Abstract
Introduction: Osteogenesis imperfecta (OI) is a rare mendelian skeletal dysplasia with autosomal dominant or recessive inheritance pattern, and almost the most common primary osteoporosis in prenatal settings. The diversity of clinical presentation and genetic etiology in prenatal OI cases presents a challenge to counseling yet has seldom been discussed in previous studies. Methods: Ten cases with suspected fetal OI were enrolled and submitted to a genetic detection using conventional karyotyping, chromosomal microarray analysis (CMA), and whole-exome sequencing (WES). Sanger sequencing was used as the validation method for potential diagnostic variants. In silico analysis of specific missense variants was also performed. Results: The karyotyping and CMA results of these cases were normal, while WES identified OI-associated variants in the COL1A1/2 genes in all ten cases. Six of these variants were novel. Additionally, four cases here exhibited distinctive clinical and/or genetic characteristics, including the situations of intrafamilial phenotypic variability, parental mosaicism, and “dual nosogenesis” (mutations in collagen I and another gene). Conclusion: Our study not only expands the spectrum of COL1A1/2-related OI, but also highlights the complexity that occurs in prenatal OI and the importance of clarifying its pathogenic mechanisms.
1. Introduction
Osteogenesis imperfecta (OI), also known as “brittle bonedisease”, is a spectrum of rare inherited connective-tissue disorders mainly characterized by fractures with minimal or absent trauma, variable dentinogenesis imperfecta (DI), and, in adult years, hearing loss [1]. The incidence of OI is approximately 6–7 per 100,000 births [2]. OI is phenotypically and genetically heterogeneous with clinical severity ranges from perinatal lethal to nearly asymptomatic [2]. Based on clinical presentations, radiographic features, family history, and natural history, OI was initially classified into four common types by Sillence et al. [3], and has recently been re-categorized into 18 fine subtypes according to its genetic pathogenesis [4].
To date, at least 20 genes with autosomal dominant or recessive inheritance pattern have been recognized to be responsible for OI, including COL1A1, COL1A2, BMP1, SERPINF1, SERPINH1, CRTAP, P3H1, PPIB, TMEM38B, WNT1, FKBP10, PLOD2, IFITM5, MBTPS2, CREB3L1, SP7, SPARC, P4HB, PLS3, and SEC24D [1], of which the COL1A1/2 (MIM *120150/120160) associated subtypes account for ~90% cases [2]. As the main etiological components, COL1A1/2 encode the alpha 1 and alpha 2 chains of type I collagen, the major protein component of the extracellular matrix inbone, skin, and tendon. Untilnow, over 3000 variants in COL1A1/2 have been indexed in the osteogenesis imperfecta variant database, yet the phenotype–genotype correlation in this disorder is still not meticulously established (http://www.le.ac.uk/ge/collagen/; accessed on 25 September 2020).
In the clinical practice of prenatal diagnosis, ultrasonographic evaluation is the first-line screening method for skeletal dysplasias (SD) during both early and late gestational stages [5]. Yet, it has obvious shortcomings in the defining of disease types and possible prognosis, which requires the facilitation of genetic techniques. In our previous study, we established a trio whole-exome sequencing (trio-WES, meaning to submit “proband–father–mother” trios simultaneously for sequencing and mutation screening analysis) detection strategy, introduced it into the diagnosis of prenatal SD cases, and achieved a considerable detection efficiency [6,7]. We also reported a rare autosomal recessive OI case and identified a novel compound heterozygous variation in the PLOD2 gene, which encodes the lysine hydroxylase responsible for the proper cross-linking of type I collagen pro-fibrils [8]. Continuous effort has been made to better understand the various conditions of prenatal OI.
Here in the present study, 10 cases with prenatal skeletal dysplasia were enrolled and underwent a comprehensive clinical and genetic investigation. All the patients were positive with COL1A1 or COL1A2 variants; however, distinct situations existed in different cases, including the intrafamilial phenotypic variability, parental mosaicism, and “dual nosogenesis” with variants in other genes. Our study highlights the complexity of COL1A1/COL1A2 associated OI, especially in the prenatal cases.
2. Materials and Methods
The present study was approved by the Ethics Committee of Beijing Obstetrics and Gynecology Hospital (No. 2018-KY-003-01). Informed consent wasprovided by all the participants.Allprocedures performed in the present studywere in accordance withtheDeclaration of Helsinki 1964 and its later amendments orcomparable ethical standards.
2.1. Subjects and Clinical Evaluation
Between December 2018 and January 2021, a total of 10 cases with fetuses displaying in utero skeletal dysplasia were enrolled in our centers. Clinical evaluation was made via the combination of ultrasound monitoring at mid-trimester X-ray examination after fetus’ abortion, and a comprehensive survey on family history.
2.2. Karyotyping and Copy Number Variation (CNV) Analysis
To conduct a prenatal genetic diagnosis, amniocentesis was performed to obtain the amniotic fluid (AF) samples according to the routineprocess. Conventional chromosomal karyotyping by G-binding was performed to detect overall chromosomal anomalies [9].
