Abstract
This study provides new data on the whole-exome sequencing of a cohort of children with autistic spectrum disorders (ASD) from an underexplored Russian population. Using both a cross-sectional approach involving a control cohort of the same ancestry and an annotation-based approach involving relevant public databases, we explored exonic single nucleotide variants and copy-number variation potentially involved in the manifestation of ASD. The study results reveal new potential ASD candidate-variants found in the studied Russian cohort and show a high prevalence of common ASD-associated genomic variants, especially those in the genes known to be associated with the manifestation of intellectual disabilities. Our screening of an ASD cohort from a previously understudied population allowed us to flag at least a few novel genes (IGLJ2, FAM21A, OR11H12, HIP1, PRAMEF10, and ZNF717) regarding their potential involvement in ASD.
1. Introduction
Autism spectrum disorders (ASDs) represent a broad spectrum of neurodevelopmental conditions characterized by severe impairment in social interactions and communication skills and the manifestation of restricted, stereotypical behaviors. In 50–70 percent of cases, an ASD co-occurs with intellectual disability [1,2,3]. In addition, multiple other comorbid medical, psychiatric, neurological, and psychological conditions are commonly observed in ASD [3,4,5]. Emerging in early childhood, ASD are lifespan disorders that can be highly disabling [2]. ASD are high-incidence disorders. With slight regional variations, a worldwide average of ASD prevalence was estimated at ~7.6 per 1000 or one in 132 persons in 2010 [6]. As new practices for identifying ASD continue to be developed, these estimates tend to increase. For example, in the USA, the 2016 prevalence estimate of 18.5 per 1000 children was 2.8 times higher than that reported in 2000–2002 [7].
The etiology of ASD is not fully understood. Similar to many other developmental disorders, genetic risk, environmental exposure, and their interplay appears to contribute to the causal landscape of ASD [2,8,9,10]. The genetic contribution to ASD is strongly supported by twin and family studies [9,11,12,13]. A recent large-scale multinational population-based investigation has demonstrated a remarkably high heritability of ASD of 80% [10]. The empirical literature provides a growing body of evidence that the genetic etiology of ASD might be linked to a complex architecture of rare, often pathogenic, de novo mutations of large effects and/or the cumulative impact of multiple common variants with small effects [14,15,16]. Both the rare occurrence of the former and the small effect sizes of the latter underlie the main challenges in detecting ASD-associated genetic factors; addressing these challenges would require a considerable increase in sample size, an extension of the research geography, and the involvement of multinational patient cohorts.
Over the past decade, research on ASD—ASD prevalence, the spectrum variability, and the contribution of environment and genetics to ASD etiology—has actively extended to previously less outreached minority groups: various racial, ethnical, and cultural communities. This research has revealed reduced differences in ASD prevalence for different ethnic cohorts destroying the stereotype of the significant prevalence of ASD in white populations and attributing this difference in prevalence mainly to limited access to healthcare services and screenings for minority communities; see, e.g., [17,18]. While the ASD prevalence can be relatively similar across various ethnic and cultural groups, the frequencies of different syndromes and disorders falling under the ASD umbrella, the disorders’ severity, and comorbid health conditions seem to vary across populations. While the ASD prevalence can be relatively similar across various ethnic and cultural groups, the frequencies of different syndromes and disorders falling under the ASD umbrella, the disorders’ severity, and the patterns of comorbid health conditions seem to vary across populations [19,20,21]. Collectively, the studies reporting on such phenotypic variability in ASD among various ethnic groups have attributed this to differences in both the cultural environment and the genetic background. The latter may shape the population-specific pattern of ASD via (1) elevated frequencies of some common ASD-related genetic variants, e.g., as has been shown for several SNPs in the SCN2A, FOXP1, and SYNGAP1 genes in indigenous American populations in the Amazon [22], rare population-specific causal variants, and (3) additive effects of common disease-associated variants and rare, likely pathogenic variants in the genetic background [23].
Thus, exploring ASD-associated genetic variants in various ethnic and geographic populations may increase diversity in reference genetic databases connecting specific variants to ASD and comorbid conditions that, in turn, may significantly improve genetic testing and its interpretation. To fill a ‘geographical’ gap, here we report data from a genomic study of a Russian cohort of children with ASD using exome sequencing.
2. Materials and Methods
2.1. Participants
The study participants were 193 children with ASD (mean age = 7 ± 4 years; 18 girls and 175 boys), ascertained through the ‘Genetico’ Center for Genetics and Reproductive Medicine (Moscow, Russia). The children were enrolled in a project focused on the genetic screening of ASD in a west-Russian population, based on ASD diagnoses established by child psychiatrists. The diagnostics of ASD in Russia are carried out in accordance with the Clinical Recommendations for Autism Spectrum Disorders (ID:594) issued by the Association of Psychiatrists and Psychologists for Evidence-Based Practice and approved by the Ministry of Health of the RF (https://cr.minzdrav.gov.ru/recomend/594_1, accessed on 15 May 2022). The diagnosis of ASD was established following the Autism Diagnostic Observation Schedule Second Edition (ADOS-II) based on the clinical picture and developmental history characteristics of ASD—a combination of symptoms of qualitative disorders of social interaction, communication, and limited, stereotyped, repetitive behavior. According to the Clinical Guidelines, the following tools to screen for the ASD symptoms were used: Checklist for Autism Spectrum Disorders (CASD), Social Communication Questionnaire (SCQ), and Autism Diagnostic Interview-Revised (ADI-R).
The children’s demographics and comorbid diagnoses, along with the participants’ family histories, are shown in Supplementary Table S1. The data on the clinical features in ASD individuals are summarized in Table 1. These data were collected from the children’s medical records and interviews with parents. In addition, 51 individuals without ASD from the west-Russian general population enrolled via the Almazov National Medical Research Centre (St. Petersburg, Russia) were involved in the study as a comparison group; hereafter, they are referred to as nonASD. The nonASD comparison group included children and adults (mean age = 23 ± 16 years; 29 females and 23 males) who did not have a lifetime or current medical history of ASD, and any neurodevelopmental and psychiatric disorders, as per the interview with the participants and their medical records.

Table 1.
Summarized data on clinical features of ASD individuals derived from the participants’ medical records.
2.2. Exome Library Preparation and WES Data Processing
For the sequencing library preparation, three different exome capture assays were utilized. For the ASD cohort, the TruSeq DNA Exome and the SureSelect Human All Exon V7 were used for 73 and 120 individual samples, respectively. Whole-exome sequencing (WES) was performed using the Illumina HiSeq 4000 platform. The library preparation and sequencing were conducted at the ‘Genetico’ Center. WES data for the controls—51 nonASD individuals—were obtained using the SureSelect V6 r2 (hg19 build) capture assay for library preparation followed by sequencing on the Illumina platform. The sequencing was performed at the Almazov National Medical Research Centre.
Sequencing data quality control was performed using the FastQC v. 0.11.9 [24]. Sequencing reads were aligned using the BWA v. 0.7.17-r1188 [25] to a reference genome depending on the exome enrichment panel utilized: the hg19 (UCSC) was used for the samples prepared with the TruSeq DNA Exome and the SureSelect V6 panels, and the hg38 (UCSC) for the samples prepared with the SureSelect V7 exome capture assay. Subsequently, the aligned sequences were subjected to sorting, deduplication, and base quality score recalibration as per the GATK Best Practices [26].
