Abstract
Phenylalanine ammonia-lyase (PAL) is an essential enzyme in the phenylpropanoid pathway, in which numerous aromatic intermediate metabolites play significant roles in plant growth, adaptation, and disease resistance. Cultivated peanuts are highly susceptible to Aspergillus flavus L. infection. Although PAL genes have been characterized in various major crops, no systematic studies have been conducted in cultivated peanuts, especially in response to A. flavus infection. In the present study, a systematic genome-wide analysis was conducted to identify PAL genes in the Arachis hypogaea L. genome. Ten AhPAL genes were distributed unevenly on nine A. hypogaea chromosomes. Based on phylogenetic analysis, the AhPAL proteins were classified into three groups. Structural and conserved motif analysis of PAL genes in A. hypogaea revealed that all peanut PAL genes contained one intron and ten motifs in the conserved domains. Furthermore, synteny analysis indicated that the ten AhPAL genes could be categorized into five pairs and that each AhPAL gene had a homologous gene in the wild-type peanut. Cis-element analysis revealed that the promoter region of the AhPAL gene family was rich in stress- and hormone-related elements. Expression analysis indicated that genes from Group I (AhPAL1 and AhPAL2), which had large number of ABRE, WUN, and ARE elements in the promoter, played a strong role in response to A. flavus stress.
1. Introduction
The phenylpropanoid pathway, one of the important secondary metabolic ways, widely exists in higher plants and some microorganisms [1]. All substances containing the phenylpropanoid skeleton are direct or indirect products of this pathway. This route produces a wide range of aromatic metabolites, including phytoalexin, lignin, flavonoids, iso-flavonoids, anthocyanins, and phytohormones [2], all of which are crucial for plant growth, development, and their resilience to biotic and abiotic stressors [3,4,5]. Phenylalanine ammonia-lyase (PAL, EC4.3.1.5), part of the ammonia-lyase superfamily, is the initial enzyme of this pathway and serves as a pivotal rate-limiting catalyst. It catalyzes the deamination of L-phenylalanine to trans-cinnamic acid, which is responsible for diverting carbon flux from the primary metabolic pathway to the trans-cinnamic acid synthesis pathway, and then further converts into different secondary metabolites in plants [2,6].
Numerous studies have indicated that the count of PAL family members is relatively limited. However, this number exhibits significant variation across different plant species. In Arabidopsis thaliana, four PAL genes have been identified [7], while Scutellaria baicalensis and Coffea canephora each have three [8,9], and poplar possesses five [10]. Twelve PAL members have been identified in rice and watermelon, 13 in cucumber [11,12], 16 in grapevines, and more than 20 in potato and tomato [13,14,15,16]. Functional differentiation results from duplication events in the PAL genes of plants. PAL families in plants likely originate from gene duplications, such as tandem, segmental, and whole-genome duplications [11]. And PAL isoforms play unique roles in plant development. For instance, Arabidopsis thaliana AtPAL1, AtPAL2, and AtPAL4 are strongly expressed in inflorescent stems, a tissue rich in lignifying cells, whereas the AtPAL3 transcript is expressed at an exceptionally low level, implying a role for AtPAL1, AtPAL2, and AtPAL4 in tissue-specific lignin synthesis [7]. Phenotypic analysis of single and multiple mutants of AtPAL genes revealed that AtPAL1 and AtPAL2 might have redundant roles in Arabidopsis thaliana flavonoid biosynthesis. However, AtPAL1 and AtPAL2 double mutants are more tolerant to drought conditions than wild-type plants [3]. In raspberry (Rubus idaeus) plants, RiPAL1 is associated with early fruit ripening, whereas RiPAL2 expression correlates more with later stages of flower and fruit development, although the two genes are similar in sequence [17]. In Salvia miltiorrhiza, SmPAL1 and SmPAL3 are highly expressed in roots and leaves, whereas SmPAL2 is predominantly expressed in stems and flowers, indicating that SmPAL1 and SmPAL3 function redundantly in rosmarinic acid biosynthesis [18]. What is more, it has been reported that PAL genes play a role in response to pathogenic infections. For example, of the four Arabidopsis PAL genes, AtPAL1 and AtPAL2 are co-expressed in different plant organs in response to inductive treatment with phytopathogenic Pseudomonas. Additionally, in response to nitrogen depletion, AtPAL1 and AtPAL4 transcript levels significantly increase [19]. In beans, GmPAL1 and GmPAL3 can be induced by fungal infection [20], and in pepper, CaPAL1 acts as a positive regulator of salicylic acid-dependent (SA-dependent) defense signaling to combat microbial pathogens [4].
Cultivated peanuts (Arachis hypogaea L.) are one of the crops most susceptible to A. flavus infection, and aflatoxins with strong toxicity and carcinogenicity are produced rapidly after infection, seriously threatening food safety and human health. Mining of regulatory genes related to A. flavus stress was considered as an important way to fundamentally solve the problem. It was reported that the fungal components, Aspergillus flavus L., induce a defense response in peanut varieties, resulting in increased PAL activity [21]. And polyhydroxylated flavonoids and intermediate products of the phenylpropanoid pathway were found to inhibit covalent adduct formation between the aflatoxin B1 and DNA, suggesting that PAL plays a unique role in resistance to A. flavus [22]. However, there are no reports on the potential regulatory role of PAL in peanut response to A. flavus infection. And the genome-wide bioinformatic analysis of the PAL gene family in cultivated peanut is still yet to be carried out. The genome sequence of A. hypogaea has recently been completed and released [23,24], providing an important resource for genome-wide analysis of disease-related genes. In the present study, ten AhPAL genes were identified and analyzed using bioinformatic approaches. Furthermore, the expression levels of AhPAL genes in cultivated peanut kernels were calculated at different time points under A. flavus stress using available RNA-seq datasets (accession number: PRJNA825125) and RT-qPCR. The findings of this research offer valuable perspectives on the evolutionary trajectory of PAL genes and elucidate their adaptive roles when confronting A. flavus-induced stress in peanut kernels.
