A Novel 90-kbp Deletion of RUNX2 Associated with Cleidocranial Dysplasia
Abstract
1. Introduction
2. Materials and Methods
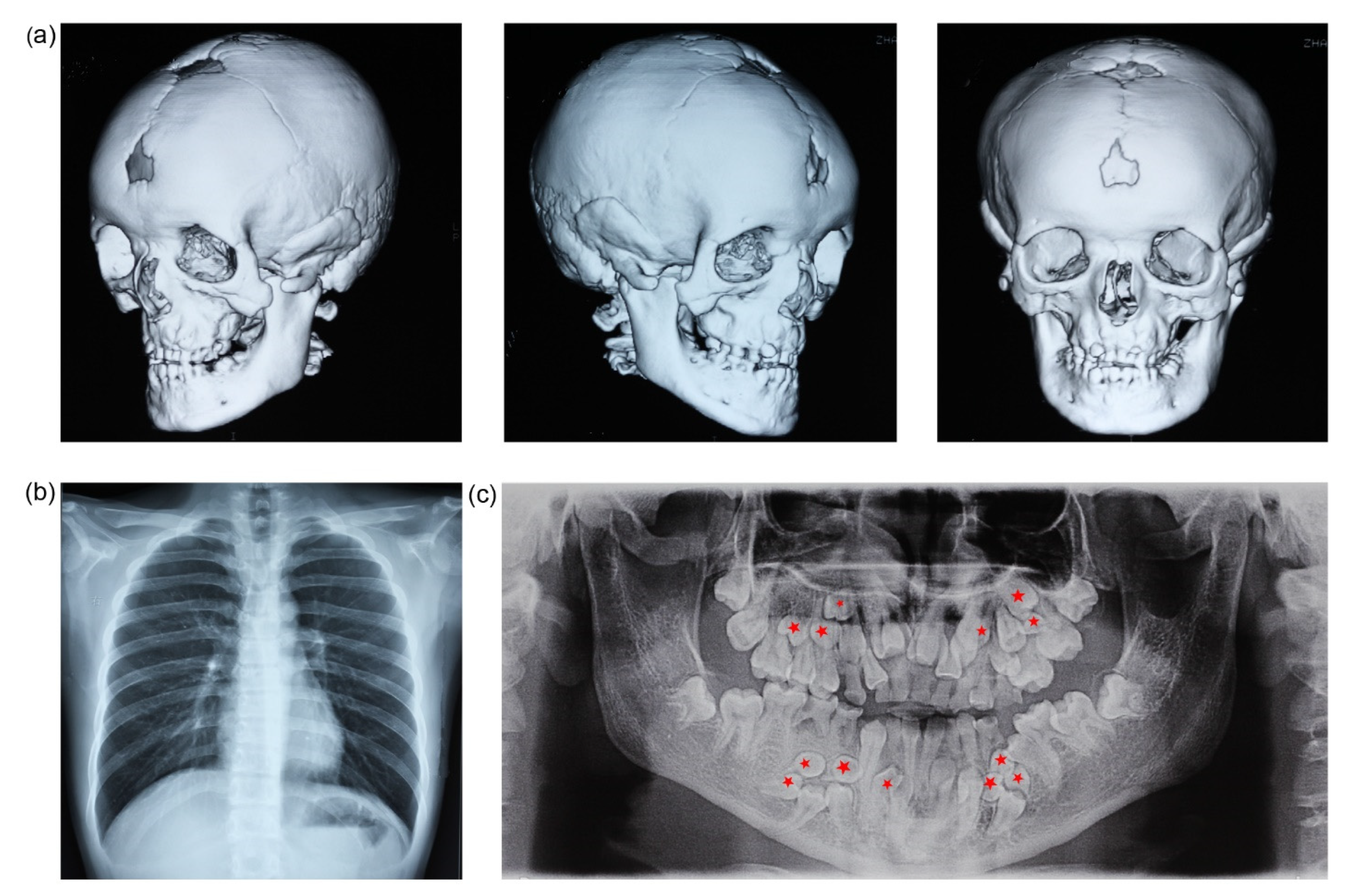
2.1. Clinical Manifestations
2.2. Whole-Exome Sequencing (WES) and Identification of Variants
2.3. Polymerase Chain Reaction (PCR) and Sanger Sequencing
2.4. Quantitative PCR Analysis (Q-PCR)
2.5. Predictions of the Protein Structure
2.6. Retrospective Study
3. Results
3.1. Clinical Findings
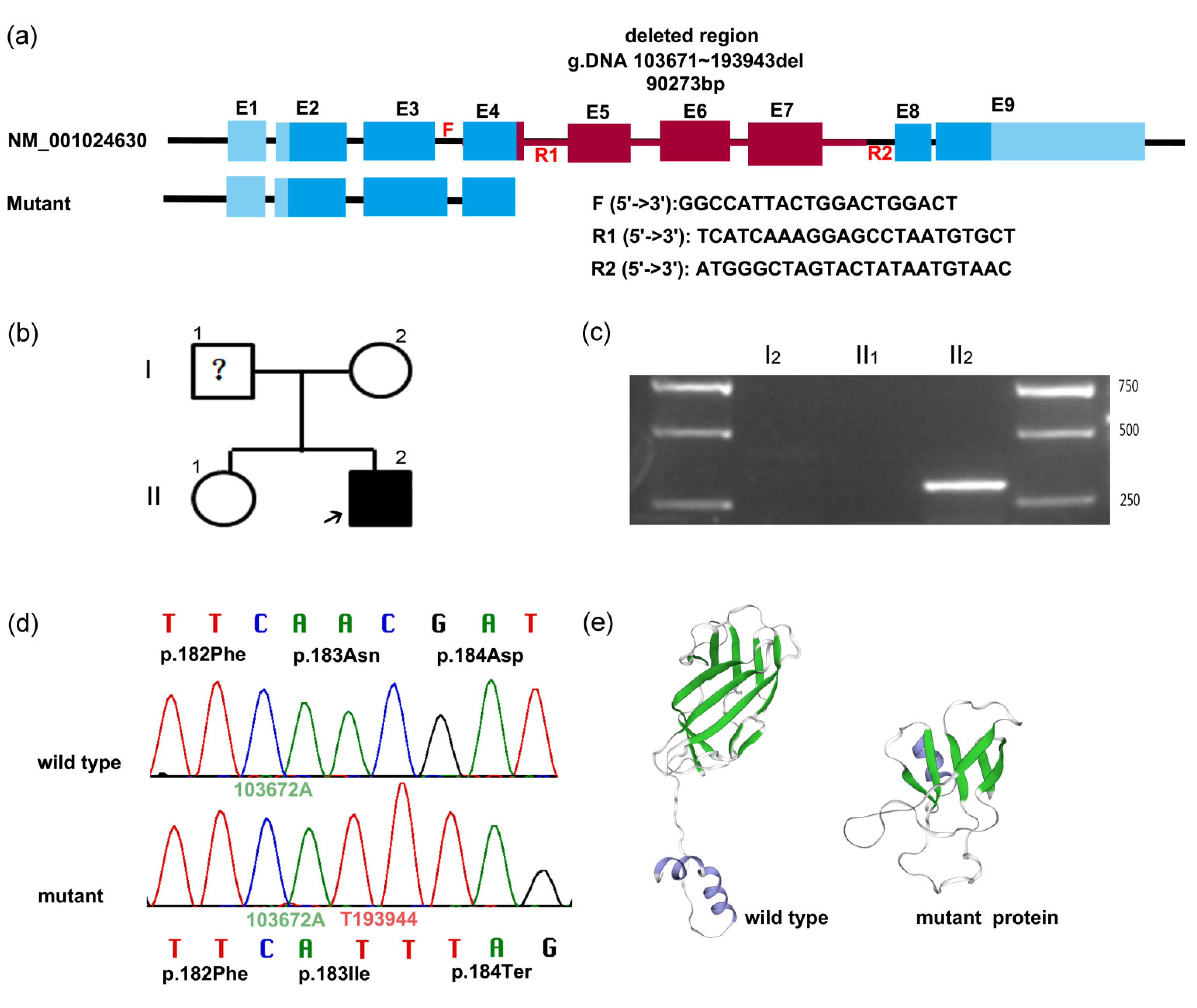
3.2. Genetic Findings
3.3. PCR and Sanger Sequencing to Determine Mutation
3.4. Bioinformatics Analysis
3.5. RUNX2 Expression Analysis
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zhang, X.; Liu, Y.; Wang, X.; Sun, X.; Zhang, C.; Zheng, S. Analysis of novel RUNX2 mutations in Chinese patients with cleidocranial dysplasia. PLoS ONE 2017, 12, e181653. [Google Scholar] [CrossRef] [PubMed]
- Quack, I.; Vonderstrass, B.; Stock, M.; Aylsworth, A.S.; Becker, A.; Brueton, L.; Lee, P.J.; Majewski, F.; Mulliken, J.B.; Suri, M.; et al. Mutation analysis of core binding factor A1 in patients with cleidocranial dysplasia. Am. J. Hum. Genet. 1999, 65, 1268–1278. [Google Scholar] [CrossRef] [PubMed]
- Hsueh, S.J.; Lee, N.C.; Yang, S.H.; Lin, H.I.; Lin, C.H. A limb-girdle myopathy phenotype of RUNX2 mutation in a patient with cleidocranial dysplasia: A case study and literature review. BMC Neurol. 2017, 17, 2. [Google Scholar] [CrossRef] [PubMed]
- Cardoso, B.M.; Dupont, J.; Castanhinha, S.; Ejarque-Albuquerque, M.; Pereira, S.; Miltenberger-Miltenyi, G.; Oliveira, G. Cleidocranial dysplasia with severe parietal bone dysplasia: A new (p.Val124Serfs) RUNX2 mutation. Clin. Dysmorphol. 2010, 19, 150–152. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, T.; Kanegane, H.; Osato, M.; Yanagida, M.; Miyawaki, T.; Ito, Y.; Shigesada, K. Functional analysis of RUNX2 mutations in Japanese patients with cleidocranial dysplasia demonstrates novel genotype-phenotype correlations. Am. J. Hum. Genet. 2002, 71, 724–738. [Google Scholar] [CrossRef]