Genomic DNA was extracted from fetal samples and their parents’ peripheral blood using QIAamp DNA Midi Kit (Qiagen, Dusseldorf, Germany). Chromosomal microarray analysis (CMA) tests with CytoScan 750K SNP-array (Affymetrix Inc., Santa Clara, CA, USA) were carried out accordingto the manufacturer’s manual workflow on all fetal specimensin order to investigate genomic CNVs with clinical significance.
2.3. Whole-Exome Sequencing
Whole-exome sequencing (WES) was employed to detect thesequence variants inthe probands’ fetal samples, as describedin our previous study [10]. Briefly, the target-region sequence enrichment was performed using the Agilent Sure Select Human Exon Sequence Capture Kit (Agilent, Palo Alto, CA, USA).DNA libraries were tested by quantitative PCR, where the size, distribution and concentration were determined using Agilent Bioanalyzer 2100 (Agilent, Palo Alto, CA, USA). Along with ~150 bp pair-endreads, the NovaSeq-6000 platform (Illumina, Inc., San Diego, CA, USA), was used for sequencing of DNA with~300 pM per sample with NovaSeq Reagent kit. Sequencing raw reads (quality level Q30%>90%; and the quality criteria was listed at https://www.illumina.com/science/technology/next-generation-sequencing/plan-experiments/quality-scores.html; accessed on 25 September 2020) were aligned to the human reference genome (accession No. hg19/GRCh37) using the Burrows Wheeler Aligner tool, and PCR duplicates were removed using Picardv1.57. Variant calling was performed with the Verita Trekker® Variants Detection system (v2.0; Berry Genomics, Beijing, China) and Genome Analysis Tool Kit (https://software.broadinstitute.org/gatk/; accessed on 25 September 2020). Then, variants were annotated and interpreted using ANNOVAR (v2.0) [11] and Enliven® Variants Annotation Interpretation systems (Berry Genomics), based on the common guidelines by ACMG (American College of Medical Genetics and Genomics) [12].To assist withthe interpretation of variant pathogenicity, we referred to three frequency databases (ExAC_EAS, http://exac.broadinstitute.org, accessed on 25 September 2020; gnomAD_exome_EAS, http://gnomad.broadinstitute.org, accessed on 25 September 2020; 1000G_2015aug_eas, https://www.internationalgenome.org, accessed on 25 September 2020) and HGMD (Human Gene Mutation Database) pro v2019; Revel score (a combined method of pathogenicity prediction) [13] and pLI score (representing the tolerance for truncating variants) were also employed.
Sanger sequencing was performed as a validation method with the 3500DX Genetic Analyzer (Applied Biosystems, Thermofisher, LA, CA, USA).
2.4. Conservatism and Structure Analysis
The evolutionary conservatism of all affected amino acid (AA) residues by corresponding missense variants was analyzed using the online tool, MEGA7 (http://www.megasoftware.net/previousVersions.php; accessed on 25 September 2020), with default parameters. Structural analysis and molecular dynamics simulation of a novel missense variant detected in the CANT1 gene were presented in Supplementary Material 1.
3. Results
3.1. Clinical Manifestations
In the 10 recruited families, the average maternal age was 31.2 (ranging from 26 to 38), and the gestational weeks with initial SD diagnosis ranged from 16W to 23W4D. All the couples were non-consanguineous and claimed to have no family history of genetic disorders. Most fetuses showed OI-like characteristics in the second or third gestational trimester, e.g., the bending or angulation of the limb long bones. However, there were a few cases with unique phenotypes, such as Case 7 and Case 8. In Case 7, the fetus presented with tibiofibula and ulnar flexure dysplasia, poor skull ossification, knee bendingabnormity, bilateral short choroid plexus; in Case 8, the fetus additionally showed thickened NT (nuchal translucency = 0.6 cm), anasarca, abnormal ankle joint and foot posture at the early second trimester. Empirically, these symptoms are highly unusual in common OI cases. According to our pregnancy tracking, all the fetus probands were aborted.
The detailed clinical manifestations and information of the 10 cases were listed in Table 1. The representative clinical images of affected fetuses were demonstrated in Figure 1, and the diagram of each case pedigree was shown in Figure 2 and Figure 3.

Table 1.
Clinical information of the ten recruited cases.
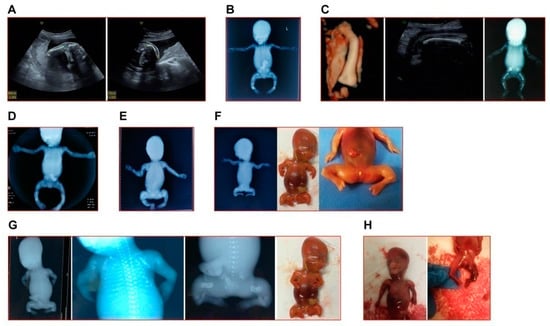
Figure 1.
