Abstract
Mitochondria are responsible for energy generation, as well as key metabolic and signaling pathways, and thus affect the entire developmental process of plants as well as their responses to stress. In metazoans, mitochondrial transcription termination factors (mTERFs) are known to regulate mitochondrial transcription. mTERFs have also been discovered in plants, but only a few of these proteins have been explored for their biological functions. Here, we report a role in root growth for mitochondria-associated protein AhmTERF1 in peanut (Arachis hypogaea L.). Overexpressing AhmTERF1 significantly stimulated the growth of peanut hairy roots and transgenic Arabidopsis. Surprisingly, AhmTERF1 is predominantly expressed in the root meristem where it increases mitochondrial abundance. AhmTERF1 binding to mtDNA was enriched in the RRN18 and RRN26 regions, suggesting it is related to the accumulation of mitochondrial ribosomes. Peanut is one of the main oil crops and the important source of edible oil and AhmTERF1 likely affects agronomic traits related to root growth in different peanut cultivars. We propose that peanut AhmTERF1 is an important protein for root growth due to its role in regulating mitochondrial abundance.
1. Introduction
Plant roots anchor the plant in the soil and are responsible for the uptake, storage and transport of minerals and water. They also communicate and interact with the soil microbiome and other plants, and sense biotic and abiotic stresses in the soil [1]. Overall, plants depend on root development, growth and function for their survival.
Mitochondria originate from endosymbiotic α-proteobacteria and possess their own vestigial genome, but most mitochondrial proteins are encoded by the nuclear genome and then are transported into the organelles. Mitochondria are essential for energy production [2], maintenance of calcium homeostasis [3] and the regulation of various intracellular signaling pathways; hence, they play crucial roles in plant growth, development and stress responses, such as seed germination [4,5], plant immunity [6] and the touch response [7,8].
A number of mitochondria-associated proteins that are involved in root growth and its regulation have been described. For example, AtPHB3 is a conserved mitochondrial type I prohibitin, which is reported to maintain the mitochondrial morphology and cell division in the root meristem of Arabidopsis [9]. This finding also provides evidence of a role for mitochondria in regulating cell division in the root meristem. The RETARDED ROOT GROWTH (RRG) gene, encoding a mitochondrial protein, is predominantly expressed in the root meristem to control cell division [10]. Arabidopsis slow growth3 (slo3) is a menber of a large family called pentatricopeptide repeat (PPR) proteins; when slo3 mutates, the generation of meristem cells is hindered, resulting in reduced root apical meristem area [11]. These proteins all affect root growth and development by impacting cell division in the root meristem.
In view of the small size of and number of genes in their genomes, mitochondria seem to have an unexpectedly complex transcriptional regulatory network. The mitochondrial transcription termination factor (mTERF) proteins share repeated conserved sequences of 30-aa mTERF motif [12]. They were identified in human mitochondria three decades ago as important control factors of transcription termination [13]. In animals, only four mTERF members have been described [12], but there are more examples in plants, and they are predicted to be targeted to nucleus, chloroplasts, mitochondria or other subcellular locations [14], implying a range of complex and diverse functions. Nevertheless, in contrast to mammalian mTERFs, reports about the roles of plant mTERFs are still scarce. To date, these studies mostly focus on Arabidopsis and maize, which have 35 and 31 members of this mTERF family, respectively [12,15].
The first mTERF gene identified in higher plants, SOLDAT10, is localized to chloroplasts; in Arabidopsis, entirely inactivation of SOLDAT10 lead to lethal mutation [16]. For other Arabidopsis mTERF mutants, including loss-of-function alleles of BSM/RUG2 (BELAYA SMERT/RUGOSA2) [17,18], MDA1 (mTERF DEFECTIVE IN Arabidopsis1) [19], mTERF6 [20], TWR-1/mTERF9 (TWIRT1/mTERF9) [21,22], mTERF15 [23] and SHOT1 (SUPPRESSOR OF HOT1-4) [24], stunted growth and altered organelle genes expression have been reported. Recently, molecular functions have been proposed for some of the mTERF genes characterised, all of which are related to post-transcriptional regulation of chloroplast and/or mitochondrial gene expression. BSM/RUG2 is necessary for the splicing of the second group II intron of the plastid clpP gene [17]. mTERF6 interacts with an RNA sequence in the chloroplast isoleucine transfer RNA gene (trnI.2) of the rRNA operon and promotes its maturation [20]. mTERF15 works in mitochondria as a splicing factor for nad2 intron 3 splicing [23].
Peanut (Arachis hypogaea L.), an important economic and oil crop, possesses a complex allotetraploid genome due to its hybridization from two diploid species [25]. This complexity results in the existence of many genes, including the mTERF family, of unknown function. Our previous study indicated that, although the mTERF gene family underwent large changes as a result of the polyploidization event, some mTERFs were conserved. Among these, AhmTERF1 (Genebank of USA No. MG957109) is highly conserved between the cultivated allotetraploid species and its wild progenitors. Subcellular localization analysis indicates that the AhmTERF1 protein is closely associated with mitochondria [26], consistent with it having a mitochondrial function.
Here, we report the function of AhmTERF1. AhmTERF1 is predominantly expressed in the root meristem. In consideration of its subcellular localization, we further figure out the relationship of AhmTERF1 with mitochondria and the regulation for peanut root growth by modulating mitochondrial abundance. In conclusion, our results reveal an unexpected role of AhmTERF1 and highlight the importance of this protein in peanut development.
2. Materials and Methods
2.1. Vector Construction
For the pAhmTERF1::GUS fusion construct, a 1.531-kb upstream sequence from the AhmTERF1 gene was amplified from peanut genomic DNA using the primers shown in Supplementary Table S1. The PCR product was then cloned into the HY107 vector [27]. To construct the 35S::AhmTERF1-GFP and 35S::AhmTERF1-mcherry plasmids, the AhmTERF1 coding sequence was amplified from peanut cDNA (for the primers used, see Supplementary Table S1). The PCR products were then cloned into the 35S::GFP vector or the 35S::mcherry vector. The AhmTERF1 RNAi construct was made by associate research fellow Xu Liu of South China Botanical Garden, Chinese Academy of Sciences according to the method [28].
