OsGSK5/OsSK41 on Wx [7,8]. These findings lay the foundation for a deeper understanding of the molecular network involved in amylose formation.
In recent years, gene editing technologies, particularly the CRISPR/Cas9 system, have provided powerful tools for achieving precise targeting and modification of the Wx gene. Research has shown that by editing the Wx gene promoter [3,9] or coding region [5], a variety of new rice materials with different amylose contents and physicochemical properties (such as the gelatinization temperature and resistant starch content) can be created [3,5,6,8,9]. For example, editing the Wx promoter (such as reducing the expression of Wxa) can reduce chalkiness and improve appearance quality [9]; creating specific Wx alleles or combining them with other starch synthesis-related genes (such as SBEIIb, SSIIa, and FLO2) can result in rice materials with high amylose content and high-resistant starch characteristics [10,11,12]. These successful cases highlight the core role of the Wx gene locus in the breeding of rice with specific qualities, including high RS.
To more effectively apply these Wx gene-based improvements in breeding practices, the development of closely linked functional molecular markers is crucial. Functional markers (FMs) are molecular markers developed from allele variants that cause phenotypic variations in genes or regulatory regions, such as SNPs and Indels. Compared with conventional markers, these markers significantly increase the precision and efficiency of marker-assisted selection (MAS). In crop breeding, functional markers are widely recognized as key tools for accelerating the integration of target traits, such as disease resistance and quality traits [13,14,15,16,17,18,19,20,21,22]. In the context of rice-resistant starch breeding, developing high-precision functional molecular markers at key functional sites of the Wx gene (including its promoter, 5′-UTR, and coding region) can accurately distinguish different alleles, predict amylose content and related starch characteristics (such as RS potential), and enable rapid and precise tracking and aggregation of superior alleles in breeding populations, thus significantly shortening the breeding cycle and improving selection efficiency [13,19,20].
In summary, the Wx gene is the core switch regulating the amylose content and physicochemical properties of the rice endosperm, and its diversity is the key genetic basis for differences in rice quality, including RS content [3,5,6]. A deep study of the regulatory mechanisms of the Wx gene and the development of new functional molecular markers based on key functional sites will provide strong technical support for the efficient breeding of new rice varieties with excellent taste qualities and high RS contents [1,2,9,11,13]. This study aims to introduce methods for the development of new functional molecular markers around the Wx gene locus, their validation, and their practical application in the breeding of high-RS rice.
2. Materialsand Methods
2.1. Plant Materials
Fourteen rice varieties and seven F4 populations were provided by the Rice Research Institute of the Chongqing Academy of Agricultural Sciences. The rice varieties were Li, Lan, HR52, QR, ML, BR50, BR19, CN-9, Sx, BR18, Ky43, CN-5, BR89, and BR28, and the F4 hybrid populations were Lan/CN-5, Lan/CN-9, Lan/Sx, QR/Sx, QR/Ky43, Li/BR50, and QR/BR50. The seeds were sown in a seedling bed after disinfection and germination. The 25-day-old seedlings were then transplanted into the Dianjiang experimental field (Chongqing, China, 30°25′31″ N 107°24′1″ E, with a typical subtropical monsoon climate and purplish clay soil) of the Rice Research Institute of Chongqing Academy of Agricultural Sciences. Field management followed local high-yielding practices: transplanting at a spacing of 20 cm × 26.7 cm with two seedlings per hill, applying fertilizer at the rate of 150 kg N/ha, 75 kg P2O5/ha, and 100 kg K2O/ha, and maintaining a shallow water layer except during the mid-season drainage period for about 10 days to control ineffective tillering.
2.2. Determination of Apparent Amylose and RS Contents
The apparent amylose content (AAC) was determined according to the Chinese National Agricultural Industry Standard NY/T2639-2014. The four AAC standard samples (1.2, 11.2, 16.8 and 26.8%) were provided by the Rice Product Quality Inspection and Supervision Testing Center of the China National Rice Research Institute of the Ministry of Agriculture. In rice, AAC can be classified into the following grades: waxy (0–2%), very low (5–12%), low (12–20%), medium (20–25%), or high (25–33%) [23]. The RS content was determined according to the ministry standard NY/T2638-2014 and calculated using the following formula: RS (%) = Sample/Standard glucose × 0.9 × 0.1 × 10.3/0.1 × 100/w (where w represents the dry weight of the sample in mg). Each measurement was performed with three technical replicates. On the basis of previous studies and our own data, we classified the rice RS content into three grades: low (<3%), medium (3–6%), and high (>6%).
