Identification and Characterization of Circular RNAs (circRNAs) Using RNA-Seq in Two Breeds of Cashmere Goats
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethics Statement
2.2. Sample Collection
2.3. RNA Extraction and Sequencing
2.4. RNA-Seq Data Analysis
2.5. Validation of the Authenticity of circRNAs Using RT-PCR and Sanger Sequencing
2.6. Validation of the Reliability of RNA-Seq by RT-qPCR
2.7. GO and KEGG Enrichment Analysis of the Parent Genes of Differentially Expressed circRNAs
2.8. Construction of circRNA-miRNA Regulatory Network
3. Results
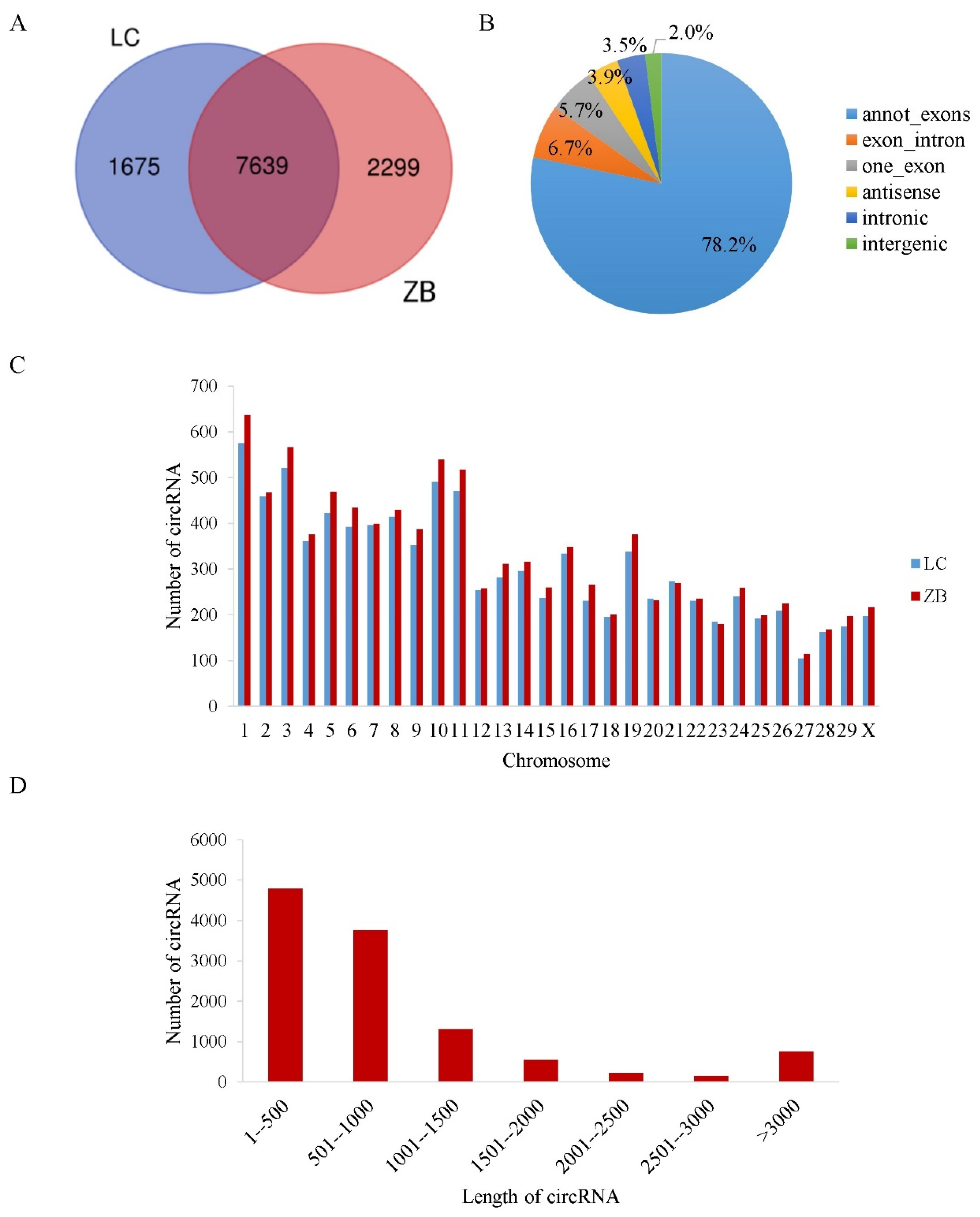
3.1. Identification and Characterization of circRNAs in Skin Tissue of Cashmere Goats
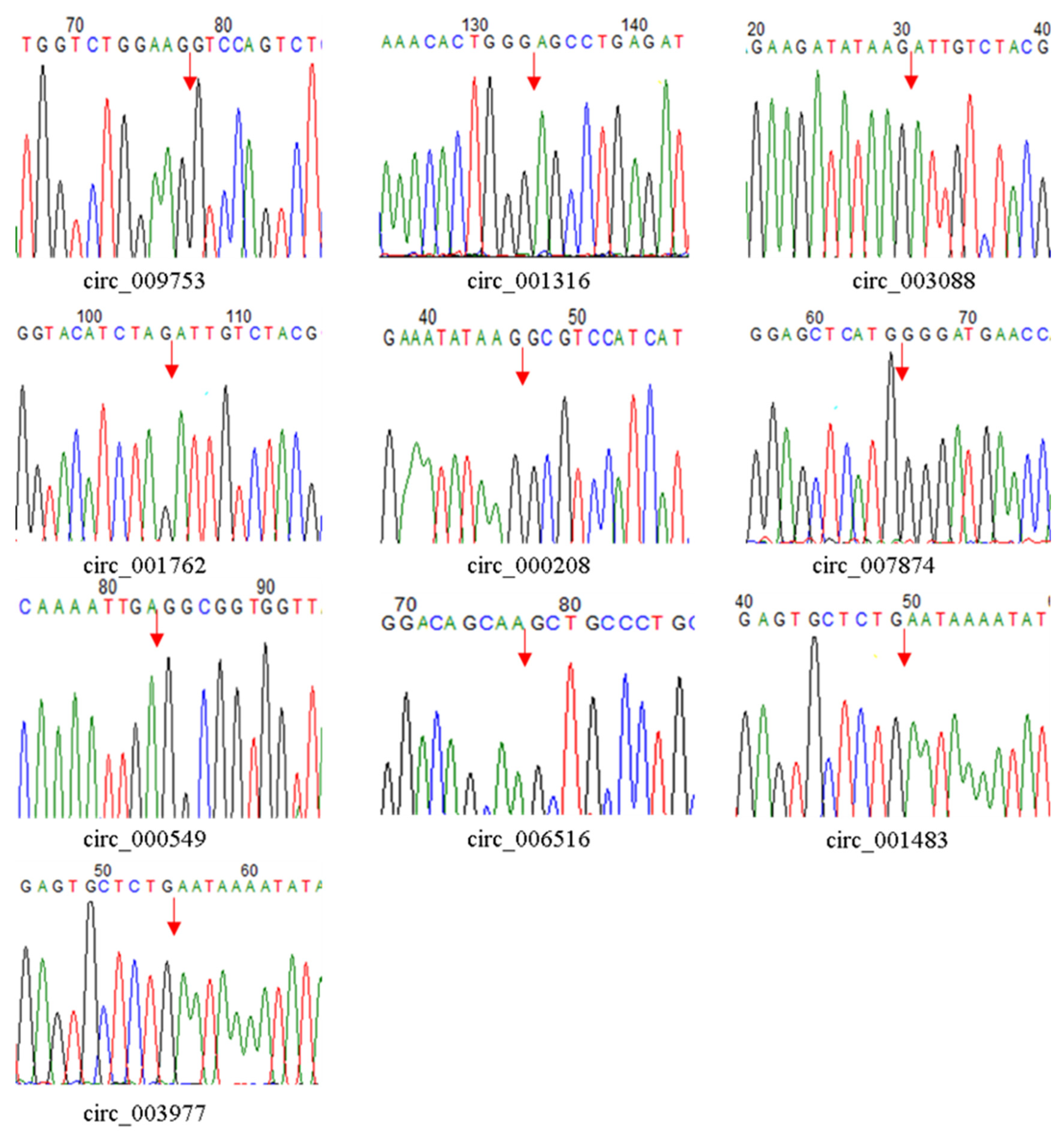
3.2. Validation of the Authenticity of circRNAs Identified in the Caprine Skin Tissue
3.3. Analysis and Validation of Differentially Expressed circRNAs between the Two Caprine Breeds
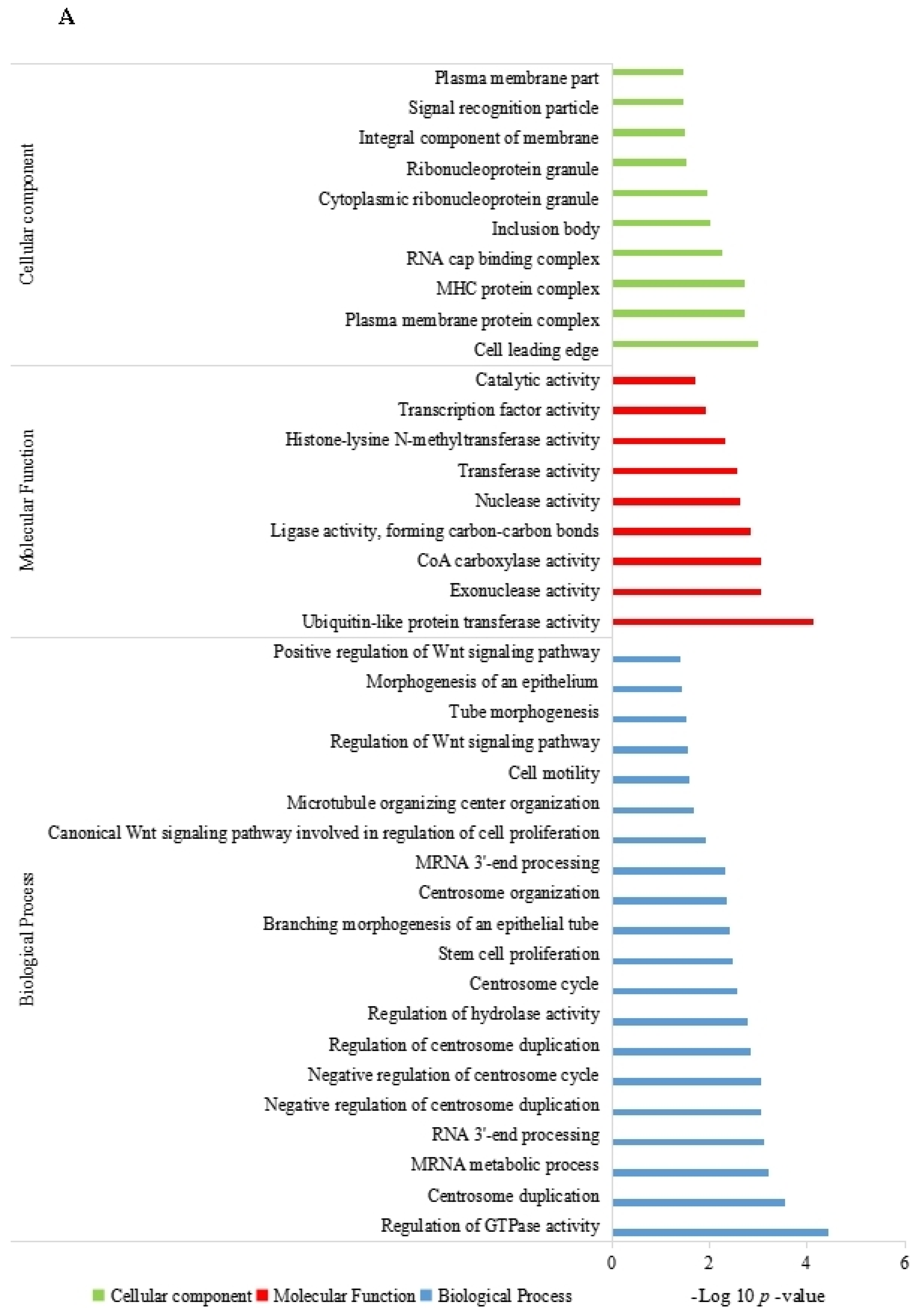
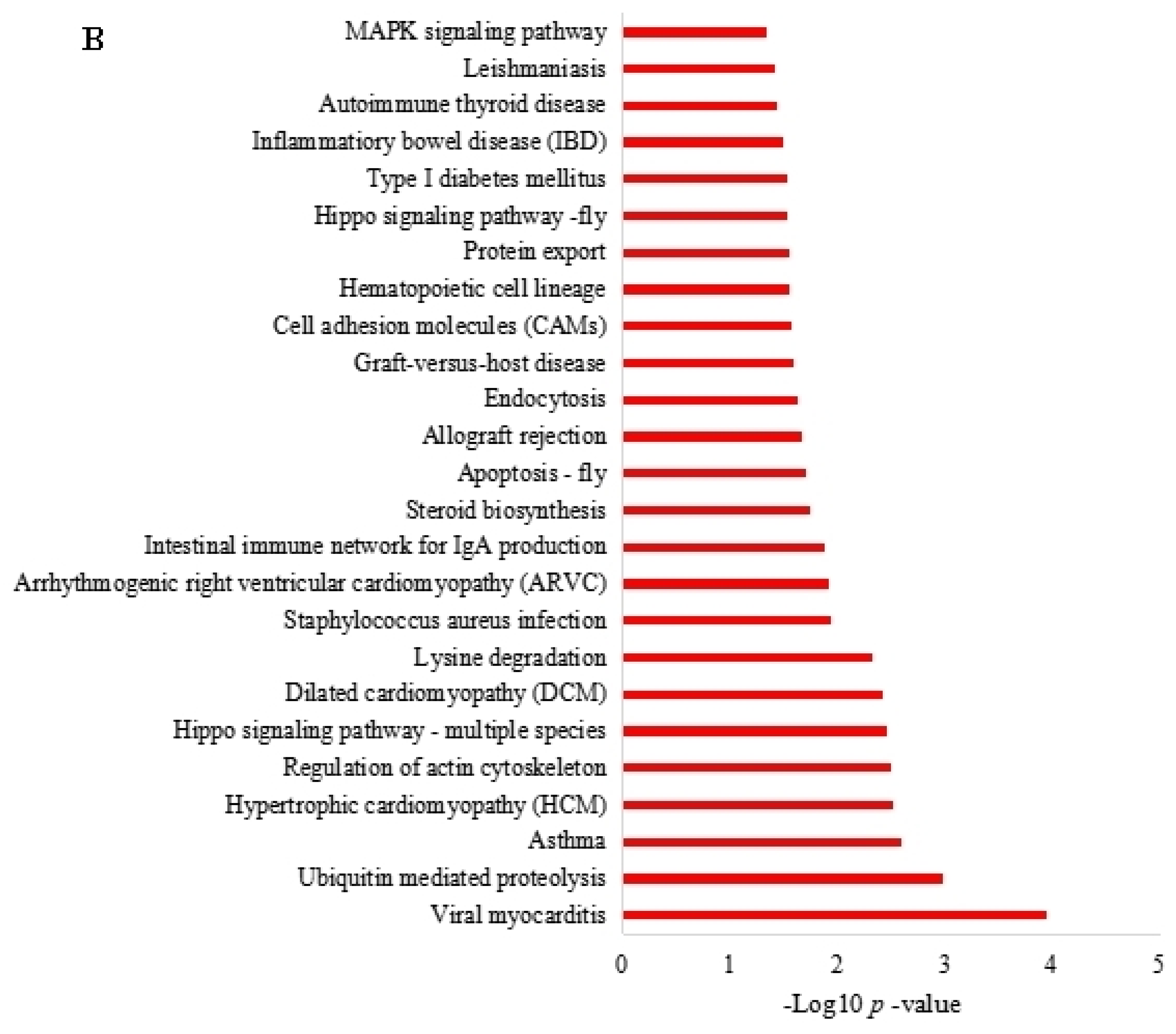
3.4. Function Enrichment Analysis of the Parent Genes of Differentially Expressed circRNAs
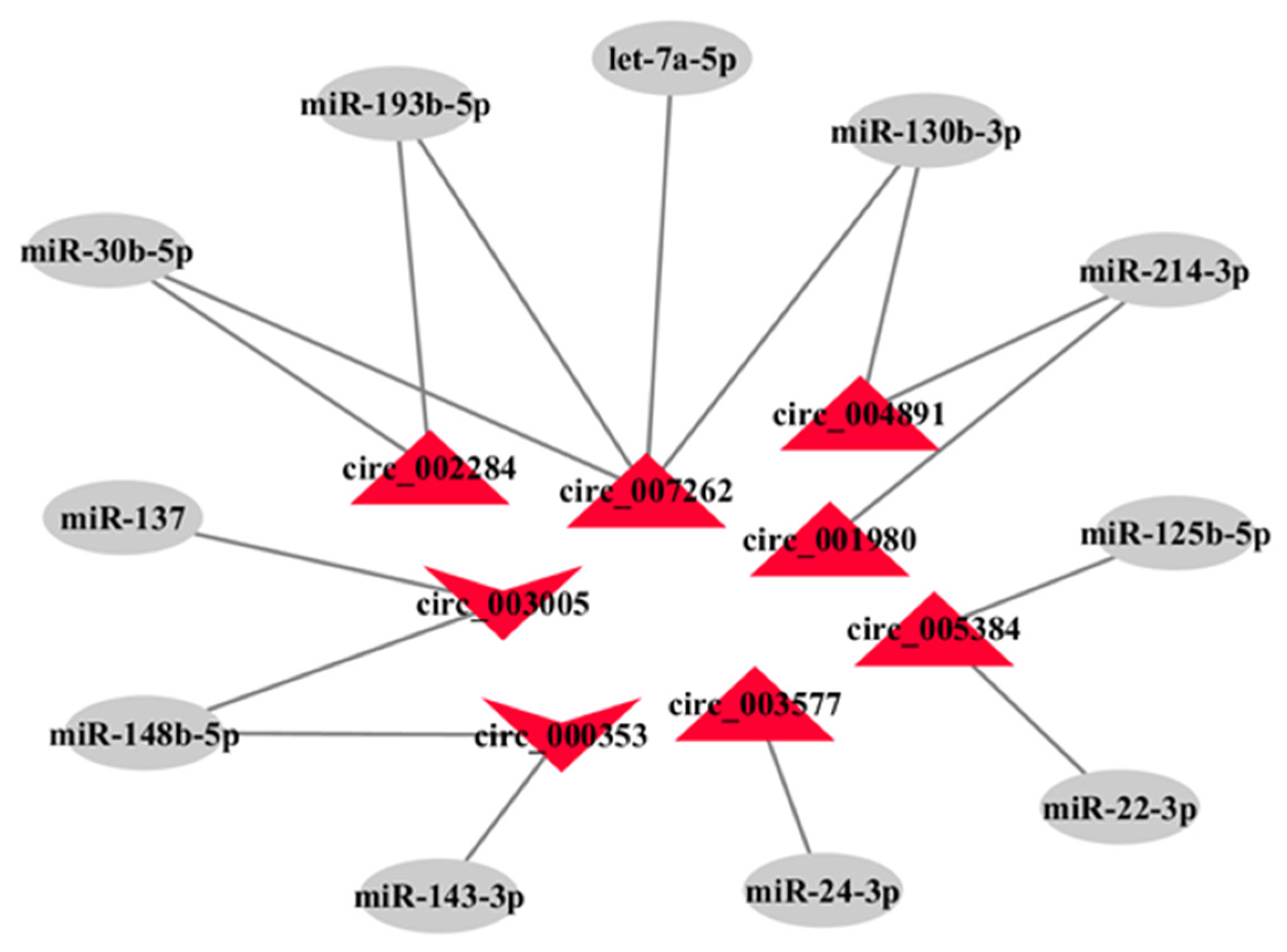
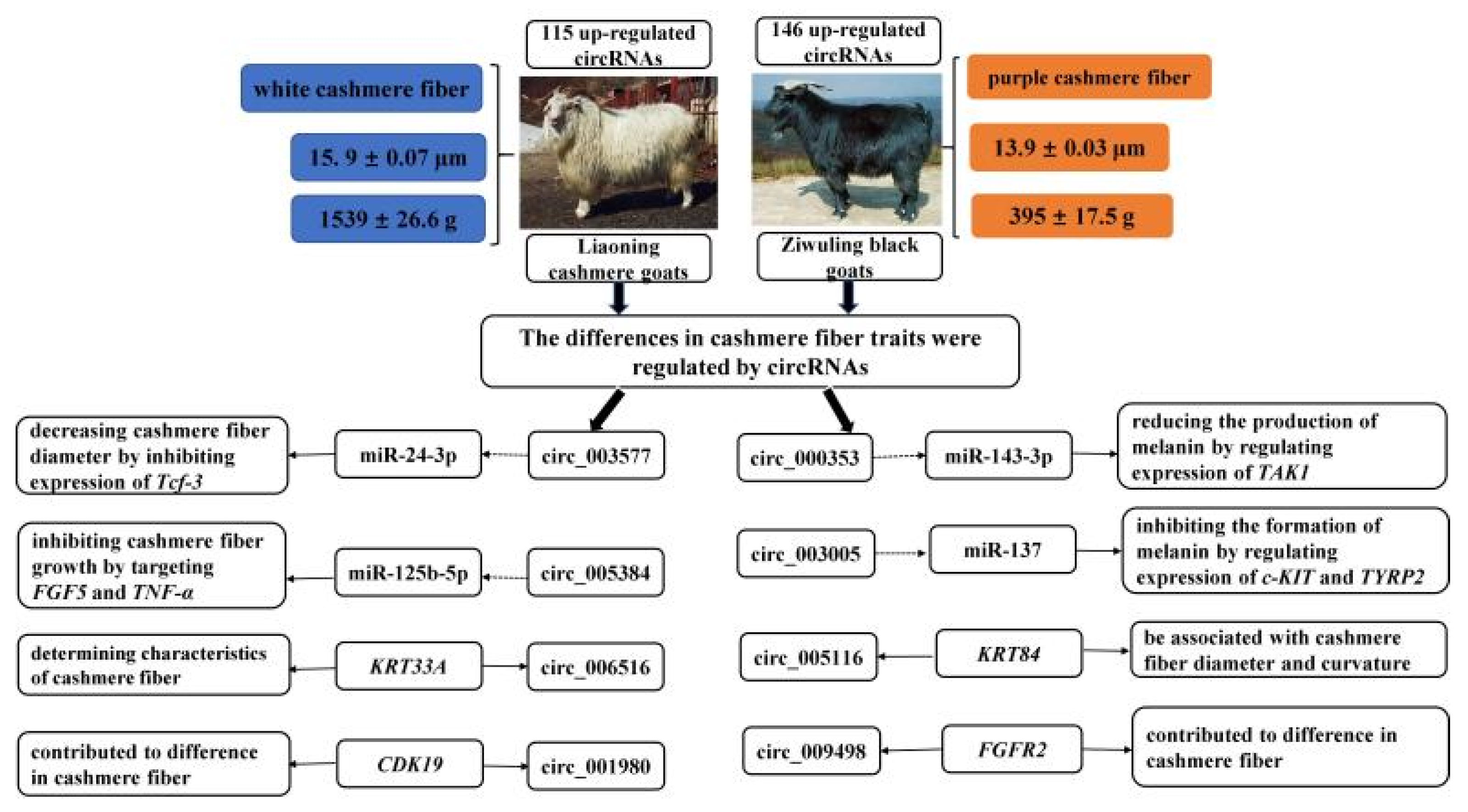
3.5. The miRNA Sponges Analysis of Differentially Expressed circRNAs
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Jeck, W.R.; Sharpless, N.E. Detecting and characterizing circular RNAs. Nat. Biotechnol. 2014, 32, 453–461. [Google Scholar] [CrossRef]
- Salzman, J.; Gawad, C.; Wang, P.L.; Lacayo, N.; Brown, P.O. Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PLoS ONE 2012, 7, e30733. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Zhang, X.O.; Chen, T.; Xiang, J.F.; Yin, Q.F.; Xing, Y.H.; Zhu, S.; Yang, L.; Chen, L.L. Circular intronic long noncoding RNAs. Mol. Cell 2013, 51, 792–806. [Google Scholar] [CrossRef] [PubMed]