2.3. Single Nucleotide Variant Calling
Detection of single nucleotide polymorphisms (SNPs) was performed as follows. First, base recalibrated BAM files were submitted to the Germline short variant (SNPs and indels) discovery workflow implemented in the GATK v. 4.1.6. Second, each sample was submitted to the HaplotypeCaller separately with an appropriate BED file of intervals. Since three different capture assays were used for the sequencing library preparation, the corresponding three datasets were subjected to joint-genotyping separately. Third, the consolidated GVCF files were submitted to an exact test of the heterozygosity excess, applying the hard filtering parameter of ExcessHet > 54.69. The subsequent filtering procedures were performed using the VariantRecalibrator algorithm implemented in the GATK with the following truth sensitivity level parameters: 99.95 and 99.90 for the SureSelect and Truseq ASD-data subsets, respectively, and 99.50 for the nonASD subset. The established thresholds were chosen empirically to increase the possibility of detecting rare variants. Specifically, this allowed the detection of the maximum number of true-positive calls with minimal risk for false-positive calls, as can be seen in the tranches plots presented in Supplementary Figures S1 and S2).
The hg38 SNP call-sets were converted to the hg19 build using the Assembly Converter. Three SNP call-sets were merged using the bcftools v. 1.10.2 [27]; the ‘include-non-variant-sites flag’ function implemented in the GenotypeGVCF pipeline was applied to account for the missing and reference genotypes. The indels were excluded, and the final merged SNP call-set was used in the downstream analysis. The SNP call-set was annotated to several databases: refGene, cytoBand, ClinVar, ExAC, avSNP147, dbNFSP v. 3.0a, and gnomAD Exome, using the ANNOVAR tools [28].
2.4. CNV Detection in WES Data
The detection of copy number variations (CNVs) in the WES data was performed by the ‘exomecn.mops’ function implemented in the cn.mops R package v. 1.32.0 [29]. The following parameters were adjusted: priorImpact = 100, upperThreshold = 0.59, lowerThreshold = −0.99, and useMedian = TRUE. Both segmentation algorithms, fastseg [29] and DNAcopy [30], were applied. Sixteen ASD samples were excluded: one sample prepared with the SureSelect V7 panel, due to a low genome coverage; and 15 samples prepared with the TruSeqExome panel, due to a batch-effect related to the use of IDT adapters. The CNV calling was performed in three separate runs for the subsets of samples processed with different exome capture assays for the library preparation—the TruSeq Exome (N = 58 ASD), the SureSelect V7 (N = 119 ASD), and the SureSelect V6 (N = 51 nonASD).
2.5. Analysis of ASD-Associated Variants
The association analysis was performed using the PLINK v1.9 toolset [31]. Prior to the association analysis, the SNP call-set underwent linkage disequilibrium (LD) pruning and clumping procedures. The LD pruning was conducted according to the following parameters of the ‘indep’ command: window size = 50, window shift = 5, and VIF threshold = 2. The SNP call-set was adjusted based on the following QC parameters: –maf 0.01, –geno 0.05, –hwe 0.001. After relatedness testing, nine individuals from closely related pairs (PI HAT ≥ 0.125) in the ASD cohort were removed. A case-control association analysis was performed using Fisher’s exact test to generate significance values adjusted for multiple testing using Bonferroni correction. Established candidate SNP variants had to meet inclusion criteria based on the predicted pathogenicity score thresholds: a SIFT score < 0.05 [32] and PolyPhen2 HDIV score ≥ 0.453 [33]. The polymorphisms with unknown pathogenicity were also considered as potential candidate variants. In addition, the SNP and CNV call-sets were intersected with an autism gene database, AutDB [34]. For the CNV call-sets, the intersection with the list of CNVs reported in the AutDB (validated) was performed using the following command: ‘bedtools intersect -a test.bed -b autdb.bed -f 0.70 -r -wa -wb > output.’ For a successful query, a test region had to overlap at least 70% of an AutDB record.
3. Results
3.1. Genome-Wide SNP Association Analysis
A summary of the variant calling statistics across the comparison groups and the WES-data subsets is shown in Supplementary Table S2. Altogether we detected 237,019 SNPs across all individuals from both comparison cohorts, ASD and nonASD. The mean number of the detected SNPs per individual varied from 23 to 30K depending on the exome enrichment assay applied, which corresponds to the value of ~25K SNPs per individual expected for the exome data [35]. The transition/transversion (Ti/Tv) ratio varied from 2.73 to 2.93, which corresponds to the value of 3.0 observed in the exonic regions [36].
After variant filtering and LD pruning, 22,249 remaining SNPs were included in the case-control association analysis. The association analysis identified ten variants related to eight genes that surpassed the genome-wide significance threshold (Table 2). These ten SNPs were novel variants not previously reported in association with ASD. Moreover, the eight genes harboring these variants also were not found among the over a thousand human genes implicated in ASD as per records in relevant databases—the SFARI [37] and AutDB [34]. According to the SNP functional annotation, the following variants might be highlighted: two likely pathogenic synonymous substitutions in the IGLJ2 (rs8033) and HIP1 (rs1167801) genes and five missense variants with moderate or high deleterious effects located in the PRAMEF10, ZNF717, FAM21A, and OR11H12 genes. The annotation of these genes against the databases on human diseases MalaCards [38] and OMIM [39], and human phenotype ontologies HPO [40], did not reveal associations with ASD and ASD-related phenotypes (Table 2).

Table 2.
The results of the case-control, ASD vs. nonASD, association analysis of SNPs. Ten genome-wide significant ASD-associated SNPs are shown along with their pathogenicity scores and genomic annotations.
3.2. CNV Burden in the ASD Cohort Compared to nonASD
Altogether 4991 CNVs across all individuals from both comparison cohorts were detected: 4084 and 907 CNV events were identified in the ASD and nonASD cohorts, respectively (Supplementary Table S3). As seen in Figure 1, the distribution of the CNV sizes differed between the comparison cohorts; the results of the Kolmogorov–Smirnov test indicated a statistical significance of this difference (D = 0.0934, p = 4.749 × 10−6). In particular, we observed a wider range in CNV length with a lower prevalence of smaller CNVs and a higher prevalence of larger CNVs in the ASD group compared to nonASD. In addition, we found a remarkable difference in the occurrence of different CNV-types between the comparison groups. In both comparison groups, deletions predominated over duplications: the deletions/duplications ratio was 2.22 and 1.48 for the ASD and nonASD cohorts, respectively. However, the predominance of deletions was more profound in the ASD group, where a significantly greater proportion of deletions was found (OR = 1.5; z = 5.365; p = 1.222 × 10−7).
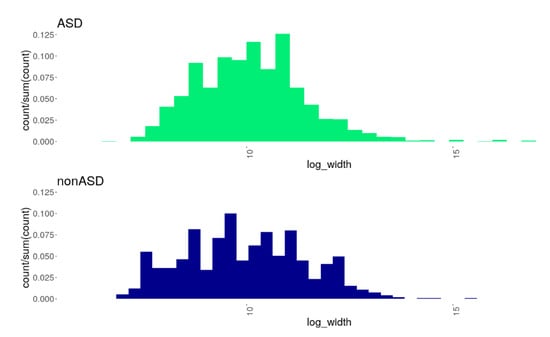
Figure 1.