2.2. Plant Materials and Growth Conditions
Peanut was grown in pots with a mixed soil consisting of vermiculite, perlite and soil (1:1:1), and cultivated at the condition of 28 °C and 16 h light photoperiod for 3 days. The 35S::eGFP, 35S::AhmTERF1-GFP, 35S::AhmTERF1 RNAi and pAhmTERF1::GUS constructs were transformed into Agrobacterium rhizogenes K599 to induce hairy roots, according to the protocol [29].
Arabidopsis plants were grown in potting soil with 16 h light/8 h dark photoperiod. To obtain AhmTERF1 overexpression and AhmTERF1::GUS Arabidopsis plants, constructs were transformed into the Col-0 strain of Arabidopsis by the floral dip method as previous reported [30]. AhmTERF1 overexpressing Arabidopsis plants were selected on 1/2 MS medium containing 50 mg/L kanamycin. AhmTERF1::GUS Arabidopsis plants were selected using the herbicide.
2.3. Quantitative Real-Time PCR (qRT-PCR)
RNA was extracted from peanut roots as described by Wan and Li [31]. Reverse transcription process was carried out using the HiScript® III 1st Strand cDNA Synthesis Kit (+gDNA wiper) (Vazyme). ChamQ Universal SYBR qPCR Master Mix (Vazyme) was used according to the manufacturer’s instructions with a BioRad CFX96 Real-Time PCR detection system. Primers for qRT-PCR are listed in Supplementary Table S1.
2.4. Histochemical Staining of GUS Activity
According to a previous reported methodology [32], histochemical staining of GUS activity was carried out. To remove chlorophyll, stained samples were soaked with 70% ethanol for 24 h. Nomarski DIC images of GUS expression were taken as mentioned in Xu and Scheres [32].
2.5. Subcellular Localization Analysis
To visualise fluorescent fusion proteins, leaves of 2-week old Col-0 Arabidopsis grown on soil were incubated for 3 h at 26 °C in the dark in a protoplasting solution (1.5% cellulase, 0.75% macerozyme, 0.5 M mannitol, 10 mM MES pH 5.7, 10 mM CaCl2 and 0.1% BSA). The isolation and transformation of protoplasts were performed as described [33], and then protoplasts were observed with a confocal laser scanning microscopy (LSM800, Carl Zeiss, Germany).
2.6. Western Blotting
According to the reported protocol [5] with minor modifications, the extraction of a crude preparation of mitochondria was conducted. Approximately 10 g of young roots was ground at 4 °C in extraction buffer (10 mM KH2PO4, pH 7.5, 0.3 M sucrose, 2 mM EDTA, 5 mM tetrasodium pyrophosphate, 5 mM cysteine, 20 mM ascorbic acid, 1% polyvinylpyrrolidone 40, 1% BSA and 10% glycerin). Two layers of Miracloth were used for filtering. And then the homogenate was centrifuged in a process of 5 min at 3000× g. Mitochondria were extracted from the clear supernatant by centrifugation at 20,000× g for 10 min. The AhmTERF1 antibody was made by Willget Biotech Co., Ltd. (Shanghai, China). Western blotting was conducted as described previously [33].
2.7. Ultrastructural Analyses
The ultrastructural analyses were carried out according to the method previous reported [34]. 2.5% glutaraldehyde solution was used to fix the hairy root tips at 4 °C for 12 h. Then the glutaraldehyde solution was discarded and phosphate-buffered saline (PBS; 0.1 M, pH 7.0) was added and allowed to stand for 15 min. The hairy root tips were washed three times with PBS after which 1% osmic acid was added and the roots were soaked for 2 h. The osmic acid was discarded and PBS was added and allowed to stand for another 15 min. Hairy root tips were washed three times with PBS and then the hairy root tips were dehydrated with alcohol step by step (30%→50%→70%→80%→90%→100%; 15 min at each concentration). Then the roots were treated with 100% alcohol for 20 min, and finally with acetone for 20 min.
The roots were submerged in osmotic solution Ⅰ (Spurr embedding agent mixed with acetone (v/v = 1/1)) for 1 h, then removed to osmotic solution Ⅱ (Spurr embedding agent mixed with acetone (v/v = 3/1)) for 3 h. The samples were then transferred to Spurr embedding agent overnight. The permeated samples were then heated overnight in a 70 °C oven to complete the embedding treatment. The samples were processed using a Leica EM UC7 ultrathin slicer to obtain 70–90 nm ultrathin slices, which were imaged using a Hitachi H-7650 transmission electron microscope.
2.8. ChIP-qPCR Assay
The ChIP assay was conducted as previous described [35]. Primers for qPCR are listed in Supplementary Table S1. An intergenic region (atp9) that does not bind AhmTERF1 was used as a negative control.
2.9. mtDNA Copy Number Analysis
The mtDNA copy number was determined by reference to Mondal et al. [36] and Mei et al. [37]. The relative mtDNA copy number is defined as the ratio of mtDNA to nuclear DNA. mtDNA was labeled with RRN18 and RRN26, and nuclear DNA was labeled with a single-copy gene, Arahy.U6ZXMA.
2.10. Statistical Analysis
Results are expressed as mean values ± standard deviation (SD). Data were evaluated by one-way analysis of variance (ANOVA) or the Student t-test using SPSS19.0 software. p < 0.05 were considered as significance.
3. Results
3.1. Overexpression of AhmTERF1 Promotes Root System Architecture
To investigate the function of AhmTERF1, we first transformed peanut hairy roots with a vector carrying either a fusion of the CaMV35S promoter and AhmTERF1 cDNA or a AhmTERF1 RNAi construct to obtain AhmTERF1 overexpression (AhmTERF1-OX) and AhmTERF1-silenced hairy root lines. It was found that the number of primary and lateral roots increased in AhmTERF1-OX lines. However, 70% of AhmTERF1-RNAi hairy roots were thinner and shorter (<3 cm) and exhibited growth retardation (Figure 1). This suggests a role for AhmTERF1 in hairy root formation in peanut. In other words, hairy roots were stimulated to develop when AhmTERF1 was overexpressed, but roots were stunted when AhmTERF1 expression was silenced. We obtained similar results in transgenic Arabidopsis lines overexpressing AhmTERF1: two AhmTERF1 overexpression lines with increased root length and number are shown in Figure S1.
