Abstract
Mitochondrial dynamics, including fission and fusion processes, are essential for heart health. Mitochondria, the powerhouses of cells, maintain their integrity through continuous cycles of biogenesis, fission, fusion, and degradation. Mitochondria are relatively immobile in the adult heart, but their morphological changes due to mitochondrial morphology factors are critical for cellular functions such as energy production, organelle integrity, and stress response. Mitochondrial fusion proteins, particularly Mfn1/2 and Opa1, play multiple roles beyond their pro-fusion effects, such as endoplasmic reticulum tethering, mitophagy, cristae remodeling, and apoptosis regulation. On the other hand, the fission process, regulated by proteins such as Drp1, Fis1, Mff and MiD49/51, is essential to eliminate damaged mitochondria via mitophagy and to ensure proper cell division. In the cardiac system, dysregulation of mitochondrial dynamics has been shown to cause cardiac hypertrophy, heart failure, ischemia/reperfusion injury, and various cardiac diseases, including metabolic and inherited cardiomyopathies. In addition, mitochondrial dysfunction associated with oxidative stress has been implicated in atherosclerosis, hypertension and pulmonary hypertension. Therefore, understanding and regulating mitochondrial dynamics is a promising therapeutic tool in cardiac diseases. This review summarizes the role of mitochondrial morphology in heart diseases for each mitochondrial morphology regulatory gene, and their potential as therapeutic targets to heart diseases.
Keywords:
mitochondrial dynamics; fission and fusion; heart failure; cardiovascular diseases; Drp1; Mfn1; Mfn2; Opa1 1. Introduction
Mitochondria are essential organelles for adenosine triphosphate (ATP) production and are closely linked to cellular health and energy demand. On the other hand, mitochondria release reactive oxygen species (ROS) that cause oxidative damage. These mitochondrial functions are regulated by two distinct systems: fission and fusion. Fission, facilitated by proteins such as dynamin-related protein 1 (Drp1), fission 1 (Fis1), mitochondrial fission factor (Mff), and mitochondrial division (MiD) 49 and 51, leads to mitochondrial fragmentation, which is essential for cell division and removal of damaged mitochondria. Fusion, orchestrated by mitofusin 1 (Mfn1), mitofusin 2 (Mfn2), and optic atrophy 1 (Opa1), results in interconnected networks that help repair mitochondrial DNA and distribute energy. While the roles of fission proteins are primarily associated with morphological changes, fusion proteins show diverse roles influencing mitophagy, apoptosis, and energy production. Consequently, mitochondrial morphology is strictly regulated by several key genes. Unlike other cells, the adult heart contains relatively static mitochondria that are segregated into three functional subpopulations. Despite this fixed arrangement, emerging evidence suggests that mitochondrial morphological factors play an important role in cardiac health and pathology.
Mitochondrial dysfunction has been implicated in a spectrum of cardiac diseases. Defective fission has been associated with severe cardiac abnormalities, highlighting the importance of these dynamics in cardiac pathologies, including cardiomyopathies and ischemic conditions. A profound understanding of mitochondrial dynamics opens a new avenue for therapeutic approaches to heart disease. The balance between fission and fusion processes offers a promising strategy to preserve cardiac health, especially under stress.
In this review, we summarize the relationship between mitochondrial morphology regulatory genes and heart diseases. We discuss the molecular biology underlying mitochondrial dynamics, clarify how defects in mitochondrial dynamics are associated with heart damage during stress, and detail the importance of considering mitochondrial dynamics as a new therapeutic target for mitochondrial morphology regulatory genes.
2. Mitochondrial Morphology
Mitochondrial dynamics involves a continuous process of mitochondrial fission and fusion. Mitochondrial dynamics regulate mitochondrial turnover and the intracellular environment [1]. Here, we summarize the basic knowledge of the regulation of mitochondrial morphology.
2.1. Mitochondrial Fission
Mitochondrial fission depends on the activation of Drp1 in the cytoplasm, which causes Drp1 to translocate into the mitochondria and bind to receptors on the mitochondrial outer membrane (OMM). Mitochondrial fission produces daughter mitochondria with different membrane potentials (Δψm). This function supports the selective degradation of damaged mitochondria by autophagy and maintains cellular homeostasis [2,3]. During mitosis, fission ensures the distribution of damaged mitochondria to daughter cells [4]. Drp1 mainly regulates mitochondrial fragmentation in mammals and interacts with several organelles [5]. Dynamin 2 is involved in mitochondrial fragmentation [6], but its deletion does not stop mitochondrial fragmentation [7,8]. Thus, Drp1 is essential for membrane contraction and separation [9]. Once translocated into the mitochondria, Drp1 aggregates and surrounds the mitochondria, causing mitochondrial fission [10,11,12,13]. The endoplasmic reticulum (ER) influences and constricts the sites that will align Drp1 rings to divide mitochondria [10,14,15]. As the ER membrane wraps around mitochondria, actin associates primarily with Formin 2 on the ER surface. Formin 2 cooperates with Spire1C to promote actin assembly at the mitochondrial surface [16] and also communicates with myosin II [17,18,19]. When fission is triggered, actin groups on the OMM turn on Drp1, which is translocated to the mitochondria. Fission requires actin to form chains and apply tension to the membrane [20,21].
Drp1, a key fission protein, is regulated by multiple post-translational modifications, including phosphorylation [22,23,24], ubiquitination [25], SUMOylation [26], O-GlcNAcylation [27], and nitrosylation [14,28]. Phosphorylation, particularly at S637 and S616, plays an important role. Protein kinase A phosphorylates S637 in response to cAMP and inhibits fission by blocking its GTPase activity [23,24]. The phosphorylation of Drp1 at the Ser 616 (S616) site, mediated by cyclin-dependent kinase (CDK) 1/Cyclin B or CDK5, facilitates mitochondrial fission during mitosis [29,30]. Drp1 is regulated by transcription and protein degradation. Small RNA molecules such as miR can alter the Drp1 gene expression. During apoptosis, RNA known as miR-30 is reduced, and Drp1 expression is increased [31]. In myocardial infarction, miR-499 regulates calcineurin and acts to protect the heart [32]. Drp1 degradation affects mitochondrial fragmentation. When Parkin is deleted, Drp1 increases, and fission is accelerated [33]. Mitochondrial ubiquitin ligase (MITOL/MARCH5) interacts with Drp1 to affect fragmentation [34,35]. MITOL/MARCH5 deficiency in the heart causes heart failure with increased mitochondrial fragmentation in mice [36].
Receptors Required for Drp1 Recruitment
In mammals, Drp1 recruitment to the OMM depends on key adaptors: Fis1, Mff, and MiD 49 and 51 [37,38,39]. These adaptors are essential for mitochondrial fragmentation by Drp1. Fis1 is a protein anchored at its C-terminus to the OMM, exposing a 15 kDa soluble domain to the cytosol [40,41,42] (Figure 1). Fis1 was first identified as a Drp1 adaptor in mammalian cells. Although Fis1 recruits Drp1 in specific cells or situations, its overexpression consistently induces mitochondrial fragmentation in mammalian cells [38,42,43,44,45,46,47]. This may be due to its ability to bind and repress fusion proteins or to mitochondrial dysfunction and Ca2+ overload [48,49]. It has been proposed that Fis1 not only acts as a Drp1 receptor but mainly promotes lysosomal recruitment of Drp1 for peripheral fission and mitophagy [3,45,50]. This is consistent with the role of Fis1 in regulating mitochondria-lysosome contact, as exemplified by its association with Tbc1d15 [51,52].
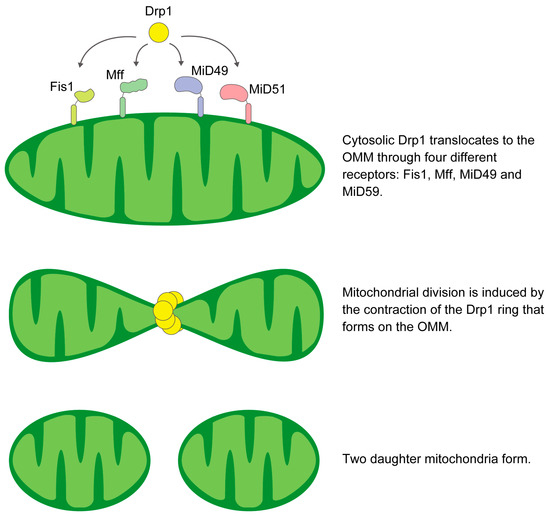
Figure 1.
Schematic illustration of the mechanism of mitochondrial fission.
Mff, which is anchored to the OMM, has a putative Drp1 binding domain at its N-terminus [37,38]. Inhibition of Mff expression attenuates both Drp1 recruitment and subsequent fission events [37,38]. Mff mediates the recruitment of Drp1 primarily in the mitochondrial midzone [3]. Under conditions of mitochondrial dysfunction and elevated AMP levels, protein kinase AMP-activated (AMPK) phosphorylates Mff and enhances its pro-fission activity [53].
MiD49 and MiD51 are mitochondrial adaptors of Drp1 that promote fission [54]. However, high levels of these adaptors can trap Drp1 and inhibit mitochondrial fragmentation [55,56]. The cryo-EM structure of the Drp1-MiD49 complex shows that after co-assembly, detachment of the MiD receptor occurs due to guanosine triphosphate (GTP) hydrolysis and exchange, resulting in a shortened filament and a contracted Drp1 ring. Therefore, an abundance of these receptors may not enhance fission, but their effect could be modulated by the precise timing of assembly and disassociation [57].
2.2. Mitochondrial Fusion
The fusion process of the OMM is orchestrated by a GTPase-driven mechanism that is predominantly mediated by the homo- or hetero-oligomeric interactions of the mitofusins [58,59,60]. The subsequent inner mitochondrial membrane (IMM) fusion is mediated by Opa1, allowing the exchange of matrix components such as mtDNA, lipids and proteins between fused mitochondria [61,62]. Mfns are prevalent at contact points between neighboring mitochondria, facilitating tethering, docking, and extension of OMM contacts through either homotypic or more effective heterotypic complexes [63] (Figure 2). The presence of these heterotypic complexes means that overexpression of one protein can compensate for the loss of the other [64,65]. An increase in either protein leads to extensive mitochondrial fusion, resulting in elongated mitochondria. Among the Mfns, Mfn1 exhibits greater efficiency in docking and tethering, which is attributed to its superior GTPase activity compared to Mfn2 [60,63]. Mfn2 is essential for tethering mitochondria to the ER and other ER-derived organelles, crucial for Ca2+ and phosphatidylserine (PS) transfer [66,67]. This transfer supports phospholipid synthesis and influences membrane fission and fusion dynamics, particularly Drp1-mediated processes [8].
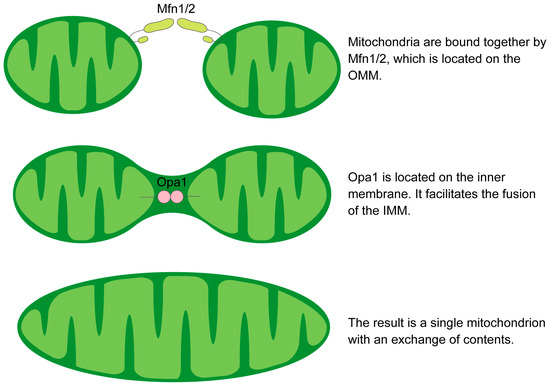
Figure 2.
Schematic illustration of the mechanism of mitochondrial fusion.
Mfns are regulated at multiple levels to control fusion. The peroxisome proliferator-activated receptor γ coactivator 1-α (Pgc-1α), estrogen-related receptor-α (Err-α), and Pgc-1β increase Mfn2 expression [68], whereas the transcription factor Mef2 degradation during neuronal excitotoxicity decreases Mfn2 expression [69]. Several miRNAs, including MiR-214 and MiR-106b, affect mitochondrial morphology by targeting Mfns [70,71]. Mitofusins undergo post-translational modifications such as oxidation, leading to Mfn oligomerization [72]. MITOL/MARCH5 ubiquitinates Mfn1 during stress [73], and histone deacetylase 6 (HDAC6) supports Mfn1 function during glucose shortage [74]. Various phosphorylation events can either support or inhibit the role of Mfns in fusion, such as ERK-mediated modification of Mfn1 [75,76]. These regulatory processes underscore the critical nature of mitochondrial dynamics in cellular function.
Opa1 is the only dynamin-like protein in the IMM and is essential for mitochondrial elongation [77]. Opa1 has eight splice variants. It’s processed at three sites by the metalloproteases YME1, like 1 ATPase (YME1L1) and M-AAA protease 1 (OMA1). Further cleavage by PARL produces a soluble intermembrane space (IMS) fraction that, in combination with IMM-anchored l-Opa1, maintains tight cristae junctions [78]. The human OPA1 gene contains more than 30 exons and eight mRNAs that differ in biological function, such as maintaining mitochondrial fusion [79,80]. Mammalian Opa1 is mainly regulated by protease cleavage by Oma1 and Yme1l. Excess s-Opa1 inhibits fusion, whereas l-Opa1 can maintain fusion under stress [81,82]. The role of s-Opa1 in fission remains controversial, but it is known that both need to work simultaneously for effective fusion [83].
3. Dynamics of Mitochondria in Cardiomyocytes
In cultured cells and neonatal cardiomyocytes, mitochondria are dynamic organelles that spread throughout the cytoplasm in a network-like pattern. In adult cardiomyocytes, however, mitochondria have a different arrangement, being more compact and tightly interconnected (Figure 3). Thus, the general hypothesis of mitochondrial morphology that mitochondrial fragmentation is detrimental and mitochondrial fusion is beneficial should be carefully applied to adult cardiomyocytes under physiological conditions [84]. Mitochondria in adult cardiomyocytes can be divided into three main types based on their location and function. Most mitochondria, called interfibrillar mitochondria (IFM), are located near the myofibrils and are essential for calcium signaling and as a source of energy for contraction. A smaller group, the sarcoplasmic submembrane mitochondria (SSM), are located near the sub-coronary artery membrane and are essential for ion channel energy and cell signaling. Another group, the perinuclear mitochondria (PNM), are located near the nucleus and support the energy required for transcription [85,86]. Recent studies have shown that despite their unique structure, mitochondria can change shape in adult cardiomyocytes [87,88,89,90,91,92]. This was observed by briefly tracking the movement of various fluorescent markers using tools such as electron microscopy and confocal microscopy [93,94,95,96,97]. These markers range from light-activated proteins to pH-sensitive probes [98,99,100,101,102,103,104,105,106]. Although many observational techniques are revealing mitochondrial dynamics in cardiomyocytes, these results differ from the basic mitochondrial dynamics obtained from cultured cells, and more studies in the heart and adult cardiomyocytes are needed to study mitochondrial dynamics in cardiomyocytes.