Histograms showing the distributions of CNVs of different sizes (X-axis: log-width) in the comparison groups, ASD and nonASD. A statistically significant difference in the distributions was found (the Kolmogorov–Smirnov D = 0.0934, p = 4.749 × 10−6): the ASD cohort was characterized by a wider range in CNV length with a lower prevalence of smaller CNVs and a higher prevalence of larger CNVs compared to the nonASD cohort.
3.3. Genome-Wide Screening of Common ASD-Associated Variants, SNPs and CNVs
The total unfiltered call-set of 237K SNPs identified in both comparison cohorts, ASD and nonASD, was intersected with the list of 891 common ASD-associated variants derived from the AutDB repository [34]. We found 138 of such SNPs in the studied groups (Supplementary Table S4). The distribution of these SNPs across the groups is shown in Figure 2. Although we did not find a significant overrepresentation of the common candidate variants in the ASD cohort compared to the nonASD controls, a greater number of such SNPs were identified in the discovery group—137 SNPs in ASD vs. 102 SNPs in nonASD. Additionally, Fisher’s exact test did not reveal a significant difference in the frequency of the ASD-associated SNPs between the comparison groups.
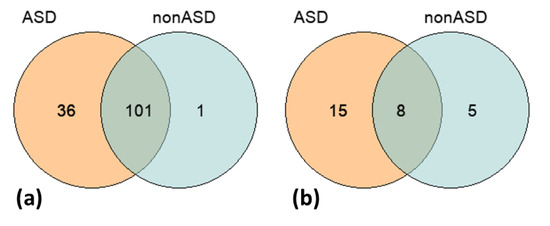
Figure 2.
Venn diagrams represent the distributions of common ASD-associated SNPs (a) and CNVs (b) across the two comparison groups, ASD and nonASD. Both diagrams reflect a greater number of the common ASD-associated genomic variants in the ASD cohort compared to the nonASD controls.
The CNV-call sets (ASD and nonASD) were also intersected with relevant data on the common ASD-related variations from the AutDB repository as per the procedures described in the Methods section. The list of overlapping CNVs was filtered according to the following criteria: localization within the same chromosome band, the same type of variation (either deletions or duplications), and the same gene content. The CNVs located on sex chromosomes and the CNVs containing HLA (human leukocyte antigen) genes, having the most extensive variability, were removed from the analysis. Detailed results of the intersection analysis are reported in Supplementary Table S5, and a summary is shown in Table 3. Altogether 29 CNVs previously associated with ASD were detected in the studied cohorts—23 and 13 CNVs in the ASD and nonASD groups, respectively, including 8 CNVs that overlapped between the comparison groups (Table 3; Figure 2). Similar to the SNP-based analysis results, despite the lack of significance in the CNV frequencies between the comparison groups, we observed a greater number of the common ASD-associated CNV-events in the ASD cohort than in the nonASD controls.

Table 3.
The distribution of 29 common ASD-associated CNVs in the studied ASD and nonASD cohorts. Despite a lack of significant differences in the variants’ frequencies between the comparison groups, a greater number of the ASD-associated CNVs were detected in the ASD cohort compared to the nonASD controls, 23 vs. 13 CNVs.
In addition, the CNV call-sets were compared with the CNV morbidity map [59,60], or the list of structural genomic variants that have been linked to severe pediatric diseases, including developmental delays, intellectual disability, and ASD. The development delay track derived using the UCSC Genome Browser tools consists of over 29 thousand individual entries for case subjects. An intersection of these records with the ASD and non-ASD CNV call-sets resulted in 52 overlapping entries (Supplementary Table S6). It was remarkable that only four of 51 nonASD individuals (7.8%) had those CNVs; in contrast, among ASD participants, 21 of 148 individuals (14.2%) harbored CNVs linked to a developmental disorder, including the deletion CNVR6294.56 on chromosome 14, known as a very common variant [61].
3.4. Gene-Based and Gene Ontology-Based Analyses
Concerning the gene content of the loci harboring CNVs, we identified 1562 and 990 genes having CNVs in the ASD and nonASD cohorts, respectively (Supplementary Table S7). We performed several overrepresentation analyses (ORA) to identify particular gene ontology (GO) and human phenotype ontology (HPO) terms enriched in these gene sets. It is necessary to note that both the ASD and nonASD sets of genes harboring CNVs were extremely enriched in those encoding olfactory receptors (OR), which were assigned to the GO: olfactory receptor activity. Specifically, 173 of 1562 genes in the ASD gene-set (Fold Enrichment = 5.45; FDR = 5.61 × 10−59) and 167 of 990 genes in the nonASD gene-set (Fold Enrichment = 8.35; FDR = 1.88 × 10−84) were OR genes. Consequently, this superfamily of highly polymorphic OR genes was removed from the gene-sets prior to the ORAs.
The ORA results are reported in the Supplementary Table S8. A graphical summary showing a top list of the most overrepresented (at an Enrichment FDR < 10−5) GO terms is shown in Figure 3. As can be seen in Figure 3, at the established significance threshold, the only functional category overrepresented among the genes having CNVs in the nonASD group is the JAK-STAT pathway. This signaling pathway mediates cellular transcriptional responses to cytokines and, as a consequence, is related, first of all, to the immune response. Notably, STAT protein-related pathways were also found among the GO terms overrepresented in the set of genes having CNVs in the ASD cohort (Figure 3). In turn, in comparison to the nonASD gene-set, the ASD gene-set was remarkably enriched in genes related to meiotic processes, in particular chromosome segregation, and in genes involved in the primary cilium assembly and organization (Figure 3).
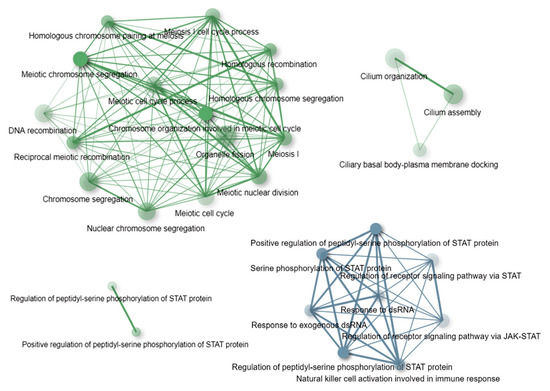
Figure 3.
The plot shows functional categories (GO: biological process terms) significantly (at an Enrichment FDR < 10−5) enriched in the sets of genes harboring CNVs in the ASD cohort (green) and nonASD cohort (blue). A network indicates GO terms sharing 30% or more genes; thicker edges represent more overlapped genes. Bigger nodes correspond to larger gene sets, and darker nodes correspond to more significant enrichment FDR-values. The enrichment tests and the network constructions were performed using ShinyGO tools [62].