2. Material and Methods
2.1. Database Searching and Identification of PAL Genes in Cultivated Peanut Genome
The genome sequences of cultivated peanuts (A. hypogaea cv. Tifrunner) were retrieved from the National Center for Biotechnology Information (NCBI) database under the accession number GCA_003086295.2 (available at: https://www.ncbi.nlm.nih.gov/assembly (accessed on 10 January 2023)). We used the established PAL sequences from Arabidopsis thaliana and Cucumis sativus as references, which were procured from TAIR at https://www.arabidopsis.org/index.jsp (accessed on 12 January 2023)and the CuGenDBv2 http://cucurbitgenomics.org/v2/ (accessed on 13 January 2023), respectively. These sequences served as probes for the Basic Local Alignment Search Tool (BLAST) to identify homologous PAL genes within the ‘Tifrunner’ genome. Proteins lacking MIO domains were removed via manual examination [25]. Physicochemical profiling of PAL genes was performed using ExPASy [26]. Subcellular localization analysis of all identified PAL proteins was conducted using the Plant-Ploc server (http://www.csbio.sjtu.edu.cn/bioinf/plant/ (accessed on 18 January 2023)).
2.2. Phylogenetic Analysis of PAL Proteins
The PAL amino acid sequences were retrieved from the reference genome of the cultivated peanut, A. hypogaea cv. Tifrunner, accessible at https://www.peanutbase.org/ (accessed on 20 January 2023). We performed multiple sequence alignments utilizing DNAMAN V9 and ClustalX 2.1 software [27,28]. Phylogenetic trees were then generated via MEGA 7.0 software, employing the neighbor-joining model with 1000 bootstrap replications [29]. The following PAL amino acid sequences in dicotyledonous and monocot plants were used for phylogenetic analysis: Arabidopsis thaliana (NP_181241, NP_190894, NP_196043, NP_187645), Nicotiana tabacum (BAA22963, ACJ66298, AAA34122, CAA55075), Cucumis sativus (Csa1M590300.1, Csa4M008760.1, Csa4M008780.1, Csa6M147460.1, Csa6M405960.1, Csa6M445240.1, Csa6M445750.1, Csa6M445760.1, Csa6M445770.1, Csa6M446290.1), Vitis vinifera (XP_002285277, XP_002281799, XP_003633986, CAN77065), Oryza sativa (BAD23149, BAD23155, CBC40318, CBC40320), and Zea mays (AAL40137, NP_001147922, NP_001147433, NP_001151482) [11,12,15].
2.3. Chromosome Localization, Gene Structure, Conserved Motif, and Duplication Event Analysis of AhPALs
Information regarding the chromosome localization and the exon–intron structure was obtained from the peanut reference genome (A. hypogaea cv. Tifrunner, https://www.peanutbase.org/ (accessed on 25 January 2023)). The conserved domains of AhPALs proteins were extracted using NCBI-Batch-CDD software. Chromosome localization and conserved motifs were identified and presented by the methods described by Cui et al. [30]. Representation of tandem and segmentally duplicated gene pairs and circos figures of PAL duplication links were conducted using TBtools software [31].
2.4. Cis-Acting Element Analysis of AhPALs
Approximately 2000 bp sequences upstream of the transcription start site (TSS) of the AhPAL genes were analyzed to identify potential cis-regulatory elements. The analysis utilized the Plant CARE online resource (available at http://bioinformatics.psb.ugent.be/webtools/plantcare/html/, accessed on 25 November 2022) for the identification of these elements [32]. Visualization of the findings was accomplished through TBtools.
2.5. In Silico Expression Analysis of AhPAL Genes Using RNA-seq Data
RNA-seq data from A. flavus-infected cultivated peanut samples were retrieved from the NCBI’s Sequence Read Archive (SRA) database (Accession No. PRJNA825125) [33]. We quantified the transcript levels of the AhPAL genes across different samples using the Fragments Per Kilobase Million (FPKM). Heat maps generated via TBtools illustrated the differential expression patterns of the AhPAL genes.
2.6. Plant Materials and Quantitative RT-PCR-Based Expression Assays of AhPALs
In our preceding study [33], we collected samples from kernels of “J-11” (a resistance cultivar) and “Zhonghua 12” (a susceptible cultivar) at various time intervals (0, 1, 3, 5, and 7 days) following inoculation with A. flavus spores. Total RNA was isolated from the collected kernels using the RNAprep Pure Plant Plus Kit (Tiangen Biotech, Co., Beijing, China) according to the manufacturer’s protocol. First-strand cDNA was obtained using the PrimeScrit™ RT Kit with gDNA eraser (perfect real-time, Takara Biomedical Technology, Ltd., Beijing, China). RT-qPCR primers were designed by Primer 3.0 online software (https://bioinfo.ut.ee/primer3/ (accessed on 18 February 2023)), and the alcohol dehydrogenase class III (AhADH3, Arahy. VYWU26.2) was selected as the internal reference control [33] (Table S1). Quantitative RT-qPCR and expression levels of AhPALs were conducted using the methods presented in the study by Cui et al. [30].
3. Results
3.1. Genome-Wide Identification of PAL Genes in the Cultivated Peanut Genome
To accurately retrieve the complete PAL proteins in peanuts, all chromosomes and scaffold sequences in the cultivated peanut genome database (A. hypogaea cv. Tifrunner, https://www.peanutbase.org/ (accessed on 10 January 2023)) were searched using PAL proteins from Arabidopsis (AtPALs, NP_181241) and cucumber (CsPALs, Csa6G445760.1) as queries. In this study, we identified 10 MIO domain-containing proteins as PAL members in cultivated peanuts, with their lengths ranging from 695 to 729 amino acids. Analysis of molecular properties revealed that the molecular weights of these PAL members span between 75.75 and 79.30 KDa, while their isoelectric points vary from 5.93 to 6.68. Significantly, subcellular localization predications predicted that all 10 PAL genes in cultivated peanuts are cytoplasmic (refer to Table 1).

Table 1.
Phenylalanine ammonia-lyase (PAL) gene family information in cultivated peanut (A. hypogaea).
The ten AhPAL genes were mapped to nine chromosomes of A. hypogaea, corresponding to their physical positions as depicted in Figure 1. According to their order on the chromosomes, PAL genes were renamed as AhPAL1–AhPAL10 (Table 1). Only chromosome 17 contained two PAL genes (AhPAL7 and AhPAL8), whereas all other chromosomes contained one PAL member.