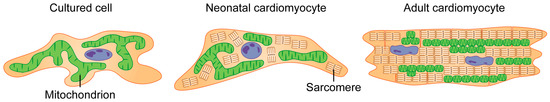
Figure 3.
Mitochondrial morphology variation among cells. In cultured cells such as HEK293, neonatal cardiomyocytes and adult cardiomyocytes, mitochondrial morphology is different in the cells. Mitochondrial morphology in neonatal cardiomyocytes is similar to cultured cells. In contrast, adult cardiomyocytes are characterized by smaller, segmented mitochondria.
4. Animal Models of Mitochondrial Dynamics
Mitochondrial dynamics during heart development and in the resting state have been studied using whole-body or heart-specific knockout models targeting mitochondrial fission and fusion (Table 1). Here, we show the phenotypes of the heart driven by each genetic engineering of mitochondrial morphology factors.
4.1. Drp1
Deletion of the Drp1 gene results in embryonic death by day E12.5 [107]. In the adult heart, loss of Drp1 disrupts mitophagy and causes cardiomyopathy [108]. This underscores the essential function of Drp1 in ensuring a robust mitochondrial network. In mice lacking Drp1, specifically in cardiomyocytes, there is a marked reduction in lifespan accompanied by mitochondrial respiratory dysfunction and accumulation of ubiquitinated proteins [109,110]. Furthermore, postnatal ablation of myocardial Drp1 leads to increased mortality [111]. In adult cardiomyocytes, deletion of Drp1 leads to upregulation of Parkin, resulting in increased mitophagy. This overactivity contributes to the onset of lethal cardiomyopathy. Simultaneous cardiac-specific deletion of both Drp1 and Parkin ameliorates cardiac remodeling and improves survival, underscoring the critical role of Parkin in regulating baseline mitophagic quality control [112]. In adult mice subjected to inducible cardiomyocyte-specific Drp1 deletion, dilated cardiomyopathy is developed under unstressed conditions, leading to mortality within 13 days. Cellular analyses reveal the presence of elongated and damaged mitochondria, reduced autophagy and increased cell death [108,111]. Overexpression of Drp1 results in mitochondrial fragmentation, while other proteins involved in mitochondrial dynamics remain unaffected. These fragmented mitochondria retain a typical cristae structure and maintain regular mitochondrial respiration. Thus, persistent Drp1-induced hyper-fragmentation does not inherently damage cardiomyocyte mitochondria or the mammalian heart [113]. Taken together, these findings suggest that a state of mitochondrial hyperactivity closely associated with Drp1 abnormalities is a strong predisposing factor for the development of hypertrophic cardiomyopathy.
4.2. Mfn1 and Mfn2
In mice, combined cardiac deletion of Mfn1 and Mfn2 results in spherical and functionally normal mitochondrial function, eventually leading to cardiac dysfunction by postnatal day 7 and death within 16 days of birth [114]. Combined ablation of tamoxifen-induced Mfn1/2 in adult cardiomyocytes causes mitochondrial fragmentation and dysfunction, leading to eccentric hypertrophy and lethal dilated cardiomyopathy [111,115,116]. In mice with cardiomyocyte-specific deletion of Mfn1, cardiac function and mitochondrial respiration remain intact, though spherical mitochondria are observed [117,118]. In contrast, mice with cardiomyocyte-specific Mfn2 ablation develop dilated cardiomyopathy [118], preceded by an initial phase of mild hypertrophy [119]. Cardiomyocyte-specific knockout models of Mfn1 and Mfn2 show different phenotypic outcomes, which can be explained by several factors. Primarily, Mfn2 functions as a Parkin receptor in addition to its overlapping roles with Mfn1. In addition, only Mfn2 is involved in tethering the endoplasmic reticulum (ER) to mitochondria, a critical interaction for mitochondrial energy metabolism and calcium regulation [66]. Therefore, in addition to mitophagy perturbations, Mfn2 knockout cardiomyocytes exhibit disruptions in calcium-related pathways due to decreased mitochondrial-sarcoplasmic reticulum (SR) tethering and impaired calcium uptake [67,120].
In Drosophila, knockdown of the cardiac genes Marf (human Mfn) and Opa1 results in cardiomyopathy and reduces contractility. This effect is rescued by overexpression of human mitofusins [121]. In contrast to cardiomyocytes with fusion defects due to Mfn1/Mfn2 knockout or fission defects due to Drp1 knockout, the Mfn1/Mfn2/Drp1 triple knockout cardiomyocytes exhibit prolonged survival and overt cardiac hypertrophy. The simultaneous disruption of mitochondrial fission and fusion leads to the accumulation of mitochondria, which compromises the sarcomeric structure of the cardiomyocytes [113]. Taken together, these findings indicate that Mfn1/Mfn2 double knockout results in mitochondrial fragmentation and stronger cardiotoxicity than deletion of only one of the Mfns.
4.3. Mff
Mff-deficient mice exhibit impaired mitochondrial function and increased mitophagy, leading to dilated cardiomyopathy and heart failure by 13 weeks of age. This cascade leads to a decrease in cardiac ATP concentration, which predisposes cardiomyocytes to apoptosis, culminating in fibrotic changes and heart failure. Surprisingly, concomitant deletion of Mfn1 attenuates these pathological changes, preserved cardiac function and extended lifespan [122].
4.4. Opa1
Cardiomyocyte-specific heterozygous Opa1-deficient mice do not show cardiac abnormalities under non-stress conditions, but they show enlarged mitochondria. Furthermore, in vitro studies on these cardiomyocytes reveal a reduced sensitivity of the opening of the mitochondrial permeability transition pore (mPTP) to calcium accumulation [123].

Table 1.
Animal models of mitochondrial morphology factors.
Table 1.
Animal models of mitochondrial morphology factors.
| Gene | Model | Phenotype | Mitochondrial Morphology | Reference |
|---|---|---|---|---|
| Drp1 | Drp1 deletion homo | Embryonically lethal | Reduced mitochondrial fission | [107] |
| Drp1 deletion hetero | Similar to WT | |||
| Cardiac Drp1 deletion (early postnatal) | Lethal | Enlarged, heterogeneity | [109] | |
| Drp1 deletion (muscle-specific) | Lethal, Dilated heart | [110] | ||
| Cardiac Drp1 KO (postnatal) | Lethal, Dilated heart | - | [111] | |
| Cardiac Drp1 KO using tamoxifen (ad 8 weeks) | DCM, Fibrosis | Enlarged | ||
| Cardiac Drp1 KO using tamoxifen (ad 15 weeks) | Hypertrophy, Fibrosis | Elongated | [108] | |
| Cardiac Drp1 KO using tamoxifen (ad 8 weeks) | DCM | Enlarged | [112] | |
| Overexpression of Drp1 (Transgenic mice) | Not deleterious | Fragmented | [113] | |
| Mfn1/2 | Mfn1/2 KO | Embryonically lethal | - | [124] |
| Cardiac Mfn1/2 KO (mid-gestational and postnatal) | Normal at birth Lethal DCM | Spherical, heterogeneity | [114] | |
| Cardiac Mfn1/2 KO (embryonic) | Cardiac development failure | Fragmented | [125] | |
| Cardiac Mfn1/2 KO using tamoxifen (early embryonic) | Lethal | Fragmented | [115] | |
| Cardiac Mfn1/2 KO using tamoxifen (within 8 weeks) | Lethal, DCM | |||
| Cardiac Mfn1/2 KO (8 weeks–10 weeks) | Impaired myocardial function | Fragmented | [116] | |
| Marf (Drosophila) | DCM | Spherical, Fragmented, heterogeneity | [121] | |
| Mfn1 | Cardiac Mfn1 deletion | Normal | Spherical, small | [117,118] |
| Cardiac Mfn1 deletion | Normal | Large | [126] | |
| Mfn2 | Cardiac Mfn2 deletion | DCM | Enlarged | [118,119] |
| Cardiac Mfn2 deletion | Normal | Heterogeneity | [126] | |
| Drp1/Mfn1/Mfn2 | Cardiac triple knockout Mfn1/Mfn2/Drp1 | HCM | Fragmented | [113] |
| Mff | Mff deletion | DCM | No changes, heterogeneity | [122] |
| Opa1 | Opa1 KO | Embryonically lethal | - | [127] |
| Heterozygous Opa1 KO | Normal | Enlarged | [123] |
Abbreviations: DCM—Dilated cardiomyopathy; HCM—Hypertrophic cardiomyopathy.
5. Cardiomyopathy
Several mitochondrial morphology factors have been implicated in the complex relationship between mitochondrial dynamics and cardiomyopathy, heart failure, and reperfusion injury. Here, we focus on five key molecules—Drp1, Mfn1/2, Mff and Opa1.
5.1. Drp1
Mitochondrial dynamics between fission and fusion are finely regulated in response to nutrient availability and metabolic demands. C57BL/6 mice develop hyperlipidemia and hyperglycemia when exposed to a high-fat diet. This condition is caused by activation of Drp1 at serine 616, which leads to myocardial insulin resistance, reduces contractile efficiency, and ultimately cardiomyocyte death [128]. The accumulation of lipids results in an increase in mitochondrial ROS and affects the activity of Drp1. This is characterized by a decrease in phosphorylation at serine 637, along with an increase in phosphorylation at serine 616 [129]. In addition, hyperglycemia causes Drp1-mediated mitochondrial fragmentation, resulting in increased ROS production, which adversely affects mitochondrial energy production [130]. Hyperglycemia activates Drp1 phosphorylation at serine 616 and induces mitochondrial fission through Ca2+-mediated ERK1/2 signaling in cardiac myoblast cells [131]. In cardiomyocytes from Zucker diabetic rats, the expression of Opa1 and Mfn2 is decreased, and the phosphorylation of Drp1 at serine 637 is reduced. Conversely, Drp1 phosphorylation at serine 616 is increased, resulting in cardiomyocyte hypertrophy with abnormal mitochondrial dynamics and calcium handling due to activation of the ORAI calcium release-activated calcium modulator 1 (Orai1) calcium channel [132]. In neonatal rat cardiomyocytes, hyperglycemia downregulates Opa1 and Mfn1 but upregulates Drp1 and Mfn2, affecting mitochondrial potential and increasing apoptosis [133]. In human cardiomyocytes, diabetes-associated glycation end products stimulate ERK1/2 and O-linked-N-acetylglucosamine glycosylation [134]. This glycosylation modulates Opa1 expression and Drp1 phosphorylation, which promotes mitochondrial fragmentation in diabetic mouse cardiomyocytes [27,135]. In cardiomyocytes, Drp1-regulated mitochondrial fragmentation is associated with insulin resistance. Drp1 knockdown in H9C2 cells attenuates H2O2-induced mitochondrial dysfunction [136]. In addition, lipotoxic cardiomyopathy, exacerbated by fatty acid overload, is associated with insulin resistance through ceramide accumulation and upregulation of Drp1 and Mff [137]. In advanced stages of dilated cardiomyopathy, the association with abnormal mitochondrial fragmentation is significant, highlighting the important role of mitochondrial dynamics in the pathological state of the heart [138]. Sepsis reduces myocardial mitochondrial respiration and membrane potential. In addition, the interaction between Drp1 and Fis1 induces ROS production and excessive mitochondrial fragmentation [139].
5.2. Mfn1/2
Mitofusins, Mfn1 and Mfn2, are GTPase proteins essential for mediating fusion of the OMM. The prognosis for severe heart failure remains poor. Some heart failure patients do not respond to established multidisciplinary therapy and are classified as “non-responders”. Mfn1 is significantly reduced in non-responders [140]. In patients with diabetic cardiomyopathy, mitochondria in the heart become smaller. This morphological change is associated with decreased hemoglobin A1C levels and associated Mfn1 expression, suggesting that hyperglycemia promotes mitochondrial remodeling [141].
5.3. Mff
Mff acts as a receptor for Drp1 on the OMM, facilitating the fission process. Its role is highlighted in lipotoxic cardiomyopathy, where fatty acid overload coupled with ceramide accumulation increases the expression of both Drp1 and Mff, perpetuating mitochondrial dysfunction [137].
5.4. Opa1
Opa1 is critical for fusion of the inner mitochondrial membrane. Cardiomyocytes from Zucker diabetic rats show reduced Opa1 expression, which affects hypertrophy and calcium handling via the Orai1 channel [132]. Hyperglycemia decreases Opa1 in neonatal rat cardiomyocytes, reducing mitochondrial potential and increasing apoptosis [133]. Glycation end products generated in human cardiomyocytes increase Opa1 expression and cause mitochondrial fragmentation in diabetic mouse cardiomyocyte models [27,135]. In addition, doxorubicin treatment in FVB/N mice increases Opa1 expression and affects mitochondrial function [142].
6. Heart Failure
6.1. Drp1 and Mfn2
Cardiac-specific overexpression of miR-122, elevated in heart failure patients, induces mitochondria-dependent cardiomyocyte apoptosis and accelerates heart failure through the activation of Drp1 by inhibiting Hand2 [143]. Drp1 plays a pivotal role in the induction of mitophagy. Drp1-mediated mitochondrial fission is crucial for the onset of mitophagy in the heart [109,144]. The Drp1 C452F mutation in mice exhibits increased Drp1 GTPase activity as well as confers resistance to oligomer degradation, ultimately leading to impaired mitophagy, mitochondrial depolarization, abnormal calcium handling, impaired ATP synthesis, and activation of sterile myocardial inflammation [145]. In mice with heart failure with transverse aortic constriction (TAC), mitochondrial autophagy associated with Drp1 is transiently activated and subsequently downregulated in the mouse heart in response to pressure overload [146]. In a rat heart failure model, cardiomyocytes demonstrate augmented production of reactive oxygen species (ROS) from mitochondria. Notably, the expression levels of Mfn2 and Drp1 are diminished by approximately 50%. Such downregulation leads to an accumulation of mitochondrial Parkin and subsequent induction of mitophagy. This observed regulation of mitochondrial dynamics and mitophagy, mediated through Mfn2 and Drp1, seems to play a pivotal role in cardioprotection, possibly via modulation of ketone body dynamics [147]. HFrEF patient exhibits increased Drp1 levels. Clinically, Mfn2 levels are stable in HFrEF patient samples [148].
6.2. Mfn2
Xbp1 expression, a sarcoplasmic reticulum stress-responsive transcription factor, enhances the stress-responsive capacity of the sarcoplasmic reticulum and rescues cardiomyopathy caused by mitofusin/MARF deficiency without ameliorating cardiomyopathy caused by Opa1 deletion [149]. Mitochondrial health in cardiomyocytes is maintained not only by mitochondrial dynamics but also by mitophagy (selective isolation of damaged mitochondria by autophagy). Mitophagy in cardiomyocytes is mediated by Parkin and PTEN-induced kinase 1 (PINK1). PINK1 is stabilized in depolarized mitochondria, allowing its accumulation and facilitating the recruitment of parkin from the cytosol to depolarized mitochondria by Mfn2, an important mediator of mitophagy through the PINK1-Mfn2-perkin signaling pathway [150]. PINK1 phosphorylates Mfn2, and Mfn2 activates parkin [151,152]. Parkin then ubiquitinates Mfn2 [153]. P62 interacts with ubiquitinated substrates marked by Parkin, connecting them to LC3 [112]. Following the elongation of the isolation membrane, mitochondria become encased by this extended membrane, culminating in the formation of an autophagosome. This leads to the subsequent degradation of the ensnared mitochondria [112,144].