2.3. DNA Extraction from Rice Leaves
Fresh leaves of the different rice varieties and F4 populations were collected at the tillering stage, and genomic DNA was extracted via the CTAB method [24].
2.4. Sequence Comparison, Sequencing and Primer Design
The full-length DNA sequence of the Wx gene (chromosome 6, nc_029261.1 (1765524.1770644)) was downloaded from the NCBI database. The PCR primers used were designed with Primer3 (https://bioinfo.ut.ee/primer3-0.4.0/ (accessed on 27 May 2020). The genomic DNA of 14 rice varieties with different RS contents was amplified to create templates for the PCR products. The PCR products were sent to Sangon Biotech (Shanghai) Co., Ltd. (Shanghai, China), for sequencing. The sequencing results were compared with those of Vector NTI Advance 11.5.1 to determine the differences between high-RS- and low-RS-content rice varieties. Primers for tetra-primer amplification refractory mutation system polymerase chain reaction (ARMS-PCR) were designed on the basis of these differences and synthesized by Sangon Biotech (Shanghai) Co., Ltd., using the online four-primer analysis software PRIMER1 (http://primer1.soton.ac.uk/primer1.html (accessed on 11 June 2021) reported by Ye et al. [25].
2.5. PCR Amplification and Recovery of the Wx Allele Fragment
The Wx allele fragment was amplified using a Phanta Max Super-Fidelity DNA Polymerase Kit (P505-d1; Nanjing Vazyme Biotech Co., Ltd., Nanjing, China). The 50 μL PCR system included 2.0 μL of template DNA (20 ng/µL), 2.0 μL each of upstream and downstream primers (10 μM), 25.0 μL of 2× Phanta Max buffer, 1.0 μL of dNTP mixture (10 mM each), 1.0 μL of Phanta Max Super-Fidelity DNA Polymerase, and 17.0 μL of ddH2O. The PCR procedure was as follows: predenaturation at 95 °C for 3 min; 35 cycles of 95 °C for 15 s, 54 °C for 15 s, and 72 °C for 3 min; extension at 72 °C for 5 min; and cooling at 10 °C for 10 min. A bromophenol blue indicator was added to the amplification product for later use. The reaction product was electrophoresed on an agarose gel with 1.5% DuRed nucleic acid dye at 150 V for 30 min. The target band was removed on a UV transmission table and sent to Sangon Biotech (Shanghai) Co., Ltd., for sequencing.
2.6. Genotype Identification
The 20 μL PCR reaction mixture included 1.0 μL of template DNA (20 ng/µL), 0.5 μL of each of the four primers (10 μM), 10.0 μL of 2× 3G Taq Master Mix for PAGE (red dye) (P115-01, Nanjing Vazyme Biotech Co., Ltd.), and 7.0 μL of ddH2O. The PCR procedure was as follows: predenaturation at 95 °C for 3 min; 35 cycles of 95 °C for 15 s, 61 °C for 15 s, and 72 °C for 30 s; extension at 72 °C for 5 min; and cooling at 10 °C for 10 min. The PCR products were analyzed via 10% polyacrylamide gel electrophoresis to detect polymorphisms. Offspring with genotypes consistent with the low-RS parent were marked with “−”, those with genotypes consistent with the high-RS parent were marked with “+”, and those with heterozygous genotypes were marked with “H”.
3. Results
3.1. AAC and RS Contents
The AACs and RS contents of the 14 different rice varieties are shown in Table 1. The AACs of Li, QR, Lan and HR52 ranged from 14.86 to 15.67%, indicating low-amylose varieties, and the RS contents ranged from 0.44 to 0.57%, indicating low-RS varieties. The AACs of BR18, BR19, BR50, BR28, Sx, Ky43, BR89 and CN-5 ranged from 21.26 to 24.99%, indicating medium-amylose varieties, and the RS contents ranged from 6.25 to 9.10%, indicating high-RS varieties. The AACs of CN-9 and ML were 25.35 and 25.78%, respectively, indicating high-amylose varieties, and the RS contents were 10.66 and 6.68%, respectively, indicating high-RS varieties. Correlation analysis revealed a significant positive correlation between the RS content and AAC of each variety (r = 0.913, p ≤ 0.01).