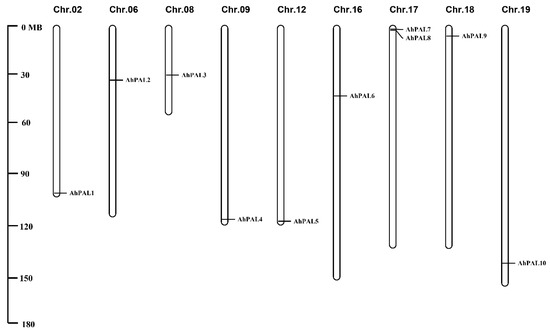
Figure 1.
Chromosome distributions of AhPAL genes in cultivated peanut with the chromosome number indicated at the top of each representation.
3.2. Sequence Characterization and Phylogenetic Analysis of PAL Family Proteins in Cultivated Peanut
Detailed sequence alignment was performed for all deduced AhPAL proteins. As has been reported for PAL proteins in cucumber, melon, and watermelon [11,12,15], all AhPAL proteins contained four conserved domains (Figure 2 and Figure S1): the N-terminus (residues 1–22, numbered according to AhPAL1), MIO (residues 23–259), core (residues 260–525 and 643–714), and inserted shielding (residues 526–642). Importantly, the N-terminus of all AhPAL proteins showed the greatest divergence, consistent with PAL families in other plants [11,12,15,17,34]. Additionally, the conserved enzymatic active site, -200Ala-Ser-Gly (numbered according to AhPAL1), in the MIO domain and residues that were essential for PAL enzymatic activity (349Tyr and 492Gly) in the core domain were found in all AhPAL proteins [11,12,15,35]. Another significant residue is 547Thr, located within the shielding domain, identified as a potential phosphorylation site [12]. As depicted in Figure 2B, this site is conserved across only six AhPALs isoforms; in contrast, three out of the remaining four PAL proteins exhibit substitutions with Asn and Ser. This pattern suggests that the AhPAL family may exhibit enzymatic activity, albeit with varying degrees of substrate specificity.
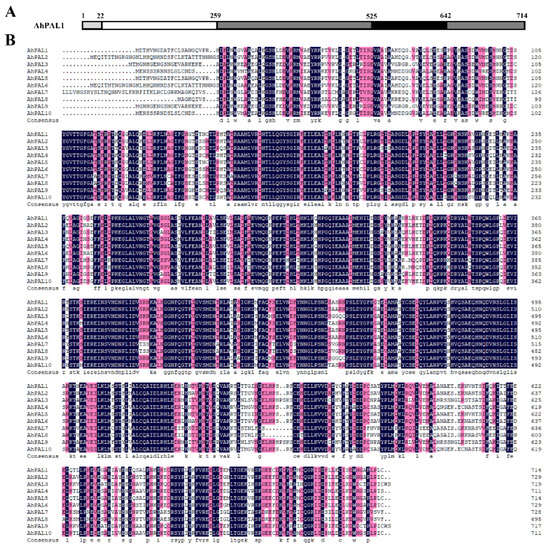
Figure 2.
Sequence analysis of AhPAL proteins. A schematic diagram of AhPAL amino acid sequence is presented in panel (A). AhPAL1 is depicted as a representative of the PAL protein families. The diagram indicates the four functional domains: the N-terminal domain (light gray), MIO domain (white), core domain (dark gray), and inserting shielding domain (black). Panel (B) shows the sequence alignment of AhPAL proteins, which was performed using DNAMAN V6. Gaps in the alignment are represented by dashes.
To determine the evolutionary relationship among plant PAL proteins, sequences of the PAL family from dicot (Arabidopsis, tobacco, cucumber, and grape) and monocot plants (rice and maize) were analyzed using a neighbor-joining phylogenetic tree. As shown in Figure 3, the corresponding tree categorizes these PALs into monocot and dicot groups. The ten AhPAL proteins were classified into three groups (AhPAL3-AhPAL7-AhPAL8-AhPAL9, AhPAL4-AhPAL10, and AhPAL1-AhPAL2-AhPAL5-AhPAL6) and members of the dicot group (Figure 3 and Figure S2). Moreover, the plant PALs within one species were more closely related to each other than to the homologs in other plants, indicating the functional conservation of PALs in the same plant.
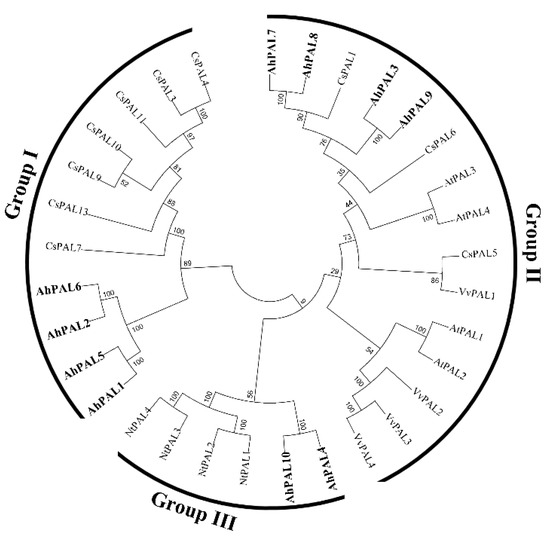
Figure 3.
Phylogenetic tree of PAL proteins in plants. The conserved PAL proteins from Arabidopsis (AtPAL), tobacco (NtPAL), cucumber (CsPAL), grape (VvPAL), and cultivated peanut (AhPAL) were aligned using Clustal X. The phylogenic tree was constructed using the neighbor-joining model (1000 replicates) with the MEGA 7.0 program. The AhPAL proteins are highlighted in bold.
3.3. Exon–Intron and Conserved Motif Analysis of AhPALs
The exon–intron structures and conserved motifs of AhPALs in A. hypogaea were investigated using the Gene Structure Display Serve (GSDS) online software and MEME tool, respectively. Notably, all AhPAL genes had one intron in the middle and ten motifs in the conserved domains (Figure 4), indicating that AhPAL genes are conserved in A. hypogaea.
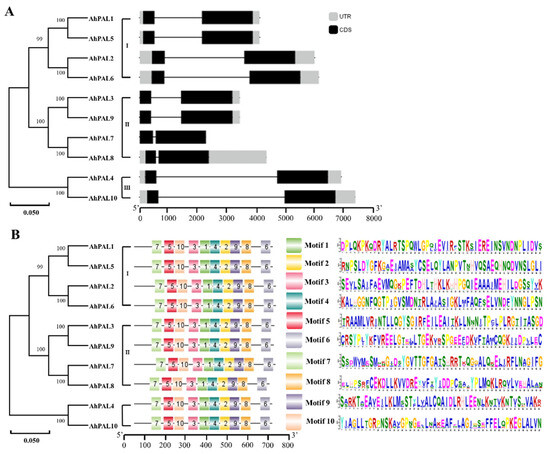
Figure 4.