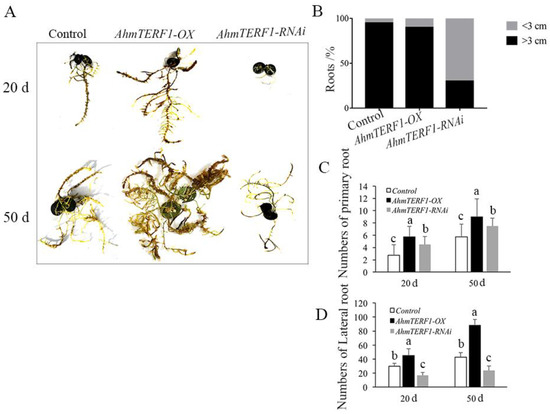
Figure 1.
AhmTERF1 affects the growth of peanut hairy roots. (A): Phenotype of 20 d-old and 50 d-old peanut hairy roots in control, AhmTERF1-overexpression (AhmTERF1-OX) and AhmTERF1-silenced (AhmTERF1-RNAi) lines. (B): Proportion of roots of different length, i.e., <3 cm or >3 cm. (C,D): Number of primary roots and lateral roots in 20 d-old and 50 d-old seedlings. Lower case letters (a, b, c) indicate significantly different groups (p < 0.05).
3.2. AhmTERF1 Is Preferentially Expressed in the Root Meristem
To understand the role of AhmTERF1 in root system architecture, we fused the AhmTERF1 promoter to the GUS reporter gene and transformed the construct into peanut hairy roots and Col Arabidopsis. The hairy roots containing pAhmTERF1:GUS displayed a GUS expression pattern in the primary and lateral root tips (Figure 2A,B). Arabidopsis transgenic plants showed GUS expression signal in the mucilage of imbibed seeds, root hairs and seedling stems, and especially in the meristematic zone of primary and lateral root tips (Figure 2C–E), consistent with the results obtained with transgenic peanut hairy roots. To further explore AhmTERF1 expression, we next investigated the expression pattern of AhmTERF1 in peanut roots at different development stages. Q-PCR results showed that AhmTERF1 was upregulated as peanut roots develop (Figure 3), indicating that AhmTERF1 is involved in the root system architecture of peanut.
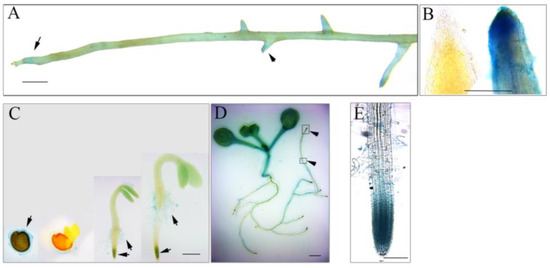
Figure 2.
Tissue distribution of AhmTERF1 gene expression. (A): GUS staining of pAhmTERF1:GUS hairy roots. Scale bar: 0.5 cm. (B): Root tips of hairy roots (Left: Control hariy root; Right: pAhmTERF1:GUS hairy roots). Scale bar: 200 µm. (C): GUS staining of transgenic pAhmTERF1:GUS Arabidopsis seeds and seedlings (images from left): seed imbibing water; seed at germination stage; seedlings 3 and 5 d after the onset of germination, respectively. Scale bar: 0.5 cm. (D): GUS staining of transgenic pAhmTERF1:GUS Arabidopsis seedling 7 d after the onset of germination. The boxes and the arrowheads indicate root tips. Scale bar: 0.5 cm. (E): Larger version of root tip. Scale bar: 100 µm.

Figure 3.
Expression pattern of AhmTERF1 at various peanut root growth stages. (A): Phenotype at various peanut root growth stages. Scale bar: 1 cm. (B): AhmTERF1 expression level at the corresponding root growth stages. Lower case letters (a, b, c) indicate significantly different groups (p < 0.05).
3.3. AhmTERF1 Overexpression Increases the Number of Mitochondria in Hairy Roots
AhmTERF1 has been localized within cells to a position close to mitochondria [26]. To further confirm the role of AhmTERF1 in mitochondria and its function in root growth, we investigated the size and number of mitochondria in AhmTERF1-OX hairy root tips by transmission electron microscopy. The size and morphology of mitochondria in AhmTERF1-OX hairy roots were similar to those of controls. However, the number of mitochondria was significantly higher in AhmTERF1-OX hairy roots (Figure 4).
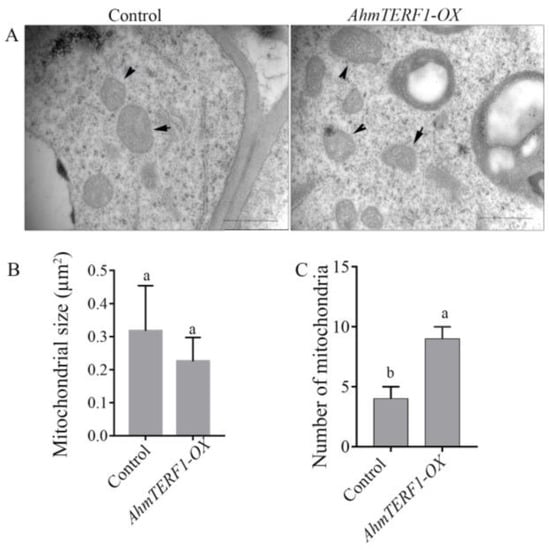
Figure 4.
Mitochondria in the tips of Control and AhmTERF1-OX hairy roots shown by transmission electron microscopy. (A): Images of mitochondria in the root tip of 20 d-old Control and AhmTERF1-OX hairy roots. Scale bars: 1 µm. (B,C): The size and number of mitochondria in the root tips of 20 d-old Control and AhmTERF1-OX hairy roots. Lower case letters (a, b) indicate significantly different groups (p < 0.05).