- Zhou, C.; Molinie, B.; Daneshvar, K.; Pondick, J.V.; Wang, J.; Van Wittenberghe, N.; Xing, Y.; Giallourakis, C.C.; Mullen, A.C. Genome-Wide Maps of m6A circRNAs Identify Widespread and Cell-Type-Specific Methylation Patterns that Are Distinct from mRNAs. Cell Rep. 2017, 20, 2262–2276. [Google Scholar] [CrossRef]
- Yang, Y.; Fan, X.; Mao, M.; Song, X.; Wu, P.; Zhang, Y.; Jin, Y.; Yang, Y.; Chen, L.L.; Wang, Y.; et al. Extensive translation of circular RNAs driven by N6-methyladenosine. Cell Res. 2017, 27, 626–641. [Google Scholar] [CrossRef]
- Zhu, Y.; Wang, Y.; Zhao, J.; Shen, J.; Wang, Z.; Bai, M.; Fan, Y.; Yin, R.; Mao, Y.; Bai, W. CircRNA-1967 participates in the differentiation of goat SHF-SCs into hair follicle lineage by sponging miR-93-3p to enhance LEF1 expression. Anim. Biotechnol. 2021, 22, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Shen, J.; Wang, Z.; Bai, M.; Fan, Y.; Zhu, Y.; Bai, W. CircRNA-0100 positively regulates the differentiation of cashmere goat SHF-SCs into hair follicle lineage via sequestering miR-153-3p to heighten the KLF5 expression. Arch. Anim. Breed. 2022, 65, 55–67. [Google Scholar] [CrossRef]
- Yin, R.H.; Zhao, S.J.; Jiao, Q.; Wang, Z.Y.; Bai, M.; Fan, Y.X.; Zhu, Y.B.; Bai, W.L. CircRNA-1926 Promotes the Differentiation of Goat SHF Stem Cells into Hair Follicle Lineage by miR-148a/b-3p/CDK19 Axis. Animals 2020, 10, 1552. [Google Scholar] [CrossRef]
- Hui, T.; Zheng, Y.; Yue, C.; Wang, Y.; Bai, Z.; Sun, J.; Cai, W.; Zhang, X.; Bai, W.; Wang, Z. Screening of cashmere fineness-related genes and their ceRNA network construction in cashmere goats. Sci. Rep. 2021, 11, 21977. [Google Scholar] [CrossRef]