Exon–intron structure and conserved motif analysis of AhPAL genes. (A) The exon–intron structure of AhPAL genes. (B) The distribution of conserved motifs in AhPAL proteins.
3.4. Duplication Events in AhPAL Genes and Synteny Analysis
Segmental and tandem duplications lead to duplicated gene pairs and play a major role in the expansion and evolution of gene families in various plant species [36]. Ten segmental duplicated gene pairs involving eight AhPAL gene members were identified in the A. hypogaea genome (Figure 5 and Table S2). However, no tandem duplications occurred, indicating that segmental duplication may be the major driving force for the expansion of the PAL gene family in the A. hypogaea genome. Similar findings have been reported for the AP2/ERF [30], WRKY [37], GRFs [38], MST [39], and bHLH [40] gene families of A. hypogaea. It is noteworthy that all duplicated gene pairs were clustered in the same group based on phylogenetic analysis, implying that the gene structure was relatively conserved during evolution.
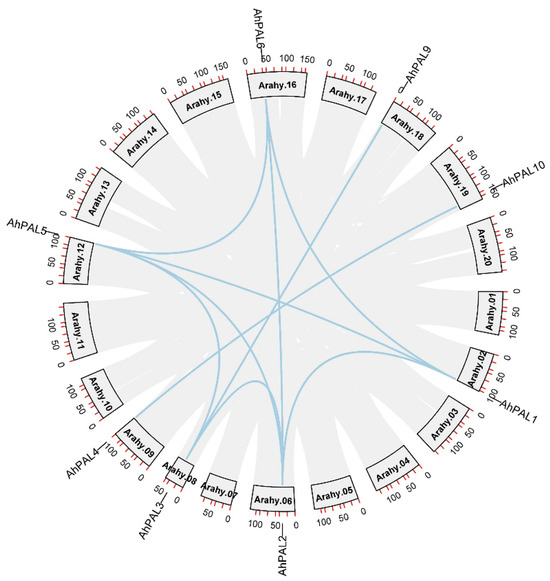
Figure 5.
Circos figures for chromosome locations with AhPAL segmental duplication links. The blue lines indicate segmental duplicated gene pairs.
Furthermore, we investigated the synteny of PAL genes in A. hypogaea, A. duranensis, A. ipaensis, Medicago truncatula, and soybean genomes to characterize the evolutionary patterns of AhPAL genes within the Leguminosae species (Figure 6 and Figure S3). Ten and eleven gene pairs were identified between cultivated peanuts and wild-type A. duranensis and A. ipaensis, respectively (Tables S3 and S4). It appeared that most genomes of A. duranensis and A. ipaensis might have more than one ortholog in the cultivated peanuts (Figure S3). A. hypogaea contained twice the number of PAL members observed for A. duranensis and A. ipaensis, which is consistent with the fact that A. hypogaea, a tetraploid plant, is formed by chromosome doubling after the natural hybridization of two diploid wild species [23,24,41]. Similarly, 11 and 13 synteny gene pairs were detected between A. hypogaea and M. truncatula and the common bean, respectively (Tables S5 and S6). Additionally, nine AhPAL genes were orthologous in M. truncatula, the dicot plant, whereas seven AhPAL genes were found in the common bean, the monocot plant, suggesting that cultivated peanuts have close genetic relationships with other dicotyledonous plants.

Figure 6.
Synteny of AhPAL genes in different genomes of A. duranensis, A. ipaensis, Glycine max L., and A. hypogaea cv. Tifrunner.
3.5. Cis-Acting Elements of AhPALs Promoter Region
To further ascertain the potential role of peanut PAL genes in various biological processes, the promoter region upstream of 2000 bp of 10 AhPALs was investigated [42,43]. Based on the PLANTCARE results, we obtained a total of 43 known cis-acting elements that could be classified into four types: tissue-specific elements (4), stress-related elements (6), hormone-related elements (9), and light-related elements (24) (Table S7). As shown in Figure 7A, except for AhPAL2, AhPAL3, AhPAL7, AhPAL8, and AhPAL9, which lack tissue-specific elements, all other AhPALs contained the four types of elements in their promoter region. To better explain the regulatory roles of AhPALs in plant stress responses, the number and types of hormone- and stress-related elements were counted and are presented in Figure 7B. Among them, the ABRE element responded to abscisic acid; the CGTCA-motif and TGACG-motif were responsive to MeJA; the AuxRR-core and TGA-element responded to auxin; the GARE-motif and TATC-box were responsive to gibberellin; and the TCA-element responded to salicylic acid. The cis-elements related to stress include drought inducibility (MBS), wound response (WUN), low-temperature (LTR), anaerobic induction (ARE), and TC-rich elements. Among these cis-elements, ABRE and MeJA were the two most frequently identified elements, followed by MBS, ARE, and WUN, indicating that most AhPAL genes participate in the regulation of plant stress processes by responding to hormones.
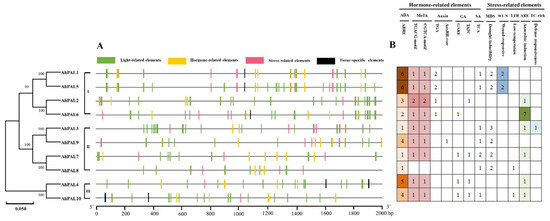
Figure 7.
Predicted cis-acting elements of AhPAL gene promoters in A. hypogaea. (A) Promoter regions 2000 bp upstream of ten AhPALs were analyzed using PlantCARE. The different colored rectangles represent the four types of cis-elements. (B) The number of hormone- and stress-related elements of AhPALs in A. hypogaea. Different colors represent numbers of various cis-acting elements.