The lists of genes with CNVs were also submitted to the HPO-based ORA. No significant enrichment was found for the nonASD gene-set. In contrast, the ASD gene-set was significantly enriched in a number of phenotypes related, first of all, to intellectual disability and developmental delays (Table 4). Tracking HPO-related genes in the data on the CNV distribution across individuals (Supplementary Table S9), we observed that several highlighted phenotypes had been reported in the participants’ medical records. In particular, a representative number of individuals (N = 79) harbored CNVs in the genes associated with intellectual disability. One of the ASD individuals harboring CNVs in genes related to hypertelorism manifested this phenotype according to his medical history. Two of four participants who had records on the epicanthic fold carried CNVs in the genes associated with the epicanthus. Two ASD participants were recorded as having hypotonia and one with confirmed microcephaly; however, they did not have CNVs in the genes linked to these phenotypes as per the HPO.

Table 4.
The human phenotype ontology (HPO) terms that were significantly overrepresented among those related to genes harboring CNVs in the ASD cohort.
Important to note that enrichment of functional groups of genes does not equate to the presence of phenotypes; thus, not necessary for every patient with perturbations in particular genes to develop the corresponding phenotype. However, the top list of HPO terms in Table 4 indicates a burden of CNVs in the genes directly related to the manifestation of severe developmental issues mostly known in disorders with autosomal recessive inheritance. Remarkable, in contrast to microcephaly, we did not observe an enrichment in the HPO Macrocephaly—a clinical feature also overrepresented in individuals with ASD, which reported rates are 10–20%; see, e.g., [63,64,65]. We tend to attribute this to a power insufficiency of the enrichment analysis for this particular HPO term, for which a limited amount of associated genes (about ten) are known compared to the HPO Microcephaly, which has been reliably linked to hundreds of various genes.
4. Discussion
In summarizing the results of this study exploring exonic variations in a Russian cohort of children with ASD, several findings and observations need to be pointed out and discussed. First, based on the data aggregated in relevant repositories, such as AutDB [34], SFARI [37], and the CNV morbidity map of developmental delay [59,60], we tracked the genomic variants known to be implicated in ASD and related developmental disorders in the studied ASD cohort compared to the nonASD individuals from the general population of the same origin. Although below the threshold of statistical significance, an elevated prevalence of common ASD-associated genomic alterations, both SNPs and CNVs, was observed in the ASD compared to the nonASD cohorts.
Second, in comparing the CNV metrics (length, prevalence, and distribution) between the comparison groups, we observed that the ASD cohort is characterized by a higher CNV burden. Specifically, we noted a higher prevalence of larger CNV events and a remarkable predominance of deletions over duplications (about 1.5 times) in the ASD compared to the nonASD group. These observations are consistent with the consensus in the literature that CNVs are one of the most prominent sources of genetic risks for ASD. Specifically, it has been reported that CNVs, as a genomic event, are highly prevalent (i.e., observed in up to 20%) in individuals with an ASD [66]. Additionally, it has been shown that de novo CNV events occur almost five times more frequently in individuals with ASD than in unaffected siblings and controls (5–10% vs. 1–2%, respectively), and large CNVs were consistently observed in the cases with developmental delays or intellectual disability [49,51]. A recent study exploring the effect sizes of the CNV types on the development of multiple cognitive domains and overall ASD risk suggested a differential effect of deletions and duplications on different phenotypic features of ASD. Specifically, whereas both CNV types may equally affect motor skills, IQ-related cognitive deficits in ASD have been predominantly attributed to haploinsufficiency due to deletions [66]. Remarkably, our phenotype-focused enrichment tests revealed a significant overrepresentation of the comorbid phenotypes related to intellectual disabilities and developmental delays among those associated with genes having CNVs, predominantly deletions, in the studied ASD cohort. It is important to note that, in addition to these two major phenotypes, several other comorbid conditions and health problems were highlighted by the CNV gene-set enrichment analysis; for example, conditions of hypertelorism and epicanthic fold were tracked in participants’ medical records. Altogether, these observations provide additional evidence supporting the essential role of structural genomic variations in the etiology of diverse ASD phenotypes often accompanied by multiple comorbid developmental conditions.
Third, we explored potential functional outcomes of the CNV burden across the comparison groups based on tests of gene-set enrichment in the specific biological pathways that these genes control. In contrast to the nonASD controls, the list of genes harboring CNVs in the genomes of individuals with ASD showed a significant overrepresentation of gene ontologies related to meiosis and chromosome segregation, and to the biological pathways involved in primary cilium assembly and organization. Both of these biological pathways might be potentially linked to the ASD phenotype. Specifically, the former may indicate an aggravated risk of chromosomal rearrangements in ASD genomes; such rearrangements, both de novo and inherited, are known to be involved in the etiology of ASD [67,68,69]. In fact, defects and deficits in primary cilia, known to impact brain development and maturation [70,71,72,73], have been demonstrated, directly and indirectly, to contribute to ASD [74] and ASD-related phenotypes, such as Asperger syndrome [75] and Fragile X syndrome [76].
Fourth, our case-control association analysis allowed us to identify seven SNPs with a predicted deleterious effect, which showed a significantly higher prevalence in the ASD cohort compared to controls. Six genes harboring these variants: IGLJ2, FAM21A, OR11H12, HIP1, PRAMEF10, and ZNF717, have not been previously linked to ASD or ASD-related phenotypes. However, several studies have shown that ZNF717, HIP1, and several genes from the PRAME (Preferentially Expressed Antigen In Melanoma) family might be involved in cognitive development, learning disorders, and developmental disorders with autistic features [77,78]. Specifically, mutations in PRAMEF5 and PRAMEF7 were described in patients with delayed speech and language development, hearing deficits, and reading disability [77]. In the same study [77], a mutation in the ZNF717 gene has been identified among 16 other rare homozygous variants in at least two families, those of patients with Joubert syndrome—a disorder characterized by autistic behavior and intellectual disability. Two reciprocal microduplications inclusive of HIP1 have been described in three children from two unrelated families who had neurobehavioral problems: one child had an expressive language disorder, and two children had attention deficit hyperactivity disorder and manifested aggressive behavior [78]. The detection of variants within the OR11H12 gene encoding an olfactory receptor is also not surprising, as rare and common variants in at least several OR genes have been reported in association with autism, such as OR2M4 [79], OR2T10 [80,81], and OR52M1 [82].
In conclusion, despite the main limitation—a relatively small sample size—the study’s findings and observations are consistent with the growing body of evidence supporting the genetic bases of such heterogeneous disorders as ASD. We also report on several suggestive candidate genes that might be associated with ASD. Undoubtedly, follow-up research involving additional extended cohorts from the population is required to confirm the involvement of these genes in ASD; we consider these findings preliminary. To our knowledge, our study is one of the first attempts to investigate genome-wide polymorphic variants, SNPs and CNVs, in a previously understudied cohort of ASD from the Russian Federation, using a whole-exome sequencing technique.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/genes13050920/s1, Figure S1: Tranches plot (SureSelect V7), Figure S2: Tranches plot (TruSeqExome), Table S1: Participants. Demographics and phenotypic data, Table S2: Summary statistics on genotyping results, provided separately for three WES data subsets prepared with three exome capture assays, Table S3: CNVs identified across all individuals from both comparison groups, ASD and nonASD, Table S4: The distribution of common ASD-associated SNPs in the studied ASD and nonASD cohorts, as per the results of an intersection analysis with relevant data sets from the AutDB repository, Table S5: Common ASD-associated CNVs detected in the studied cohorts, as per the results of an intersection analysis with relevant data sets from the AutDB repository, Table S6: CNV dataset and Developmental delay morbidity map intersection results, Table S7: List of genes located within the CNVs’ loci detected in the studied cohorts, ASD and nonASD, Table S8: Top-30 list of GO terms ranked by Enrichment P-value, which were significantly overrepresented in the sets of genes harboring CNVs in ASD and nonASD, respectively, Table S9: HPO tracking in individual data.