6.3. Opa1
Cardiomyocyte-specific Yme1L deficient mice exhibit reduced Opa1 levels via activation of Oma1, leading to a shift in cardiac metabolism and, ultimately, heart failure [81]. Ischemic conditions lead to a reduction in Opa1 expression both in vivo and in vitro [154]. In the rat hearts followed for 12 to 18 weeks after myocardial infarction, Mfn2 was reduced, and Fis1 was increased, but Opa1 expression was unchanged [155]. Furthermore, Opa1 mutant heart tissues show increased ROS levels and mitochondrial dysfunction [156]. Ventricular hypertrophy model mice have been shown to have increased expression of Drp1 and reduced expression of Opa1. This indicates that modulating the optimal balance of mitochondrial dynamics can improve mitochondrial function and delay the onset of right ventricular dysfunction [157].
7. Ischemia-Reperfusion Injury and Cardioprotection
7.1. Drp1
Dynamin-related protein 1 (Drp1) plays a crucial role as an inducer of mitochondrial fragmentation in the detrimental effects of cardiomyocytes after ischemia-reperfusion (I/R) injury. In I/R injury, Drp1 translocates into the OMM and causes mitochondrial fragmentation [158]. Mitochondrial fission inhibitor-1 (mdivi-1), a specific inhibitor of Drp1, administered prior to I/R, inhibits mitochondrial fragmentation, prevents the opening of mitochondrial permeability transition pore, and showed a reduction in cell death and infarct size in a mouse I/R model [93]. Under I/R conditions, both mitochondrial calcium overload and oxidative stress promote mitochondrial fragmentation [159]. After reperfusion, elevated Ca2+ strongly regulates Drp1 activity. Notably, miR-499 attenuates the effects of Ca2+ overload on Drp1 function [32]. Drp1 has five phosphorylation sites located at serines 585, 616, 637, 656, and 693. Of these, serines 616, 637, and 656 are especially involved in the regulation of Drp1 activation and inactivation under I/R conditions [159] [24,30,160,161,162]. Under conditions of oxidative stress, Drp1 is phosphorylated at Ser 579 by PKCδ, leading to its translocation to the mitochondria [163].
7.2. Mfn1 and Mfn2
Mitofusin 1 and 2 (Mfn1/2) are elongation factors of mitochondrial dynamics, and their expression levels are strongly associated with the pathophysiology of I/R injury. Mitochondrial fragmentation induced by I/R also results from decreased expression of Mfn1, Mfn2, or Opa1, leading to impaired respiratory function. Consequently, it promotes I/R-induced cardiomyocyte apoptosis [164,165,166]. I/R stress increases miR-140, inhibits Mfn1 expression in cardiomyocytes, blocks mitochondrial networks, and exacerbates apoptosis in cardiomyocytes [167]. In vitro experiments with cardiomyocytes, hypoxia upregulates Mfn2 expression [168]. In other in vivo experiments, heart-specific Mfn2 KO Mice exhibit a large reduction in mitochondrial membrane potential and significantly reduced viability under I/R conditions [169]. Additionally, heart-specific Mfn1/Mfn2 double KO Mice exhibit a smaller myocardial infarction size in response to I/R [170]. Overexpression of Mfn1 significantly improves microvascular function under hypoxic conditions, and overexpression of Mfn2 protects cardiomyocytes from I/R injury [171,172].
7.3. Opa1
Opa1 has been identified as playing a protective role in cardiomyocytes following I/R injury. A notable downregulation of Opa1 expression is observed in cardiomyocytes subjected to I/R injury in vivo, as well as in hypoxia-treated cardiomyocytes in vitro [154]. Opa1 is reduced in rat heart failure models after MI and in tissue samples of human dilated and ischemic cardiomyopathy [173]. Opa1 heterozygous KO mice display an increased propensity for cardiomyocyte death under I/R conditions, resulting in a larger infarct size post-I/R exposure than their wild-type counterparts [174]. Opa1 overexpression has been linked to the induction of mitophagy, presenting a potential therapeutic avenue for improving cardiomyocyte damage and viability under hypoxic conditions [154].
8. Therapeutic Targeting of Mitochondrial Fusion and Fission Proteins
This section summarizes the therapeutic potential for mitochondrial morphological abnormalities. Numerous investigations have explored various agents targeting each mitochondrial regulatory factor (Table 2). Although still in the preliminary stages of experimentation, continued research may further establish mitochondrial morphogenesis as a promising therapeutic target.
8.1. Drp1
Melatonin inactivates I/R injury-induced phosphorylation of Drp1 at serine 616. Long-term melatonin treatment attenuates the dilated cardiomyopathy progression and reduces myocardial vulnerability to I/R injury by maintaining mitochondrial quality control. Melatonin membrane receptors are associated with regulating the SIRT6-AMPK-PGC-1α-AKT axis in this effect [175]. Hydralazine reduces myocardial infarct size in cardiac I/R injury by inactivating Drp1 [176]. However, excessive inhibition of mitochondrial fragmentation, as observed with high doses of mdivi-1, which inhibits Drp1 activation, may exacerbate myocardial injury through inhibition of mitophagy and accumulation of abnormal mitochondria [108]. In the area of Drp1 inhibition, P110 and Dynasore emerge as promising therapeutic compound [177,178]. Donepezil inactivates Drp1 phosphorylation at serine 616, indicating its therapeutic potential after I/R injury [179]. Inhibition of Ca2+ entry into mitochondria via Orai1 reduces Drp1-mediated fission during hyperglycemia, providing a therapeutic hint for diabetic cardiomyopathy [132]. Klotho inhibits the phosphorylation of Drp1 at serine 616 and attenuates doxorubicin-related cardiomyopathy [180]. Sevoflurane protects the heart from I/R injury by increasing Drp1 and Parkin and stabilizes ATP levels by ameliorating mitochondrial damage in the rat heart [181]. For arterial calcification, Mdivi-1, melatonin and irisin inhibit arterial calcification by inhibiting mitochondrial fragmentation [182,183,184]. Quercetin, a polyphenol, also improves arterial calcification by inactivating Drp1 phosphorylation at serine 616 [185]. In hypertension, Mdivi-1 is shown to be effective [186,187,188]. As a treatment for pulmonary Hypertension (PAH), dichloroacetic acid, a pyruvate dehydrogenase kinase inhibitor, improves fibrosis and hypertrophy of the right ventricular in rats treated with monocrotaline. This therapeutic effect is mediated through the DNA methyltransferase-1/HIF-1α/PDK/Drp1 pathway [189]. Trimetazidine inhibits hypoxia-induced pulmonary artery smooth muscle cell (PASMC) proliferation by modulating Drp1 and Mfn2 [190]. Liraglutide, a glucagon-like peptide-1 receptor agonist, inhibits PASMC proliferation in PAH via inactivation of the Drp1/NADPH oxidase pathway and by LC3-dependent autophagy [191]. AT1R inhibition also reverses premature aging in VSMC exposed to ox-LDL and in the arteries of ApoE KO mice [192]. In a Huntington’s disease model, the Drp1 inhibitor P110 improves mitochondrial structure in the heart [193].
8.2. Mfn1 and Mfn2
SAMβA attenuates cell death through Mfn1 enhancement in ischemic heart failure [194]. In the Wistar rat I/R model, aerobic exercise increases Mfn1 and Mfn2 expression and improves infarct size [195]. Cordycepin also reduces infarct size in diabetic model mice by mitochondrial fusion through the AMPK/Mfn2 pathway [196]. The angiotensin II type I receptor inhibitor and adiponectin mitigate VSMC proliferation via Mfn2-mediated Ras/Raf/ERK signaling [197,198]. Donepezil activates Mfn2 and Opa1, inducing mitophagy, which results in the improvement of ROS production, mitochondrial dysfunction, and cardiac apoptosis under I/R conditions [179]. In hypertension, pomegranate extract mitigates oxidative stress and improves mitochondrial function, coinciding with reduced Mfn2 [199]. Resveratrol increases Mfn1 and Mfn2 expression and reduces oxidative stress damage in human umbilical vein endothelial cells [200]. Fish oil supplementation boosts Mfn2 and Opa1, preventing endothelial dysfunction in high-fat-fed ApoE KO mice [201]. Ferulic acid may restore Mfn1 and Mfn2 levels and attenuate oxidative stress in high-fat-fed ApoE KO mice [202]. In spontaneously hypertensive rat hearts, Drp1 is notably upregulated while Opa1 and Mfn2 decrease. Calhex231, a calcium sensing receptor inhibitor, counters these alterations in mitochondrial dynamics, subsequently reducing apoptosis in hypertensive hearts [203].
8.3. Opa1
Irisin administration induces Opa1-mediated mitophagy and protects myocardial cells from post-myocardial infarction damage [154]. Remote ischemic preconditioning enhances Opa1 expression, decreasing myocardial infarct size [166]. Melatonin activates the AMPK/Opa1 axis, promoting mitochondrial fusion and mitophagy, which ameliorates cardiomyocyte death and mitochondrial dysfunction after I/R injury [204]. Paeonol activates Opa1-mediated fusion through the CK2α-STAT3 pathway during diabetes, both in vitro and in vivo [205]. Sevoflurane postconditioning reduces Opa1 expression, protects against I/R injury, stabilizes ATP levels, and rescues mitochondrial damage in the rat heart [181]. Nicorandil suppresses fission and boosts fusion by downregulating Drp1 and upregulating Opa1 and Mfn1 in ischemic cardiomyopathy rats [206]. Coenzyme Q10 promotes mitochondrial function and energy metabolism by activating the AMPK/YAP/Opa1 pathway, attenuating atherosclerosis [207].

Table 2.
Targeting mitochondrial morphology factors for cardiac disease.
Table 2.
Targeting mitochondrial morphology factors for cardiac disease.
| Target Up (+) or Down (−) | Method | Model | Result | Reference | |
|---|---|---|---|---|---|
| Drp1 (−) | Melatonin | I/R | Diabetic rat with I/R injury | Decrease in MI size and apoptosis | [175] |
| Hydralazine | C57BL/6N with I/R injury | MI size reduction | [176] | ||
| P110 | Wistar rat, Ex vivo | Improvement of mitochondrial morphology and mitochondrial respiratory function. | [177] | ||
| Dynasore | C6/Black, Ex vivo | Increase in cardiomyocyte viability | [178] | ||
| BTP2 | Diabetic CM | Zucker diabetic fat | Improvement of cardiomyocyte hypertrophy | [132] | |
| Klotho | Dox-CM | C57BL/6 treated with doxorubicin | Inhibition of apoptosis | [180] | |
| Sevoflurane postconditioning | Ischemic HF | Sprague-Dawley rat with I/R injury | Improvement of ATP production with mitophagy | [181] | |
| Mdivi-1 | ATS | Human VSMC | Suppression of VSMC calcification | [182] | |
| Melatonin | Rat VSMC | Suppression of arterial calcification | [183] | ||
| Irisin | High-phosphorus-diet C57BL/6 | Suppression of arterial calcification | [184] | ||
| Quercetin | Adenine-rich diet rat | Suppression of arterial calcification | [185] | ||
| Mdivi-1 | HTN | C57BL/6 treated with AngII | Inhibition of AngII-mediated phenotypic switch | [186] | |
| Mdivi-1 | High-salt-fed rat | Reduction of cardiac hypertrophy and fibrosis | [187] | ||
| Mdivi-1 | Rat VSMC | Suppression of arterial calcification | [188] | ||
| Dichlorpacetate | PH | Monocrotaline-treated rat | Inhibition of right ventricular fibrosis and hypertrophy | [189] | |
| Liraglutide | Rat PASMC | Inhibition of cell proliferation | [191] | ||
| ARB | Aging | Human VSMC and ApoE KO mice | Reduction of hyperlipidemic aging | [192] | |
| P110 | Huntington Disease (HD) model mice | Reduction of pathological mitochondrial fission | [193] | ||
| Mfn1 (+) | SAMβA | Ischemic HF | Rat treated with AngII | Inhibition of apoptosis | [194] |
| Mfn2 (+) | Cordycepin | I/R | Diabetic mice with I/R injury | MI size reduction | [196] |
| ARB | ATS | Rat VSMC | Inhibition of cell proliferation | [197] | |
| Adiponectin | ATS | Human VSMC | Inhibition of cell proliferation | [198] | |
| Mfn2 (−) | Pomegranate | HTN | SHR | Reduction of oxidative stress | [199] |
| Mfn1 and Mfn2 (+) | Aerobic exercise | I/R | Wistar rat with I/R injury | MI size reduction | [195] |
| Ferulic acid | ATS | High-fat-fed ApoE KO mice and Human mononuclear cell | Reduction of oxidative stress | [202] | |
| Mfn1 and Opa1 (+) | Fish oil | ATS | High-fat-fed ApoE KO mice | Improvement of endothelial dysfunction | [201] |
| Mfn1/2 and Opa1 (+) | Resveratrol | ATS | HUVEC treated with palmitic acid | Improvement of cell viability and reduction of oxidative stress | [200] |
| Drp1 (−), Mfn2 (+) | Trimetazidine | PH | Human PASMC | Inhibition of hypoxia-induced cell proliferation | [190] |
| Drp1 (−), Mfn2 and Opa1 (+) | Donepezil | I/R | Wistar rat with I/R injury | Amelioration of apoptosis and mitochondrial dysfunction | [179] |
| Calhex231 | HTN | SHR | Inhibition of apoptosis | [203] | |
| Drp1 (−), Mfn1 and Opa1 (+) | Nicorandil | Ischemic HF | Rat with I/R injury | Increased mitochondrial ATP-sensitive potassium channel opening | [206] |
| Drp1 (−), Opa1 (+) | Sevoflurane postconditioning | Ischemic HF | Sprague-Dawley rat with I/R injury | Induction of mitophagy and improvement of myocardial ATP production | [181] |
| Opa1 (+) | RIPC | I/R | Wistar rat with I/R injury | MI size reduction | [166] |
| Irisin | Hypoxia-treated cardiomyocyte | Inhibition of apoptosis | [154] | ||
| Melatonin | C57BL/6 with I/R injury | Amelioration of apoptosis and mitochondrial dysfunction | [204] | ||
| Paeonol | Diabetic CM | Sprague-Dawley rat cardiomyocytes under high glucose condition | Improvement of cardiomyocyte hypertrophy and interstitial fibrosis | [205] | |
| Coenzyme Q10 | ATS | High-fat-fed ApoE KO mice | Inhibition of oxidative stress and promotion of energy metabolism | [207] | |
Abbreviations: +—upregulation; −—downregulation (inhibition); ARB—Angiotensin II type I receptor inhibitor; MI—myocardial infarct; HUVEC—human umbilical vein endothelial cell; RIPC—remote ischemic preconditioning; CM—cardiomyopathy; Dox-CM—doxorubicin-associated cardiomyopathy; ATS—Atherosclerosis; HTN—hypertension; HF—Heart failure; SHR—spontaneously hypertensive rat. The table was modified from Yoshihiro et al. [208].