3.6. Expression Patterns of AhPAL Genes in Response to A. flavus Stress in Peanut Seeds
To better investigate the possible regulatory roles of AhPALs in peanuts’ response to A. flavus infection, the expression patterns of AhPAL genes were analyzed at different time points under A. flavus stress in cultivated peanut kernels using the available RNA-seq datasets (accession number: PRJNA825125). Among them, the expression levels of six AhPAL genes (four genes from group I and two from group II) were higher in the S genotype than in the R genotype, implying a main role in the response to A. flavus infection (Figure 8A). As shown in Figure S4, profuse mycelial growth and sporulation was observed in the S genotype on the 7th day after inoculation, whereas very little mycelial growth occurred on the R seed coat. According to the methods described by Cui et al. [33], the infection index was calculated on the 7th day after inoculation and was nearly 5-times higher in S (92.22) than that in R (17.00). Furthermore, two genes from each group, AhPAL1 and AhPAL2 from Group I, AhPAL3 and AhPAL8 from Group II, and AhPAL4 and AhPAL10 from Group III, were selected to detect whether their expression levels would be induced under A. flavus stress by RT-qPCR. As shown in Figure 8, the expression levels of all AhPALs genes were higher in S than in R, suggesting that all AhPALs genes were significantly induced by A. flavus infection, which may be due to more serious damage in S. Moreover, AhPAL1 and AhPAL2 from Group I and AhPAL4 from Group IIII exhibited continuous increasing trends in overall time points in both R and S kernels. And the expression level of AhPAL1 and AhPAL2 in S were nearly 10-times higher than that in R, while AhPAL4 only 5-times, implying that genes from Group I functioned strongly upon A. flavus stress.
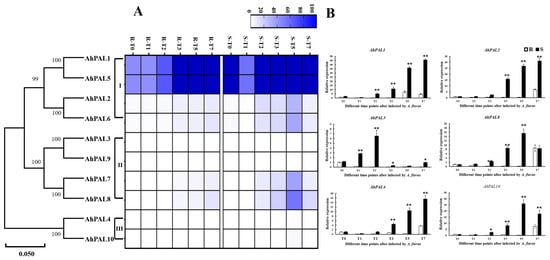
Figure 8.
Expression patterns of AhPAL genes in A. hypogaea cv. Tifrunner. (A) The expression patterns of AhPAL genes in peanut seeds under A. flavus stress. (B) Relative expression level of AhPAL genes at different time points after infection by A. flavus. Error bars were obtained from three biological replicates. Values are means ± standard errors (SEs) of three independent biological replicates (n = 3). Asterisks indicate a significant difference between R and S at each time point as determined by Student’s t-test (* p < 0.05; ** p < 0.01).
4. Discussion
PAL, a member of the ammonia-lyase superfamily, is widely found in higher plants. The characterization of PAL genes in certain plants is crucial, as these genes have been shown to be closely related to biotic and abiotic resistance in other studies [3,4]. The released genome sequence of A. hypogaea facilitated identification and isolation of the PAL gene in cultivated peanuts. In the present study, ten PAL genes in cultivated peanuts were identified using BLAST. The analysis of PAL proteins translated from these genes showed basic properties, such as isoelectric point and amino acid length varying within a small range, implying that PAL genes in the cultivated peanut were conserved and had similar important functions. The subcellular localization predicted that PAL proteins are localized in the cytoplasm, which is consistent with reports in other plants [44,45]. Phylogenetic analysis showed that PAL genes from plants were categorized into monocot and dicot groups. Ten AhPAL proteins were classified into three groups, similar to that found in walnut [46], whereas most woody plants have only two groups [12,45,47,48], suggesting that the PAL gene was relatively conserved in dicot plant evolution. Moreover, gene structure analysis showed that all AhPAL genes contained one intron in the middle and ten motifs in the conserved domains (Figure 4), indicating that AhPAL genes are conserved in A. hypogaea.
Gene duplication events include whole-genome fragment duplication, small-scale fragment replication, partial tandem replication, or a combination of these [49]. The ten PAL genes can be divided into five pairs, of which four pairs correspond one-to-one in their two chromosome groups (chr02-chr12, chr06-chr16, chr08-chr18, and chr09-chr19). Excluding one gene pair (chr8-chr18), the other three gene pairs were also similar in position and may have been produced by the duplication of whole-genome segments. AhPAL3 and AhPAL9 are located in the middle and top of their chromosomes, respectively, which could be due to the replication of whole-genome fragments, followed by chromosome recombination. Cultivated peanuts included two sub-genomes: A and B. In the peanut reference genome, chr01 to chr10 correspond to the A sub-genome and chr11 to chr20 correspond to the B sub-genome [41]. These four gene pairs were equivalent to a one-to-one correspondence in the A and B sub-genomes. Studies have shown that cultivated peanuts may have resulted from the fusion of two diploid wild-type peanuts [50]. Comparing the cultivated peanut PAL with that of the wild-type peanut, it was observed that each PAL gene of the cultivated peanut corresponded to the PAL from either of the wild-type peanut PAL genes (Figure 6 and Figure S3).
The phenylpropanoid pathway produces a vast number of aromatic metabolites, including flavonoids, isoflavonoids, anthocyanins, plant hormones, phytoalexins, and lignins, and PAL activity is related to the productivity of these products. Transcriptome research showed that PAL gene expression in resistant seeds differed from that in susceptible seeds during A. flavus infection [45,51,52]. It was also found that phenylpropanoid biosynthesis pathways are triggered by transcription factors, such as WRKY, bHLH, and MYB, to evoke defense response mechanisms against A. flavus [53]. The expression profile of AhPAL genes in peanuts infected with A. flavus showed that all AhPALs genes exhibited a higher level in S than that in R, suggesting that all PALs genes were significantly induced by A. flavus infection, which may due to more serious damage and more mycelia and spores in S. Moreover, the spores on the surface of the kernel increased during A. flavus infection process, combing the fact that expression levels of AhPALs exhibited continuous increasing trends in overall time points, implying that expression of PAL genes is closely related to mycelial growth. Meanwhile, the expression levels of AhPAL1 and AhPAL2 in S were nearly 10-times higher than that in R, suggesting its critical role in response to A. flavus infection. The promoter sequence was regarded as a major factor determining whether and when the transcription of a gene will be initiated and possessing cis-acting elements, which might imply the potential functions of genes [30,54]. Here, genes from Group I showed the largest number of ABRE, WUN, and ARE in the promoter region, in combination with the phylogenetic tree, illustrating that the phylogenetically similar genes shared identical cis-elements, and increased number of ABRE, WUN, and ARE elements correspond to an increased responsiveness to A. flavus infection.