Author Contributions
CConceptualization: E.L.G. and E.A.P.; Funding acquisition: E.L.G., E.A.P. and A.K.; Research: E.A.G., P.V.D., A.K., I.E., S.Y.R., O.V.Z. and O.Y.N.; Data analyses and manuscript preparation: E.A.G., P.V.D. and O.Y.N.; Writing—original draft preparation: E.A.G., P.V.D. and E.A.P. Writing—review and editing: E.A.G., P.V.D., E.A.P., E.V.M., A.K., I.E., S.Y.R., O.V.Z., O.Y.N. and E.L.G. All authors have read and agreed to the published version of the manuscript.
Funding
This work was financially supported by the Ministry of Science and Higher Education of the Russian Federation (Agreement No. 075-15-2022-301).
Institutional Review Board Statement
All procedures performed in the study abided by the ethical standards of the institutional and national research committees and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards. All protocols and procedures of this study were approved by the Almazov National Medical Research Centre Ethical Committee (Approval Code: 0101-22-01C/2014; Date: 14 June 2014) and the Saint-Petersburg State University Research Ethics Board (Approval Code: 02-155; Date: 20 June 2018).
Informed Consent Statement
Informed written consent was obtained from the study’s participants for the data collection and the publication of the study results.
Data Availability Statement
The data that support the findings of this study are available from the authors [E.A.P., E.A.G., P.V.D., and A.K.] upon reasonable request.
Acknowledgments
We are grateful to all the families and children who participated in this study, who provided information from their medical records and family histories, and donated biosamples for genetic analysis. We thank the Sirius University and the ‘Genetico’ Center for resource support of this study. Also, we would like to thank Mei Tan for her editorial assistance.
Conflicts of Interest
The authors declare that they have no competing interest.
References
- Yeargin-Allsopp, M.; Rice, C.; Karapurkar, T.; Doernberg, N.; Boyle, C.; Murphy, C. Prevalence of autism in a US metropolitan area. JAMA 2003, 289, 49–55. [Google Scholar] [CrossRef] [PubMed]
- Newschaffer, C.J.; Croen, L.A.; Daniels, J.; Giarelli, E.; Grether, J.K.; Levy, S.E.; Mandell, D.S.; Miller, L.A.; Pinto-Martin, J.; Reaven, J. The epidemiology of autism spectrum disorders. Annu. Rev. Public Health 2007, 28, 235–258. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mpaka, D.M.; Okitundu, D.L.; Ndjukendi, A.O.; N’Situ, A.M.; Kinsala, S.Y.; Mukau, J.E.; Ngoma, V.M.; Kashala-Abotnes, E.; Ma-Miezi-Mampunza, S.; Vogels, A.; et al. Prevalence and comorbidities of autism among children referred to the outpatient clinics for neurodevelopmental disorders. Pan. Afr. Med. J. 2016, 25, 82. [Google Scholar] [CrossRef]
- Doshi-Velez, F.; Ge, Y.; Kohane, I. Comorbidity clusters in autism spectrum disorders: An electronic health record time-series analysis. Pediatrics 2014, 133, e54–e63. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mannion, A.; Leader, G. Comorbidity in autism spectrum disorder: A literature review. Res. Autism Spectr. Disord. 2013, 7, 1595–1616. [Google Scholar] [CrossRef] [Green Version]
- Baxter, A.J.; Brugha, T.S.; Erskine, H.E.; Scheurer, R.W.; Vos, T.; Scott, J.G. The epidemiology and global burden of autism spectrum disorders. Psychol. Med. 2015, 45, 601–613. [Google Scholar] [CrossRef] [PubMed]
- Maenner, M.J.; Shaw, K.A.; Baio, J.; Washington, A.; Patrick, M.; DiRienzo, M.; Christensen, D.L.; Wiggins, L.D.; Pettygrove, S.; Andrews, J.G.; et al. Prevalence of autism spectrum disorder among children aged 8 years—Autism and developmental disabilities monitoring network, 11 sites, United States, 2016. Morb. Mortal. Wkly. Rep. Surveill. Summ. 2020, 69, 1–12. [Google Scholar] [CrossRef]
- Lyall, K.; Croen, L.; Daniels, J.; Fallin, M.D.; Ladd-Acosta, C.; Lee, B.K.; Park, B.Y.; Snyder, N.W.; Schendel, D.; Volk, H.; et al. The changing epidemiology of autism spectrum disorders. Annu. Rev. Public Health 2017, 38, 81–102. [Google Scholar] [CrossRef] [Green Version]
- Hallmayer, J.; Cleveland, S.; Torres, A.; Phillips, J.; Cohen, B.; Torigoe, T.; Miller, J.; Fedele, A.; Collins, J.; Smith, K.; et al. Genetic heritability and shared environmental factors among twin pairs with autism. Arch. Gen. Psychiatry 2011, 68, 1095–1102. [Google Scholar] [CrossRef]
- Bai, D.; Yip, B.H.K.; Windham, G.C.; Sourander, A.; Francis, R.; Yoffe, R.; Glasson, E.; Mahjani, B.; Suominen, A.; Leonard, H.; et al. Association of genetic and environmental factors with autism in a 5-country cohort. JAMA Psychiatry 2019, 76, 1035–1043. [Google Scholar] [CrossRef]
- Colvert, E.; Tick, B.; McEwen, F.; Stewart, C.; Curran, S.R.; Woodhouse, E.; Gillan, N.; Hallett, V.; Lietz, S.; Garnett, T.; et al. Heritability of autism spectrum disorder in a UK population-based twin sample. JAMA Psychiatry 2015, 72, 415–423. [Google Scholar] [CrossRef] [PubMed]
- Sandin, S.; Lichtenstein, P.; Kuja-Halkola, R.; Larsson, H.; Hultman, C.M.; Reichenberg, A. The familial risk of autism. Jama 2014, 311, 1770–1777. [Google Scholar] [CrossRef] [PubMed]
- Tick, B.; Bolton, P.; Happé, F.; Rutter, M.; Rijsdijk, F. Heritability of autism spectrum disorders: A meta-analysis of twin studies. J. Child Psychol. Psychiatry 2016, 57, 585–595. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grove, J.; Ripke, S.; Als, T.D.; Mattheisen, M.; Walters, R.K.; Won, H.; Pallesen, J.; Agerbo, E.; Andreassen, O.A.; Anney, R.; et al. Identification of common genetic risk variants for autism spectrum disorder. Nat. Genet. 2019, 51, 431–444. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Havdahl, A.; Niarchou, M.; Starnawska, A.; Uddin, M.; van der Merwe, C.; Warrier, V. Genetic contributions to autism spectrum disorder. Psychol. Med. 2021, 51, 2260–2273. [Google Scholar] [CrossRef] [PubMed]