Table 1.
Amylose and RS contents of different rice varieties.
3.2. Identification and Primer Design of Specific SNPs in the Wx Gene of High-RS Rice Varieties
The primers WL3 and WR (Table 2) were designed according to the rice Wx gene sequence in the NCBI database, and the genomic DNA of 14 rice varieties with different RS contents was amplified as a template. A 3043 bp fragment was obtained by sequencing. The sequence similarities and differences between the high-RS- and low-RS-content rice varieties were analyzed. Compared with that of low-RS-content rice, the Wx gene of high-RS-content rice was missing 4 bases; a T → C base mutation at position 202 of exon 9 (Figure 1) and a C → T mutation at position 115 of exon 10 (Figure 2) were detected in high-RS-content rice, in addition to several base mutations in introns.

Table 2.
Sequencing primers and related markers for detecting specific SNP variations in the rice Wx gene.

Figure 1.
Partial nucleotide sequences of exon 9 of the Wx gene in different rice varieties. The gray letters represent the same nucleotide sequence, and the black and red letters indicate base mutations in T → C at position 202.
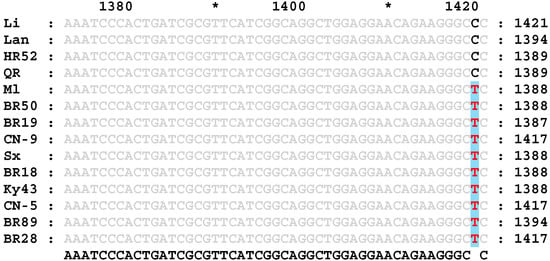
Figure 2.
Partial nucleotide sequences of exon 10 of the Wx gene in different rice varieties. The gray letters represent identical nucleotide sequences, while the black and red letters indicate base mutations in C → T located at position 115.
Two sets of primers, T-Wx9-RS1 and T-Wx9-RS2 (Table 2), were designed using online software (http://primer1.soton.ac.uk/primer1.html (accessed on 11 June 2021)). These primers were designed to detect the base mutation at position 202 of exon 9 of the Wx gene in rice, which is closely related to high RS content.
3.3. Validation of Molecular Markers
The molecular markers of 40 individual plants in the F4 population were verified. The results of nucleotide electrophoresis of representative plants are shown in Figure 3 and Figure 4. The results of amplification using the primer group T-Wx9-RS1 are shown in Figure 3. Two bands of 350 and 190 bp indicate a low-RS-content rice variety; two bands of 350 and 215 bp indicate the homozygous genotype of a high-RS-content rice variety (T → C base mutation at position 202 of the ninth exon of the Wx gene in both strands), whereas three bands of 350, 190 and 215 bp indicate the heterozygous genotype of a high-RS-content rice variety (T → C base mutation in one strand).
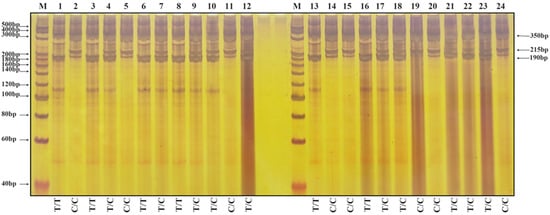
Figure 3.
Nucleic acid electrophoresis results after amplification of the genomic DNA of F4 generation rice plants using the primer set T-Wx9-RS1. The expected PCR products are labeled on the right. Two bands of 350 and 190 bp indicate a low-RS-content rice variety, two bands of 350 and 215 bp indicate the homozygous genotype of a high-RS-content rice variety, whereas three bands of 350, 190 and 215 bp indicate the heterozygous genotype of a high-RS-content rice variety. Lane M: 20 bp DNA Ladder Marker from TaKaRa, Lane 1: Parent Li, Lane 2: Parent BR50, Lanes 3–12: F4 plants from cross Li/BR50 (7553 2-1 to 7553 2-10). Lane 13: Parent QR, Lane 14: Parent BR50, Lanes 15–24: F4 plants from cross QR/BR50 (7586 2-1 to 7586 2-10).