5. Conclusions
This study conducted a comprehensive identification and characterization of the peanut PAL gene family at the genome level. We identified ten PAL genes in cultivated peanuts, which were unevenly distributed across nine chromosomes. Phylogenetic analysis revealed that dicot PAL genes segregate into three distinct clades. The pattern of chromosome distribution and synteny analysis suggest that segmental duplications played a significant role in the proliferation of the peanut PAL gene repertoire. Notably, genes within Group I (AhPAL1 and AhPAL2), which possess a high density of ABRE, WUN, and ARE elements within their promoter regions, exhibited pronounced activity in response to A. flavus stress.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/genes15030265/s1, Figure S1: Sequence analysis of PAL proteins. A, scheme diagram of AhPAL amino acid sequence. AhPAL1 is shown as a representative of the PAL protein families. The four functional domains, including the N-terminal domain (light gray), MIO domain (white), core domain (dark gray), and inserting shielding domain (black), are indicated. B, sequence alignment of PAL proteins in A. hypogaea and C. sativus. The alignment was performed by DNAMAN V6. The gaps are indicated as dashes. Figure S2: Phylogenetic tree of plant PAL proteins. The conserved PAL proteins from Arabidopsis (AtPAL), tobacoo (NtPAL), cucumber (CsPAL), grape (AtPAL), cultivated peanut (AhPAL), rice (OsPAL), and maize (ZmPAL) were aligned using Clustal X, and the phylogenic tree was constructed using the neighbor-joining model (1000 replicates) with the program MEGA 7.0. The AhPAL proteins are shown in bold. Figure S3: Synteny of AhPAL genes in different genomes of A. duranensis, A. ipaensis, G. max, and A. hypogaea cv. Tifrunner. Figure S4: Comparison of mycelia growth on the surface of resistance cultivar “J-11” and susceptible cultivar “Zhonghua 12”. Table S1: Primers for RT-qPCR. Table S2: Duplicated gene pairs of AhPALs. Table S3: Synteny gene pairs between A. hypogaea cv. Tifrunner and A. duranensis. Table S4: Synteny gene pairs between A. hypogaea cv. Tifrunner and A. ipaeansis. Table S5: Synteny gene pairs between A. hypogaea cv. Tifrunner and M. truncatula. Table S6: Synteny gene pairs between A. hypogaea cv. Tifrunner and G. max. Table S7: Cis-acting elements of AhPAL promoter.
Author Contributions
Conceptualization, M.C. and P.D.; data curation, J.G. and Z.Z.; funding acquisition, X.Z.; investigation, P.C. and C.W.; methodology, P.C., T.G. and J.X.; software, P.C., M.C. and Q.Z.; supervision, B.H. and X.Z.; validation, M.C., L.C. and W.D.; visualization, H.L.; writing—original draft, M.C. and P.C.; writing—review & editing, M.C., S.H. and X.Z. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the National Key R&D Plan (2022YFD1200400), the National Natural Science Foundation of China (32301851; 31871663), First Class Project of Shennong Laboratory (SN01-2002-03), and the China Agriculture Research System (CARS-13).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Data are contained within the article or Supplementary Materials.
Conflicts of Interest
The authors declare that they have no conflict of interest.
References
- MacDonald, M.J.; Godwin, B.D. A modern view of phenylalanine ammonia lyase. Biochem. Cell Biol. 2007, 85, 273–282. [Google Scholar] [CrossRef]
- Vogt, T. Phenylpropanoid biosynthesis. Mol. Plant 2010, 3, 2–20. [Google Scholar] [CrossRef]
- Huang, J.L.; Gu, M.; Lai, Z.B.; Fan, B.; Shi, K.; Zhou, Y.H.; Yu, J.Q.; Chen, Z. Functional analysis of the Arabidopsis PAL gene family in plant growth, development, and response to environmental stress. Plant Physiol. 2010, 153, 1526–1538. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.S.; Hwang, B.K. An important role of the pepper phenylalanine ammonia-lyase gene (PAL1) in salicylic acid-dependent signaling of the defense response to microbial pathogens. J. Exp. Bot. 2014, 65, 2295–2306. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Liu, C.J. Multifaceted regulations of gateway enzyme phenylalanine ammonia-lyase in the biosynthesis of phenylpropanoids. Mol. Plant 2015, 8, 17–27. [Google Scholar] [CrossRef]
- Ferrer, J.L.; Austin, M.B.; Stewart, C.; Noel, J. Structure and function of enzymes involved in the biosynthesis of phenylpropanoids. Plant Physiol. Biochem. 2008, 46, 356–370. [Google Scholar] [CrossRef]
- Raes, J.; Rohde, A.; Christensen, J.H.; Peer, Y.; Boerjan, W. Genome-wide characterization of the lignification toolbox in Arabidopsis. Plant Physiol. 2003, 133, 1051–1071. [Google Scholar] [CrossRef]
- Xu, H.; Park, N.I.; Li, X.; Kim, Y.; Lee, S.; Park, S. Molecular cloning and characterization of phenylalanine ammonia-lyase, cinnamate 4-hydroxylase and genes involved in flavone biosynthesis in Scutellaria baicalensis. Bioresour. Technol. 2010, 101, 9715–9722. [Google Scholar] [CrossRef]
- Lepelley, M.; Mahesh, V.; McCarthy, J.; Rigoreau, M.; Crouzillat, D.; Chabrillange, N.; Kochko, A.D.; Campa, C. Characterization, high-resolution mapping and differential expression of three homologous PAL genes in Coffea canephora Pierre (Rubiaceae). Planta 2012, 236, 313–326. [Google Scholar] [CrossRef]