- Gao, X.; Li, K.; Fan, Y.; Sun, Y.; Luo, X.; Wang, L.; Liu, H.; Gong, Z.; Wang, J.; Wang, Y.; et al. Identification of RUNX2 variants associated with cleidocranial dysplasia. Hereditas 2019, 156, 31. [Google Scholar] [CrossRef]
- Ma, D.; Wang, X.; Guo, J.; Zhang, J.; Cai, T. Identification of a novel mutation of RUNX2 in a family with supernumerary teeth and craniofacial dysplasia by whole-exome sequencing: A case report and literature review. Medicine 2018, 97, e11328. [Google Scholar] [CrossRef]
- Gong, L.; Odilov, B.; Han, F.; Liu, F.; Sun, Y.; Zhang, N.; Zuo, X.; Yang, J.; Wang, S.; Hou, X.; et al. Identification a novel de novo RUNX2 frameshift mutation associated with cleidocranial dysplasia. Genes Genomics 2022, 44, 683–690. [Google Scholar] [CrossRef]
- Lee, M.T.; Tsai, A.C.; Chou, C.H.; Sun, F.M.; Huang, L.C.; Yen, P.; Lin, C.C.; Liu, C.Y.; Wu, J.Y.; Chen, Y.T.; et al. Intragenic microdeletion of RUNX2 is a novel mechanism for cleidocranial dysplasia. Genom. Med. 2008, 2, 45–49. [Google Scholar] [CrossRef][Green Version]
- Duan, X.; Yang, S.; Zhang, H.; Wu, J.; Zhang, Y.; Ji, D.; Tie, L.; Boerkoel, C.F. A Novel AMELX Mutation, Its Phenotypic Features, and Skewed X Inactivation. J. Dent. Res. 2019, 98, 870–878. [Google Scholar] [CrossRef]
- Farrow, E.; Nicot, R.; Wiss, A.; Laborde, A.; Ferri, J. Cleidocranial Dysplasia: A Review of Clinical, Radiological, Genetic Implications and a Guidelines Proposal. J. Craniofac. Surg. 2018, 29, 382–389. [Google Scholar] [CrossRef]
- Puvabanditsin, S.; February, M.; Mayne, J.; Mcconnell, J.; Mehta, R. Cleidocranial Dysplasia with 6p21.1-p12.3 Microdeletion: A Case Report and Literature Review. Cleft Palate-Craniofacial J. 2018, 55, 891–894. [Google Scholar] [CrossRef]
- Xin, Y.; Liu, Y.; Liu, D.; Li, J.; Zhang, C.; Wang, Y.; Zheng, S. New Function of RUNX2 in Regulating Osteoclast Differentiation via the AKT/NFATc1/CTSK Axis. Calcif. Tissue Int. 2020, 106, 553–566. [Google Scholar] [CrossRef]