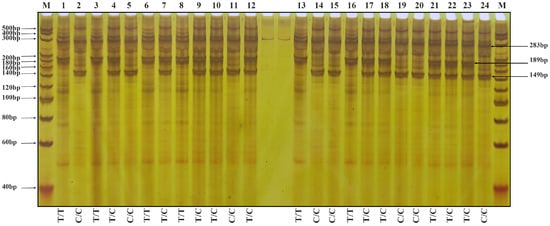
Figure 4.
Nucleic acid electrophoresis results after amplification of the genomic DNA of F4 generation rice plants using the primer set T-Wx9-RS2. The expected PCR products are labeled on the right. Two bands of 283 and 189 bp indicate a low-RS-content rice variety, two bands of 283 and 149 bp indicate the homozygous genotype of a high-RS-content rice variety, whereas three bands of 283, 189 and 149 bp indicate the heterozygous genotype of a high-RS-content rice variety. Lane M: 20 bp DNA Ladder Marker from TaKaRa, Lane 1: Parent Li, Lane 2: Parent BR50, Lanes 3–12: F4 plants from cross Li/BR50 (7553 2-1 to 7553 2-10). Lane 13: Parent QR, Lane 14: Parent BR50, Lanes 15–24: F4 plants from cross QR/BR50 (7586 2-1 to 7586 2-10).
The results of amplification using the primer group T-Wx9-RS2 are shown in Figure 4. Two bands of 283 and 189 bp indicate a low-RS-content rice variety; two bands of 283 and 149 bp indicate the homozygous genotype of a high-RS-content rice variety (mutation in both strands), whereas three bands of 283, 189 and 149 bp indicate the heterozygous genotype of a high-RS-content rice variety (mutation in one strand). Individual plants from the F4 generation were analyzed with the two primer sets, and the results were consistent.
The genotypes and RS contents of 40 individual plants are shown in Table 3 and Figure 5. Among the 40 F4 generation plants, 23 genotypes were consistent with those of the low–RS-content parent (without the mutant Wx gene, indicated by the symbol “−”), and the RS content was 0.41–1.99%, with an average of 0.88%. The genotypes of 8 plants were consistent with those of the high–RS-content parent (homozygous for the mutant Wx gene, indicated by the symbol “+”), and the RS content was 3.10–11.10%, with an average of 5.91%. The genotypes of 9 plants were heterozygous (inherited the mutant Wx gene from one parent, indicated by the letter “H”), and the RS content was 1.42–4.00%, with an average of 3.00%. These findings indicate that the RS content of a rice variety can be accurately predicted according to its genotype.

Table 3.
Genotypes and corresponding RS contents of 40 individual plants.
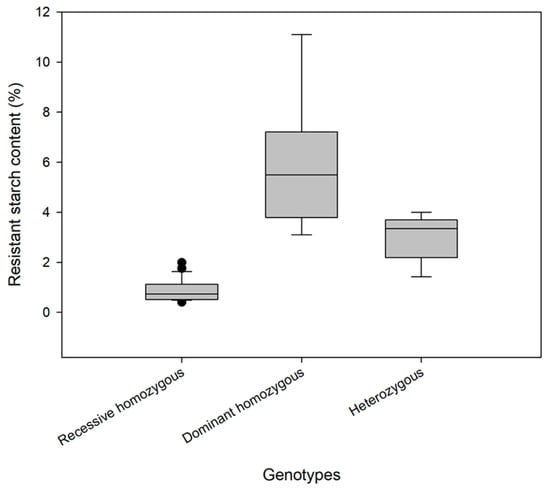
Figure 5.
Box plot of resistant starch content in 40 individual plants of different genotypes.
4. Discussion
4.1. Core Findings and Marker Utility
This study identified a novel, synonymous T → C SNP within exon 9 of the rice Wx gene that exhibits a perfect diagnostic association with elevated resistant starch (RS) content across our biparental populations. The key achievement is the successful development and validation of two robust, codominant tetra-primer ARMS-PCR markers (T-Wx9-RS1 and T-Wx9-RS2) precisely targeting this SNP. These markers efficiently discriminated among all three possible genotypes (homozygous for the low-RS allele, homozygous for the high-RS allele, and heterozygous) within parental lines and F4 segregating populations. The observed complete concordance between marker genotypes and chemically quantified RS levels (Table 3, Figure 5) underscores their immediate, high practical value for marker-assisted selection (MAS). This breakthrough equips breeders with a cost-effective, rapid, and accurate tool to screen for the high-RS trait in early generations, dramatically accelerating the breeding pipeline compared to reliance on time-consuming, cumbersome phenotypic assays alone.