Mitochondrial defects in the Arabidopsis mterf14 mutant result in a dwarf phenotype. Therefore, we hypothesized that AhmTERF1 might restore the dwarf phenotype of the mterf14 mutant. Indeed, AhmTERF1mterf14 plants exhibited a normal Col phenotype, confirming our hypothesis (Supplemental Figure S2). By examining the root tips, we showed that the size of the meristem region and the number of cells decreased in the mterf14 mutant, but were higher than controls in the AhmTERF1-OX line. In 35S:AhmTERF1/mterf14 Arabidopsis plants, the short-root phenotype of the mterf14 mutant was restored to that of controls (Figure 5), implying that AhmTERF1 regulates root growth by promoting the activity of the root tip meristem.
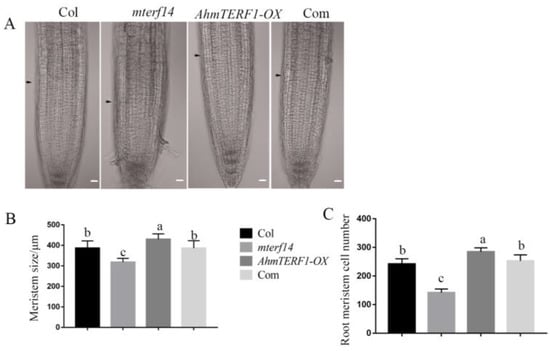
Figure 5.
AhmTERF1 accelerates root meristem growth in Arabidopsis plants. (A): Images of root meristem in AhmTERF1-OX transgenic Arabidopsis. Scale bars: 20 µm. Arrows indicate the boundary between the root meristem and the elongation zone. (B,C): Meristem size and cell number of roots in A. Com indicates 35S:AhmTERF1/mterf14 Arabidopsis plants. Lower case letters (a, b, c) indicate significantly different groups (p < 0.05).
We next treated the root tip with MitoTracker Red CMXRos to fluorescently mark mitochondria. Compared with Col and mterf14 mutant plants, the intensity of fluorescence in AhmTERF1-OX root tips was higher, which is consistent with an elevated number of mitochondria in AhmTERF1-OX plants (Figure 6A,B). ATP content correlated with fluorescence levels (Figure 6C). Overall, these results suggest that AhmTERF1 is crucial for facilitating mitochondrial accumulation.
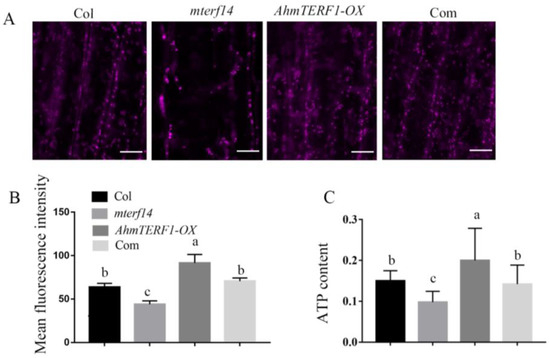
Figure 6.
Mitochondria and ATP content are increased in AhmTERF1-OX Arabidopsis. (A): Images of mitochondria in the root tip of 3-d old Arabidopsis taken by confocal laser scanning microscope (LSM800, Carl Zeiss, Germany). Scale bars: 5 µm. MitoTracker was used as a mitochondrial marker. (B): Fluorescence intensity of A. (C): ATP content of the roots of 3-d old Arabidopsis seedlings. ATP content was calculated as µmol/g. Com indicates 35S:AhmTERF1/mterf14 Arabidopsis plants. Lower case letters (a, b, c) indicate significantly different groups (p < 0.05).
3.4. Identification of the Target Genes of AhmTERF1
We supposed that the target genes of AhmTERF1 would be located in the mitochondrial genome. ChIP-qPCR indicated that AhmTERF1 was enriched in regions of the RRN18 and RRN26 rRNA genes (Figure 7A). Furthermore, Q-PCR results suggested that the expression levels of RRN18 and RRN26 were enhanced in AhmTERF1-OX hairy roots (Figure 7B). 18S RNA and 26S RNA, the products of RRN18 and RRN26, are important components of mitochondrial ribosomes and are involved in their assembly. Presumably, AhmTERF1 promotes RRN18 and RRN26 expression to affect accumulation of mitochondrial ribosomes, and thus to influence the number of mitochondria in cells.
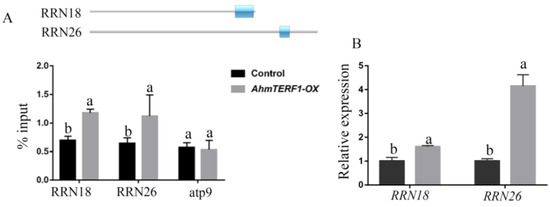
Figure 7.
AhmTERF1 binds specifically to and regulates its target rRNA genes, RRN18 and RRN26. (A): Enrichment of AhmTERF1 binding to RRN18 and RRN26 DNA sequences shown by ChIP-qPCR, with atp9 as negative control. Cox is used as an internal reference. The upper figure represents gene sequences. Blue rectangles represent the enrichment region. (B): RRN18 and RRN26 expression in Control and AhmTERF1-OX hairy roots. Lower case letters (a, b) indicate significantly different groups (p < 0.05).
3.5. AhmTERF1 Expression in Different Cultivars of Peanut
Different cultivars of peanut exhibit different agronomic traits, for example, varying in the characteristics of their roots. Fuhua9, which has a well-developed root system, showed stronger expression of AhmTERF1 than Zhonghua16, which has a relatively underdeveloped root system at the same stage (5 d) of seedling development (Figure 8A–H). The target genes of AhmTERF1, RRN18 and RRN26, were also variably expressed in these two peanut cultivars. Thus, RRN18 and RRN26 showed higher transcript levels in Fuhua9 than in Zhonghua16 (Figure 8I). We also measured the abundance of mitochondrial DNA relative to a nuclear gene to evaluate mtDNA copy number. In Fuhua9, the mtDNA copy number was higher than in Zhonghua16 (Figure 8J). These results indicate a role for AhmTERF1 in regulating agronomic traits. In particular, it seems likely that those peanut cultivars with strong AhmTERF1 expression have a more extensive root system.