- Zheng, Y.; Hui, T.; Yue, C.; Sun, J.; Guo, D.; Guo, S.; Guo, S.; Li, B.; Wang, Z.; Bai, W. Comprehensive analysis of circRNAs from cashmere goat skin by next generation RNA sequencing (RNA-seq). Sci. Rep. 2020, 10, 516. [Google Scholar] [CrossRef]
- Memczak, S.; Jens, M.; Elefsinioti, A.; Torti, F.; Krueger, J.; Rybak, A.; Maier, L.; Mackowiak, S.D.; Gregersen, L.H.; Munschauer, M.; et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 2013, 495, 333–338. [Google Scholar] [CrossRef]
- Lv, X.; Chen, W.; Sun, W.; Hussain, Z.; Chen, L.; Wang, S.; Wang, J. Expression profile analysis to identify circular RNA expression signatures in hair follicle of Hu sheep lambskin. Genomics 2020, 112, 4454–4462. [Google Scholar] [CrossRef]
- Cai, B.; Li, M.; Zheng, Y.; Yin, Y.; Jin, F.; Li, X.; Dong, J.; Jiao, X.; Liu, X.; Zhang, K.; et al. EZH2-mediated inhibition of microRNA-22 promotes differentiation of hair follicle stem cells by elevating STK40 expression. Aging 2020, 12, 12726–12739. [Google Scholar] [CrossRef]
- Amelio, I.; Lena, A.M.; Bonanno, E.; Melino, G.; Candi, E. miR-24 affects hair follicle morphogenesis targeting Tcf-3. Cell Death Dis. 2013, 4, e922. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.L.; Li, J.P.; Chen, Y.; Chang, Q.; Li, Y.M.; Yao, J.Y.; Jiang, H.Z.; Zhao, Z.H.; Guo, D. Growth and viability of Liaoning Cashmere goat hair follicles during the annual hair follicle cycle. Genet. Mol. Res. 2014, 13, 4433–4443. [Google Scholar] [CrossRef] [PubMed]
- Dai, B.; Liang, H.; Guo, D.D.; Bi, Z.W.; Yuan, J.L.; Jin, Y.; Huan, L.; Guo, X.D.; Cang, M.; Liu, D.J. The Overexpression of Tβ4 in the Hair Follicle Tissue of Alpas Cashmere Goats Increases Cashmere Yield and Promotes Hair Follicle Development. Animals 2019, 10, 75. [Google Scholar] [CrossRef]