4.2. Interpretation of the Synonymous Mutation and Its Potential Mechanisms
A central finding demanding careful interpretation is the robust association of a synonymous SNP with a distinct phenotypic effect. While our data unequivocally establishes this SNP as an outstanding molecular marker, its direct functional role remains elusive. We consider several non-exclusive possibilities: (1) Linkage Disequilibrium (LD): The T202C SNP likely resides in extremely tight LD with the true, yet unidentified causal variant. This causal mutation could be a cis-regulatory element influencing Wx expression levels or a non-synonymous change within another gene inside the haplotype block. (2) Effects on mRNA Processing: Synonymous mutations can profoundly impact mRNA splicing efficiency, stability, or secondary structure. The exon 9 SNP may disrupt an exonic splicing enhancer/silencer motif or alter the co-translational folding of the GBSSI protein, potentially compromising its activity or abundance. (3) Translational Kinetics: Altered codon usage, even for the identical amino acid, can modulate translation speed, thereby influencing protein folding and function.
We emphasize that our current experimental design—association analysis within a mapping population—cannot definitively distinguish between these scenarios. Therefore, we present the T202C SNP primarily as a highly reliable and tightly linked diagnostic marker for breeding applications. Definitive proof of causality necessitates future functional studies, such as in vitro splicing assays, allele-specific expression analysis, or the creation of isogenic lines differing solely at this SNP via precise gene editing.
4.3. Marker Development, Application, and Technical Considerations
The Wx locus stands as a well-documented major regulator of amylose and RS content. Our work introduces a novel, practical tool to the arsenal of Wx-based markers. Our deliberate choice of tetra-primer ARMS-PCR offered an optimal balance of accuracy, speed, and affordability, perfectly suited for routine breeding applications. However, we must acknowledge the technique’s inherent limitation: its success critically depends on specific primer design. Our inability to develop a functional ARMS-PCR marker for the exon 10 SNP, despite its established association with gel consistency [26], underscores this critical limitation. The probable cause is the complex genomic context surrounding that SNP, such as high sequence similarity to other regions or stable secondary structures, which prevented the design of four specific primers that meet all stringent requirements for robust amplification.
The two successful markers developed here precisely convert genotype data into a clear breeding decision: select plants harboring the homozygous “C” allele (or potentially heterozygotes for further advancement) to enrich for high RS content. This direct genotype-to-phenotype link, rigorously validated in our populations, forms the cornerstone of their practical value.
4.4. Limitations, Generalizability, and Future Perspectives
While our results are promising, several limitations must be noted to frame the appropriate scope of the markers’ application: (1) Genetic Background and Generalizability: Our validation occurred within specific crosses. The utility of these markers across the broad diversity of rice germplasm (e.g., indica vs. japonica, landraces vs. elite lines) hinges critically upon the conservation of the causal haplotype tagged by our SNP. We strongly recommend preliminary screening of target breeding panels to confirm the marker-trait association before large-scale deployment. (2) Environmental Effects and G × E: This study was conducted in a single environment. RS content, like many quality traits, is susceptible to environmental influences such as temperature during grain filling. The potential for genotype-by-environment (G × E) interaction remains unassessed. Therefore, implementing rigorous multi-location and multi-season trials constitutes an essential next step. These trials are crucial to evaluate the stability of the phenotype predicted by these markers and validate their effectiveness across diverse growing conditions. (3) Statistical Analysis: The statistical correlations presented, while clear and significant, are initial. Future studies utilizing larger, more diverse populations should employ more sophisticated models to account for additional genetic or environmental covariates.
In conclusion, the functional markers developed herein offer a rigorously validated, highly efficient tool for MAS in high-RS rice breeding within analogous genetic backgrounds. Future work must prioritize: (1) rigorous validation of these markers across independent and diverse germplasm collections; (2) expanding trials to multiple environments to critically assess stability; and (3) initiating fine-mapping or functional studies to definitively elucidate the precise biological mechanism underpinning the observed perfect association. In the breeding program, parallel selection using these markers alongside functional markers of key genes regulating starch quality, such as SSIIa and SBEIIb, will provide an efficient approach to achieve strategic accumulation of superior alleles and develop new materials with improved nutritional quality.