- Zhang, T.; Wu, J.; Zhao, X.; Hou, F.; Ma, T.; Wang, H.; Zhang, X.; Zhang, X. Whole-exome sequencing identification of a novel splicing mutation of RUNX2 in a Chinese family with cleidocranial dysplasia. Arch. Oral Biol. 2019, 100, 49–56. [Google Scholar] [CrossRef]
- Ott, C.E.; Leschik, G.; Trotier, F.; Brueton, L.; Brunner, H.G.; Brussel, W.; Guillen-Navarro, E.; Haase, C.; Kohlhase, J.; Kotzot, D.; et al. Deletions of the RUNX2 gene are present in about 10% of individuals with cleidocranial dysplasia. Hum. Mutat. 2010, 31, E1587–E1593. [Google Scholar] [CrossRef]
- Hansen, L.; Riis, A.K.; Silahtaroglu, A.; Hove, H.; Lauridsen, E.; Eiberg, H.; Kreiborg, S. RUNX2 analysis of Danish cleidocranial dysplasia families. Clin. Genet. 2011, 79, 254–263. [Google Scholar] [CrossRef]
- El-Gharbawy, A.H.; Peeden, J.J.; Lachman, R.S.; Graham, J.J.; Moore, S.R.; Rimoin, D.L. Severe cleidocranial dysplasia and hypophosphatasia in a child with microdeletion of the C-terminal region of RUNX2. Am. J. Med. Genet. A 2010, 152, 169–174. [Google Scholar] [CrossRef]
- Takenouchi, T.; Sato, W.; Torii, C.; Kosaki, K. Progressive cognitive decline in an adult patient with cleidocranial dysplasia. Eur. J. Med. Genet. 2014, 57, 319–321. [Google Scholar] [CrossRef]
- Zhou, G.; Chen, Y.; Zhou, L.; Thirunavukkarasu, K.; Hecht, J.; Chitayat, D.; Gelb, B.D.; Pirinen, S.; Berry, S.A.; Greenberg, C.R.; et al. CBFA1 mutation analysis and functional correlation with phenotypic variability in cleidocranial dysplasia. Hum. Mol. Genet. 1999, 8, 2311–2316. [Google Scholar] [CrossRef]
- Cunningham, M.L.; Seto, M.L.; Hing, A.V.; Bull, M.J.; Hopkin, R.J.; Leppig, K.A. Cleidocranial dysplasia with severe parietal bone dysplasia: C-terminal RUNX2 mutations. Birth Defects Res. Part A Clin. Mol. Teratol. 2006, 76, 78–85. [Google Scholar] [CrossRef]
- Qi, Z.; Yang, W.; Meng, Y.; Liu, Y. Identification of three novel frameshift mutations in the RUNX2 gene in three sporadic Chinese cases with cleidocranial dysplasia. Chin. J. Med. Genet. 2014, 31, 415–419. [Google Scholar] [CrossRef]