9. Conclusions
Mitochondrial dynamics, including fission and fusion processes, play an essential role in heart health and disease. Recent research has highlighted the potential of targeting these dynamics to treat various cardiac diseases, including acute myocardial infarction, hypertrophy, PAH, ischemic diseases and heart failure. However, it is important to recognize the multifaceted role of mitochondrial morphology factors, particularly fusion proteins such as Mfn2 and Opa1. Acute and chronic modulation of these proteins may have therapeutic effects or unintended adverse consequences. Although most of the current knowledge is derived from transgenic animal models, early pharmacological interventions have yielded promising results. It is essential to extend this research to human samples and to improve the diagnostic and therapeutic toolkit by integrating biopsy analysis and indirect markers of mitochondrial dynamics. This would help to refine patient risk assessment and tailor treatment strategies for cardiovascular disease.
Furthermore, there is a perception that mitochondrial fusion is beneficial and fission is detrimental, but this binary view may be too simplistic. Fission plays a fundamental role in separating damaged mitochondria for removal by mitophagy, and excessive inhibition of this process may exacerbate myocardial injury. Thus, striking the right balance in the regulation of mitochondrial dynamics is critical for potential clinical applications.
Author Contributions
Conceptualization, T.T. and S.Y.; writing—original draft preparation, T.T.; writing—review and editing, T.T. and S.Y.; visualization, T.T.; supervision, T.T. and S.Y.; funding acquisition, T.T. and S.Y. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the Japan Society for the Promotion of Science KAKENHI (Grant Nos. 22K15397 and 23H02691).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Suen, D.F.; Norris, K.L.; Youle, R.J. Mitochondrial dynamics and apoptosis. Genes Dev. 2008, 22, 1577–1590. [Google Scholar] [CrossRef] [PubMed]
- Twig, G.; Elorza, A.; Molina, A.J.; Mohamed, H.; Wikstrom, J.D.; Walzer, G.; Stiles, L.; Haigh, S.E.; Katz, S.; Las, G.; et al. Fission and selective fusion govern mitochondrial segregation and elimination by autophagy. EMBO J. 2008, 27, 433–446. [Google Scholar] [CrossRef]
- Kleele, T.; Rey, T.; Winter, J.; Zaganelli, S.; Mahecic, D.; Perreten Lambert, H.; Ruberto, F.P.; Nemir, M.; Wai, T.; Pedrazzini, T.; et al. Distinct fission signatures predict mitochondrial degradation or biogenesis. Nature 2021, 593, 435–439. [Google Scholar] [CrossRef] [PubMed]
- Moore, A.S.; Coscia, S.M.; Simpson, C.L.; Ortega, F.E.; Wait, E.C.; Heddleston, J.M.; Nirschl, J.J.; Obara, C.J.; Guedes-Dias, P.; Boecker, C.A.; et al. Actin cables and comet tails organize mitochondrial networks in mitosis. Nature 2021, 591, 659–664. [Google Scholar] [CrossRef]
- Tabara, L.C.; Prudent, J. The last wall of defense to prevent extreme and deleterious mitochondrial fusion. EMBO J. 2020, 39, e107326. [Google Scholar] [CrossRef]
- Lee, J.E.; Westrate, L.M.; Wu, H.; Page, C.; Voeltz, G.K. Multiple dynamin family members collaborate to drive mitochondrial division. Nature 2016, 540, 139–143. [Google Scholar] [CrossRef]
- Fonseca, T.B.; Sanchez-Guerrero, A.; Milosevic, I.; Raimundo, N. Mitochondrial fission requires DRP1 but not dynamins. Nature 2019, 570, E34–E42. [Google Scholar] [CrossRef] [PubMed]
- Kraus, F.; Roy, K.; Pucadyil, T.J.; Ryan, M.T. Function and regulation of the divisome for mitochondrial fission. Nature 2021, 590, 57–66. [Google Scholar] [CrossRef]
- Kamerkar, S.C.; Kraus, F.; Sharpe, A.J.; Pucadyil, T.J.; Ryan, M.T. Dynamin-related protein 1 has membrane constricting and severing abilities sufficient for mitochondrial and peroxisomal fission. Nat. Commun. 2018, 9, 5239. [Google Scholar] [CrossRef]
- Ingerman, E.; Perkins, E.M.; Marino, M.; Mears, J.A.; McCaffery, J.M.; Hinshaw, J.E.; Nunnari, J. Dnm1 forms spirals that are structurally tailored to fit mitochondria. J. Cell Biol. 2005, 170, 1021–1027. [Google Scholar] [CrossRef]
- Legesse-Miller, A.; Massol, R.H.; Kirchhausen, T. Constriction and Dnm1p recruitment are distinct processes in mitochondrial fission. Mol. Biol. Cell 2003, 14, 1953–1963. [Google Scholar] [CrossRef] [PubMed]
- Smirnova, E.; Shurland, D.L.; Ryazantsev, S.N.; van der Bliek, A.M. A human dynamin-related protein controls the distribution of mitochondria. J. Cell Biol. 1998, 143, 351–358. [Google Scholar] [CrossRef] [PubMed]
- Smirnova, E.; Griparic, L.; Shurland, D.L.; van der Bliek, A.M. Dynamin-related protein Drp1 is required for mitochondrial division in mammalian cells. Mol. Biol. Cell 2001, 12, 2245–2256. [Google Scholar] [CrossRef]
- van der Bliek, A.M.; Shen, Q.; Kawajiri, S. Mechanisms of mitochondrial fission and fusion. Cold Spring Harb. Perspect. Biol. 2013, 5, a011072. [Google Scholar] [CrossRef] [PubMed]
- Mears, J.A.; Lackner, L.L.; Fang, S.; Ingerman, E.; Nunnari, J.; Hinshaw, J.E. Conformational changes in Dnm1 support a contractile mechanism for mitochondrial fission. Nat. Struct. Mol. Biol. 2011, 18, 20–26. [Google Scholar] [CrossRef]
- Manor, U.; Bartholomew, S.; Golani, G.; Christenson, E.; Kozlov, M.; Higgs, H.; Spudich, J.; Lippincott-Schwartz, J. A mitochondria-anchored isoform of the actin-nucleating spire protein regulates mitochondrial division. eLife 2015, 4, 08828. [Google Scholar] [CrossRef]
- Korobova, F.; Ramabhadran, V.; Higgs, H.N. An actin-dependent step in mitochondrial fission mediated by the ER-associated formin INF2. Science 2013, 339, 464–467. [Google Scholar] [CrossRef] [PubMed]
- Moore, A.S.; Wong, Y.C.; Simpson, C.L.; Holzbaur, E.L. Dynamic actin cycling through mitochondrial subpopulations locally regulates the fission-fusion balance within mitochondrial networks. Nat. Commun. 2016, 7, 12886. [Google Scholar] [CrossRef] [PubMed]
- Korobova, F.; Gauvin, T.J.; Higgs, H.N. A role for myosin II in mammalian mitochondrial fission. Curr. Biol. 2014, 24, 409–414. [Google Scholar] [CrossRef] [PubMed]
- Mahecic, D.; Carlini, L.; Kleele, T.; Colom, A.; Goujon, A.; Matile, S.; Roux, A.; Manley, S. Mitochondrial membrane tension governs fission. Cell. Rep. 2021, 35, 108947. [Google Scholar] [CrossRef]
- Ji, W.K.; Hatch, A.L.; Merrill, R.A.; Strack, S.; Higgs, H.N. Actin filaments target the oligomeric maturation of the dynamin GTPase Drp1 to mitochondrial fission sites. eLife 2015, 4, e11553. [Google Scholar] [CrossRef] [PubMed]
- Cho, S.G.; Du, Q.; Huang, S.; Dong, Z. Drp1 dephosphorylation in ATP depletion-induced mitochondrial injury and tubular cell apoptosis. Am. J. Physiol. Renal Physiol. 2010, 299, F199–F206. [Google Scholar] [CrossRef]
- Chang, C.R.; Blackstone, C. Cyclic AMP-dependent protein kinase phosphorylation of Drp1 regulates its GTPase activity and mitochondrial morphology. J. Biol. Chem. 2007, 282, 21583–21587. [Google Scholar] [CrossRef]
- Cribbs, J.T.; Strack, S. Reversible phosphorylation of Drp1 by cyclic AMP-dependent protein kinase and calcineurin regulates mitochondrial fission and cell death. EMBO Rep. 2007, 8, 939–944. [Google Scholar] [CrossRef]
- Nakamura, N.; Kimura, Y.; Tokuda, M.; Honda, S.; Hirose, S. MARCH-V is a novel mitofusin 2- and Drp1-binding protein able to change mitochondrial morphology. EMBO Rep. 2006, 7, 1019–1022. [Google Scholar] [CrossRef] [PubMed]
- Figueroa-Romero, C.; Iniguez-Lluhi, J.A.; Stadler, J.; Chang, C.R.; Arnoult, D.; Keller, P.J.; Hong, Y.; Blackstone, C.; Feldman, E.L. SUMOylation of the mitochondrial fission protein Drp1 occurs at multiple nonconsensus sites within the B domain and is linked to its activity cycle. FASEB J. 2009, 23, 3917–3927. [Google Scholar] [CrossRef] [PubMed]
- Gawlowski, T.; Suarez, J.; Scott, B.; Torres-Gonzalez, M.; Wang, H.; Schwappacher, R.; Han, X.; Yates, J.R., 3rd; Hoshijima, M.; Dillmann, W. Modulation of dynamin-related protein 1 (DRP1) function by increased O-linked-beta-N-acetylglucosamine modification (O-GlcNAc) in cardiac myocytes. J. Biol. Chem. 2012, 287, 30024–30034. [Google Scholar] [CrossRef]
- Cho, D.H.; Nakamura, T.; Fang, J.; Cieplak, P.; Godzik, A.; Gu, Z.; Lipton, S.A. S-nitrosylation of Drp1 mediates beta-amyloid-related mitochondrial fission and neuronal injury. Science 2009, 324, 102–105. [Google Scholar] [CrossRef] [PubMed]
- Liesa, M.; Palacin, M.; Zorzano, A. Mitochondrial dynamics in mammalian health and disease. Physiol. Rev. 2009, 89, 799–845. [Google Scholar] [CrossRef]
- Taguchi, N.; Ishihara, N.; Jofuku, A.; Oka, T.; Mihara, K. Mitotic phosphorylation of dynamin-related GTPase Drp1 participates in mitochondrial fission. J. Biol. Chem. 2007, 282, 11521–11529. [Google Scholar] [CrossRef]
- Li, J.; Donath, S.; Li, Y.; Qin, D.; Prabhakar, B.S.; Li, P. miR-30 regulates mitochondrial fission through targeting p53 and the dynamin-related protein-1 pathway. PLoS Genet. 2010, 6, e1000795. [Google Scholar] [CrossRef]
- Wang, J.X.; Jiao, J.Q.; Li, Q.; Long, B.; Wang, K.; Liu, J.P.; Li, Y.R.; Li, P.F. miR-499 regulates mitochondrial dynamics by targeting calcineurin and dynamin-related protein-1. Nat. Med. 2011, 17, 71–78. [Google Scholar] [CrossRef] [PubMed]
- Lutz, A.K.; Exner, N.; Fett, M.E.; Schlehe, J.S.; Kloos, K.; Lammermann, K.; Brunner, B.; Kurz-Drexler, A.; Vogel, F.; Reichert, A.S.; et al. Loss of parkin or PINK1 function increases Drp1-dependent mitochondrial fragmentation. J. Biol. Chem. 2009, 284, 22938–22951. [Google Scholar] [CrossRef]
- Yonashiro, R.; Ishido, S.; Kyo, S.; Fukuda, T.; Goto, E.; Matsuki, Y.; Ohmura-Hoshino, M.; Sada, K.; Hotta, H.; Yamamura, H.; et al. A novel mitochondrial ubiquitin ligase plays a critical role in mitochondrial dynamics. EMBO J. 2006, 25, 3618–3626. [Google Scholar] [CrossRef] [PubMed]
- Karbowski, M.; Neutzner, A.; Youle, R.J. The mitochondrial E3 ubiquitin ligase MARCH5 is required for Drp1 dependent mitochondrial division. J. Cell Biol. 2007, 178, 71–84. [Google Scholar] [CrossRef] [PubMed]
- Tokuyama, T.; Uosaki, H.; Sugiura, A.; Nishitai, G.; Takeda, K.; Nagashima, S.; Shiiba, I.; Ito, N.; Amo, T.; Mohri, S.; et al. Protective roles of MITOL against myocardial senescence and ischemic injury partly via Drp1 regulation. iScience 2022, 25, 104582. [Google Scholar] [CrossRef]
- Gandre-Babbe, S.; van der Bliek, A.M. The novel tail-anchored membrane protein Mff controls mitochondrial and peroxisomal fission in mammalian cells. Mol. Biol. Cell 2008, 19, 2402–2412. [Google Scholar] [CrossRef]
- Otera, H.; Wang, C.; Cleland, M.M.; Setoguchi, K.; Yokota, S.; Youle, R.J.; Mihara, K. Mff is an essential factor for mitochondrial recruitment of Drp1 during mitochondrial fission in mammalian cells. J. Cell Biol. 2010, 191, 1141–1158. [Google Scholar] [CrossRef] [PubMed]
- Palmer, C.S.; Elgass, K.D.; Parton, R.G.; Osellame, L.D.; Stojanovski, D.; Ryan, M.T. Adaptor proteins MiD49 and MiD51 can act independently of Mff and Fis1 in Drp1 recruitment and are specific for mitochondrial fission. J. Biol. Chem. 2013, 288, 27584–27593. [Google Scholar] [CrossRef]
- James, D.I.; Parone, P.A.; Mattenberger, Y.; Martinou, J.C. hFis1, a novel component of the mammalian mitochondrial fission machinery. J. Biol. Chem. 2003, 278, 36373–36379. [Google Scholar] [CrossRef]