- Bai, W.L.; Dang, Y.L.; Wang, J.J.; Yin, R.H.; Wang, Z.Y.; Zhu, Y.B.; Cong, Y.Y.; Xue, H.L.; Deng, L.; Guo, D.; et al. Molecular characterization, expression and methylation status analysis of BMP4 gene in skin tissue of Liaoning cashmere goat during hair follicle cycle. Genetica 2016, 144, 457–467. [Google Scholar] [CrossRef] [PubMed]
- Müller-Röver, S.; Handjiski, B.; van der Veen, C.; Eichmüller, S.; Foitzik, K.; McKay, I.A.; Stenn, K.S.; Paus, R. A comprehensive guide for the accurate classification of murine hair follicles in distinct hair cycle stages. J. Investig. Dermatol. 2001, 117, 3–15. [Google Scholar] [CrossRef]
- Yuan, C.; Wang, X.; Geng, R.; He, X.; Qu, L.; Chen, Y. Discovery of cashmere goat (Capra hircus) microRNAs in skin and hair follicles by Solexa sequencing. BMC Genom. 2013, 14, 511. [Google Scholar] [CrossRef]
- Wang, S.; Ge, W.; Luo, Z.; Guo, Y.; Jiao, B.; Qu, L.; Zhang, Z.; Wang, X. Integrated analysis of coding genes and non-coding RNAs during hair follicle cycle of cashmere goat (Capra hircus). BMC Genom. 2017, 18, 767. [Google Scholar] [CrossRef]
- Shang, F.; Wang, Y.; Ma, R.; Di, Z.; Wu, Z.; Hai, E.; Rong, Y.; Pan, J.; Liang, L.; Wang, Z.; et al. Expression Profiling and Functional Analysis of Circular RNAs in Inner Mongolian Cashmere Goat Hair Follicles. Front. Genet. 2021, 12, 678825. [Google Scholar] [CrossRef] [PubMed]
- Hao, Z.; Zhou, H.; Hickford, J.G.H.; Gong, H.; Wang, J.; Hu, J.; Liu, X.; Li, S.; Zhao, M.; Luo, Y. Identification and characterization of circular RNA in lactating mammary glands from two breeds of sheep with different milk production profiles using RNA-Seq. Genomics 2020, 112, 2186–2193. [Google Scholar] [CrossRef]
- Wang, J.; Zhou, H.; Hickford, J.G.H.; Hao, Z.; Gong, H.; Hu, J.; Liu, X.; Li, S.; Shen, J.; Ke, N.; et al. Identification and characterization of circular RNAs in mammary gland tissue from sheep at peak lactation and during the nonlactating period. J. Dairy Sci. 2021, 104, 2396–2409. [Google Scholar] [CrossRef] [PubMed]