- Chen, C.P.; Lin, S.P.; Liu, Y.P.; Chern, S.R.; Wu, P.S.; Chen, Y.T.; Su, J.W.; Lee, C.C.; Wang, W. 6p21.2-p12.3 deletion detected by aCGH in an 8-year-old girl with cleidocranial dysplasia and developmental delay. Gene 2013, 523, 99–102. [Google Scholar] [CrossRef]
- Franceschi, R.; Maines, E.; Fedrizzi, M.; Piemontese, M.R.; De Bonis, P.; Agarwal, N.; Bellizzi, M.; Di Palma, A. Familial cleidocranial dysplasia misdiagnosed as rickets over three generations. Pediatr. Int. 2015, 57, 1003–1006. [Google Scholar] [CrossRef]
- Bedeschi, M.F.; Bonarrigo, F.; Manzoni, F.; Milani, D.; Piemontese, M.R.; Guez, S.; Esposito, S. Ehlers-Danlos syndrome versus cleidocranial dysplasia. Ital. J. Pediatr. 2014, 40, 49. [Google Scholar] [CrossRef]
- Chen, M.; Lin, S.M.; Li, N.; Li, Y.; Li, Y.; Zhang, L. An induced pluripotent stem cell line (GZHMCi003-A) derived from a fetus with exon 3 heterozygous deletion in RUNX2 gene causing cleidocranial dysplasia. Stem Cell Res. 2021, 51, 102166. [Google Scholar] [CrossRef]
- Dincsoy, B.F.; Dinckan, N.; Guven, Y.; Bas, F.; Altunoglu, U.; Kuvvetli, S.S.; Poyrazoglu, S.; Toksoy, G.; Kayserili, H.; Uyguner, Z.O. Cleidocranial dysplasia: Clinical, endocrinologic and molecular findings in 15 patients from 11 families. Eur. J. Med. Genet. 2017, 60, 163–168. [Google Scholar] [CrossRef]
- Hashmi, J.A.; Almatrafi, A.; Latif, M.; Nasir, A.; Basit, S. An 18 bps in-frame deletion mutation in RUNX2 gene is a population polymorphism rather than a pathogenic variant. Eur. J. Med. Genet. 2019, 62, 124–128. [Google Scholar] [CrossRef]
- Zeng, L.; Wei, J.; Zhao, N.; Sun, S.; Wang, Y.; Feng, H. A novel 18-bp in-frame deletion mutation in RUNX2 causes cleidocranial dysplasia. Arch. Oral Biol. 2018, 96, 243–248. [Google Scholar] [CrossRef]
- Qian, Y.; Zhang, Y.; Wei, B.; Zhang, M.; Yang, J.; Leng, C.; Ge, Z.; Xu, X.; Sun, M. A novel Alu-mediated microdeletion in the RUNX2 gene in a Chinese patient with cleidocranial dysplasia. J. Genet. 2018, 97, 137–143. [Google Scholar] [CrossRef]
- Berkay, E.G.; Elkanova, L.; Kalayci, T.; Uludag, A.D.; Altunoglu, U.; Cefle, K.; Mihci, E.; Nur, B.; Tasdelen, E.; Bayramoglu, Z.; et al. Skeletal and molecular findings in 51 Cleidocranial dysplasia patients from Turkey. Am. J. Med. Genet. A 2021, 185, 2488–2495. [Google Scholar] [CrossRef]