- Stojanovski, D.; Koutsopoulos, O.S.; Okamoto, K.; Ryan, M.T. Levels of human Fis1 at the mitochondrial outer membrane regulate mitochondrial morphology. J. Cell Sci. 2004, 117, 1201–1210. [Google Scholar] [CrossRef] [PubMed]
- Yoon, Y.; Krueger, E.W.; Oswald, B.J.; McNiven, M.A. The mitochondrial protein hFis1 regulates mitochondrial fission in mammalian cells through an interaction with the dynamin-like protein DLP1. Mol. Cell Biol. 2003, 23, 5409–5420. [Google Scholar] [CrossRef]
- Yamano, K.; Fogel, A.I.; Wang, C.; van der Bliek, A.M.; Youle, R.J. Mitochondrial Rab GAPs govern autophagosome biogenesis during mitophagy. eLife 2014, 3, e01612. [Google Scholar] [CrossRef]
- Gomes, L.C.; Scorrano, L. High levels of Fis1, a pro-fission mitochondrial protein, trigger autophagy. Biochim. Biophys. Acta 2008, 1777, 860–866. [Google Scholar] [CrossRef] [PubMed]
- Shen, Q.; Yamano, K.; Head, B.P.; Kawajiri, S.; Cheung, J.T.; Wang, C.; Cho, J.H.; Hattori, N.; Youle, R.J.; van der Bliek, A.M. Mutations in Fis1 disrupt orderly disposal of defective mitochondria. Mol. Biol. Cell 2014, 25, 145–159. [Google Scholar] [CrossRef]
- Loson, O.C.; Liu, R.; Rome, M.E.; Meng, S.; Kaiser, J.T.; Shan, S.O.; Chan, D.C. The mitochondrial fission receptor MiD51 requires ADP as a cofactor. Structure 2014, 22, 367–377. [Google Scholar] [CrossRef]
- Koirala, S.; Guo, Q.; Kalia, R.; Bui, H.T.; Eckert, D.M.; Frost, A.; Shaw, J.M. Interchangeable adaptors regulate mitochondrial dynamin assembly for membrane scission. Proc. Natl. Acad. Sci. USA 2013, 110, E1342–E1351. [Google Scholar] [CrossRef] [PubMed]
- Alirol, E.; James, D.; Huber, D.; Marchetto, A.; Vergani, L.; Martinou, J.C.; Scorrano, L. The mitochondrial fission protein hFis1 requires the endoplasmic reticulum gateway to induce apoptosis. Mol. Biol. Cell 2006, 17, 4593–4605. [Google Scholar] [CrossRef]
- Yu, R.; Jin, S.B.; Lendahl, U.; Nister, M.; Zhao, J. Human Fis1 regulates mitochondrial dynamics through inhibition of the fusion machinery. EMBO J. 2019, 38, e99748. [Google Scholar] [CrossRef]
- Xian, H.; Yang, Q.; Xiao, L.; Shen, H.M.; Liou, Y.C. STX17 dynamically regulated by Fis1 induces mitophagy via hierarchical macroautophagic mechanism. Nat. Commun. 2019, 10, 2059. [Google Scholar] [CrossRef]
- Boutry, M.; Kim, P.K. ORP1L mediated PI(4)P signaling at ER-lysosome-mitochondrion three-way contact contributes to mitochondrial division. Nat. Commun. 2021, 12, 5354. [Google Scholar] [CrossRef] [PubMed]
- Wong, Y.C.; Ysselstein, D.; Krainc, D. Mitochondria-lysosome contacts regulate mitochondrial fission via RAB7 GTP hydrolysis. Nature 2018, 554, 382–386. [Google Scholar] [CrossRef]
- Toyama, E.Q.; Herzig, S.; Courchet, J.; Lewis, T.L., Jr.; Loson, O.C.; Hellberg, K.; Young, N.P.; Chen, H.; Polleux, F.; Chan, D.C.; et al. Metabolism. AMP-activated protein kinase mediates mitochondrial fission in response to energy stress. Science 2016, 351, 275–281. [Google Scholar] [CrossRef]
- Schrader, M.; Costello, J.L.; Godinho, L.F.; Azadi, A.S.; Islinger, M. Proliferation and fission of peroxisomes—An update. Biochim. Biophys. Acta 2016, 1863, 971–983. [Google Scholar] [CrossRef] [PubMed]
- Elgass, K.D.; Smith, E.A.; LeGros, M.A.; Larabell, C.A.; Ryan, M.T. Analysis of ER-mitochondria contacts using correlative fluorescence microscopy and soft X-ray tomography of mammalian cells. J. Cell Sci. 2015, 128, 2795–2804. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Liu, T.; Jin, S.; Wang, X.; Qu, M.; Uhlen, P.; Tomilin, N.; Shupliakov, O.; Lendahl, U.; Nister, M. Human MIEF1 recruits Drp1 to mitochondrial outer membranes and promotes mitochondrial fusion rather than fission. EMBO J. 2011, 30, 2762–2778. [Google Scholar] [CrossRef] [PubMed]
- Kalia, R.; Wang, R.Y.; Yusuf, A.; Thomas, P.V.; Agard, D.A.; Shaw, J.M.; Frost, A. Structural basis of mitochondrial receptor binding and constriction by DRP1. Nature 2018, 558, 401–405. [Google Scholar] [CrossRef]
- Koshiba, T.; Detmer, S.A.; Kaiser, J.T.; Chen, H.; McCaffery, J.M.; Chan, D.C. Structural basis of mitochondrial tethering by mitofusin complexes. Science 2004, 305, 858–862. [Google Scholar] [CrossRef] [PubMed]
- Song, Z.; Ghochani, M.; McCaffery, J.M.; Frey, T.G.; Chan, D.C. Mitofusins and OPA1 mediate sequential steps in mitochondrial membrane fusion. Mol. Biol. Cell 2009, 20, 3525–3532. [Google Scholar] [CrossRef]
- Ishihara, N.; Eura, Y.; Mihara, K. Mitofusin 1 and 2 play distinct roles in mitochondrial fusion reactions via GTPase activity. J. Cell Sci. 2004, 117, 6535–6546. [Google Scholar] [CrossRef] [PubMed]
- Nunnari, J.; Marshall, W.F.; Straight, A.; Murray, A.; Sedat, J.W.; Walter, P. Mitochondrial transmission during mating in Saccharomyces cerevisiae is determined by mitochondrial fusion and fission and the intramitochondrial segregation of mitochondrial DNA. Mol. Biol. Cell 1997, 8, 1233–1242. [Google Scholar] [CrossRef]
- Jakobs, S.; Martini, N.; Schauss, A.C.; Egner, A.; Westermann, B.; Hell, S.W. Spatial and temporal dynamics of budding yeast mitochondria lacking the division component Fis1p. J. Cell Sci. 2003, 116, 2005–2014. [Google Scholar] [CrossRef] [PubMed]
- Eura, Y.; Ishihara, N.; Yokota, S.; Mihara, K. Two mitofusin proteins, mammalian homologues of FZO, with distinct functions are both required for mitochondrial fusion. J. Biochem. 2003, 134, 333–344. [Google Scholar] [CrossRef] [PubMed]
- Detmer, S.A.; Chan, D.C. Complementation between mouse Mfn1 and Mfn2 protects mitochondrial fusion defects caused by CMT2A disease mutations. J. Cell Biol. 2007, 176, 405–414. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Detmer, S.A.; Ewald, A.J.; Griffin, E.E.; Fraser, S.E.; Chan, D.C. Mitofusins Mfn1 and Mfn2 coordinately regulate mitochondrial fusion and are essential for embryonic development. J. Cell Biol. 2003, 160, 189–200. [Google Scholar] [CrossRef]
- de Brito, O.M.; Scorrano, L. Mitofusin 2 tethers endoplasmic reticulum to mitochondria. Nature 2008, 456, 605–610. [Google Scholar] [CrossRef]
- Chen, Y.; Csordas, G.; Jowdy, C.; Schneider, T.G.; Csordas, N.; Wang, W.; Liu, Y.; Kohlhaas, M.; Meiser, M.; Bergem, S.; et al. Mitofusin 2-containing mitochondrial-reticular microdomains direct rapid cardiomyocyte bioenergetic responses via interorganelle Ca2+ crosstalk. Circ. Res. 2012, 111, 863–875. [Google Scholar] [CrossRef] [PubMed]
- Liesa, M.; Borda-d’Agua, B.; Medina-Gomez, G.; Lelliott, C.J.; Paz, J.C.; Rojo, M.; Palacin, M.; Vidal-Puig, A.; Zorzano, A. Mitochondrial fusion is increased by the nuclear coactivator PGC-1beta. PLoS ONE 2008, 3, e3613. [Google Scholar] [CrossRef]
- Martorell-Riera, A.; Segarra-Mondejar, M.; Munoz, J.P.; Ginet, V.; Olloquequi, J.; Perez-Clausell, J.; Palacin, M.; Reina, M.; Puyal, J.; Zorzano, A.; et al. Mfn2 downregulation in excitotoxicity causes mitochondrial dysfunction and delayed neuronal death. EMBO J. 2014, 33, 2388–2407. [Google Scholar] [CrossRef]
- Wu, H.; Li, Z.; Wang, Y.; Yang, P.; Li, Z.; Li, H.; Wu, C. MiR-106b-mediated Mfn2 suppression is critical for PKM2 induced mitochondrial fusion. Am. J. Cancer Res. 2016, 6, 2221–2234. [Google Scholar]
- Bucha, S.; Mukhopadhyay, D.; Bhattacharyya, N.P. Regulation of mitochondrial morphology and cell cycle by microRNA-214 targeting Mitofusin2. Biochem. Biophys. Res. Commun. 2015, 465, 797–802. [Google Scholar] [CrossRef] [PubMed]
- Mattie, S.; Riemer, J.; Wideman, J.G.; McBride, H.M. A new mitofusin topology places the redox-regulated C terminus in the mitochondrial intermembrane space. J. Cell Biol. 2018, 217, 507–515. [Google Scholar] [CrossRef]
- Park, Y.Y.; Nguyen, O.T.; Kang, H.; Cho, H. MARCH5-mediated quality control on acetylated Mfn1 facilitates mitochondrial homeostasis and cell survival. Cell Death Dis. 2014, 5, e1172. [Google Scholar] [CrossRef]
- Lee, J.Y.; Kapur, M.; Li, M.; Choi, M.C.; Choi, S.; Kim, H.J.; Kim, I.; Lee, E.; Taylor, J.P.; Yao, T.P. MFN1 deacetylation activates adaptive mitochondrial fusion and protects metabolically challenged mitochondria. J. Cell Sci. 2014, 127, 4954–4963. [Google Scholar] [CrossRef] [PubMed]
- Leboucher, G.P.; Tsai, Y.C.; Yang, M.; Shaw, K.C.; Zhou, M.; Veenstra, T.D.; Glickman, M.H.; Weissman, A.M. Stress-induced phosphorylation and proteasomal degradation of mitofusin 2 facilitates mitochondrial fragmentation and apoptosis. Mol. Cell 2012, 47, 547–557. [Google Scholar] [CrossRef] [PubMed]
- Pyakurel, A.; Savoia, C.; Hess, D.; Scorrano, L. Extracellular regulated kinase phosphorylates mitofusin 1 to control mitochondrial morphology and apoptosis. Mol. Cell 2015, 58, 244–254. [Google Scholar] [CrossRef] [PubMed]
- Meeusen, S.; DeVay, R.; Block, J.; Cassidy-Stone, A.; Wayson, S.; McCaffery, J.M.; Nunnari, J. Mitochondrial inner-membrane fusion and crista maintenance requires the dynamin-related GTPase Mgm1. Cell 2006, 127, 383–395. [Google Scholar] [CrossRef]
- Sprenger, H.G.; Langer, T. The Good and the Bad of Mitochondrial Breakups. Trends Cell Biol. 2019, 29, 888–900. [Google Scholar] [CrossRef]
- Olichon, A.; Elachouri, G.; Baricault, L.; Delettre, C.; Belenguer, P.; Lenaers, G. OPA1 alternate splicing uncouples an evolutionary conserved function in mitochondrial fusion from a vertebrate restricted function in apoptosis. Cell Death Differ. 2007, 14, 682–692. [Google Scholar] [CrossRef]
- Delettre, C.; Lenaers, G.; Griffoin, J.M.; Gigarel, N.; Lorenzo, C.; Belenguer, P.; Pelloquin, L.; Grosgeorge, J.; Turc-Carel, C.; Perret, E.; et al. Nuclear gene OPA1, encoding a mitochondrial dynamin-related protein, is mutated in dominant optic atrophy. Nat. Genet. 2000, 26, 207–210. [Google Scholar] [CrossRef]
- Wai, T.; Garcia-Prieto, J.; Baker, M.J.; Merkwirth, C.; Benit, P.; Rustin, P.; Ruperez, F.J.; Barbas, C.; Ibanez, B.; Langer, T. Imbalanced OPA1 processing and mitochondrial fragmentation cause heart failure in mice. Science 2015, 350, aad0116. [Google Scholar] [CrossRef] [PubMed]
- Anand, R.; Wai, T.; Baker, M.J.; Kladt, N.; Schauss, A.C.; Rugarli, E.; Langer, T. The i-AAA protease YME1L and OMA1 cleave OPA1 to balance mitochondrial fusion and fission. J. Cell Biol. 2014, 204, 919–929. [Google Scholar] [CrossRef]
- Song, Z.; Chen, H.; Fiket, M.; Alexander, C.; Chan, D.C. OPA1 processing controls mitochondrial fusion and is regulated by mRNA splicing, membrane potential, and Yme1L. J. Cell Biol. 2007, 178, 749–755. [Google Scholar] [CrossRef] [PubMed]
- Song, M.; Dorn, G.W., 2nd. Mitoconfusion: Noncanonical functioning of dynamism factors in static mitochondria of the heart. Cell Metab. 2015, 21, 195–205. [Google Scholar] [CrossRef] [PubMed]
- Hoppel, C.L.; Tandler, B.; Fujioka, H.; Riva, A. Dynamic organization of mitochondria in human heart and in myocardial disease. Int. J. Biochem. Cell Biol. 2009, 41, 1949–1956. [Google Scholar] [CrossRef] [PubMed]
- Amchenkova, A.A.; Bakeeva, L.E.; Chentsov, Y.S.; Skulachev, V.P.; Zorov, D.B. Coupling membranes as energy-transmitting cables. I. Filamentous mitochondria in fibroblasts and mitochondrial clusters in cardiomyocytes. J. Cell Biol. 1988, 107, 481–495. [Google Scholar] [CrossRef] [PubMed]
- Ong, S.B.; Hall, A.R.; Hausenloy, D.J. Mitochondrial dynamics in cardiovascular health and disease. Antioxid. Redox Signal. 2013, 19, 400–414. [Google Scholar] [CrossRef]
- Ong, S.B.; Gustafsson, A.B. New roles for mitochondria in cell death in the reperfused myocardium. Cardiovasc. Res. 2012, 94, 190–196. [Google Scholar] [CrossRef]