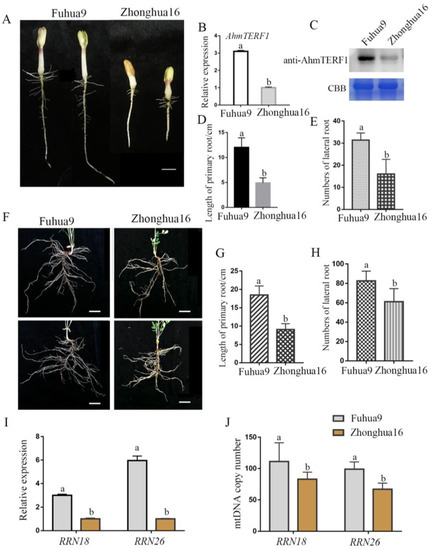
Figure 8.
The effect of AhmTERF1 expression on root growth and mtDNA copy number in Fuhua9 and Zhonghua16 peanut cultivars. (A): Phenotype of 5 d-old Fuhua9 and Zhonghua16 cultivars. (B,C): AhmTERF1 transcript and protein expression, respectively, in the roots of 5 d-old Fuhua9 and Zhonghua16. (D,E): Length of primary root and number of lateral roots of 5 d-old Fuhua9 and Zhonghua16. (F): Phenotype of 25 d-old Fuhua9 and Zhonghua16 cultivars. (G,H): Length of primary root and number of lateral roots, respectively, of 25 d-old Fuhua9 and Zhonghua16. (I): RRN18 and RRN26 expression in the roots of 5 d-old Fuhua9 and Zhonghua16. (J): mtDNA copy number in the roots of 5 d-old Fuhua9 and Zhonghua16. mtDNA was labeled with RRN18 and RRN26. Lower case letters (a, b) indicate significantly different groups (p < 0.05).
3.6. Mechanism of AhmTERF1 to Regulate Root Growth
In order to determine accurately the location of AhmTERF1 within cells, we futher conducted the subcellular localization assay and it is revealed the co-localization of AhmTERF1 with the ER and mtDNA (Supplemental Figure S3). To sum up, we can speculate that AhmTERF1 is located at ER-mitochondrion contract sites to regulate root growth by modulating mitochondrial abundance (Figure 9).
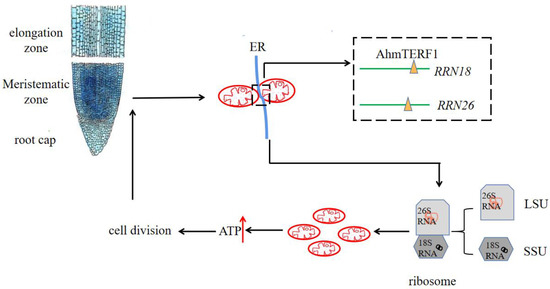
Figure 9.
Working model of AhmTERF1 function.
AhmTERF1 is preferentially expressed in the root meristem. At mitochondrion-ER contact sites, AhmTERF1 combines with mtDNA, and is enriched in the RRN18 and RRN26 loci. The respective transcripts 18S RNA and 26S RNA are involved in the assembly of mitochondrial ribosomes, which promotes an increase in the number of mitochondria. Thus, energy production is increased and can then be used for cell division, which will increase the extent of the meristematic zone and result in peanut root growth.
4. Discussion
The mitochondrion is one of the most malleable organelles in the cell. It can change its abundance, structure and distribution in the cytoplasm according to specific energy requirements. In plants, when levels of energy demand and metabolic flux are high, this is reflected in an increase in the number or area of mitochondria, as well as in higher levels of mitochondrial electron transport chain (mETC) components [38,39,40]. During the seed germination, development of pollen and ripening of fruit stages, mitochondria show the highest respiration rate [41]. Pollen respiration increases 10-fold, and the number of mitochondria increases 20 to 40 times, in meiotic and tapetal cells [38]. The doubling of mitochondrial volume and number, as well as a boost in mitochondrial dynamics, occur in germinated seedlings to provide energy for seedling growth and development [4]. In the root apical meristem of Arabidopsis, along with an increasing of respiratory rate, an approximately 3-fold increase in mitochondrial number have been found [41]. Within the root meristem, cell division is important to maintain the root meristem activity and support root growth [42]. Thus, presumably, our observations of an increase in the number of mitochondria in peanut root tips indicate an acceleration in respiratory activity, providing energy for root growth.
In comparison with other organelles, biosynthesis displays a particular level of complexity in mitochondria, because they contain their own genomes and specialized ribosomes. The mitochondrial translation process seems to be a key point to monitor mitochondrial homeostasis and may play a part in establishing the abundance of mitochondria within the cells.
Previous research has shown that mTERFs are involved in ribosome biogenesis. Lack of DmMTERF3 was found in Drosophila to be related with a reducing of the amount of 16S rRNA and the assembly of mitochondrial ribosomes was hindered [43]. MTERF4 and NSUN4 RNA methyltransferase join together to form a complex, which is necessary to target NSUN4 to the mammalian mitochondrial ribosome [44]. We found that binding of AhmTERF1 to mtDNA was enriched in the region containing the rRNA genes, RRN18 and RRN26. 18S RNA and 26S RNA, the products of these genes, are essential components of mitochondrial ribosomes and are crucial for their assembly. In the mitochondrial genome of higher plants, three ribosomal RNA (rRNA) genes, including rrn26, rrn18 and rrn5, encode 26S, 18S and 5S rRNAs, respectively [45]. Post-transcriptional modifications of rRNAs are essential for ribosome biogenesis and translation [46]. In spite of their greater diversity in plants, knowledge of mTERF functions in mitochondrial ribosomal biogenesis remains sparse. For example, levels of 16S and 23S rRNAs were decreased in Zm-mterf4 mutants of maize, showing the important role of Zmmterf4 in the accumulation of plastid ribosomes [47]. In this paper, we speculate that the expression of RRN18 and RRN26 accelerates the accumulation of mitochondrial ribosomes, thereby increasing the number of mitochondria and supporting the growth of roots.