- Bourgeron, T. Current knowledge on the genetics of autism and propositions for future research. Comptes Rendus Biol. 2016, 339, 300–307. [Google Scholar] [CrossRef]
- Christensen, D.L.; Baio, J.; Van Naarden Braun, K.; Bilder, D.; Charles, J.; Constantino, J.N.; Daniels, J.; Durkin, M.S.; Fitzgerald, R.T.; Kurzius-Spencer, M.; et al. Prevalence and characteristics of autism spectrum disorder among children aged 8 years—Autism and developmental disabilities monitoring network, 11 Sites, United States, 2012. Morb. Mortal. Wkly. Rep. Surveill. Summ. 2016, 65, 1–23. [Google Scholar] [CrossRef] [Green Version]
- Tromans, S.; Chester, V.; Gemegah, E.; Roberts, K.; Morgan, Z.; Yao, G.L.; Brugha, T. Autism identification across ethnic groups: A narrative review. Adv. Autism 2021, 7, 241–255. [Google Scholar] [CrossRef]
- Schott, W.; Tao, S.; Shea, L. Co-occurring conditions and racial-ethnic disparities: Medicaid enrolled adults on the autism spectrum. Autism Res. 2022, 15, 70–85. [Google Scholar] [CrossRef]
- Becerra, T.A.; von Ehrenstein, O.S.; Heck, J.E.; Olsen, J.; Arah, O.A.; Jeste, S.S.; Rodriguez, M.; Ritz, B. Autism spectrum disorders and race, ethnicity, and nativity: A population-based study. Pediatrics 2014, 134, e63–e71. [Google Scholar] [CrossRef] [Green Version]
- Morinaga, M.; Rai, D.; Hollander, A.-C.; Petros, N.; Dalman, C.; Magnusson, C. Migration or ethnic minority status and risk of autism spectrum disorders and intellectual disability: Systematic review. Eur. J. Public Health 2020, 31, 304–312. [Google Scholar] [CrossRef] [PubMed]
- Da Costa, G.E.; Fernandes, G.L.; Rodrigues, J.C.G.; da VB Leal, D.F.; Pastana, L.F.; Pereira, E.E.B.; Assumpção, P.P.; Burbano, R.M.R.; dos Santos, S.E.B.; Guerreiro, J.F.; et al. Exome evaluation of autism-associated genes in amazon american populations. Genes 2022, 13, 368. [Google Scholar] [CrossRef] [PubMed]
- Pizzo, L.; Jensen, M.; Polyak, A.; Rosenfeld, J.A.; Mannik, K.; Krishnan, A.; McCready, E.; Pichon, O.; Le Caignec, C.; Van Dijck, A.; et al. Rare variants in the genetic background modulate cognitive and developmental phenotypes in individuals carrying disease-associated variants. Genet. Med. 2019, 21, 816–825. [Google Scholar] [CrossRef] [Green Version]
- Andrews, S. FastQC: A Quality Control Tool for High Throughput Sequence Data. 2010. Available online: http://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 20 March 2022).
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van der Auwera, G.A.; Carneiro, M.O.; Hartl, C.; Poplin, R.; Del Angel, G.; Levy-Moonshine, A.; Jordan, T.; Shakir, K.; Roazen, D.; Thibault, J.; et al. From FastQ data to high confidence variant calls: The genome analysis toolkit best practices pipeline. Curr. Protoc. Bioinform. 2013, 43, 11.10.1–11.10.33. [Google Scholar] [CrossRef]
- Li, H. A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics 2011, 27, 2987–2993. [Google Scholar] [CrossRef] [Green Version]
- Wang, K.; Li, M.; Hakonarson, H. ANNOVAR: Functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010, 38, e164. [Google Scholar] [CrossRef]
- Klambauer, G.; Schwarzbauer, K.; Mayr, A.; Clevert, D.-A.; Mitterecker, A.; Bodenhofer, U.; Hochreiter, S. MOPS: Mixture of Poissons for discovering copy number variations in next-generation sequencing data with a low false discovery rate. Nucleic Acids Research 2012, 40, e69. [Google Scholar] [CrossRef]
- Seshan, V.E.; Olshen, A.; DNAcopy: DNA Copy Number Data Analysis. R Package Version 1.66.0. 2021. Available online: https://bioconductor.org/packages/release/bioc/html/DNAcopy.html (accessed on 20 March 2022).
- Purcell, S.; Neale, B.; Todd-Brown, K.; Thomas, L.; Ferreira, M.A.R.; Bender, D.; Maller, J.; Sklar, P.; de Bakker, P.I.W.; Daly, M.J.; et al. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007, 81, 559–575. [Google Scholar] [CrossRef] [Green Version]
- Kumar, P.; Henikoff, S.; Ng, P.C. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat. Protoc. 2009, 4, 1073–1081. [Google Scholar] [CrossRef]
- Adzhubei, I.; Jordan, D.M.; Sunyaev, S.R. Predicting functional effect of human missense mutations using PolyPhen-2. Curr. Protoc. Hum. Genet. 2014. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Basu, S.N.; Kollu, R.; Banerjee-Basu, S. AutDB: A gene reference resource for autism research. Nucleic Acids Res. 2009, 37, D832–D836. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- 1000 Genomes Project Consortium; Auton, A.; Brooks, L.D.; Durbin, R.M.; Garrison, E.P.; Kang, H.M.; Korbel, J.O.; Marchini, J.L.; McCarthy, S.; McVean, G.A.; et al. A global reference for human genetic variation. Nature 2015, 526, 68–74. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, J.; Raskin, L.; Samuels, D.C.; Shyr, Y.; Guo, Y. Genome measures used for quality control are dependent on gene function and ancestry. Bioinformatics 2014, 31, 318–323. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Banerjee-Basu, S.; Packer, A. SFARI Gene: An evolving database for the autism research community. Dis. Models Mech. 2010, 3, 133–135. [Google Scholar] [CrossRef] [Green Version]
- Rappaport, N.; Nativ, N.; Stelzer, G.; Twik, M.; Guan-Golan, Y.; Stein, T.I.; Bahir, I.; Belinky, F.; Morrey, C.P.; Safran, M.; et al. MalaCards: An integrated compendium for diseases and their annotation. Database 2013, 2013, bat018. [Google Scholar] [CrossRef] [Green Version]
- Hamosh, A.; Scott, A.F.; Amberger, J.S.; Bocchini, C.A.; McKusick, V.A. Online Mendelian Inheritance in Man (OMIM), a knowledgebase of human genes and genetic disorders. Nucleic Acids Res. 2005, 33, D514–D517. [Google Scholar] [CrossRef]
- Robinson, P.N.; Köhler, S.; Bauer, S.; Seelow, D.; Horn, D.; Mundlos, S. The human phenotype ontology: A tool for annotating and analyzing human hereditary disease. Am. J. Hum. Genet. 2008, 83, 610–615. [Google Scholar] [CrossRef] [Green Version]