The representative clinical images of fetuses in this study. (A) The left humerus (left image) and right femur (right image) of fetus in Case 1 appeared curved at angled. (B) The X-ray image of induced fetus in Case 2 showed typical OI indications (short and curved femur, tibia and fibula). (C) Case 3: left and middle, the 3D reconstruction of the curved femur and the in utero curve of femur of the second fetus; right, the X-ray image of the first induced fetus in this family. (D) The X-ray image of induced fetus in Case 5. (E) The X-ray image of induced fetus in Case 6. (F) The X-ray image (left) and appearance (middle and right) of induced fetus in Case 7. (G) The X-ray images and appearance (farthest to the right) of induced fetus in Case 8. (H) The appearance of induced fetus in Case 9. Note: In accordance with the wishes of the affected families, clinical images of Cases 4 and 10 are not shown.
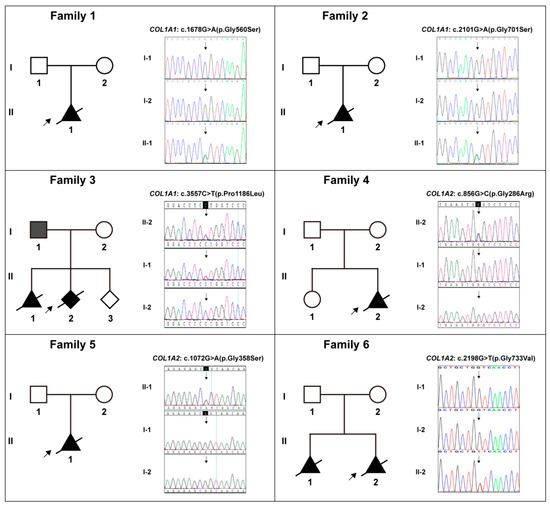
Figure 2.
The pedigree diagrams of Cases 1–6 (Families 1–6) and the variants in collagen genes (COL1A1/2) in Sanger sequencing form and their corresponding carrying status in each family. Note: Solid shapes represent mutation carriers; the arrow points to the probands; the diagonal line means dead.
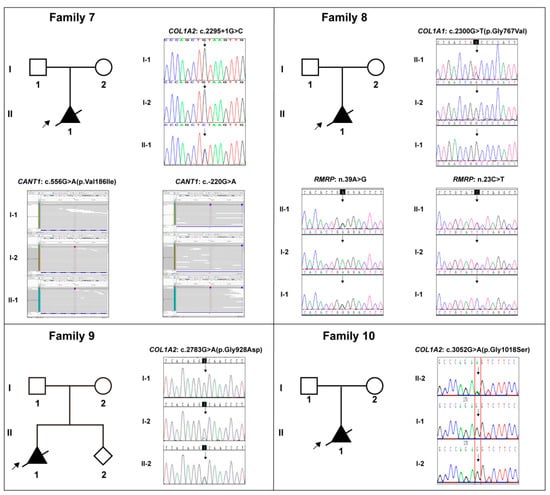
Figure 3.
The pedigree diagrams of Cases 7–10 (Families 7–10) and the variants in collagen genes (COL1A1/2), CANT1 gene, and RMRP gene in Sanger sequencing or Bam form, and their corresponding carrying status in each family. Note: Solid shapes represent mutation carriers; the arrow points to the probands; the diagonal line means dead.
3.2. Genetic Findings
All 10 cases were negative for G-banding karyotyping and CMA detection. Subsequently, WES identified 10 specific diagnostic variants in these cases, which were distributed in the COL1A1 and COL1A2 genes, namely COL1A1: c.1678G>A (p.Gly560Ser) in Case 1, COL1A1: c.2101G>A (p.Gly701Ser) in Case 2, COL1A1: c.3557C>T (p.Pro1186Leu) in Case 3, COL1A2: c.856G>C (p.Gly286Arg) in Case 4, COL1A2: c.1072G>A (p.Gly358Ser) in Case 5, COL1A2: c.2198G>T (p.Gly733Val) in Case 6, COL1A2: c.2295+1(IVS37)G>C in Case 7, COL1A1: c.2300G>T (p.Gly767Val) in Case 8, COL1A2: c.2783G>A (p.Gly928Asp) in Case 9, and COL1A2: c.3052G>A (p.Gly1018Ser) in Case 10. Table 2 included the details of all these COL1A1/2 variants. Sanger sequencing verified these variants and their familial carrying status, and the results were demonstrated in Figure 2 and Figure 3. Except for the variant in Case 3, all the variants were de novo. Six novel variants amongst them were reported, which were COL1A1: c.3557C>T, COL1A2: c.856G>C, COL1A2: c.2295+1(IVS37)G>C, COL1A1: c.2300G>T, COL1A2: c.2783G>A, and COL1A2: c.3052G>A.

Table 2.
Information of the genetic variations identified in this study.