5. Conclusions
The results of the present study identified a significant association between the T → C base mutation at position 202 of exon 9 of the rice Wx gene and the formation of highly resistant starch, and the two sets of molecular markers, T-Wx9-RS1 and T-Wx9-RS2, which were developed for the base mutation at this position, can effectively distinguish among the three genotypes. Moreover, the genotypes are in complete agreement with the results of the chemical method. These two sets of molecular markers can be used to achieve rapid and accurate detection of resistant starch genotypes and accelerate the selection of new rice varieties with high starch resistance.
While the developed Wx gene functional markers show great promise in high-RS rice breeding, it is important to note that their performance may vary across different genetic backgrounds and environmental conditions. Future research should focus on validating these markers in diverse rice populations and under various field conditions, as well as optimizing breeding strategies to maintain high RS content while improving yield and disease resistance. Additionally, efforts should be made to simplify and reduce the cost of marker detection techniques to enhance their applicability in large-scale breeding programs.
6. Patents
Patent number: ZL 2022 1 0541772.0.
Author Contributions
Conceptualization, methodology, writing—original draft, writing—review and editing, funding acquisition, J.O.; supervision, writing—review and editing, Z.Z.; validation, software, writing—review and editing, Y.G.; formal analysis, project administration, writing—review and editing, Q.H.; resources, writing—review and editing, T.H.; investigation, writing—review and editing, S.Z.; data curation, writing—review and editing, C.P. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Chongqing Academy of Agricultural Sciences Municipal Financial Science and Technology Innovation Projects, China, [grant number KYLX20240500090], the Chongqing Research Institutions Performance Incentive Guide Special, China, [grant number cstc2021jxjl80014], and the Chongqing Technology Innovation and Application Development Key Special Project, China, [grant number CSTB2022TIAD-KPX0017].
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data sets supporting the conclusions of this article are available in the China National GeneBank Database (CNGBdb) at https://db.cngb.org/, with the following accession numbers: CNP0008416.
Conflicts of Interest
Authors Jie Ouyang, Zichao Zhu, Yusheng Guan and Qianlong Huang are the inventors of patent ZL2022 1 0541772.0. Author Chuxiang Pan is an employee of Hunan Shennong Dafeng Seed Industry Technology Co., Ltd. Authors Tao Huang and Shun Zang declare that they have no conflicts of interest.
Abbreviations
The following abbreviations are used in this manuscript:
| ARMS-PCR | Amplification refractory mutation system polymerase chain reaction |
| RS | Resistant starch |
| GBSSI | Granule-bound starch synthase I |
| GT | Gelatinization temperature |
| ECQ | Eating and cooking quality |
| FMs | Functional markers |
| MAS | Marker-assisted selection |
| AAC | Apparent amylose content |
| LD | Linkage Disequilibrium |
References
- Praphasanobol, P.; Purnama, P.R.; Junbuathong, S.; Chotechuen, S.; Moung-Ngam, P.; Kasettranan, W.; Paliyavuth, C.; Comai, L.; Pongpanich, M.; Buaboocha, T.; et al. Genome-Wide Association Study of Starch Properties in Local Thai Rice. Plants 2023, 12, 3290. [Google Scholar] [CrossRef]