- Hamberger, B.; Ellis, M.; Friedmann, M.; Clarice, D.; Barbazuk, B.; Douglas, C. Genome-wide analyses of phenylpropanoid-related genes in Populus trichocarpa, Arabidopsis thaliana, and Oryza sativa: The Populus lignin toolbox and conservation and diversification of angiosperm gene families. Can. J. Bot. 2007, 85, 1182–1201. [Google Scholar] [CrossRef]
- Shang, Q.M.; Li, L.; Dong, C.J. Multiple tandem duplication of the phenylalanine ammonia-lyase genes in Cucumis sativus L. Planta 2012, 236, 1093–1105. [Google Scholar] [CrossRef]
- Dong, C.J.; Cao, N.; Zhang, Z.; Shang, Q. Phenylalanine ammonia-lyase gene families in cucurbit species: Structure, evolution, and expression. J. Integr. Agric. 2016, 15, 1239–1255. [Google Scholar] [CrossRef]
- Joos, H.J.; Hahlbrock, K. Phenylalanine ammonia-lyase in potato (Solanum tuberosum L.). Genomic complexity, structural comparison of two selected genes and modes of expression. Eur. J. Biochem. 1992, 204, 621–629. [Google Scholar] [CrossRef]
- Chang, A.; Lim, M.H.; Lee, S.W.; Robb, J.; Nazar, R. Tomato phenylalanine ammonia-lyase gene family, highly redundant but strongly underutilized. J. Biol. Chem. 2008, 283, 33591–33601. [Google Scholar] [CrossRef]
- Dong, C.J.; Shang, Q.M. Genome-wide characterization of phenylalanine ammonia-lyase gene family in watermelon (Citrullus lanatus). Planta 2013, 238, 35–49. [Google Scholar] [CrossRef]
- Rawal, H.C.; Singh, N.K.; Sharma, T.R. Conservation, divergence, and genome-wide distribution of PAL and POX A gene families in plants. Int. J. Genom. 2013, 2013, 678969–678974. [Google Scholar]
- Kumar, A.; Ellis, B.E. The phenylalanine ammonia-lyase gene family in raspberry. Structure, expression, and evolution. Plant Physiol. 2001, 127, 230–239. [Google Scholar] [CrossRef]
- Hou, X.M.; Shao, F.J.; Ma, Y.M.; Lu, S. The phenylalanine ammonia-lyase gene family in Salvia miltiorrhiza: Genome-wide characterization, molecular cloning and expression analysis. Mol. Biol. Rep. 2013, 40, 4301–4310. [Google Scholar] [CrossRef]
- Olsen, K.M.; Lea, U.S.; Slimestad, R.; Verheul, M.; Lillo, C. Differential expression of four Arabidopsis PAL genes: PAL1 and PAL2 have functional specialization in abiotic environmental-triggered flavonoid synthesis. J. Plant Physiol. 2008, 165, 1491–4199. [Google Scholar] [CrossRef] [PubMed]
- Liang, X.W.; Dron, M.; Cramer, C.L.; Dixon, R.A.; Lamb, C. Differential regulation of phenylalanine ammonia-lyase genes during plant development and by environmental cues. J. Biol. Chem. 1989, 264, 14486–14492. [Google Scholar] [CrossRef]
- Nandini, D.; Bariya, H. Induction of systemic acquired resistance in Arachis hypogaea L. by aspergillus flavus derived elicitors. J. Cell Tissue Res. 2014, 14, 4395–4403. [Google Scholar]
- Firozi, P.; Bhattacharya, R. Effects of natural polyphenols on aflatoxin B1 activation in a reconstituted microsomal monooxygenase system. J. Biochem. Toxicol. 1995, 10, 25–31. [Google Scholar]
- Bertioli, D.J.; Jenkins, J.; Clevenger, J.; Dudchenko, O.; Gao, D.; Seijo, G.; Leal, B.; Soraya, C.M.; Ren, L.; Farmer, A.D.; et al. The genome sequence of segmental allotetraploid peanut Arachis hypogaea. Nat. Genet. 2019, 51, 877–884. [Google Scholar] [CrossRef]
- Zhuang, W.J.; Chen, H.; Yang, M.; Wang, J.; Pandey, M.; Zhang, C.; Chang, W.C.; Zhang, L.; Zhang, X.; Tang, R.; et al. The genome of cultivated peanut provides insight into legume karyotypes, polyploid evolution and crop domestication. Nat. Genet. 2019, 51, 865–876. [Google Scholar] [CrossRef]
- Holger, R.; Georg, E.S. Structural basis for the entrance into the phenylpropanoid metabolism catalyzed by phenylalanine ammonia-lyase. Plant Cell 2004, 16, 3426–3436. [Google Scholar]
- Gasteiger, E.; Hoogland, C.; Gattiker, A.; Duvaud, S.; Wilkins, M.R.; Appel, R.D.; Bairoch, A. Protein identification and analysis tools on the ExPASy server. In The Proteomics Protoc Handbook; Humana Press: Totowa, NJ, USA, 2005; pp. 571–607. [Google Scholar]
- Thompson, J.D.; Higgins, D.G.; Gibson, T.J. CLUSTALW: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994, 22, 4673–4680. [Google Scholar] [CrossRef]
- Zhu, X.; Leng, X.; Sun, X.; Mu, Q.; Wang, B.; Li, X.; Wang, C.; Fang, J. Discovery of conservation and diversification of miR171 genes by phylogenetic analysis based on global genomes. Plant Genome 2015, 8, plantgenome2014-10. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef]
- Cui, M.; Haider, M.S.; Chai, P.; Guo, J.; Du, P.; Li, H.; Dong, W.; Huang, B.; Zheng, Z.; Shi, L.; et al. Genome-wide identification and expression analysis of AP2/ERF transcription factor related to drought stress in cultivated peanut (Arachis hypogaea L.). Front. Genet. 2021, 12, 750761. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Xia, R. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Magali, L.; Patrice, D.; Gert, T.; Kathleen, M.; Yves, M.; Yves, V.; Pierre, R.; Stephane, R. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar]
- Cui, M.; Han, S.; Wang, D.; Haider, M.S.; Guo, J.; Zhao, Q.; Du, P.; Sun, Z.; Qi, F.; Zheng, Z.; et al. Gene co-expression network analysis of the comparative transcriptome identifies hub genes associated with resistance to Aspergillus flavus L. in cultivated peanut (Arachis hypogaea L.). Front. Plant Sci. 2022, 13, 899177. [Google Scholar] [CrossRef] [PubMed]