There were four cases with special circumstances that required special attention. Case 3: Three subjects (the father and two fetuses) harboring the identical COL1A1: c.3557C>T variant showed a strong variable expressivity. The father was asymptomatic, while both fetuses had typical OI characteristics. Case 7: An additional heterozygous compound variation in the CANT1 gene, consisting of c.556G>A and c.-220G>A variants inherited from the parents, respectively, was identified and considered to be with contribution to the fetus’ phenotype (Figure 3, Family 7; Table 2). Case 8: A heterozygous compound variation in the RMRP gene, consisting of n.39A>G and n.23C>T variants inherited from the parents, respectively, was identified and considered to contribute to the fetus’ phenotype (Figure 3, Family 8; Table 2). Case 9: The mother, who was asymptomatic, was a mosaic carrier of the COL1A2: c.2783G>A variant (Figure 3, Family 9).
3.3. Analysis of the Missense Variants
In this study, a total of nine missense variants in COL1A1/2 genes were detected. In addition, according to the analysis results, the AA residues they affected maintained a high degree of evolutionary conservatism among species (Figure 4). Since the pathogenicity of the CANT1: Val186Ile variant was not certain, we conducted the structural and molecular dynamics analysis. It was demonstrated that this variant probably impacted the protein stability and secondary structure (detailed results see Supplementary Material).
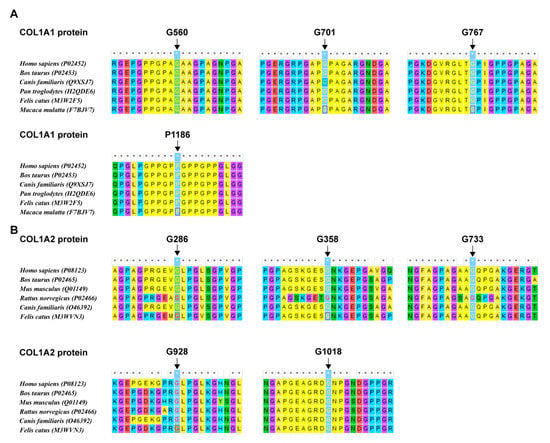
Figure 4.
The evolutionaryconservatism of AA residues affected by nine missense variants in the COL1A1/2 genes identified in this study. (A) Conservatism of the residues in COL1A1 affected by missense variants detected in this study. (B) Conservatism of the residues in COL1A2 affected by missense variants detected in this study.
4. Discussion
Osteogenesis imperfecta (OI) is a rare genetic disorder with an estimated prevalence between 1/13,500 and 1/9700(14), yet the most common form of primary osteoporosis in prenatal and perinatal circumstances [7,14,15]. There were numerous studies using molecular genetic techniques to achieve the prenatal diagnosis on OI [8,16,17,18,19]. A great deal of mutations in the COL1A1/2 genes, which confirms to the autosomal dominant pattern and contributed to a major part of OI cases, have been reported and indexed; as a result, researchers have begun to develop methods to assess the prognosis caused by specific types of mutations in recent years [20]. However, in the prenatal settings, OI cases may present with more complexity, which still warrants more sufficient attention. In the present study, we retrospectively summarized the clinical and genetic characteristics of 10 cases with COL1A1/2-related OI. Generally speaking, the affected fetuses were diagnosed by ultrasonography at middle to late trimester with characteristic long bone curvature. Among all the COL1A1/2 variations, missense variants resulting in the replacement of Gly residue in the triple helical domain occupied the major part, up to eight in total, which indicates the predominant role of these variations in OI pathogenesis. Besides, the conservatism of AA residues affected by each missense variant strongly supports their pathogenicity. On the other hand, some individual cases displayed unique clinical presentations, which could likely be related to their specific genetic backgrounds.
In Case 3, the asymptomatic father and two affected fetuses all carried the missense variant COL1A1: p.Pro1186Leu, which reflects the strong intra-familial phenotypic variability of OI. According to the large cohort studied by Zhytniket al., 32.81% families had increasing or decreasing OI phenotype severity across generations, and higher intrafamilial variability of phenotypes correlated with the collagen I dominant negative variants [21]. In other words, the father in the family may have cryptic mild skeletal symptoms that went undetected by the clinician. As for this novel variant, the pathogenicity of another missense variation (p.Pro1186Ala) affecting the identical residue has been reported [22], thus supporting it to be deleterious. It was reported that dominant-negative mutations in COL1A1 were more likely to result in phenotypic variability, but functional experiments are needed to test this theory and provide evidence of whether specific variations are more suitable for particular treatment [21,23].