- Shen, J.; Zhen, H.; Li, L.; Zhang, Y.; Wang, J.; Hu, J.; Liu, X.; Li, S.; Hao, Z.; Li, M.; et al. Identification and characterization of circular RNAs in Longissimus dorsi muscle tissue from two goat breeds using RNA-Seq. Mol. Genet. Genom. 2022, 297, 817–831. [Google Scholar] [CrossRef]
- Wilusz, J.E. A 360° view of circular RNAs: From biogenesis to functions. Wiley Interdiscip. Rev. RNA 2018, 9, e1478. [Google Scholar] [CrossRef]
- Coulombe, P.A.; Omary, M.B. ‘Hard’ and ‘Soft’ principles defining the structure, function and regulation of keratin intermediate filaments. Curr. Opin. Cell Biol. 2002, 14, 110–122. [Google Scholar] [CrossRef]
- Powell, B.C.; Rogers, G.E. The role of keratin proteins and their genes in the growth, structure and properties of hair. EXS 1997, 78, 59–148. [Google Scholar] [CrossRef] [PubMed]
- Yu, Z.; Wildermoth, J.E.; Wallace, O.A.; Gordon, S.W.; Maqbool, N.J.; Maclean, P.H.; Nixon, A.J.; Pearson, A.J. Annotation of sheep keratin intermediate filament genes and their patterns of expression. Exp. Dermatol. 2011, 20, 582–588. [Google Scholar] [CrossRef]
- Langbein, L.; Schweizer, J. Keratins of the human hair follicle. Int. Rev. Cytol. 2005, 243, 1–78. [Google Scholar] [CrossRef]
- Gao, Y.; Wang, X.; Yan, H.; Zeng, J.; Ma, S.; Niu, Y.; Zhou, G.; Jiang, Y.; Chen, Y. Comparative Transcriptome Analysis of Fetal Skin Reveals Key Genes Related to Hair Follicle Morphogenesis in Cashmere Goats. PLoS ONE 2016, 11, e0151118. [Google Scholar] [CrossRef]
- Yu, Z.; Gordon, S.W.; Nixon, A.J.; Bawden, C.S.; Rogers, M.A.; Wildermoth, J.E.; Maqbool, N.J.; Pearson, A.J. Expression patterns of keratin intermediate filament and keratin associated protein genes in wool follicles. Differentiation 2009, 77, 307–316. [Google Scholar] [CrossRef] [PubMed]