- Wu, D.; Li, T.; Hou, Q.; Huo, X.; Wang, X.; Wang, T.; Yang, Y.; Liu, H.; Liao, S. Genetic analysis of a child with cleidocranial dysplasiaand 6q21-q22.31 microdeletion. Chin. J. Med. Genet. 2018, 35, 253–256. [Google Scholar] [CrossRef]
- Ogawa, E.; Maruyama, M.; Kagoshima, H.; Inuzuka, M.; Lu, J.; Satake, M.; Shigesada, K.; Ito, Y. PEBP2/PEA2 represents a family of transcription factors homologous to the products of the Drosophila runt gene and the human AML1 gene. Proc. Natl. Acad. Sci. USA 1993, 90, 6859–6863. [Google Scholar] [CrossRef] [PubMed]
- Mundlos, S.; Otto, F.; Mundlos, C.; Mulliken, J.B.; Aylsworth, A.S.; Albright, S.; Lindhout, D.; Cole, W.G.; Henn, W.; Knoll, J.H.; et al. Mutations involving the transcription factor CBFA1 cause cleidocranial dysplasia. Cell 1997, 89, 773–779. [Google Scholar] [CrossRef]
- Lou, Y.; Javed, A.; Hussain, S.; Colby, J.; Frederick, D.; Pratap, J.; Xie, R.; Gaur, T.; van Wijnen, A.J.; Jones, S.N.; et al. A Runx2 threshold for the cleidocranial dysplasia phenotype. Hum. Mol. Genet. 2009, 18, 556–568. [Google Scholar] [CrossRef]

| Number | Description | Predicted Changes in Protein or RNA | Methods | Location | Reference |
|---|---|---|---|---|---|
| 1 | ~7.3 Mb incl entire gene + VEGFA + NFKBIE | Microarry | entire areas | [12] | |
| 2 | ~70 kbp incl entire gene | aCGH | entire areas | [17] | |
| 3 | 11.87 kbp incl ex. 8–9 | Microarry | PST | [18] | |
| 4 | nt.220 del173 | Sanger sequencing | QA | [19] | |
| 5 | 2.1 Mb, entire gene +~20 additional genes | Q-PCR, aCGH | entire areas | [15] | |
| 6 | nt.950-971del22 | p.Leu317ThrfsX483 | Sanger sequencing | PST | [20] |
| 7 | 500 kbp incl ex. 1–5 | Q-PCR, FISH | QA + RHD + NLS + PST | [16] | |
| 8 | 750 kbp incl ex. 1–5 | Q-PCR, FISH | QA + RHD + NLS + PST | [16] | |
| 9 | c.227_306del80 | p.Ala76GlyfsX58 | QA | [21] | |
| 10 | 9.7 Mb incl. entire gene + CUL7, VEGFA and NFKBIE | aCGH | entire areas | [22] | |