In Case 7, the affected fetus presented with a very complex and extensive skeletal dysplasia, while a suspected causative compound heterozygous variations in the CANT1 gene was detected, along with the splicing site variant in COL1A2. The CANT1 gene (Calcium-activated nucleotidase 1, MIM *613165), encoding an extracellular protein that functions as a nucleotide tri- and diphosphatase [24], is responsible for a spectrum of skeletal dysplasia with diverse indications, including the Desbuquois dysplasia 1 (MIM #251450) [25], the multiple epiphyseal dysplasia 7 (MED, MIM #617719) [26], and the pseudodiastrophic dysplasia (PDD, MIM #264180) [27]. Among them, PDD is the rarest subtype and associated with prenatal manifestation and early lethality, characterized by short-limbed short stature, facial dysmorphism, and distinctive skeletal abnormalities including short ribs, mild tomoderate platyspondyly, broad ilia with flaring, increased acetabular angle, shortened long boneswith metaphyseal flaring, elongation of the proximal and middle phalanges with subluxation of theproximal interphalangeal joints, subluxation of the elbow, and talipes equinovarus [28,29]. These descriptions from previous reports fit well with the fetal phenotype in Case7, so it was supposed that this variation could also most likely contribute to its manifestation. In that case, fertility counseling for the couple would face greater challenges. Ideally, the couple would need to find a way to avoid the c.2295+1(IVS37)G>C variant and the co-existing of both variants in CANT1 in their future pregnancies, which would be difficult to achieve.
In Case 8, the situation was similar. Besides the Glycine substitution in COL1A1, another heterozygous compound variation in the RMRP gene was identified. RMRP (RNA component of mitochondrial RNA processing endoribonuclease, MIM *157660) can be transcribed into a long non-coding RNA, which is associated with a wide spectrum of autosomal recessive skeletal conditions, ranging from the mild metaphyseal dysplasia without hypotrichosis (MDWH, MIM #250460) [30] and cartilage-hairhypoplasia (CHH, MIM #250250) [31] to the severe anauxetic dysplasia 1 (ANXD1, MIM #607095) [32]. This clinical spectrum includes different degrees of short stature, hair hypoplasia, defective erythrogenesis, and immunodeficiency, and the mutant RMRP may affect both messenger RNA (mRNA) and ribosomal RNA (rRNA) cleavage and thus cell-cycle regulation and protein synthesis [33]. In terms of morphology, the fetus in Case 8 had similar characteristics to ANXD1, such as short stature and multiple skeletal dysplasia; meanwhile, it also exhibited unique prenatal indications at early pregnancy, including NT thickening and systemic edema. Admittedly, the two variants of RMRP are novel and need to be validated by functional experiments; however, we do support the idea that variations in both COL1A1 and RMRP contribute to the fetal phenotype. In a large cohort study, Posey et al. pointed out that cases involving two-gene pathogenic variants were not uncommon, and that the most typical case is a compound heterozygous variationin a recessive gene plus a de novo variation in a dominant gene [34]. We speculate that Case7 and Case8 should fit this scenario. Therefore, with regard to their future pregnancies, special care should be taken, and interventional assisted reproduction may be best recommended.
In Case 9, a novel missense variant leading to the Glycine substitution in COL1A2 was detected, and Sanger sequencing revealed that this variant was inherited from the asymptomatic mosaic mother. Parental mosaicism, whether confined to the genital system or not, has been reported in OI from time to time and should be noted in reproductive counseling [35,36,37]. According to our follow-up, the couple chose preimplantation diagnosis and selected a wild-type embryo. At present, the pregnancy has reached 38 weeks and the prenatal phenotype is normal.
The main limitation of this study is the lack of adequate functional experimental verification for several specific mutations with uncertain clinical significance at genetic level. In conclusion, this pilot study presents a landscape of complicated prenatal COL1A1/2-OI cases. We identified six novel variants in these two collagen genes, as well as compound variations in two other genes associated with skeletal dysplasia. However, most importantly, our findings suggest the need to be particularly aware of these specific conditions during prenatal and reproductive counseling in OI case.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/genes13091578/s1, Figure S1: Structural and MD results of the CANT1V186I variant, Supplementary Material 1: Structural and molecular dynamics analysis on the CANT1-V186I variant, refs [38,39,40].
Author Contributions
C.-h.Y. and Q.-q.W. designed and supervised this study. Y.L., J.Z., J.W. and W.-q.C. recruited the cases and conducted the clinical evaluation. L.-j.S. and Y.-q.S. performed the ultrasonic examination. K.Y., J.Z., Y.-s.Y. and H.-y.H. performed the genetic analysis. S.-f.Y. and L.-j.S. carried out the in silico analysis. K.Y. wrote the manuscript, and all co-authors reviewed it and approved its submission. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the Project Funded by China Postdoctoral Science Foundation (No. 2022T150445), and the Beijing Hospitals Authority Youth Programme (No. QML20211401).
Data Availability Statement
All the data involved in this study were provided in the manuscript, tables and figures.
Acknowledgments
We acknowledge all the participants in this study.
Conflicts of Interest
Kai Yang, Yan Liu, Jing Zhang, Jue Wu, Hua-ying Hu, You-sheng Yan, Wen-qi Chen, Shu-fa Yang, Li-juan Sun, Yong-qing Sun, Qing-qing Wu and Cheng-hong Yin declare that they have no conflict of interest.