In addition, this paper revealed the co-localization of AhmTERF1 with the ER and mtDNA (Supplemental Figure S3). Mitochondria have a close relationship with the ER; ER-mitochondrion contact sites is important for mitochondrial fission, lipid transfer, calcium signaling [48] and the synchronization of mtDNA synthesis [49]. The majority (84%) of mitochondrial fission events happen at this location [50]. The unit of mtDNA inheritance, nucleoids contain lots of copies of mtDNA and display distribution in mitochondrial networks. In mammalian cells, these nucleoids linked to a small subset of ER-mitochondrion contact points spatially and temporally, involve in mtDNA synthesis. Before mitochondrial constriction and division. mtDNA replication occurs. ER-mitochondrion contact sites regulate mtDNA replication and distribute the newly replicated mtDNA to progeny mitochondria [49]. In our previous study, it was found that AhmTERF1 was closely surrounded by mitochondria [26]. This might suggest that AhmTERF1 is located at ER-mitochondrion contract sites and that the mitochondria around AhmTERF1 are daughter mitochondria produced by mitochondrial division at those sites (Figure 9).
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/genes14010209/s1, Figure S1: AhmTERF1 promotes root growth in Arabidopsis; Figure S2: AhmTERF1 restores the phenotype of mterf14 in Arabidopsis; Figure S3: AhmTERF1 co-localizes with mtDNA and ER; Table S1: Primers used in this study.
Author Contributions
L.L. (Limei Li) and X.L. performed most of the experiments; C.Y. prepared material for the assay; L.L. (Limei Li) wrote the manuscript; X.L. and L.L. (Ling Li) conceived the project. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the National Natural Science Foundation of China, grant number 32071924, the Scientific Research Starting Foundation for PhD of Zhaoqing University, grant number 210073, and Youth Foundation of Zhaoqing University, grant number QN202221. And The APC was funded by the National Natural Science Foundation of China, grant number 32071924.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data that support the findings of this study are available from the authors, upon reasonable request.
Acknowledgments
We are grateful to Xingliang Hou and associate research fellow Xu Liu of South China Botanical Garden, Chinese Academy of Sciences for supporting vector construction and ultrastructural analyses. We also grateful to Caiji Gao of South China Normal University for providing ER-mCherry plasmid.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Karlova, R.; Boer, D.; Hayes, S.; Testerink, C. Root plasticity under abiotic stress. Plant Physiol. 2021, 187, 1057–1070. [Google Scholar] [CrossRef] [PubMed]
- Snyder, C.; Stefano, G.B. Mitochondria and chloroplasts shared in animal and plant tissues: Significance of communication. Med. Sci. Monit. 2015, 21, 1507–1511. [Google Scholar] [PubMed]
- Sarasija, S.; Norman, K.R. A γ-Secretase Independent Role for Presenilin in Calcium Homeostasis Impacts Mitochondrial Function and Morphology in Caenorhabditis elegans. Genetics 2015, 201, 1453–1466. [Google Scholar] [CrossRef] [PubMed]
- Paszkiewicz, G.; Gualberto, J.M.; Benamar, A.; Macherel, D.; Logan, D.C. Arabidopsis Seed Mitochondria Are Bioenergetically Active Immediately upon Imbibition and Specialize via Biogenesis in Preparation for Autotrophic Growth. Plant Cell 2017, 29, 109–128. [Google Scholar] [CrossRef] [PubMed]
- Ma, W.; Guan, X.; Li, J.; Pan, R.; Wang, L.; Liu, F.; Ma, H.; Zhu, S.; Hu, J.; Ruan, Y.L.; et al. Mitochondrial small heat shock protein mediates seed germination via thermal sensing. Proc. Natl. Acad. Sci. USA 2019, 116, 4716–4721. [Google Scholar] [CrossRef]
- Huang, Y.; Chen, X.; Liu, Y.; Roth, C.; Copeland, C.; McFarlane, H.E.; Huang, S.; Lipka, V.; Wiermer, M.; Li, X. Mitochondrial AtPAM16 is required for plant survival and the negative regulation of plant immunity. Nat. Commun. 2013, 4, 2558. [Google Scholar] [CrossRef]
- Xu, Y.; Berkowitz, O.; Narsai, R.; De Clercq, I.; Hooi, M.; Bulone, V.; Van Breusegem, F.; Whelan, J.; Wang, Y. Mitochondrial function modulates touch signalling in Arabidopsis thaliana. Plant J. 2019, 97, 623–645. [Google Scholar] [CrossRef]
- Fernie, A.R. Making sense of the complex role of the mitochondria in mediating the plant touch response. Plant J. 2019, 97, 621–622. [Google Scholar] [CrossRef]
- Van Aken, O.; Pecenková, T.; van de Cotte, B.; De Rycke, R.; Eeckhout, D.; Fromm, H.; De Jaeger, G.; Witters, E.; Beemster, G.T.; Inzé, D.; et al. Mitochondrial type-I prohibitins of Arabidopsis thaliana are required for supporting proficient meristem development. Plant J. 2007, 52, 850–864. [Google Scholar] [CrossRef]
- Zhou, X.; Li, Q.; Chen, X.; Liu, J.; Zhang, Q.; Liu, Y.; Liu, K.; Xu, J. The Arabidopsis RETARDED ROOT GROWTH gene encodes a mitochondria-localized protein that is required for cell division in the root meristem. Plant Physiol. 2011, 157, 1793–1804. [Google Scholar] [CrossRef]