| 11 | c.230_276del47 | p.Ala77ValfsX68 | Sanger sequencing | QA | [15] |
| 12 | c.718_721del4 | p.Ser240CysfsX9 | Sanger sequencing | PST | [15] |
| 13 | c.879_885del7 | p.Ser294ArgfsX12 | Sanger sequencing | PST | [15] |
| 14 | entire gene | Q-PCR, aCGH | entire areas | [15] | |
| 15 | ex. 1–3 | Q-PCR, MLPA | QA + RHD | [23] | |
| 16 | ex. 2 | MLPA | RHD | [24] | |
| 17 | 125.6 kbp incl ex. 2–6 | Sanger sequencing | QA + RHD + NLS + PST | [9] | |
| 18 | ex. 2–8 | Q-PCR | QA + RHD + NLS + PST | [15] | |
| 19 | ex. 3 | Q-PCR | RHD | [15,25] | |
| 20 | ex. 3–6 | Q-PCR | RHD + NLS + PST | [15] | |
| 21 | ex. 6–8 | Q-PCR | PST | [15] | |
| 22 | ex. 7 | Q-PCR | PST | [15] | |
| 23 | ex. 7–8 | Q-PCR | PST | [15] | |
| 24 | c.241_258del18 | p. 84_89del | Sanger sequencing, MLPA | QA | [26,27] |
| 25 | c.243-260del18 | P.85_90 del | WES, Sanger sequencing | QA | [27,28] |
| 26 | ~500 kbp incl ex. 1–4 + SUPT3H | Q-PCR CNV analysis by microarrays | QA + RHD + NLS | [29] | |
| 27 | c.443_454del12insG | p.Val148GlyfsX9 | sequencing | RHD | [30] |
| 28 | ex. 2–9 | unmentioned | QA + RHD + NLS + PST | [30] | |
| 29 | ex. 6–9 | unmentioned | PST | [30] | |
| 30 | ~800 kbp incl entire gene | aCGH | entire areas | [31] | |
| 31 | ~90 kbp incl ex. 4–7 | p.Asn294LlefsX1 | WES, PCR, Sanger sequencing | RHD + NLS + PST | This paper |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, Y.; Duan, X. A Novel 90-kbp Deletion of RUNX2 Associated with Cleidocranial Dysplasia. Genes 2022, 13, 1128. https://doi.org/10.3390/genes13071128
Zhang Y, Duan X. A Novel 90-kbp Deletion of RUNX2 Associated with Cleidocranial Dysplasia. Genes. 2022; 13(7):1128. https://doi.org/10.3390/genes13071128
Chicago/Turabian StyleZhang, Yanli, and Xiaohong Duan. 2022. "A Novel 90-kbp Deletion of RUNX2 Associated with Cleidocranial Dysplasia" Genes 13, no. 7: 1128. https://doi.org/10.3390/genes13071128
APA StyleZhang, Y., & Duan, X. (2022). A Novel 90-kbp Deletion of RUNX2 Associated with Cleidocranial Dysplasia. Genes, 13(7), 1128. https://doi.org/10.3390/genes13071128