- Yan, C.; Meng, H.; Pei, Y.; Sun, W.; Zhang, J. Breeding by Design for Functional Rice with Genome Editing Technologies. JoVE 2025, 215, 67336. [Google Scholar] [CrossRef] [PubMed]
- Zhao, P.; Liu, Y.; Deng, Z.; Liu, L.; Yu, T.; Ge, G.; Chen, B.; Wang, T. Creating of Novel Wx Allelic Variations Significantly Altering Wx Expression and Rice Eating and Cooking Quality. J. Plant Physiol. 2024, 303, 154384. [Google Scholar] [CrossRef]
- Zhu, M.; Liu, Y.; Jiao, G.; Yu, J.; Zhao, R.; Lu, A.; Zhou, W.; Cao, N.; Wu, J.; Hu, S.; et al. The Elite Eating Quality Alleles Wx(b) and ALK(b) Are Regulated by OsDOF18 and Coordinately Improve Head Rice Yield. Plant Biotechnol. J. 2024, 22, 1582–1595. [Google Scholar] [CrossRef]
- Fu, Y.; Hua, Y.; Luo, T.; Liu, C.; Zhang, B.; Zhang, X.; Liu, Y.; Zhu, Z.; Tao, Y.; Zhu, Z.; et al. Generating Waxy Rice Starch with Target Type of Amylopectin Fine Structure and Gelatinization Temperature by Waxy Gene Editing. Carbohydr. Polym. 2023, 306, 120595. [Google Scholar] [CrossRef]
- Kumar, S.; Li, Y.; Zheng, J.; Liu, J.; Xu, Q.; Zhang, Y.; Tang, H.; Qi, P.; Deng, M.; Ma, J.; et al. The Impact of GBSSI Inactivation on Starch Structure and Functionality in EMS-Induced Mutant Lines of Wheat. BMC Genom. 2025, 26, 501. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Guo, X.; Song, J.; Xu, K.; Qin, T.; Zhang, X.; Song, Z.; He, Y.; Zhang, B.; Zhang, H.; et al. OsSPL14 and OsNF-YB9/YC8-12 Subunits Cooperate to Enhance Grain Appearance Quality by Promoting Waxy and PDIL1-1 Expression in Rice. Plant Commun. 2025, 6, 101348. [Google Scholar] [CrossRef] [PubMed]
- Hu, Z.; Niu, F.; Yan, P.; Wang, K.; Zhang, L.; Yan, Y.; Zhu, Y.; Dong, S.; Ma, F.; Lan, D.; et al. The Kinase OsSK41/OsGSK5 Negatively Regulates Amylose Content in Rice Endosperm by Affecting the Interaction between OsEBP89 and OsBP5. J. Integr. Plant Biol. 2023, 65, 1782–1793. [Google Scholar] [CrossRef]
- Li, J.; Zhang, C.; Luo, X.; Zhang, T.; Zhang, X.; Liu, P.; Yang, W.; Lei, Y.; Tang, S.; Kang, L.; et al. Fine Mapping of the Grain Chalkiness Quantitative Trait Locus qCGP6 Reveals the Involvement of Wx in Grain Chalkiness Formation. J. Exp. Bot. 2023, 74, 3544–3559. [Google Scholar] [CrossRef]
- Lu, Y.; Lv, D.; Zhou, L.; Yang, Y.; Hao, W.; Huang, L.; Fan, X.; Zhao, D.; Li, Q.; Zhang, C.; et al. Combined Effects of SSII-2RNAi and Different Wx Alleles on Rice Grain Transparency and Physicochemical Properties. Carbohydr. Polym. 2023, 308, 120651. [Google Scholar] [CrossRef]
- Hu, Y.; Zhang, Y.; Yu, S.; Deng, G.; Dai, G.; Bao, J. Combined Effects of BEIIb and SSIIa Alleles on Amylose Contents, Starch Fine Structures and Physicochemical Properties of Indica Rice. Foods 2022, 12, 119. [Google Scholar] [CrossRef]
- Ying, Y.; Hu, Y.; Liu, X.; Zhao, J.; Deng, B.; Zhang, Z.; Bao, J. Effects of Wx, SSIIa and FLO2 Alleles and Their Interactions on the Formation of Multi-Scale Structures of Rice Starch. Int. J. Biol. Macromol. 2025, 303, 140658. [Google Scholar] [CrossRef]
- Zhong, Q.; Jia, Q.; Yin, W.; Wang, Y.; Rao, Y.; Mao, Y. Advances in Cloning Functional Genes for Rice Yield Traits and Molecular Design Breeding in China. Front. Plant Sci. 2023, 14, 1206165. [Google Scholar] [CrossRef] [PubMed]
- Yang, K.; Liu, H.; Jiang, W.; Hu, Y.; Zhou, Z.; An, X.; Miao, S.; Qin, Y.; Du, B.; Zhu, L.; et al. Large Scale Rice Germplasm Screening for Identification of Novel Brown Planthopper Resistance Sources. Mol. Breed. 2023, 43, 70. [Google Scholar] [CrossRef] [PubMed]