Ethics Approval and Consent to Participate
The samples used in thisstudy were collected with appropriate informed consent and approvalof the ethics committee of Beijing Obstetrics and Gynecology Hospital. Themethods used in this study were carried out in accordance with theapproved guidelines.
References
- Marini, J.C.; Forlino, A.; Bachinger, H.P.; Bishop, N.J.; Byers, P.H.; De Paepe, A.; Fassier, F.; Fratzl-Zelman, N.; Kozloff, K.M.; Krakow, D.; et al. Osteogenesis imperfecta. Nat. Rev. Dis. Primers. 2017, 3, 17052. [Google Scholar] [CrossRef] [PubMed]
- Steiner, R.D.; Basel, D. COL1A1/2 Osteogenesis Imperfecta. In GeneReviews® [Internet]; Adam, M.P., Everman, D.B., Mirzaa, G.M., Pagon, R.A., Wallace, S.E., Bean, L.J.H., Gripp, K.W., Amemiya, A., Eds.; University of Washington: Seattle, WA, USA, 1993. [Google Scholar]
- Sillence, D.O.; Senn, A.; Danks, D.M. Genetic heterogeneity in osteogenesis imperfecta. J. Med. Genet. 1979, 16, 101–116. [Google Scholar] [PubMed]
- Mortier, G.R.; Cohn, D.H.; Cormier-Daire, V.; Hall, C.; Krakow, D.; Mundlos, S.; Nishimura, G.; Robertson, S.; Sangiorgi, L.; Savarirayan, R.; et al. Nosology and classification of genetic skeletal disorders: 2019 revision. Am. J. Med. Genet. A. 2019, 179, 2393–2419. [Google Scholar] [CrossRef]
- Ficara, A.; Syngelaki, A.; Hammami, A.; Akolekar, R.; Nicolaides, K.H. Value of routine ultrasound examination at 35-37 weeks’ gestation in diagnosis of fetal abnormalities. Ultrasound Obstet. Gynecol. 2020, 55, 75–80. [Google Scholar] [CrossRef] [PubMed]
- Yang, K.; Shen, M.; Yan, Y.; Tan, Y.; Zhang, J.; Wu, J.; Yang, G.; Li, S.; Wang, J.; Ren, Z.; et al. Genetic Analysis in Fetal Skeletal Dysplasias by Trio Whole-Exome Sequencing. Biomed. Res. Int. 2019, 2019, 2492590. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Wang, L.; Yang, Y.K.; Liang, Y.; Zhang, T.J.; Liang, N.; Yang, L.-M.; Li, S.-J.; Shan, D.; Wu, Q.-Q. Prenatal diagnosis of fetal skeletal dysplasia using targeted next-generation sequencing: An analysis of 30 cases. Diagn. Pathol. 2019, 14, 76. [Google Scholar] [CrossRef]
- Zhang, J.; Hu, H.; Mu, W.; Yu, M.; Chen, W.; Mi, D.; Yang, K.; Guo, Q. Case Report: Exome Sequencing Identified a Novel Compound Heterozygous Variation in PLOD2 Causing Bruck Syndrome Type 2. Front. Genet. 2021, 12, 619948. [Google Scholar] [CrossRef]
- Arsham, M.S.; Barch, M.J.; Lawce, H.J. The AGT Cytogenetics Laboratory Manual; John Wiley & Sons Inc.: Hoboken, NJ, USA, 2017. [Google Scholar]
- Huang, Y.X.; Gao, C.Y.; Zheng, C.Y.; Chen, X.; Yan, Y.S.; Sun, Y.Q.; Dong, X.Y.; Yang, K.; Zhang, D.L. Investigation of a Novel LRP6 Variant Causing Autosomal-Dominant Tooth Agenesis. Front. Genet. 2021, 12, 688241. [Google Scholar] [CrossRef]
- Wang, K.L.M.; Hakonarson, H. ANNOVAR: Functional annotation of genetic variants from next-generation sequencing data. Nucleic Acids Res. 2010, 38, e164. [Google Scholar] [CrossRef]
- Richards, S.; Aziz, N.; Bale, S.; Bick, D.; Das, S.; Gastier-Foster, J.; Grody, W.W.; Hegde, M.; Lyon, E.; Spector, E.; et al. Standards and guidelines for the interpretation of sequence variants: A joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. 2015, 17, 405–424. [Google Scholar] [CrossRef]
- Ioannidis, N.M.; Rothstein, J.H.; Pejaver, V.; Middha, S.; McDonnell, S.K.; Baheti, S.; Musolf, A.; Li, Q.; Holzinger, E.; Karyadi, D.; et al. REVEL: An Ensemble Method for Predicting the Pathogenicity of Rare Missense Variants. Am. J. Hum. Genet. 2016, 99, 877–885. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Pan, L.; Teng, Y.; Liang, D.; Li, Z.; Wu, L. Molecular diagnosis for 55 fetuses with skeletal dysplasias by whole-exome sequencing: A retrospective cohort study. Clin. Genet. 2021, 100, 219–226. [Google Scholar] [CrossRef] [PubMed]