- Ong, S.B.; Hausenloy, D.J. Mitochondrial morphology and cardiovascular disease. Cardiovasc. Res. 2010, 88, 16–29. [Google Scholar] [CrossRef]
- Piquereau, J.; Caffin, F.; Novotova, M.; Lemaire, C.; Veksler, V.; Garnier, A.; Ventura-Clapier, R.; Joubert, F. Mitochondrial dynamics in the adult cardiomyocytes: Which roles for a highly specialized cell? Front. Physiol. 2013, 4, 102. [Google Scholar] [CrossRef]
- Chen, H.; Chan, D.C. Mitochondrial dynamics in mammals. Curr. Top. Dev. Biol. 2004, 59, 119–144. [Google Scholar] [CrossRef] [PubMed]
- Braschi, E.; McBride, H.M. Mitochondria and the culture of the Borg: Understanding the integration of mitochondrial function within the reticulum, the cell, and the organism. Bioessays 2010, 32, 958–966. [Google Scholar] [CrossRef] [PubMed]
- Ong, S.B.; Subrayan, S.; Lim, S.Y.; Yellon, D.M.; Davidson, S.M.; Hausenloy, D.J. Inhibiting mitochondrial fission protects the heart against ischemia/reperfusion injury. Circulation 2010, 121, 2012–2022. [Google Scholar] [CrossRef]
- Shim, S.H.; Xia, C.; Zhong, G.; Babcock, H.P.; Vaughan, J.C.; Huang, B.; Wang, X.; Xu, C.; Bi, G.Q.; Zhuang, X. Super-resolution fluorescence imaging of organelles in live cells with photoswitchable membrane probes. Proc. Natl. Acad Sci. USA 2012, 109, 13978–13983. [Google Scholar] [CrossRef] [PubMed]
- Brunstein, M.; Wicker, K.; Herault, K.; Heintzmann, R.; Oheim, M. Full-field dual-color 100-nm super-resolution imaging reveals organization and dynamics of mitochondrial and ER networks. Opt. Express 2013, 21, 26162–26173. [Google Scholar] [CrossRef]
- Sherman, S.; Nachmias, D.; Elia, N. A simple, straightforward correlative live-cell-imaging-structured-illumination-microscopy approach for studying organelle dynamics. Microsc. Res. Tech. 2015, 78, 777–783. [Google Scholar] [CrossRef] [PubMed]
- Lo, C.Y.; Chen, S.; Creed, S.J.; Kang, M.; Zhao, N.; Tang, B.Z.; Elgass, K.D. Novel super-resolution capable mitochondrial probe, MitoRed AIE, enables assessment of real-time molecular mitochondrial dynamics. Sci. Rep. 2016, 6, 30855. [Google Scholar] [CrossRef] [PubMed]
- Twig, G.; Graf, S.A.; Wikstrom, J.D.; Mohamed, H.; Haigh, S.E.; Elorza, A.; Deutsch, M.; Zurgil, N.; Reynolds, N.; Shirihai, O.S. Tagging and tracking individual networks within a complex mitochondrial web with photoactivatable GFP. Am. J. Physiol. Cell Physiol. 2006, 291, C176–C184. [Google Scholar] [CrossRef]
- Magrane, J.; Cortez, C.; Gan, W.B.; Manfredi, G. Abnormal mitochondrial transport and morphology are common pathological denominators in SOD1 and TDP43 ALS mouse models. Hum. Mol. Genet. 2014, 23, 1413–1424. [Google Scholar] [CrossRef]
- Legros, F.; Lombes, A.; Frachon, P.; Rojo, M. Mitochondrial fusion in human cells is efficient, requires the inner membrane potential, and is mediated by mitofusins. Mol. Biol. Cell 2002, 13, 4343–4354. [Google Scholar] [CrossRef]
- Hernandez, G.; Thornton, C.; Stotland, A.; Lui, D.; Sin, J.; Ramil, J.; Magee, N.; Andres, A.; Quarato, G.; Carreira, R.S.; et al. MitoTimer: A novel tool for monitoring mitochondrial turnover. Autophagy 2013, 9, 1852–1861. [Google Scholar] [CrossRef] [PubMed]
- Ferree, A.W.; Trudeau, K.; Zik, E.; Benador, I.Y.; Twig, G.; Gottlieb, R.A.; Shirihai, O.S. MitoTimer probe reveals the impact of autophagy, fusion, and motility on subcellular distribution of young and old mitochondrial protein and on relative mitochondrial protein age. Autophagy 2013, 9, 1887–1896. [Google Scholar] [CrossRef]
- Huang, S.; Han, R.; Zhuang, Q.; Du, L.; Jia, H.; Liu, Y.; Liu, Y. New photostable naphthalimide-based fluorescent probe for mitochondrial imaging and tracking. Biosens. Bioelectron. 2015, 71, 313–321. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.; Choi, S.Y.; Frohman, M.A. A quantitative assay for mitochondrial fusion using Renilla luciferase complementation. Mitochondrion 2010, 10, 559–566. [Google Scholar] [CrossRef][Green Version]
- McWilliams, T.G.; Prescott, A.R.; Allen, G.F.; Tamjar, J.; Munson, M.J.; Thomson, C.; Muqit, M.M.; Ganley, I.G. mito-QC illuminates mitophagy and mitochondrial architecture in vivo. J. Cell Biol. 2016, 214, 333–345. [Google Scholar] [CrossRef] [PubMed]
- Lee, M.H.; Park, N.; Yi, C.; Han, J.H.; Hong, J.H.; Kim, K.P.; Kang, D.H.; Sessler, J.L.; Kang, C.; Kim, J.S. Mitochondria-immobilized pH-sensitive off-on fluorescent probe. J. Am. Chem. Soc. 2014, 136, 14136–14142. [Google Scholar] [CrossRef]
- Manczak, M.; Sesaki, H.; Kageyama, Y.; Reddy, P.H. Dynamin-related protein 1 heterozygote knockout mice do not have synaptic and mitochondrial deficiencies. Biochim. Biophys. Acta 2012, 1822, 862–874. [Google Scholar] [CrossRef]
- Ikeda, Y.; Shirakabe, A.; Maejima, Y.; Zhai, P.; Sciarretta, S.; Toli, J.; Nomura, M.; Mihara, K.; Egashira, K.; Ohishi, M.; et al. Endogenous Drp1 mediates mitochondrial autophagy and protects the heart against energy stress. Circ. Res. 2015, 116, 264–278. [Google Scholar] [CrossRef]
- Kageyama, Y.; Hoshijima, M.; Seo, K.; Bedja, D.; Sysa-Shah, P.; Andrabi, S.A.; Chen, W.; Hoke, A.; Dawson, V.L.; Dawson, T.M.; et al. Parkin-independent mitophagy requires Drp1 and maintains the integrity of mammalian heart and brain. EMBO J. 2014, 33, 2798–2813. [Google Scholar] [CrossRef]
- Ishihara, T.; Ban-Ishihara, R.; Maeda, M.; Matsunaga, Y.; Ichimura, A.; Kyogoku, S.; Aoki, H.; Katada, S.; Nakada, K.; Nomura, M.; et al. Dynamics of mitochondrial DNA nucleoids regulated by mitochondrial fission is essential for maintenance of homogeneously active mitochondria during neonatal heart development. Mol. Cell Biol. 2015, 35, 211–223. [Google Scholar] [CrossRef]
- Song, M.; Mihara, K.; Chen, Y.; Scorrano, L.; Dorn, G.W., 2nd. Mitochondrial fission and fusion factors reciprocally orchestrate mitophagic culling in mouse hearts and cultured fibroblasts. Cell Metab. 2015, 21, 273–286. [Google Scholar] [CrossRef] [PubMed]
- Song, M.; Gong, G.; Burelle, Y.; Gustafsson, A.B.; Kitsis, R.N.; Matkovich, S.J.; Dorn, G.W., 2nd. Interdependence of Parkin-Mediated Mitophagy and Mitochondrial Fission in Adult Mouse Hearts. Circ. Res. 2015, 117, 346–351. [Google Scholar] [CrossRef] [PubMed]
- Song, M.; Franco, A.; Fleischer, J.A.; Zhang, L.; Dorn, G.W. Abrogating Mitochondrial Dynamics in Mouse Hearts Accelerates Mitochondrial Senescence. Cell Metab. 2017, 26, 872–883.e875. [Google Scholar] [CrossRef]
- Papanicolaou, K.N.; Kikuchi, R.; Ngoh, G.A.; Coughlan, K.A.; Dominguez, I.; Stanley, W.C.; Walsh, K. Mitofusins 1 and 2 are essential for postnatal metabolic remodeling in heart. Circ. Res. 2012, 111, 1012–1026. [Google Scholar] [CrossRef]
- Chen, Y.; Liu, Y.; Dorn, G.W., 2nd. Mitochondrial fusion is essential for organelle function and cardiac homeostasis. Circ. Res. 2011, 109, 1327–1331. [Google Scholar] [CrossRef] [PubMed]
- Hall, A.R.; Burke, N.; Dongworth, R.K.; Kalkhoran, S.B.; Dyson, A.; Vicencio, J.M.; Dorn, G.W.; Yellon, D.M.; Hausenloy, D.J. Hearts deficient in both Mfn1 and Mfn2 are protected against acute myocardial infarction. Cell Death Dis. 2016, 7, e2238. [Google Scholar] [CrossRef]
- Papanicolaou, K.N.; Ngoh, G.A.; Dabkowski, E.R.; O’Connell, K.A.; Ribeiro, R.F., Jr.; Stanley, W.C.; Walsh, K. Cardiomyocyte deletion of mitofusin-1 leads to mitochondrial fragmentation and improves tolerance to ROS-induced mitochondrial dysfunction and cell death. Am. J. Physiol. Heart Circ. Physiol. 2012, 302, H167–H179. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Dorn, G.W., 2nd. PINK1-phosphorylated mitofusin 2 is a Parkin receptor for culling damaged mitochondria. Science 2013, 340, 471–475. [Google Scholar] [CrossRef]
- Papanicolaou, K.N.; Khairallah, R.J.; Ngoh, G.A.; Chikando, A.; Luptak, I.; O’Shea, K.M.; Riley, D.D.; Lugus, J.J.; Colucci, W.S.; Lederer, W.J.; et al. Mitofusin-2 maintains mitochondrial structure and contributes to stress-induced permeability transition in cardiac myocytes. Mol. Cell Biol. 2011, 31, 1309–1328. [Google Scholar] [CrossRef]
- Konstantinidis, K.; Lederer, W.J.; Rizzuto, R.; Kitsis, R.N. Mitofusin 2 joins the sarcoplasmic reticulum and mitochondria at the hip to sustain cardiac energetics. Circ. Res. 2012, 111, 821–823. [Google Scholar] [CrossRef]
- Dorn, G.W., 2nd; Clark, C.F.; Eschenbacher, W.H.; Kang, M.Y.; Engelhard, J.T.; Warner, S.J.; Matkovich, S.J.; Jowdy, C.C. MARF and Opa1 control mitochondrial and cardiac function in Drosophila. Circ. Res. 2011, 108, 12–17. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Ren, S.; Clish, C.; Jain, M.; Mootha, V.; McCaffery, J.M.; Chan, D.C. Titration of mitochondrial fusion rescues Mff-deficient cardiomyopathy. J. Cell Biol. 2015, 211, 795–805. [Google Scholar] [CrossRef]
- Piquereau, J.; Caffin, F.; Novotova, M.; Prola, A.; Garnier, A.; Mateo, P.; Fortin, D.; Huynh Le, H.; Nicolas, V.; Alavi, M.V.; et al. Down-regulation of OPA1 alters mouse mitochondrial morphology, PTP function, and cardiac adaptation to pressure overload. Cardiovasc. Res. 2012, 94, 408–417. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; McCaffery, J.M.; Chan, D.C. Mitochondrial fusion protects against neurodegeneration in the cerebellum. Cell 2007, 130, 548–562. [Google Scholar] [CrossRef] [PubMed]
- Kasahara, A.; Cipolat, S.; Chen, Y.; Dorn, G.W., 2nd; Scorrano, L. Mitochondrial fusion directs cardiomyocyte differentiation via calcineurin and Notch signaling. Science 2013, 342, 734–737. [Google Scholar] [CrossRef] [PubMed]
- Mourier, A.; Motori, E.; Brandt, T.; Lagouge, M.; Atanassov, I.; Galinier, A.; Rappl, G.; Brodesser, S.; Hultenby, K.; Dieterich, C.; et al. Mitofusin 2 is required to maintain mitochondrial coenzyme Q levels. J. Cell Biol. 2015, 208, 429–442. [Google Scholar] [CrossRef] [PubMed]
- Alavi, M.V.; Bette, S.; Schimpf, S.; Schuettauf, F.; Schraermeyer, U.; Wehrl, H.F.; Ruttiger, L.; Beck, S.C.; Tonagel, F.; Pichler, B.J.; et al. A splice site mutation in the murine Opa1 gene features pathology of autosomal dominant optic atrophy. Brain 2007, 130, 1029–1042. [Google Scholar] [CrossRef]
- Hu, Q.; Zhang, H.; Gutierrez Cortes, N.; Wu, D.; Wang, P.; Zhang, J.; Mattison, J.A.; Smith, E.; Bettcher, L.F.; Wang, M.; et al. Increased Drp1 Acetylation by Lipid Overload Induces Cardiomyocyte Death and Heart Dysfunction. Circ. Res. 2020, 126, 456–470. [Google Scholar] [CrossRef] [PubMed]
- Tsushima, K.; Bugger, H.; Wende, A.R.; Soto, J.; Jenson, G.A.; Tor, A.R.; McGlauflin, R.; Kenny, H.C.; Zhang, Y.; Souvenir, R.; et al. Mitochondrial Reactive Oxygen Species in Lipotoxic Hearts Induce Post-Translational Modifications of AKAP121, DRP1, and OPA1 That Promote Mitochondrial Fission. Circ. Res. 2018, 122, 58–73. [Google Scholar] [CrossRef] [PubMed]
- Yu, T.; Robotham, J.L.; Yoon, Y. Increased production of reactive oxygen species in hyperglycemic conditions requires dynamic change of mitochondrial morphology. Proc. Natl. Acad. Sci. USA 2006, 103, 2653–2658. [Google Scholar] [CrossRef]
- Yu, T.; Jhun, B.S.; Yoon, Y. High-glucose stimulation increases reactive oxygen species production through the calcium and mitogen-activated protein kinase-mediated activation of mitochondrial fission. Antioxid. Redox Signal. 2011, 14, 425–437. [Google Scholar] [CrossRef]