- Rentzsch, P.; Witten, D.; Cooper, G.M.; Shendure, J.; Kircher, M. CADD: Predicting the deleteriousness of variants throughout the human genome. Nucleic Acids Res. 2018, 47, D886–D894. [Google Scholar] [CrossRef]
- Niroula, A.; Vihinen, M. How good are pathogenicity predictors in detecting benign variants? PLoS Comput. Biol. 2019, 15, e1006481. [Google Scholar] [CrossRef] [Green Version]
- Kircher, M.; Witten, D.M.; Jain, P.; O’Roak, B.J.; Cooper, G.M.; Shendure, J. A general framework for estimating the relative pathogenicity of human genetic variants. Nat. Genet. 2014, 46, 310–315. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lintas, C.; Picinelli, C.; Piras, I.S.; Sacco, R.; Brogna, C.; Persico, A.M. Copy number variation in 19 Italian multiplex families with autism spectrum disorder: Importance of synaptic and neurite elongation genes. Am. J. Med. Genet. B Neuropsychiatr. Genet. 2017, 174, 547–556. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Mandell, J.D.; Kumar, Y.; Sun, N.; Morris, M.T.; Arbelaez, J.; Nasello, C.; Dong, S.; Duhn, C.; Zhao, X.; et al. De novo sequence and copy number variants are strongly associated with tourette disorder and implicate cell polarity in pathogenesis. Cell Rep. 2018, 24, 3441–3454. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Celestino-Soper, P.B.; Shaw, C.A.; Sanders, S.J.; Li, J.; Murtha, M.T.; Ercan-Sencicek, A.G.; Davis, L.; Thomson, S.; Gambin, T.; Chinault, A.C.; et al. Use of array CGH to detect exonic copy number variants throughout the genome in autism families detects a novel deletion in TMLHE. Hum. Mol. Genet. 2011, 20, 4360–4370. [Google Scholar] [CrossRef] [PubMed]
- Krumm, N.; Turner, T.N.; Baker, C.; Vives, L.; Mohajeri, K.; Witherspoon, K.; Raja, A.; Coe, B.P.; Stessman, H.A.; He, Z.X.; et al. Excess of rare, inherited truncating mutations in autism. Nat. Genet. 2015, 47, 582–588. [Google Scholar] [CrossRef] [Green Version]
- Girirajan, S.; Brkanac, Z.; Coe, B.P.; Baker, C.; Vives, L.; Vu, T.H.; Shafer, N.; Bernier, R.; Ferrero, G.B.; Silengo, M.; et al. Relative burden of large CNVs on a range of neurodevelopmental phenotypes. PLoS Genet. 2011, 7, e1002334. [Google Scholar] [CrossRef]
- Pinto, D.; Delaby, E.; Merico, D.; Barbosa, M.; Merikangas, A.; Klei, L.; Thiruvahindrapuram, B.; Xu, X.; Ziman, R.; Wang, Z.; et al. Convergence of genes and cellular pathways dysregulated in autism spectrum disorders. Am. J. Hum. Genet. 2014, 94, 677–694. [Google Scholar] [CrossRef] [Green Version]
- Yatsenko, S.A.; Hixson, P.; Roney, E.K.; Scott, D.A.; Schaaf, C.P.; Ng, Y.T.; Palmer, R.; Fisher, R.B.; Patel, A.; Cheung, S.W.; et al. Human subtelomeric copy number gains suggest a DNA replication mechanism for formation: Beyond breakage-fusion-bridge for telomere stabilization. Hum. Genet. 2012, 131, 1895–1910. [Google Scholar] [CrossRef] [Green Version]
- Pinto, D.; Pagnamenta, A.T.; Klei, L.; Anney, R.; Merico, D.; Regan, R.; Conroy, J.; Magalhaes, T.R.; Correia, C.; Abrahams, B.S.; et al. Functional impact of global rare copy number variation in autism spectrum disorders. Nature 2010, 466, 368–372. [Google Scholar] [CrossRef] [Green Version]
- AlAyadhi, L.Y.; Hashmi, J.A.; Iqbal, M.; Albalawi, A.M.; Samman, M.I.; Elamin, N.E.; Bashir, S.; Basit, S. High-resolution SNP genotyping platform identified recurrent and novel CNVs in autism multiplex families. Neuroscience 2016, 339, 561–570. [Google Scholar] [CrossRef]
- Kaminsky, E.B.; Kaul, V.; Paschall, J.; Church, D.M.; Bunke, B.; Kunig, D.; Moreno-De-Luca, D.; Moreno-De-Luca, A.; Mulle, J.G.; Warren, S.T.; et al. An evidence-based approach to establish the functional and clinical significance of copy number variants in intellectual and developmental disabilities. Genet. Med. 2011, 13, 777–784. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- O’Roak, B.J.; Vives, L.; Girirajan, S.; Karakoc, E.; Krumm, N.; Coe, B.P.; Levy, R.; Ko, A.; Lee, C.; Smith, J.D.; et al. Sporadic autism exomes reveal a highly interconnected protein network of de novo mutations. Nature 2012, 485, 246–250. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sajan, S.A.; Fernandez, L.; Nieh, S.E.; Rider, E.; Bukshpun, P.; Wakahiro, M.; Christian, S.L.; Rivière, J.B.; Sullivan, C.T.; Sudi, J.; et al. Both rare and de novo copy number variants are prevalent in agenesis of the corpus callosum but not in cerebellar hypoplasia or polymicrogyria. PLoS Genet. 2013, 9, e1003823. [Google Scholar] [CrossRef] [PubMed]
- Asadollahi, R.; Oneda, B.; Joset, P.; Azzarello-Burri, S.; Bartholdi, D.; Steindl, K.; Vincent, M.; Cobilanschi, J.; Sticht, H.; Baldinger, R.; et al. The clinical significance of small copy number variants in neurodevelopmental disorders. J. Med. Genet. 2014, 51, 677–688. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Di Gregorio, E.; Riberi, E.; Belligni, E.F.; Biamino, E.; Spielmann, M.; Ala, U.; Calcia, A.; Bagnasco, I.; Carli, D.; Gai, G.; et al. Copy number variants analysis in a cohort of isolated and syndromic developmental delay/intellectual disability reveals novel genomic disorders, position effects and candidate disease genes. Clin. Genet. 2017, 92, 415–422. [Google Scholar] [CrossRef]
- Munnich, A.; Demily, C.; Frugère, L.; Duwime, C.; Malan, V.; Barcia, G.; Vidal, C.; Throo, E.; Besmond, C.; Hubert, L.; et al. Impact of on-site clinical genetics consultations on diagnostic rate in children and young adults with autism spectrum disorder. Mol. Autism 2019, 10, 33. [Google Scholar] [CrossRef]
- Coe, B.P.; Witherspoon, K.; Rosenfeld, J.A.; van Bon, B.W.; Vulto-van Silfhout, A.T.; Bosco, P.; Friend, K.L.; Baker, C.; Buono, S.; Vissers, L.E.; et al. Refining analyses of copy number variation identifies specific genes associated with developmental delay. Nat. Genet. 2014, 46, 1063–1071. [Google Scholar] [CrossRef]