- Hsieh, W.Y.; Liao, J.C.; Hsieh, M.H. Dysfunctional mitochondria regulate the size of root apical meristem and leaf development in Arabidopsis. Plant Signal. Behav. 2015, 10, e1071002. [Google Scholar] [CrossRef] [PubMed]
- Kleine, T.; Leister, D. Emerging functions of mammalian and plant mTERFs. Biochim. Biophys. Acta 2015, 1847, 786–797. [Google Scholar] [CrossRef] [PubMed]
- Kruse, B.; Narasimhan, N.; Attardi, G. Termination of transcription in human mitochondria: Identification and purification of a DNA binding protein factor that promotes termination. Cell 1989, 58, 391–397. [Google Scholar] [CrossRef] [PubMed]
- Kleine, T. Arabidopsis thaliana mTERF proteins: Evolution and functional classification. Front. Plant Sci. 2012, 3, 233. [Google Scholar] [CrossRef]
- Zhao, Y.; Cai, M.; Zhang, X.; Li, Y.; Zhang, J.; Zhao, H.; Kong, F.; Zheng, Y.; Qiu, F. Genome-wide identification, evolution and expression analysis of mTERF gene family in maize. PLoS ONE 2014, 9, e94126. [Google Scholar] [CrossRef]
- Meskauskiene, R.; Würsch, M.; Laloi, C.; Vidi, P.A.; Coll, N.S.; Kessler, F.; Baruah, A.; Kim, C.; Apel, K. A mutation in the Arabidopsis mTERF-related plastid protein SOLDAT10 activates retrograde signaling and suppresses (1)O(2)-induced cell death. Plant J. 2009, 60, 399–410. [Google Scholar] [CrossRef]
- Babiychuk, E.; Vandepoele, K.; Wissing, J.; Garcia-Diaz, M.; De Rycke, R.; Akbari, H.; Joubès, J.; Beeckman, T.; Jänsch, L.; Frentzen, M.; et al. Plastid gene expression and plant development require a plastidic protein of the mitochondrial transcription termination factor family. Proc. Natl. Acad. Sci. USA 2011, 108, 6674–6679. [Google Scholar] [CrossRef]
- Quesada, V.; Sarmiento-Mañús, R.; González-Bayón, R.; Hricová, A.; Pérez-Marcos, R.; Graciá-Martínez, E.; Medina-Ruiz, L.; Leyva-Díaz, E.; Ponce, M.R.; Micol, J.L. Arabidopsis RUGOSA2 encodes an mTERF family member required for mitochondrion, chloroplast and leaf development. Plant J. 2011, 68, 738–753. [Google Scholar] [CrossRef]
- Robles, P.; Micol, J.L.; Quesada, V. Arabidopsis MDA1, a nuclear-encoded protein, functions in chloroplast development and abiotic stress responses. PLoS ONE 2012, 7, e42924. [Google Scholar] [CrossRef]
- Romani, I.; Manavski, N.; Morosetti, A.; Tadini, L.; Maier, S.; Kühn, K.; Ruwe, H.; Schmitz-Linneweber, C.; Wanner, G.; Leister, D.; et al. A Member of the Arabidopsis Mitochondrial Transcription Termination Factor Family Is Required for Maturation of Chloroplast Transfer RNAIle(GAU). Plant Physiol. 2015, 169, 627–646. [Google Scholar] [CrossRef]
- Mokry, M.; Nijman, I.J.; van Dijken, A.; Benjamins, R.; Heidstra, R.; Scheres, B.; Cuppen, E. Identification of factors required for meristem function in Arabidopsis using a novel next generation sequencing fast forward genetics approach. BMC Genom. 2011, 12, 256. [Google Scholar] [CrossRef]
- Robles, P.; Micol, J.L.; Quesada, V. Mutations in the plant-conserved MTERF9 alter chloroplast gene expression, development and tolerance to abiotic stress in Arabidopsis thaliana. Physiol. Plant 2015, 154, 297–313. [Google Scholar] [CrossRef] [PubMed]
- Hsu, Y.W.; Wang, H.J.; Hsieh, M.H.; Hsieh, H.L.; Jauh, G.Y. Arabidopsis mTERF15 is required for mitochondrial nad2 intron 3 splicing and functional complex I activity. PLoS ONE 2014, 9, e112360. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.; Lee, U.; Small, I.; des Francs-Small, C.C.; Vierling, E. Mutations in an Arabidopsis mitochondrial transcription termination factor-related protein enhance thermotolerance in the absence of the major molecular chaperone HSP101. Plant Cell 2012, 24, 3349–3365. [Google Scholar] [CrossRef] [PubMed]
- Zhuang, W.; Chen, H.; Yang, M.; Wang, J.; Pandey, M.K.; Zhang, C.; Chang, W.C.; Zhang, L.; Zhang, X.; Tang, R.; et al. The genome of cultivated peanut provides insight into legume karyotypes, polyploid evolution and crop domestication. Nat. Genet. 2019, 51, 865–876. [Google Scholar] [CrossRef]
- Li, L.M.; Hu, B.; Li, X.Y.; Li, L. Characterization of mTERF family in allotetraploid peanut and their expression levels in response to dehydration stress. Biotechnol. Biotec. Eq. 2020, 34, 1176–1187. [Google Scholar] [CrossRef]
- Zhou, Y.; Zeng, L.; Hou, X.; Liao, Y.; Yang, Z. Low temperature synergistically promotes wounding-induced indole accumulation by INDUCER OF CBF EXPRESSION-mediated alterations of jasmonic acid signaling in Camellia sinensis. J. Exp. Bot. 2020, 71, 2172–2185. [Google Scholar] [CrossRef] [PubMed]
- Qian, Q.; Yang, Y.; Zhang, W.; Hu, Y.; Li, Y.; Yu, H.; Hou, X. A novel Arabidopsis gene RGAT1 is required for GA-mediated tapetum and pollen development. New Phytol. 2021, 231, 137–151. [Google Scholar] [CrossRef]