- Wu, Q.R.; Zheng, D.L.; Liu, P.M.; Yang, H.; Li, L.A.; Kuang, S.J.; Lai, Y.Y.; Rao, F.; Xue, Y.M.; Lin, J.J.; et al. High glucose induces Drp1-mediated mitochondrial fission via the Orai1 calcium channel to participate in diabetic cardiomyocyte hypertrophy. Cell Death Dis. 2021, 12, 216. [Google Scholar] [CrossRef]
- Yu, J.; Maimaitili, Y.; Xie, P.; Wu, J.J.; Wang, J.; Yang, Y.N.; Ma, H.P.; Zheng, H. High glucose concentration abrogates sevoflurane post-conditioning cardioprotection by advancing mitochondrial fission but dynamin-related protein 1 inhibitor restores these effects. Acta Physiol. 2017, 220, 83–98. [Google Scholar] [CrossRef] [PubMed]
- Li, S.Y.; Sigmon, V.K.; Babcock, S.A.; Ren, J. Advanced glycation endproduct induces ROS accumulation, apoptosis, MAP kinase activation and nuclear O-GlcNAcylation in human cardiac myocytes. Life Sci. 2007, 80, 1051–1056. [Google Scholar] [CrossRef] [PubMed]
- Makino, A.; Suarez, J.; Gawlowski, T.; Han, W.; Wang, H.; Scott, B.T.; Dillmann, W.H. Regulation of mitochondrial morphology and function by O-GlcNAcylation in neonatal cardiac myocytes. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2011, 300, R1296–R1302. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, T.; Saotome, M.; Nobuhara, M.; Sakamoto, A.; Urushida, T.; Katoh, H.; Satoh, H.; Funaki, M.; Hayashi, H. Roles of mitochondrial fragmentation and reactive oxygen species in mitochondrial dysfunction and myocardial insulin resistance. Exp. Cell Res. 2014, 323, 314–325. [Google Scholar] [CrossRef] [PubMed]
- Bekhite, M.; Gonzalez-Delgado, A.; Hubner, S.; Haxhikadrija, P.; Kretzschmar, T.; Muller, T.; Wu, J.M.F.; Bekfani, T.; Franz, M.; Wartenberg, M.; et al. The role of ceramide accumulation in human induced pluripotent stem cell-derived cardiomyocytes on mitochondrial oxidative stress and mitophagy. Free Radic. Biol. Med. 2021, 167, 66–80. [Google Scholar] [CrossRef]
- Schaper, J.; Froede, R.; Hein, S.; Buck, A.; Hashizume, H.; Speiser, B.; Friedl, A.; Bleese, N. Impairment of the myocardial ultrastructure and changes of the cytoskeleton in dilated cardiomyopathy. Circulation 1991, 83, 504–514. [Google Scholar] [CrossRef]
- Haileselassie, B.; Mukherjee, R.; Joshi, A.U.; Napier, B.A.; Massis, L.M.; Ostberg, N.P.; Queliconi, B.B.; Monack, D.; Bernstein, D.; Mochly-Rosen, D. Drp1/Fis1 interaction mediates mitochondrial dysfunction in septic cardiomyopathy. J. Mol. Cell. Cardiol. 2019, 130, 160–169. [Google Scholar] [CrossRef]
- Hsiao, Y.T.; Shimizu, I.; Wakasugi, T.; Yoshida, Y.; Ikegami, R.; Hayashi, Y.; Suda, M.; Katsuumi, G.; Nakao, M.; Ozawa, T.; et al. Cardiac mitofusin-1 is reduced in non-responding patients with idiopathic dilated cardiomyopathy. Sci. Rep. 2021, 11, 6722. [Google Scholar] [CrossRef] [PubMed]
- Montaigne, D.; Marechal, X.; Coisne, A.; Debry, N.; Modine, T.; Fayad, G.; Potelle, C.; El Arid, J.M.; Mouton, S.; Sebti, Y.; et al. Myocardial contractile dysfunction is associated with impaired mitochondrial function and dynamics in type 2 diabetic but not in obese patients. Circulation 2014, 130, 554–564. [Google Scholar] [CrossRef]
- Abdullah, C.S.; Alam, S.; Aishwarya, R.; Miriyala, S.; Bhuiyan, M.A.N.; Panchatcharam, M.; Pattillo, C.B.; Orr, A.W.; Sadoshima, J.; Hill, J.A.; et al. Doxorubicin-induced cardiomyopathy associated with inhibition of autophagic degradation process and defects in mitochondrial respiration. Sci. Rep. 2019, 9, 2002. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.; Zhang, Z.; Yin, Q.; Fu, C.; Barszczyk, A.; Zhang, X.; Wang, J.; Yang, D. Cardiac-specific overexpression of miR-122 induces mitochondria-dependent cardiomyocyte apoptosis and promotes heart failure by inhibiting Hand2. J. Cell Mol. Med. 2021, 25, 5326–5334. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.; Lee, H.Y.; Hanna, R.A.; Gustafsson, A.B. Mitochondrial autophagy by Bnip3 involves Drp1-mediated mitochondrial fission and recruitment of Parkin in cardiac myocytes. Am. J. Physiol. Heart Circ. Physiol. 2011, 301, H1924–H1931. [Google Scholar] [CrossRef]
- Cahill, T.J.; Leo, V.; Kelly, M.; Stockenhuber, A.; Kennedy, N.W.; Bao, L.; Cereghetti, G.M.; Harper, A.R.; Czibik, G.; Liao, C.; et al. Resistance of Dynamin-related Protein 1 Oligomers to Disassembly Impairs Mitophagy, Resulting in Myocardial Inflammation and Heart Failure. J. Biol. Chem. 2015, 290, 25907–25919. [Google Scholar] [CrossRef]
- Shirakabe, A.; Zhai, P.; Ikeda, Y.; Saito, T.; Maejima, Y.; Hsu, C.P.; Nomura, M.; Egashira, K.; Levine, B.; Sadoshima, J. Drp1-Dependent Mitochondrial Autophagy Plays a Protective Role against Pressure Overload-Induced Mitochondrial Dysfunction and Heart Failure. Circulation 2016, 133, 1249–1263. [Google Scholar] [CrossRef]
- Thai, P.N.; Seidlmayer, L.K.; Miller, C.; Ferrero, M.; Dorn, G.W., II; Schaefer, S.; Bers, D.M.; Dedkova, E.N. Mitochondrial Quality Control in Aging and Heart Failure: Influence of Ketone Bodies and Mitofusin-Stabilizing Peptides. Front. Physiol. 2019, 10, 382. [Google Scholar] [CrossRef]
- Chaanine, A.H.; Joyce, L.D.; Stulak, J.M.; Maltais, S.; Joyce, D.L.; Dearani, J.A.; Klaus, K.; Nair, K.S.; Hajjar, R.J.; Redfield, M.M. Mitochondrial Morphology, Dynamics, and Function in Human Pressure Overload or Ischemic Heart Disease with Preserved or Reduced Ejection Fraction. Circ. Heart Fail. 2019, 12, e005131. [Google Scholar] [CrossRef] [PubMed]
- Bhandari, P.; Song, M.; Dorn, G.W., 2nd. Dissociation of mitochondrial from sarcoplasmic reticular stress in Drosophila cardiomyopathy induced by molecularly distinct mitochondrial fusion defects. J. Mol. Cell. Cardiol. 2015, 80, 71–80. [Google Scholar] [CrossRef] [PubMed]
- Gong, G.; Song, M.; Csordas, G.; Kelly, D.P.; Matkovich, S.J.; Dorn, G.W., 2nd. Parkin-mediated mitophagy directs perinatal cardiac metabolic maturation in mice. Science 2015, 350, aad2459. [Google Scholar] [CrossRef] [PubMed]
- Kane, L.A.; Lazarou, M.; Fogel, A.I.; Li, Y.; Yamano, K.; Sarraf, S.A.; Banerjee, S.; Youle, R.J. PINK1 phosphorylates ubiquitin to activate Parkin E3 ubiquitin ligase activity. J. Cell Biol. 2014, 205, 143–153. [Google Scholar] [CrossRef] [PubMed]
- Koyano, F.; Okatsu, K.; Kosako, H.; Tamura, Y.; Go, E.; Kimura, M.; Kimura, Y.; Tsuchiya, H.; Yoshihara, H.; Hirokawa, T.; et al. Ubiquitin is phosphorylated by PINK1 to activate parkin. Nature 2014, 510, 162–166. [Google Scholar] [CrossRef]
- McLelland, G.L.; Goiran, T.; Yi, W.; Dorval, G.; Chen, C.X.; Lauinger, N.D.; Krahn, A.I.; Valimehr, S.; Rakovic, A.; Rouiller, I.; et al. Mfn2 ubiquitination by PINK1/parkin gates the p97-dependent release of ER from mitochondria to drive mitophagy. eLife 2018, 7, e32866. [Google Scholar] [CrossRef] [PubMed]
- Xin, T.; Lu, C. Irisin activates Opa1-induced mitophagy to protect cardiomyocytes against apoptosis following myocardial infarction. Aging 2020, 12, 4474–4488. [Google Scholar] [CrossRef] [PubMed]
- Javadov, S.; Rajapurohitam, V.; Kilić, A.; Hunter, J.C.; Zeidan, A.; Said Faruq, N.; Escobales, N.; Karmazyn, M. Expression of mitochondrial fusion-fission proteins during post-infarction remodeling: The effect of NHE-1 inhibition. Basic Res. Cardiol. 2011, 106, 99–109. [Google Scholar] [CrossRef] [PubMed]
- Davies, V.J.; Hollins, A.J.; Piechota, M.J.; Yip, W.; Davies, J.R.; White, K.E.; Nicols, P.P.; Boulton, M.E.; Votruba, M. Opa1 deficiency in a mouse model of autosomal dominant optic atrophy impairs mitochondrial morphology, optic nerve structure and visual function. Hum. Mol. Genet. 2007, 16, 1307–1318. [Google Scholar] [CrossRef]
- Hwang, H.V.; Sandeep, N.; Nair, R.V.; Hu, D.Q.; Zhao, M.; Lan, I.S.; Fajardo, G.; Matkovich, S.J.; Bernstein, D.; Reddy, S. Transcriptomic and Functional Analyses of Mitochondrial Dysfunction in Pressure Overload-Induced Right Ventricular Failure. J. Am. Heart Assoc. 2021, 10, e017835. [Google Scholar] [CrossRef]
- Din, S.; Mason, M.; Volkers, M.; Johnson, B.; Cottage, C.T.; Wang, Z.; Joyo, A.Y.; Quijada, P.; Erhardt, P.; Magnuson, N.S.; et al. Pim-1 preserves mitochondrial morphology by inhibiting dynamin-related protein 1 translocation. Proc. Natl. Acad. Sci. USA 2013, 110, 5969–5974. [Google Scholar] [CrossRef] [PubMed]
- Sharp, W.W.; Fang, Y.H.; Han, M.; Zhang, H.J.; Hong, Z.; Banathy, A.; Morrow, E.; Ryan, J.J.; Archer, S.L. Dynamin-related protein 1 (Drp1)-mediated diastolic dysfunction in myocardial ischemia-reperfusion injury: Therapeutic benefits of Drp1 inhibition to reduce mitochondrial fission. FASEB J. 2014, 28, 316–326. [Google Scholar] [CrossRef]
- Chou, C.H.; Lin, C.C.; Yang, M.C.; Wei, C.C.; Liao, H.D.; Lin, R.C.; Tu, W.Y.; Kao, T.C.; Hsu, C.M.; Cheng, J.T.; et al. GSK3beta-mediated Drp1 phosphorylation induced elongated mitochondrial morphology against oxidative stress. PLoS ONE 2012, 7, e49112. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.D.; Chen, H.J.; Wang, D.L.; Wang, H.; Deng, Q. Pim-1 Kinase Regulating Dynamics Related Protein 1 Mediates Sevoflurane Postconditioning-induced Cardioprotection. Chin. Med. J. 2017, 130, 309–317. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Zhang, F.; Zhao, G.; Cheng, Y.; Wu, T.; Wu, B.; Zhang, Y.E. Mitochondrial PKC-epsilon deficiency promotes I/R-mediated myocardial injury via GSK3beta-dependent mitochondrial permeability transition pore opening. J. Cell. Mol. Med. 2017, 21, 2009–2021. [Google Scholar] [CrossRef]
- Qi, X.; Disatnik, M.H.; Shen, N.; Sobel, R.A.; Mochly-Rosen, D. Aberrant mitochondrial fission in neurons induced by protein kinase Cdelta under oxidative stress conditions in vivo. Mol. Biol. Cell 2011, 22, 256–265. [Google Scholar] [CrossRef]
- Yang, Y.; Zhao, L.; Ma, J. Penehyclidine hydrochloride preconditioning provides cardiac protection in a rat model of myocardial ischemia/reperfusion injury via the mechanism of mitochondrial dynamics mechanism. Eur. J. Pharmacol. 2017, 813, 130–139. [Google Scholar] [CrossRef]
- Dong, G.; Chen, T.; Ren, X.; Zhang, Z.; Huang, W.; Liu, L.; Luo, P.; Zhou, H. Rg1 prevents myocardial hypoxia/reoxygenation injury by regulating mitochondrial dynamics imbalance via modulation of glutamate dehydrogenase and mitofusin 2. Mitochondrion 2016, 26, 7–18. [Google Scholar] [CrossRef] [PubMed]
- Cellier, L.; Tamareille, S.; Kalakech, H.; Guillou, S.; Lenaers, G.; Prunier, F.; Mirebeau-Prunier, D. Remote Ischemic Conditioning Influences Mitochondrial Dynamics. Shock 2016, 45, 192–197. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Li, J.; Li, Y.; Jiao, J.; Wang, J.; Li, Y.; Qin, D.; Li, P. Mitofusin 1 is negatively regulated by microRNA 140 in cardiomyocyte apoptosis. Mol. Cell. Biol. 2014, 34, 1788–1799. [Google Scholar] [CrossRef]
- Shen, T.; Zheng, M.; Cao, C.; Chen, C.; Tang, J.; Zhang, W.; Cheng, H.; Chen, K.H.; Xiao, R.P. Mitofusin-2 is a major determinant of oxidative stress-mediated heart muscle cell apoptosis. J. Biol. Chem. 2007, 282, 23354–23361. [Google Scholar] [CrossRef]
- Zhao, T.; Huang, X.; Han, L.; Wang, X.; Cheng, H.; Zhao, Y.; Chen, Q.; Chen, J.; Cheng, H.; Xiao, R.; et al. Central role of mitofusin 2 in autophagosome-lysosome fusion in cardiomyocytes. J. Biol. Chem. 2012, 287, 23615–23625. [Google Scholar] [CrossRef]
- Hall, A.R.; Burke, N.; Dongworth, R.K.; Kalkhoran, S.B.; Dyson, A.; Vicencio, J.M.; Dorn, G.W., 2nd; Yellon, D.M.; Hausenloy, D.J. Correction: Hearts deficient in both Mfn1 and Mfn2 are protected against acute myocardial infarction. Cell. Death Dis. 2021, 12, 660. [Google Scholar] [CrossRef]