- Cooper, G.M.; Coe, B.P.; Girirajan, S.; Rosenfeld, J.A.; Vu, T.H.; Baker, C.; Williams, C.; Stalker, H.; Hamid, R.; Hannig, V.; et al. A copy number variation morbidity map of developmental delay. Nat. Genet. 2011, 43, 838–846. [Google Scholar] [CrossRef] [Green Version]
- Conrad, D.F.; Pinto, D.; Redon, R.; Feuk, L.; Gokcumen, O.; Zhang, Y.; Aerts, J.; Andrews, T.D.; Barnes, C.; Campbell, P.; et al. Origins and functional impact of copy number variation in the human genome. Nature 2010, 464, 704–712. [Google Scholar] [CrossRef] [Green Version]
- Ge, S.X.; Jung, D.; Yao, R. ShinyGO: A graphical gene-set enrichment tool for animals and plants. Bioinformatics 2020, 36, 2628–2629. [Google Scholar] [CrossRef]
- Fidler, D.J.; Bailey, J.N.; Smalley, S.L. Macrocephaly in autism and other pervasive developmental disorders. Dev. Med. Child Neurol. 2000, 42, 737–740. [Google Scholar] [CrossRef] [PubMed]
- Fombonne, E.; Rogé, B.; Claverie, J.; Courty, S.; Frémolle, J. Microcephaly and macrocephaly in autism. J. Autism Dev. Disord. 1999, 29, 113–119. [Google Scholar] [CrossRef] [PubMed]
- Lainhart, J.E.; Bigler, E.D.; Bocian, M.; Coon, H.; Dinh, E.; Dawson, G.; Deutsch, C.K.; Dunn, M.; Estes, A.; Tager-Flusberg, H.; et al. Head circumference and height in autism: A study by the collaborative program of excellence in autism. Am. J. Med. Genet. Part A 2006, 140, 2257–2274. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Douard, E.; Zeribi, A.; Schramm, C.; Tamer, P.; Loum, M.A.; Nowak, S.; Saci, Z.; Lord, M.P.; Rodríguez-Herreros, B.; Jean-Louis, M.; et al. Effect Sizes of deletions and duplications on autism risk across the genome. Am. J. Psychiatry 2021, 178, 87–98. [Google Scholar] [CrossRef]
- Sener, E.F. Association of copy number variations in autism spectrum disorders: A systematic review. Chin. J. Biol. 2014, 2014, 713109. [Google Scholar] [CrossRef]
- Tabet, A.-C.; Verloes, A.; Pilorge, M.; Delaby, E.; Delorme, R.; Nygren, G.; Devillard, F.; Gérard, M.; Passemard, S.; Héron, D.; et al. Complex nature of apparently balanced chromosomal rearrangements in patients with autism spectrum disorder. Mol. Autism 2015, 6, 19. [Google Scholar] [CrossRef] [Green Version]
- Rylaarsdam, L.; Guemez-Gamboa, A. Genetic causes and modifiers of autism spectrum disorder. Front. Cell. Neurosci. 2019, 13, 385. [Google Scholar] [CrossRef]
- Park, S.M.; Jang, H.J.; Lee, J.H. Roles of primary cilia in the developing brain. Front. Cell. Neurosci. 2019, 13, 218. [Google Scholar] [CrossRef] [Green Version]
- Guemez-Gamboa, A.; Coufal, N.G.; Gleeson, J.G. Primary cilia in the developing and mature brain. Neuron 2014, 82, 511–521. [Google Scholar] [CrossRef] [Green Version]
- Guo, J.; Higginbotham, H.; Li, J.; Nichols, J.; Hirt, J.; Ghukasyan, V.; Anton, E.S. Developmental disruptions underlying brain abnormalities in ciliopathies. Nat. Commun. 2015, 6, 7857. [Google Scholar] [CrossRef] [Green Version]
- Guo, J.; Otis, J.M.; Higginbotham, H.; Monckton, C.; Cheng, J.; Asokan, A.; Mykytyn, K.; Caspary, T.; Stuber, G.D.; Anton, E.S. Primary cilia signaling shapes the development of interneuronal connectivity. Dev. Cell 2017, 42, 286–300. [Google Scholar] [CrossRef] [PubMed]
- Trulioff, A.; Ermakov, A.; Malashichev, Y. Primary Cilia as a possible link between left-right asymmetry and neurodevelopmental diseases. Genes 2017, 8, 48. [Google Scholar] [CrossRef]
- Kondziella, D.; Lycke, J. Autism spectrum disorders: Does cilia dysfunction in embryogenesis play a role? Acta Neuropsychiatr. 2008, 20, 227–228. [Google Scholar] [CrossRef]
- Lee, B.; Panda, S.; Lee, H.Y. Primary ciliary deficits in the dentate gyrus of fragile X syndrome. Stem Cell Rep. 2020, 15, 454–466. [Google Scholar] [CrossRef] [PubMed]
- Lei, H.; Yan, Z.; Sun, X.; Zhang, Y.; Wang, J.; Ma, C.; Xu, Q.; Wang, R.; Jarvis, E.D.; Sun, Z. Axon guidance pathways served as common targets for human speech/language evolution and related disorders. Brain Lang. 2017, 174, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Ramocki, M.B.; Bartnik, M.; Szafranski, P.; Kołodziejska, K.E.; Xia, Z.; Bravo, J.; Miller, G.S.; Rodriguez, D.L.; Williams, C.A.; Bader, P.I.; et al. Recurrent distal 7q11.23 Deletion including HIP1 and YWHAG identified in patients with intellectual disabilities, epilepsy, and neurobehavioral problems. Am. J. Hum. Genet. 2010, 87, 857–865. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kuo, P.H.; Chuang, L.C.; Su, M.H.; Chen, C.H.; Chen, C.H.; Wu, J.Y.; Yen, C.J.; Wu, Y.Y.; Liu, S.K.; Chou, M.C.; et al. Genome-wide association study for autism spectrum disorder in Taiwanese Han population. PLoS ONE 2015, 10, e0138695. [Google Scholar] [CrossRef] [Green Version]
- Iossifov, I.; O’Roak, B.J.; Sanders, S.J.; Ronemus, M.; Krumm, N.; Levy, D.; Stessman, H.A.; Witherspoon, K.T.; Vives, L.; Patterson, K.E.; et al. The contribution of de novo coding mutations to autism spectrum disorder. Nature 2014, 515, 216–221. [Google Scholar] [CrossRef] [Green Version]
- Codina-Solà, M.; Rodríguez-Santiago, B.; Homs, A.; Santoyo, J.; Rigau, M.; Aznar-Laín, G.; Del Campo, M.; Gener, B.; Gabau, E.; Botella, M.P.; et al. Integrated analysis of whole-exome sequencing and transcriptome profiling in males with autism spectrum disorders. Mol. Autism 2015, 6, 21. [Google Scholar] [CrossRef] [Green Version]
- Sanders, S.J.; He, X.; Willsey, A.J.; Ercan-Sencicek, A.G.; Samocha, K.E.; Cicek, A.E.; Murtha, M.T.; Bal, V.H.; Bishop, S.L.; Dong, S.; et al. Insights into autism spectrum disorder genomic architecture and biology from 71 risk loci. Neuron 2015, 87, 1215–1233. [Google Scholar] [CrossRef] [Green Version]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).