- Firestein, R.; Bass, A.J.; Kim, S.Y.; Dunn, I.F.; Silver, S.J.; Guney, I.; Freed, E.; Ligon, A.H.; Vena, N.; Ogino, S.; et al. CDK8 is a colorectal cancer oncogene that regulates beta-catenin activity. Nature 2008, 455, 547–551. [Google Scholar] [CrossRef]
- DasGupta, R.; Fuchs, E. Multiple roles for activated LEF/TCF transcription complexes during hair follicle development and differentiation. Development 1999, 126, 4557–4568. [Google Scholar] [CrossRef]
- Ge, W.; Wang, S.H.; Sun, B.; Zhang, Y.L.; Shen, W.; Khatib, H.; Wang, X. Melatonin promotes Cashmere goat (Capra hircus) secondary hair follicle growth: A view from integrated analysis of long non-coding and coding RNAs. Cell Cycle 2018, 17, 1255–1267. [Google Scholar] [CrossRef]
- Nixon, A.J.; Choy, V.J.; Parry, A.L.; Pearson, A.J. Fiber growth initiation in hair follicles of goats treated with melatonin. J. Exp. Zool. 1993, 267, 47–56. [Google Scholar] [CrossRef] [PubMed]
- Fischer, T.W.; Slominski, A.; Tobin, D.J.; Paus, R. Melatonin and the hair follicle. J. Pineal Res. 2008, 44, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Greco, V.; Chen, T.; Rendl, M.; Schober, M.; Pasolli, H.A.; Stokes, N.; Dela Cruz-Racelis, J.; Fuchs, E. A two-step mechanism for stem cell activation during hair regeneration. Cell Stem Cell 2009, 4, 155–169. [Google Scholar] [CrossRef]
- Fernandes, K.J.; Kobayashi, N.R.; Gallagher, C.J.; Barnabé-Heider, F.; Aumont, A.; Kaplan, D.R.; Miller, F.D. Analysis of the neurogenic potential of multipotent skin-derived precursors. Exp. Neurol. 2006, 201, 32–48. [Google Scholar] [CrossRef]