- Schramm, T.; Mommsen, H. Fetal Skeletal Disorders. Ultraschall Med.-Eur. J. Ultrasound. 2018, 39, 610–634. [Google Scholar] [CrossRef] [PubMed]
- Costantini, A.; Skarp, S.; Kampe, A.; Makitie, R.E.; Pettersson, M.; Mannikko, M.; Jiao, H.; Taylan, F.; Lindstrand, A.; Mäkitie, O. Rare Copy Number Variants in Array-Based Comparative Genomic Hybridization in Early-Onset Skeletal Fragility. Front. Endocrinol. 2018, 9, 380. [Google Scholar] [CrossRef]
- Fu, F.; Li, R.; Li, Y.; Nie, Z.Q.; Lei, T.; Wang, D.; Yang, X.; Han, J.; Pan, M.; Zhen, L.; et al. Whole exome sequencing as a diagnostic adjunct to clinical testing in fetuses with structural abnormalities. Ultrasound Obstet. Gynecol. 2018, 51, 493–502. [Google Scholar] [CrossRef]
- Higuchi, Y.; Hasegawa, K.; Futagawa, N.; Yamashita, M.; Tanaka, H.; Tsukahara, H. Genetic analysis in Japanese patients with osteogenesis imperfecta: Genotype and phenotype spectra in 96 probands. Mol. Genet. Genomic. Med. 2021, 9, e1675. [Google Scholar] [CrossRef]
- Ju, M.; Bai, X.; Zhang, T.; Lin, Y.; Yang, L.; Zhou, H.; Chang, X.; Guan, S.; Ren, X.; Li, K.; et al. Mutation spectrum of COL1A1/COL1A2 screening by high-resolution melting analysis of Chinese patients with osteogenesis imperfecta. J. Bone Miner. Metab. 2020, 38, 188–197. [Google Scholar] [CrossRef]
- Yang, L.; Liu, B.; Dong, X.; Wu, J.; Sun, C.; Xi, L.; Cheng, R.; Wu, B.; Wang, H.; Tong, S.; et al. Clinical severity prediction in children with osteogenesis imperfecta caused by COL1A1/2 defects. Osteoporos. Int. 2022, 33, 1373–1384. [Google Scholar] [CrossRef]
- Zhytnik, L.; Maasalu, K.; Reimand, T.; Duy, B.H.; Koks, S.; Martson, A. Inter- and Intrafamilial Phenotypic Variability in Individuals with Collagen-Related Osteogenesis Imperfecta. Clin. Transl. Sci. 2020, 13, 960–971. [Google Scholar] [CrossRef]
- Hruskova, L.; Fijalkowski, I.; Van Hul, W.; Marik, I.; Mortier, G.; Martasek, P.; Mazura, I. Eight mutations including 5 novel ones in the COL1A1 gene in Czech patients with osteogenesis imperfecta. Biomed. Pap. Med. Fac. Univ. Palacky Olomouc Czechoslov. 2016, 160, 442–447. [Google Scholar] [CrossRef]
- Greene, B.; Russo, R.J.; Dwyer, S.; Malley, K.; Roberts, E.; Serrielo, J.; Piepenhagen, P.; Cummings, S.; Ryan, S.; Zarazinski, C.; et al. Inhibition of TGF-β Increases Bone Volume and Strength in a Mouse Model of Osteogenesis Imperfecta. JBMR Plus. 2021, 5, e10530. [Google Scholar] [CrossRef] [PubMed]
- Smith, T.M.; Hicks-Berger, C.A.; Kim, S.; Kirley, T.L. Cloning, expression, and characterization of a soluble calcium-activated nucleotidase, a human enzyme belonging to a new family of extracellular nucleotidases. Arch. Biochem. Biophys. 2002, 406, 105–115. [Google Scholar] [CrossRef]
- Huber, C.; Oules, B.; Bertoli, M.; Chami, M.; Fradin, M.; Alanay, Y.; Al-Gazali, L.I.; Ausems, M.G.; Bitoun, P.; Cavalcanti, D.P.; et al. Identification of CANT1 mutations in Desbuquois dysplasia. Am. J. Hum. Genet. 2009, 85, 706–710. [Google Scholar] [CrossRef]
- Balasubramanian, K.; Li, B.; Krakow, D.; Nevarez, L.; Ho, P.J.; Ainsworth, J.A.; Nickerson, D.A.; Bamshad, M.J.; Immken, L.; Lachman, R.S.; et al. MED resulting from recessively inherited mutations in the gene encoding calcium-activated nucleotidase CANT1. Am. J. Med. Genet. A. 2017, 173, 2415–2421. [Google Scholar] [CrossRef] [PubMed]
- Byrne, A.B.; Mizumoto, S.; Arts, P.; Yap, P.; Feng, J.; Schreiber, A.W.; Babic, M.; King-Smith, S.L.; Barnett, C.P.; Moore, L.; et al. Pseudodiastrophic dysplasia expands the known phenotypic spectrum of defects in proteoglycan biosynthesis. J. Med. Genet. 2020, 57, 454–460. [Google Scholar] [CrossRef]