- Liu, S.; Su, L.; Liu, S.; Zeng, X.; Zheng, D.; Hong, L.; Li, L. Agrobacteriumrhizogenes-mediated transformation of Arachis hypogaea: An efficient tool for functional study of genes. Biotechnol. Biotec. Eq. 2016, 30, 869–878. [Google Scholar] [CrossRef]
- Clough, S.J.; Bent, A.F. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998, 16, 735–743. [Google Scholar] [CrossRef]
- Wan, X.; Li, L. Molecular cloning and characterization of a dehydration-inducible cDNA encoding a putative 9-cis-epoxycarotenoid dioxygenase in Arachis hygogaea L. DNA Seq. 2005, 16, 217–223. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Scheres, B. Dissection of Arabidopsis ADP-RIBOSYLATION FACTOR 1 function in epidermal cell polarity. Plant Cell 2005, 17, 525–536. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Li, L.; Li, M.; Su, L.; Lian, S.; Zhang, B.; Li, X.; Ge, K.; Li, L. AhGLK1 affects chlorophyll biosynthesis and photosynthesis in peanut leaves during recovery from drought. Sci. Rep. 2018, 8, 2250. [Google Scholar] [CrossRef]
- Fang, L.; Hou, X.; Lee, L.Y.; Liu, L.; Yan, X.; Yu, H. AtPV42a and AtPV42b redundantly regulate reproductive development in Arabidopsis thaliana. PLoS ONE 2011, 6, e19033. [Google Scholar] [CrossRef] [PubMed]
- Qiu, Q.; Mei, H.; Deng, X.; He, K.; Wu, B.; Yao, Q.; Zhang, J.; Lu, F.; Ma, J.; Cao, X. DNA methylation repels targeting of Arabidopsis REF6. Nat. Commun. 2019, 10, 2063. [Google Scholar] [CrossRef]
- Mondal, R.; Ghosh, S.K.; Choudhury, J.H.; Seram, A.; Sinha, K.; Hussain, M.; Laskar, R.S.; Rabha, B.; Dey, P.; Ganguli, S.; et al. Mitochondrial DNA copy number and risk of oral cancer: A report from Northeast India. PLoS ONE 2013, 8, e57771. [Google Scholar] [CrossRef]
- Mei, H.; Sun, S.; Bai, Y.; Chen, Y.; Chai, R.; Li, H. Reduced mtDNA copy number increases the sensitivity of tumor cells to chemotherapeutic drugs. Cell Death Dis. 2015, 6, e1710. [Google Scholar] [CrossRef]
- Warmke, H.E.; Lee, S.L. Pollen Abortion in T Cytoplasmic Male-Sterile Corn (Zea mays): A Suggested Mechanism. Science 1978, 200, 561–563. [Google Scholar] [CrossRef]
- Huang, J.; Struck, F.; Matzinger, D.F.; Levings, C.S., 3rd. Flower-enhanced expression of a nuclear-encoded mitochondrial respiratory protein is associated with changes in mitochondrion number. Plant Cell 1994, 6, 439–448. [Google Scholar]
- Zabaleta, E.; Heiser, V.; Grohmann, L.; Brennicke, A. Promoters of nuclear-encoded respiratory chain complex I genes from Arabidopsis thaliana contain a region essential for anther/pollen-specific expression. Plant J. 1998, 15, 49–59. [Google Scholar] [CrossRef]
- Liberatore, K.L.; Dukowic-Schulze, S.; Miller, M.E.; Chen, C.; Kianian, S.F. The role of mitochondria in plant development and stress tolerance. Free Radic. Biol. Med. 2016, 100, 238–256. [Google Scholar] [CrossRef] [PubMed]
- Perilli, S.; Di Mambro, R.; Sabatini, S. Growth and development of the root apical meristem. Curr. Opin. Plant Biol. 2012, 15, 17–23. [Google Scholar] [CrossRef]
- Wredenberg, A.; Lagouge, M.; Bratic, A.; Metodiev, M.D.; Spåhr, H.; Mourier, A.; Freyer, C.; Ruzzenente, B.; Tain, L.; Grönke, S.; et al. MTERF3 regulates mitochondrial ribosome biogenesis in invertebrates and mammals. PLoS Genet. 2013, 9, e1003178. [Google Scholar] [CrossRef]
- Cámara, Y.; Asin-Cayuela, J.; Park, C.B.; Metodiev, M.D.; Shi, Y.; Ruzzenente, B.; Kukat, C.; Habermann, B.; Wibom, R.; Hultenby, K.; et al. MTERF4 regulates translation by targeting the methyltransferase NSUN4 to the mammalian mitochondrial ribosome. Cell Metab. 2011, 13, 527–539. [Google Scholar] [CrossRef] [PubMed]
- Petersen, G.; Cuenca, A.; Møller, I.M.; Seberg, O. Massive gene loss in mistletoe (Viscum, Viscaceae) mitochondria. Sci. Rep. 2015, 5, 17588. [Google Scholar] [CrossRef]
- Van Haute, L.; Hendrick, A.G.; D’Souza, A.R.; Powell, C.A.; Rebelo-Guiomar, P.; Harbour, M.E.; Ding, S.; Fearnley, I.M.; Andrews, B.; Minczuk, M. METTL15 introduces N4-methylcytidine into human mitochondrial 12S rRNA and is required for mitoribosome biogenesis. Nucleic Acids Res. 2019, 47, 10267–10281. [Google Scholar] [CrossRef] [PubMed]
- Hammani, K.; Barkan, A. An mTERF domain protein functions in group II intron splicing in maize chloroplasts. Nucleic Acids Res. 2014, 42, 5033–5042. [Google Scholar] [CrossRef]
- Friedman, J.R.; Voeltz, G.K. The ER in 3D: A multifunctional dynamic membrane network. Trends Cell Biol. 2011, 21, 709–717. [Google Scholar] [CrossRef] [PubMed]
- Lewis, S.C.; Uchiyama, L.F.; Nunnari, J. ER-mitochondria contacts couple mtDNA synthesis with mitochondrial division in human cells. Science 2016, 353, aaf5549. [Google Scholar] [CrossRef]
- Guo, Y.; Li, D.; Zhang, S.; Yang, Y.; Liu, J.J.; Wang, X.; Liu, C.; Milkie, D.E.; Moore, R.P.; Tulu, U.S.; et al. Visualizing Intracellular Organelle and Cytoskeletal Interactions at Nanoscale Resolution on Millisecond Timescales. Cell 2018, 175, 1430–1442.e17. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).