- Cramer, C.L.; Edwards, K.; Dron, M.; Liang, X.; Dildine, S.L.; Bolwell, G.P.; Dixon, R.A.; Lamb, C.J.; Schuch, W. Phenylalanine ammonialyase gene organization and structure. Plant Mol. Biol. 1989, 12, 367–383. [Google Scholar] [CrossRef]
- Reichert, A.I.; He, X.Z.; Dixon, R.A. Phenylalanine ammonia-lyase (PAL) from tobacco (Nicotiana tabacum): Characterization of the four tobacco PAL genes and active heterotetrameric enzymes. Biochem. J. 2009, 424, 233–242. [Google Scholar] [CrossRef]
- Baloglu, M.C. Genome-wide in silico identification and comparison of growth regulating factor (GRF) genes in Cucurbitaceae family. Plant Omics 2014, 7, 260–270. [Google Scholar]
- Song, H.; Wang, P.; Lin, J.Y.; Zhao, C.; Bi, Y.; Wang, X. Genome-wide identification and characterization of WRKY gene family in peanut. Front. Plant Sci. 2016, 26, 534. [Google Scholar] [CrossRef]
- Zhao, K.; Li, K.; Ning, L.; He, J.; Yin, D. Genome-wide analysis of the growth-regulating factor family in peanut (Arachis hypogaea L.). Int. J. Mol. Sci. 2019, 2, 4120. [Google Scholar] [CrossRef]
- Wan, L.; Ren, W.; Miao, H.; Zhang, J.; Fang, J. Genome-wide identification, expression, and association analysis of the monosaccharide transporter (MST) gene family in peanut (Arachis hypogaea L.). 3 Biotech. 2020, 10, 130. [Google Scholar] [CrossRef] [PubMed]
- Chao, G.; Sun, J.; Wang, C.; Dong, Y.; Xiao, S.; Wang, X.; Jiao, Z.; Chen, Z.H. Genome-wide analysis of basic/helix-loophelix gene family in peanut and assessment of its roles in pod development. PLoS ONE 2017, 12, e0181843. [Google Scholar]
- Bertioli, D.J.; Cannon, S.B.; Froenicke, L.; Huang, G.; Farmer, A.D.; Cannon, E.K.S.; Liu, X.; Gao, D.; Clevenger, J.; Dash, S.; et al. The genome sequences of Arachis duranensis and Arachis ipaensis, the diploid ancestors of cultivated peanut. Nat. Genet. 2016, 48, 438–446. [Google Scholar] [CrossRef] [PubMed]
- Cui, M.; Wang, C.; Zhang, W.; Pervaiz, T.; Haider, M.S.; Tang, W.; Fang, J. Characterization of Vv-miR156: Vv-SPL pairs involved in the modulation of grape berry development and ripening. Mol. Genet. Genom. 2018, 293, 1333–1354. [Google Scholar] [CrossRef]
- Meng, J.; Yang, J.; Peng, M.; Liu, X.; He, H. Genome-wide characterization, evolution, and expression analysis of the leucine-rich repeat receptor-like protein kinase (LRR-RLK) gene family in Medicago truncatula. Life 2020, 10, 176–200. [Google Scholar] [CrossRef]
- Achnine, L.; Blancaflor, E.B.; Rasmussen, S.; Dixon, R. Colocalization of L-phenylalanine ammonia-lyase and cinnamate 4-hydroxylase for metabolic channeling in phenylpropanoid biosynthesis. Plant Cell 2004, 16, 3098–3109. [Google Scholar] [CrossRef]
- Jong, F.; Hanley, S.J.; Beale, M.H.; Karp, A. Characterization of the willow phenylalanine ammonia-lyase (PAL) gene family reveals expression differences compared with poplar. Phytochemistry 2015, 117, 90–97. [Google Scholar] [CrossRef]
- Yan, F.; Li, H.; Zhao, P. Genome-wide identification and transcriptional expression of the PAL gene family in common walnut (Juglans regia L.). Genes 2019, 10, 46. [Google Scholar] [CrossRef]
- Shi, R.; Sun, Y.H.; Li, Q.; Heber, S.; Sederoff, R.; Chiang, V. Towards a systems approach for lignin biosynthesis in Populus trichocarpa: Transcript abundance and specificity of the monolignol biosynthetic genes. Plant Cell Physiol. 2010, 51, 144–163. [Google Scholar] [CrossRef]
- Wang, Z.B.; Chen, X.; Wang, W.; Cheng, K.; Kong, J. Transcriptome-wide identification and characterization of Ornithogalum saundersiae phenylalanine ammonia lyase gene family. RSC Adv. 2014, 4, 27159–27175. [Google Scholar] [CrossRef]
- Flagel, L.E.; Wendel, J.F. Gene duplication and evolutionary novelty in plants. New Phytol. 2009, 183, 557–564. [Google Scholar] [CrossRef] [PubMed]
- Song, H.; Wang, P.; Li, C.; Han, S.; Lopez-Baltazar, J.; Zhang, X.; Wang, X. Identification of lipoxygenase (LOX) genes from legumes and their responses in wild type and cultivated peanut upon Aspergillus flavus infection. Sci. Rep. 2016, 12, 35245. [Google Scholar] [CrossRef] [PubMed]
- Nayak, S.N.; Agarwal, G.; Pandey, M.K.; Sudini, H.; Jayale, A.; Purohit, S.; Desai, A.; Wan, L.; Guo, B.; Liao, B.; et al. Aspergillus flavus infection triggered immune responses and host-pathogen cross-talks in groundnut during in-vitro seed colonization. Sci. Rep. 2017, 29, 9659–9672. [Google Scholar] [CrossRef] [PubMed]
- Wanner, L.A.; Li, G.; Ware, D.; Somssich, I.; Davis, K. The phenylalanine ammonia-lyase gene family in Arabidopsis thaliana. Plant Mol. Biol. 1995, 27, 327–338. [Google Scholar] [CrossRef] [PubMed]
- Soni, P.; Nayak, S.N.; Kumar, R.; Pandey, M.; Singh, N.; Sudini, H.K.; Bajaj, P.; Fountain, J.C.; Singam, P.; Hong, Y.; et al. Transcriptome analysis identified coordinated control of key pathways regulating cellular physiology and metabolism upon Aspergillus flavus infection resulting in reduced aflatoxin production in groundnut. J. Fungi 2020, 6, 370–392. [Google Scholar] [CrossRef] [PubMed]
- Kabir, S.M.T.; Hossain, M.S.; Bashar, K.K.; Honi, U.; Ahmed, B.; Emdad, E.M. Genome-wide identification and expression profiling of AP2/ERF superfamily genes under stress conditions in dark jute (Corchorus olitorius L.). Ind. Crops Prod. 2021, 166, 113469–113487. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).