- Scheuermann, K.K.; Pereira, A. Development of a Molecular Marker for the Pi1 Gene Based on the Association of the SNAP Protocol with the Touch-up Gradient Amplification Method. J. Microbiol. Methods 2023, 214, 106845. [Google Scholar] [CrossRef] [PubMed]
- Huang, F.; He, N.; Yu, M.; Li, D.; Yang, D. Identification and Fine Mapping of a New Bacterial Blight Resistance Gene, Xa43(t), in Zhangpu Wild Rice (Oryza rufipogon). Plant Biol. 2023, 25, 433–439. [Google Scholar] [CrossRef]
- Thulasinathan, T.; Ayyenar, B.; Kambale, R.; Manickam, S.; Chellappan, G.; Shanmugavel, P.; Narayanan, M.B.; Swaminathan, M.; Muthurajan, R. Marker Assisted Introgression of Resistance Genes and Phenotypic Evaluation Enabled Identification of Durable and Broad-Spectrum Blast Resistance in Elite Rice Cultivar, CO 51. Genes 2023, 14, 719. [Google Scholar] [CrossRef]
- Chen, S.; Feng, A.; Wang, C.; Zhao, J.; Feng, J.; Chen, B.; Yang, J.; Wang, W.; Zhang, M.; Chen, K.; et al. Identification and Fine-Mapping of Xo2, a Novel Rice Bacterial Leaf Streak Resistance Gene. Theor. Appl. Genet. 2022, 135, 3195–3209. [Google Scholar] [CrossRef]
- Srivastava, A.; Pusuluri, M.; Balakrishnan, D.; Vattikuti, J.L.; Neelamraju, S.; Sundaram, R.M.; Mangrauthia, S.K.; Ram, T. Identification and Functional Characterization of Two Major Loci Associated with Resistance against Brown Planthoppers (Nilaparvata lugens (Stal)) Derived from Oryza nivara. Genes 2023, 14, 66. [Google Scholar] [CrossRef]
- Bhatia, S.K.; Vikal, Y.; Kaur, P.; Dhillon, G.S.; Kaur, G.; Neelam, K.; Malik, P.; Lore, J.S.; Khanna, R.; Singh, K. Introgression and Mapping of a Novel Bacterial Blight Resistance Gene Xa49(t) from Oryza rufipogon Acc. CR100098A into O. sativa. Phytopathology 2024, 114, 2412–2420. [Google Scholar] [CrossRef]
- Zhang, Y.; Lin, X.-F.; Li, L.; Piao, R.-H.; Wu, S.; Song, A.; Gao, M.; Jin, Y.-M. CRISPR/Cas9-Mediated Knockout of Bsr-D1 Enhances the Blast Resistance of Rice in Northeast China. Plant Cell Rep. 2024, 43, 100. [Google Scholar] [CrossRef]
- Bian, Z.; Cao, D.; Zou, Y.; Xie, D.; Zhuang, W.; Sun, Z.; Mou, N.; Sun, Y.; Zhang, C.; Li, Q.; et al. Genetic Dissection of Major Rice QTLs for Strong Culms and Fine Mapping of qWS5 for Breeding Application in Transplanted System. Rice 2024, 17, 43. [Google Scholar] [CrossRef] [PubMed]
- Bao, J.; Shen, S.; Sun, M.; Corke, H. Analysis of Genotypic Diversity in the Starch Physicochemical Properties of Nonwaxy Rice: Apparent Amylose Content, Pasting Viscosity and Gel Texture. Starch Stärke 2006, 58, 259–267. [Google Scholar] [CrossRef]
- Chen, X.; Temnykh, S.; Xu, Y.; Cho, Y.G.; McCouch, S.R. Development of a Microsatellite Framework Map Providing Genome-Wide Coverage in Rice (Oryza sativa L.). Theor. Appl. Genet. 1997, 95, 553–567. [Google Scholar] [CrossRef]
- Ye, S.; Dhillon, S.; Ke, X.; Collins, A.R.; Day, I.N.M. An Efficient Procedure for Genotyping Single Nucleotide Polymorphisms. Nucleic Acids Res. 2001, 29, e88. [Google Scholar] [CrossRef] [PubMed]
- Tran, N.A.; Daygon, V.D.; Resurreccion, A.P.; Cuevas, R.P.; Corpuz, H.M.; Fitzgerald, M.A. A Single Nucleotide Polymorphism in the Waxy Gene Explains a Significant Component of Gel Consistency. Theor. Appl. Genet. 2011, 123, 519–525. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2026 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license.