- Fischetto, R.; Causio, F.; Corso, G.; Lillo, V.; Natale, B.; Papadia, F. Pseudodiastrophic dysplasia type Burgio in a newborn. Am. J. Med. Genet. 1997, 71, 222–225. [Google Scholar] [CrossRef]
- Yap, P.; Liebelt, J.E.; Amor, D.J.; Moore, L.; Savarirayan, R. Pseudodiastrophic dysplasia: Two cases delineating and expanding the pre and postnatal phenotype. Am. J. Med. Genet. A. 2016, 170a, 1363–1366. [Google Scholar] [CrossRef]
- Bonafe, L.; Schmitt, K.; Eich, G.; Giediona, A.; Superti-Furgaa, A. RMRP gene sequence analysis confirms a cartilage-hair hypoplasia variant with only skeletal manifestations and reveals a high density of single-nucleotide polymorphisms. Clin. Genet. 2002, 61, 146–151. [Google Scholar] [CrossRef]
- Ridanpaa, M.; EenennaamHvPelin, K.; Chadwick, R.; Johnson, C.; Yuan, B.; Vanvenrooij, W.; Pruijn, G.; Salmela, R.; Rockas, S.; Mäkitie, O.; et al. Mutations in the RNA Component of RNase MRP Cause a Pleiotropic Human Disease, Cartilage-Hair Hypoplasia. Cell 2001, 104, 195–203. [Google Scholar] [CrossRef]
- Thiel, C.T.; Horn, D.; Zabel, B.; Ekici, A.B.; Salinas, K.; Gebhart, E.; Rüschendorf, F.; Sticht, H.; Spranger, J.; Müller, D.; et al. Severely Incapacitating Mutations in Patients with Extreme Short Stature Identify RNA-Processing Endoribonuclease RMRP as an Essential Cell Growth Regulator. Am. J. Hum. Genet. 2005, 77, 795–806. [Google Scholar] [CrossRef]
- Thiel, C.T.; Mortier, G.; Kaitila, I.; Reis, A.; Rauch, A. Type and level of RMRP functional impairment predicts phenotype in the cartilage hair hypoplasia-anauxetic dysplasia spectrum. Am. J. Hum. Genet. 2007, 81, 519–529. [Google Scholar] [CrossRef] [PubMed]
- Posey, J.E.; Harel, T.; Liu, P.; Rosenfeld, J.A.; James, R.A.; Coban Akdemir, Z.H.; Walkiewicz, M.; Bi, W.; Xiao, R.; Ding, Y.; et al. Resolution of Disease Phenotypes Resulting from Multilocus Genomic Variation. N. Engl. J. Med. 2017, 376, 21–31. [Google Scholar] [CrossRef] [PubMed]
- Yamada, T.; Takagi, M.; Nishimura, G.; Akaishi, R.; Furuta, I.; Morikawa, M.; Yamada, T.; Cho, K.; Sawai, H.; Ikegawa, S.; et al. Recurrence of osteogenesis imperfecta due to maternal mosaicism of a novel COL1A1 mutation. Am. J. Med. Genet. A. 2012, 158A, 2969–2971. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.P.; Lin, S.P.; Su, Y.N.; Chern, S.R.; Su, J.W.; Wang, W. Prenatal diagnosis of recurrent autosomal dominant osteogenesis imperfecta associated with unaffected parents and paternal gonadal mosaicism. Taiwan J. Obstet. Gynecol. 2013, 52, 106–109. [Google Scholar] [CrossRef][Green Version]
- Frederiksen, A.L.; Duno, M.; Johnsen, I.B.; Nielsen, M.F.; Kroigard, A.B. Asymptomatic parental mosaicism for osteogenesis imperfecta associated with a new splice site mutation in COL1A2. Clin. Case Rep. 2016, 4, 972–978. [Google Scholar] [CrossRef] [PubMed]
- Dai, J.; Liu, J.; Deng, Y.Q.; Smith, T.M.; Lu, M. Structure and protein design of a human platelet function inhibitor. Cell 2004, 116, 649–659. [Google Scholar] [CrossRef]
- Rakhshani, H.; Dehghanian, E.; Rahati, A. Enhanced GROMACS: Toward a better numerical simulation framework. J. Mol. Model. 2019, 25, 355. [Google Scholar] [CrossRef]
- Gutiérrez, I.S.; Lin, F.Y.; Vanommeslaeghe, K.; Lemkul, J.A.; Armacost, K.A.; Brooks, C.L., 3rd; MacKerell, A.D., Jr. Parametrization of halogen bonds in the CHARMM general force field: Improved treatment of ligand-protein interactions. Bioorg. Med. Chem. 2016, 24, 4812–4825. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).