- Su, R.; Gong, G.; Zhang, L.; Yan, X.; Wang, F.; Zhang, L.; Qiao, X.; Li, X.; Li, J. Screening the key genes of hair follicle growth cycle in Inner Mongolian Cashmere goat based on RNA sequencing. Arch. Anim. Breed. 2020, 63, 155–164. [Google Scholar] [CrossRef]
- Xiang, B.; Li, Y.; Li, J.; Li, J.; Jiang, H.; Zhang, Q. MiR-193b regulated the formation of coat colors by targeting WNT10A and GNAI2 in Cashmere goats. Anim. Biotechnol. 2021, 8, 330–338. [Google Scholar] [CrossRef]
- Zhu, B.; Xu, T.; Zhang, Z.; Ta, N.; Gao, X.; Hui, L.; Guo, X.; Liu, D. Transcriptome sequencing reveals differences between anagen and telogen secondary hair follicle-derived dermal papilla cells of the Cashmere goat (Capra hircus). Physiol. Genom. 2014, 46, 104–111. [Google Scholar] [CrossRef] [PubMed]
- Xu, T.; Guo, X.; Wang, H.; Hao, F.; Du, X.; Gao, X.; Liu, D. Differential gene expression analysis between anagen and telogen of Capra hircus skin based on the de novo assembled transcriptome sequence. Gene 2013, 520, 30–38. [Google Scholar] [CrossRef] [PubMed]
- Zhou, G.; Yuan, C.; He, X.; Kang, D.; Wang, X.; Chen, Y. Effect of miR-125b on dermal papilla cells of goat secondary hair follicle. Electron. J. Biotechnol. 2017, 25, 64–69. [Google Scholar] [CrossRef]
- Ji, K.; Zhang, P.; Zhang, J.; Fan, R.; Liu, Y.; Yang, S.; Hu, S.; Liu, X.; Dong, C. MicroRNA 143-5p regulates alpaca melanocyte migration, proliferation and melanogenesis. Exp. Dermatol. 2018, 27, 166–171. [Google Scholar] [CrossRef] [PubMed]
- Jiang, S.; Yu, X.; Dong, C. MiR-137 affects melanin synthesis in mouse melanocyte by repressing the expression of c-Kit and Tyrp2 in SCF/c-Kit signaling pathway. Biosci. Biotechnol. Biochem. 2016, 80, 2115–2121. [Google Scholar] [CrossRef] [PubMed]

| CircRNA/Gene | Forward (5′→3′) | Reverse (5′→3′) |
|---|---|---|