- Olmedo, I.; Pino, G.; Riquelme, J.A.; Aranguiz, P.; Diaz, M.C.; Lopez-Crisosto, C.; Lavandero, S.; Donoso, P.; Pedrozo, Z.; Sanchez, G. Inhibition of the proteasome preserves Mitofusin-2 and mitochondrial integrity, protecting cardiomyocytes during ischemia-reperfusion injury. Biochim. Biophys. Acta Mol. Basis Dis. 2020, 1866, 165659. [Google Scholar] [CrossRef]
- Chen, Y.; Li, S.; Zhang, Y.; Wang, M.; Li, X.; Liu, S.; Xu, D.; Bao, Y.; Jia, P.; Wu, N.; et al. The lncRNA Malat1 regulates microvascular function after myocardial infarction in mice via miR-26b-5p/Mfn1 axis-mediated mitochondrial dynamics. Redox Biol. 2021, 41, 101910. [Google Scholar] [CrossRef]
- Chen, L.; Gong, Q.; Stice, J.P.; Knowlton, A.A. Mitochondrial OPA1, apoptosis, and heart failure. Cardiovasc. Res. 2009, 84, 91–99. [Google Scholar] [CrossRef]
- Le Page, S.; Niro, M.; Fauconnier, J.; Cellier, L.; Tamareille, S.; Gharib, A.; Chevrollier, A.; Loufrani, L.; Grenier, C.; Kamel, R.; et al. Increase in Cardiac Ischemia-Reperfusion Injuries in Opa1+/− Mouse Model. PLoS ONE 2016, 11, e0164066. [Google Scholar] [CrossRef]
- Yu, L.M.; Dong, X.; Xue, X.D.; Xu, S.; Zhang, X.; Xu, Y.L.; Wang, Z.S.; Wang, Y.; Gao, H.; Liang, Y.X.; et al. Melatonin attenuates diabetic cardiomyopathy and reduces myocardial vulnerability to ischemia-reperfusion injury by improving mitochondrial quality control: Role of SIRT6. J. Pineal. Res. 2021, 70, e12698. [Google Scholar] [CrossRef] [PubMed]
- Kalkhoran, S.B.; Kriston-Vizi, J.; Hernandez-Resendiz, S.; Crespo-Avilan, G.E.; Rosdah, A.A.; Lees, J.G.; Costa, J.; Ling, N.X.Y.; Holien, J.K.; Samangouei, P.; et al. Hydralazine protects the heart against acute ischaemia/reperfusion injury by inhibiting Drp1-mediated mitochondrial fission. Cardiovasc. Res. 2022, 118, 282–294. [Google Scholar] [CrossRef]
- Disatnik, M.H.; Ferreira, J.C.; Campos, J.C.; Gomes, K.S.; Dourado, P.M.; Qi, X.; Mochly-Rosen, D. Acute inhibition of excessive mitochondrial fission after myocardial infarction prevents long-term cardiac dysfunction. J. Am. Heart Assoc. 2013, 2, e000461. [Google Scholar] [CrossRef] [PubMed]
- Gao, D.; Zhang, L.; Dhillon, R.; Hong, T.T.; Shaw, R.M.; Zhu, J. Dynasore protects mitochondria and improves cardiac lusitropy in Langendorff perfused mouse heart. PLoS ONE 2013, 8, e60967. [Google Scholar] [CrossRef]
- Khuanjing, T.; Palee, S.; Kerdphoo, S.; Jaiwongkam, T.; Anomasiri, A.; Chattipakorn, S.C.; Chattipakorn, N. Donepezil attenuated cardiac ischemia/reperfusion injury through balancing mitochondrial dynamics, mitophagy, and autophagy. Transl. Res. 2021, 230, 82–97. [Google Scholar] [CrossRef]
- Zhuang, X.; Sun, X.; Zhou, H.; Zhang, S.; Zhong, X.; Xu, X.; Guo, Y.; Xiong, Z.; Liu, M.; Lin, Y.; et al. Klotho attenuated Doxorubicin-induced cardiomyopathy by alleviating Dynamin-related protein 1 - mediated mitochondrial dysfunction. Mech. Ageing Dev. 2021, 195, 111442. [Google Scholar] [CrossRef] [PubMed]
- Yu, P.; Zhang, J.; Yu, S.; Luo, Z.; Hua, F.; Yuan, L.; Zhou, Z.; Liu, Q.; Du, X.; Chen, S.; et al. Protective Effect of Sevoflurane Postconditioning against Cardiac Ischemia/Reperfusion Injury via Ameliorating Mitochondrial Impairment, Oxidative Stress and Rescuing Autophagic Clearance. PLoS ONE 2015, 10, e0134666. [Google Scholar] [CrossRef] [PubMed]
- Rogers, M.A.; Maldonado, N.; Hutcheson, J.D.; Goettsch, C.; Goto, S.; Yamada, I.; Faits, T.; Sesaki, H.; Aikawa, M.; Aikawa, E. Dynamin-Related Protein 1 Inhibition Attenuates Cardiovascular Calcification in the Presence of Oxidative Stress. Circ. Res. 2017, 121, 220–233. [Google Scholar] [CrossRef]
- Chen, W.R.; Zhou, Y.J.; Sha, Y.; Wu, X.P.; Yang, J.Q.; Liu, F. Melatonin attenuates vascular calcification by inhibiting mitochondria fission via an AMPK/Drp1 signalling pathway. J. Cell. Mol. Med. 2020, 24, 6043–6054. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.W.; Pang, Q.; Zhou, T.; Song, X.Y.; Pan, Y.J.; Jia, L.P.; Zhang, A.H. Irisin alleviates vascular calcification by inhibiting VSMC osteoblastic transformation and mitochondria dysfunction via AMPK/Drp1 signaling pathway in chronic kidney disease. Atherosclerosis 2022, 346, 36–45. [Google Scholar] [CrossRef]
- Cui, L.; Li, Z.; Chang, X.; Cong, G.; Hao, L. Quercetin attenuates vascular calcification by inhibiting oxidative stress and mitochondrial fission. Vascul. Pharmacol. 2017, 88, 21–29. [Google Scholar] [CrossRef] [PubMed]
- Deng, Y.; Li, S.; Chen, Z.; Wang, W.; Geng, B.; Cai, J. Mdivi-1, a mitochondrial fission inhibitor, reduces angiotensin-II- induced hypertension by mediating VSMC phenotypic switch. Biomed. Pharmacother. 2021, 140, 111689. [Google Scholar] [CrossRef] [PubMed]
- Hasan, P.; Saotome, M.; Ikoma, T.; Iguchi, K.; Kawasaki, H.; Iwashita, T.; Hayashi, H.; Maekawa, Y. Mitochondrial fission protein, dynamin-related protein 1, contributes to the promotion of hypertensive cardiac hypertrophy and fibrosis in Dahl-salt sensitive rats. J. Mol. Cell. Cardiol. 2018, 121, 103–106. [Google Scholar] [CrossRef]
- Chen, C.; Gao, J.L.; Liu, M.Y.; Li, S.L.; Xuan, X.C.; Zhang, X.Z.; Zhang, X.Y.; Wei, Y.Y.; Zhen, C.L.; Jin, J.; et al. Mitochondrial Fission Inhibitors Suppress Endothelin-1-Induced Artery Constriction. Cell Physiol. Biochem. 2017, 42, 1802–1811. [Google Scholar] [CrossRef]
- Tian, L.; Wu, D.; Dasgupta, A.; Chen, K.H.; Mewburn, J.; Potus, F.; Lima, P.D.A.; Hong, Z.; Zhao, Y.Y.; Hindmarch, C.C.T.; et al. Epigenetic Metabolic Reprogramming of Right Ventricular Fibroblasts in Pulmonary Arterial Hypertension: A Pyruvate Dehydrogenase Kinase-Dependent Shift in Mitochondrial Metabolism Promotes Right Ventricular Fibrosis. Circ. Res. 2020, 126, 1723–1745. [Google Scholar] [CrossRef]
- Parra, V.; Bravo-Sagua, R.; Norambuena-Soto, I.; Hernández-Fuentes, C.P.; Gómez-Contreras, A.G.; Verdejo, H.E.; Mellado, R.; Chiong, M.; Lavandero, S.; Castro, P.F. Inhibition of mitochondrial fission prevents hypoxia-induced metabolic shift and cellular proliferation of pulmonary arterial smooth muscle cells. Biochim. Biophys. Acta Mol. Basis Dis. 2017, 1863, 2891–2903. [Google Scholar] [CrossRef]
- Wu, Y.C.; Wang, W.T.; Lee, S.S.; Kuo, Y.R.; Wang, Y.C.; Yen, S.J.; Lee, M.Y.; Yeh, J.L. Glucagon-Like Peptide-1 Receptor Agonist Attenuates Autophagy to Ameliorate Pulmonary Arterial Hypertension through Drp1/NOX- and Atg-5/Atg-7/Beclin-1/LC3β Pathways. Int. J. Mol. Sci. 2019, 20, 3435. [Google Scholar] [CrossRef] [PubMed]
- Uchikado, Y.; Ikeda, Y.; Sasaki, Y.; Iwabayashi, M.; Akasaki, Y.; Ohishi, M. Association of Lectin-Like Oxidized Low-Density Lipoprotein Receptor-1 With Angiotensin II Type 1 Receptor Impacts Mitochondrial Quality Control, Offering Promise for the Treatment of Vascular Senescence. Front. Cardiovasc. Med. 2021, 8, 788655. [Google Scholar] [CrossRef]
- Joshi, A.U.; Ebert, A.E.; Haileselassie, B.; Mochly-Rosen, D. Drp1/Fis1-mediated mitochondrial fragmentation leads to lysosomal dysfunction in cardiac models of Huntington’s disease. J. Mol. Cell. Cardiol. 2019, 127, 125–133. [Google Scholar] [CrossRef]
- Ferreira, J.C.B.; Campos, J.C.; Qvit, N.; Qi, X.; Bozi, L.H.M.; Bechara, L.R.G.; Lima, V.M.; Queliconi, B.B.; Disatnik, M.H.; Dourado, P.M.M.; et al. A selective inhibitor of mitofusin 1-βIIPKC association improves heart failure outcome in rats. Nat. Commun. 2019, 10, 329. [Google Scholar] [CrossRef]
- Ghahremani, R.; Damirchi, A.; Salehi, I.; Komaki, A.; Esposito, F. Mitochondrial dynamics as an underlying mechanism involved in aerobic exercise training-induced cardioprotection against ischemia-reperfusion injury. Life Sci. 2018, 213, 102–108. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.; Hong, X.; Liu, L.; Wu, Y.; Xie, X.; Fang, G.; Zhi, S. Cordycepin Decreases Ischemia/Reperfusion Injury in Diabetic Hearts via Upregulating AMPK/Mfn2-dependent Mitochondrial Fusion. Front. Pharmacol. 2021, 12, 754005. [Google Scholar] [CrossRef]
- Liao, H.; Gong, J.; Zhang, W.; Guo, X. Valsartan inhibits angiotensin II-induced proliferation of vascular smooth muscle cells via regulating the expression of mitofusin 2. J. Huazhong Univ. Sci. Technol. Med. Sci. 2012, 32, 31–35. [Google Scholar] [CrossRef]
- Zhang, W.; Shu, C.; Li, Q.; Li, M.; Li, X. Adiponectin affects vascular smooth muscle cell proliferation and apoptosis through modulation of the mitofusin-2-mediated Ras-Raf-Erk1/2 signaling pathway. Mol. Med. Rep. 2015, 12, 4703–4707. [Google Scholar] [CrossRef]
- Sun, W.; Yan, C.; Frost, B.; Wang, X.; Hou, C.; Zeng, M.; Gao, H.; Kang, Y.; Liu, J. Pomegranate extract decreases oxidative stress and alleviates mitochondrial impairment by activating AMPK-Nrf2 in hypothalamic paraventricular nucleus of spontaneously hypertensive rats. Sci. Rep. 2016, 6, 34246. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Zhou, X.; Zeng, X.; Hu, O.; Yi, L.; Mi, M. Resveratrol attenuates oxidative injury in human umbilical vein endothelial cells through regulating mitochondrial fusion via TyrRS-PARP1 pathway. Nutr. Metab. 2019, 16, 9. [Google Scholar] [CrossRef]
- Sun, R.; Wang, X.; Liu, Y.; Xia, M. Dietary supplementation with fish oil alters the expression levels of proteins governing mitochondrial dynamics and prevents high-fat diet-induced endothelial dysfunction. Br. J. Nutr. 2014, 112, 145–153. [Google Scholar] [CrossRef]
- Perez-Ternero, C.; Werner, C.M.; Nickel, A.G.; Herrera, M.D.; Motilva, M.J.; Böhm, M.; Alvarez de Sotomayor, M.; Laufs, U. Ferulic acid, a bioactive component of rice bran, improves oxidative stress and mitochondrial biogenesis and dynamics in mice and in human mononuclear cells. J. Nutr. Biochem. 2017, 48, 51–61. [Google Scholar] [CrossRef]
- Hong, S.; Zhang, X.; Zhang, X.; Liu, W.; Fu, Y.; Liu, Y.; Shi, Z.; Chi, J.; Zhao, M.; Yin, X. Role of the calcium sensing receptor in cardiomyocyte apoptosis via mitochondrial dynamics in compensatory hypertrophied myocardium of spontaneously hypertensive rat. Biochem. Biophys. Res. Commun. 2017, 487, 728–733. [Google Scholar] [CrossRef]
- Zhang, Y.; Wang, Y.; Xu, J.; Tian, F.; Hu, S.; Chen, Y.; Fu, Z. Melatonin attenuates myocardial ischemia-reperfusion injury via improving mitochondrial fusion/mitophagy and activating the AMPK-OPA1 signaling pathways. J. Pineal Res. 2019, 66, e12542. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Han, Y.; Gu, X.; Li, M.; Du, Y.; Feng, N.; Li, J.; Zhang, S.; Maslov, L.N.; Wang, G.; et al. Paeonol promotes Opa1-mediated mitochondrial fusion via activating the CK2α-Stat3 pathway in diabetic cardiomyopathy. Redox Biol. 2021, 46, 102098. [Google Scholar] [CrossRef]
- Shaoqing, L.; Ting, Z.; Hao, L.; He, Z.; Wang, Y.; Ming, Z. Nicorandil, an ATP-sensitive potassium channel activation, attenuates myocardial injury in rats with ischemic cardiomyopathy. Med. Mol. Morphol. 2022, 55, 41–46. [Google Scholar] [CrossRef] [PubMed]
- Xie, T.; Wang, C.; Jin, Y.; Meng, Q.; Liu, Q.; Wu, J.; Sun, H. CoenzymeQ10-Induced Activation of AMPK-YAP-OPA1 Pathway Alleviates Atherosclerosis by Improving Mitochondrial Function, Inhibiting Oxidative Stress and Promoting Energy Metabolism. Front. Pharmacol. 2020, 11, 1034. [Google Scholar] [CrossRef]
- Uchikado, Y.; Ikeda, Y.; Ohishi, M. Current Understanding of the Pivotal Role of Mitochondrial Dynamics in Cardiovascular Diseases and Senescence. Front. Cardiovasc. Med. 2022, 9, 905072. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).