| circ_009753 | TTCTACTGTGATAGGGCTCG | CACATTGAAGTCAGGTGGTT |
| circ_001316 | ACGGGCATGTAGTCACGG | TCAACCCCAGGGCACAGA |
| circ_003088 | GCTCAGGCAAAGATAGAAG | ACTGAAGGTCGAGGGTCT |
| circ_001762 | CAGGTGCCCAGATGACTA | CCTTTAAGCCGTAAGACG |
| circ_000208 | TGCAGGACCTGAGAACAG | TTCACCACCGAGGACAAT |
| circ_007874 | ACACCGTATGGGACAACAA | TCTCAAGGCTCAGCTTCC |
| circ_000549 | TTTAGCCTTGGATTATGAC | TCGCCAGTGTACTTGTTG |
| circ_006516 | TGACCTGGAGCGGCAGAA | GGTGACATAGGACCCAACTGAT |
| circ_001483 | AAACTGCCAATACCCTGAA | AGGTTGCTGCCATGACTT |
| circ_003977 | CTGGAAACTGCCAATACC | CTGCCGTGACTTAGGGAT |
| GAPDH | ACACTGAGGACCAGGTTGTG | GACAAAGTGGTCGTTGAGGG |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hu, L.; Wang, J.; Luo, Y.; Liu, X.; Li, S.; Hao, Z.; Zhao, F.; Li, M.; Shi, B.; Gu, Y. Identification and Characterization of Circular RNAs (circRNAs) Using RNA-Seq in Two Breeds of Cashmere Goats. Genes 2023, 14, 331. https://doi.org/10.3390/genes14020331
Hu L, Wang J, Luo Y, Liu X, Li S, Hao Z, Zhao F, Li M, Shi B, Gu Y. Identification and Characterization of Circular RNAs (circRNAs) Using RNA-Seq in Two Breeds of Cashmere Goats. Genes. 2023; 14(2):331. https://doi.org/10.3390/genes14020331
Chicago/Turabian StyleHu, Liyan, Jiqing Wang, Yuzhu Luo, Xiu Liu, Shaobin Li, Zhiyun Hao, Fangfang Zhao, Mingna Li, Bingang Shi, and Yuanhua Gu. 2023. "Identification and Characterization of Circular RNAs (circRNAs) Using RNA-Seq in Two Breeds of Cashmere Goats" Genes 14, no. 2: 331. https://doi.org/10.3390/genes14020331
APA StyleHu, L., Wang, J., Luo, Y., Liu, X., Li, S., Hao, Z., Zhao, F., Li, M., Shi, B., & Gu, Y. (2023). Identification and Characterization of Circular RNAs (circRNAs) Using RNA-Seq in Two Breeds of Cashmere Goats. Genes, 14(2), 331. https://doi.org/10.3390/genes14020331





