Abstract
Trehalose-6-phosphate phosphatase (TPP) genes take part in trehalose metabolism and also in stress tolerance, which has been well documented in many species but poorly understood in wheat. The present research has identified a family of 31 TPP genes in Triticum aestivum L. through homology searches and classified them into five clades by phylogenetic tree analysis, providing evidence of an evolutionary status with Hordeum vulgare, Brachypodium distachyon and Oryza sativa. The exon-intron distribution revealed a discrete evolutionary history and projected possible gene duplication occurrences. Furthermore, different computational approaches were used to analyze the physical and chemical properties, conserved domains and motifs, subcellular and chromosomal localization, and three-dimensional (3-D) protein structures. Cis-regulatory elements (CREs) analysis predicted that TaTPP promoters consist of CREs related to plant growth and development, hormones, and stress. Transcriptional analysis revealed that the transcription levels of TaTPPs were variable in different developmental stages and organs. In addition, qRT-PCR analysis showed that different TaTPPs were induced under salt and drought stresses and during leaf senescence. Therefore, the findings of the present study give fundamental genomic information and possible biological functions of the TaTPP gene family in wheat and will provide the path for a better understanding of TaTPPs involvement in wheat developmental processes, stress tolerance, and leaf senescence.
1. Introduction
Cereals are indeed the single most significant part of the diet for the majority of the global population, with about 60% to 80% of carbohydrates coming straightly from them in developing and under-developing nations, respectively [1]. According to the FAO’s most current predictions, global grain production in 2021 will increase by 1.7% over 2020, achieving 2817 million tons [2]. Wheat (Triticum aestivum L.) is the world’s largest extensively grown cereal crop and is among the most often eaten cereals by the world population [3]. The major abiotic stresses that decrease wheat productivity throughout the growing period include water shortages, high temperatures, and salinity [4]. Among them, salinity is a major barrier to crop production, especially in wheat, resulting in a yield loss of 65% in moderately saline soils, by influencing nearly every stage of plant growth and development, including germination, vegetative growth, and reproductive growth [5,6]. This abiotic stress condition results in a decrease in yield related traits that directly affect the yield of cereal crops. Thus, one of the most significant tasks for plant breeders right now is to uncover the genes associated with abiotic stress responses and to cultivate genetically engineered varieties with improved stress tolerance [7,8].
Plants generate various organic molecules, such as soluble sugar and free amino acids, in response to stress exposure. Trehalose is one of these non-reducing disaccharides composed of two molecules of α-glucose that may accumulate in the cell up to 12% of its dry mass to maintain its integrity and is associated with plant abiotic stress tolerance, including high and low temperature, drought, and osmotic stress tolerance [9,10,11,12]. Many species, including yeast, fungus, invertebrates, plants, bacteria, insects, green weed, and cyanobacteria synthesize this sugar substance [12,13,14,15]. Except for vertebrates, the synthesis of trehalose in plants and other organisms involves two phases with two catalytic enzymes, trehalose-6-phosphate synthase (TPS) and trehalose-6-phosphate phosphatase (TPP). TPS produces trehalose-6-phosphate (T6P), a phosphorylated intermediate, from Uridine diphosphate-glucose (UDPG) and Glucose-6-phosphate (G6P) in the first phase, and the TPP dephosphorylates T6P to produce trehalose in the second phase (Figure 1). Trehalose is then hydrolyzed by an enzyme called trehalase (TRE) to synthesize two molecules of glucose, which suggests that TPS, TPP, and TRE are the three enzymes involved in the trehalose biosynthesis pathway [16]. The TPS and TPP families encode multiple genes, but TRE is denoted by a single copy of the gene [17,18,19].
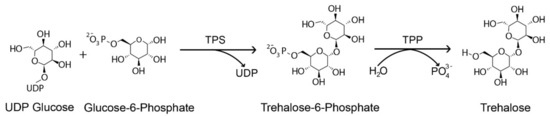
Figure 1.
Trehalose biosynthesis pathway in plants. Uridine diphosphate glucose (UDP), Trehalose-6-phosphate synthase (TPS) and Trehalose-6-phosphate phosphatase (TPP).
In addition to providing a route for the production of trehalose, TPS and TPP have been shown to serve as signaling molecules in higher plants by modulating a variety of plant metabolic and developmental processes. T6P is a signaling metabolite in plants that links growth and development to carbon metabolism and serves as a signal of sucrose status at various phases of the plant’s development [20,21,22]. TPS genes were discovered to be involved in the germination of seeds, stress signaling, vegetative phase separation, shoot branching, and flowering time regulation in Arabidopsis and rice, with TPS1 being the most studied [23,24,25,26,27]. Instead, TPP was found to inhibit SnRK1 (Sn1-related protein kinase) activity, a well-known transcriptional regulatory pathway under stress and energy metabolism [28]. The Ramosa1 (RA1) transcription factor activates the transcription of TPP to regulate flower branching, which suggests that trehalose may have a role in specific developmental processes [29]. Tobacco plant overexpressing Escherichia coli TPS gene ostA improved photosynthesis efficiency by enhancing RUBISCO concentration, although ostB, a TPP gene, exhibited the opposite impact, further suggesting the significance of trehalose in plant photosynthesis [30].
Various studies have reported trehalose enzymes to enhance abiotic and biotic stress tolerance, such as in Arabidopsis [31,32]. For example, ZxTPP (Zygophyllum xanthoxylum) or ostA and ostB containing tobacco transgenic plants were significantly tolerant to drought [33,34]. Likewise, ostA and ostB transformed rice plants showed increased trehalose levels and enhanced performance against cold, salt, and drought stresses [35]. Exogenous trehalose triggered a signal transduction pathway including calcium and reactive oxygen species (ROS) and OsTPP1 or OsTPP3 transgenic rice and maize plants induced stress-related genes that conferred drought tolerance [36,37,38]. After drought stress, vulnerable maize seedlings had lower ZmTPP1 expression, whereas resistant seedlings had higher expression [39]. TPP promoters’ Cis-regulatory elements (CREs) stimulate trehalose metabolism and improve stress response. In Arabidopsis, ABF1, ABF2, and ABF4 are ABA-responsive elements that directly influence AtTPPI expression to increase drought tolerance by changing stomatal apertures [40]. The transcription factor that responds to ABA in the presence of ABA, ABF2 binds directly to the AtTPPE promoter, triggering its expression for root elongation and stomatal movement via producing ROS [41]. DREB1A, which binds to the DRE/CR motif in the AtTPPF promoter, is thought to upregulate AtTPPF transcription in drought-stressed plants [32]. T6P role as a signal for increased carbon availability might have implications for leaf senescence control, as the accumulation of sugars has been demonstrated during leaf senescence in Arabidopsis, wheat, tobacco, and maize. The phenotype of mature otsB-overexpressing Arabidopsis plants included delayed senescence and decreased anthocyanin accumulation, suggesting that the role of TPP may perform a crucial role during leaf senescence in plants [42,43,44,45]. To date, TaTPP-6AL1 and its functional marker have been shown to improve crop yield in wheat [46]. However, the gene structure and regulatory mechanism of wheat TPPs are not well studied.
The present study intends to investigate wheat TPPs in silico by identification of TaTPPs, gene duplication analysis, phylogenetic relationship with other species, subcellular localization prediction, motif and domain analyses, proteins 3-D structure modeling, investigation of CREs, and gene transcription analysis that have all been performed to better understand TaTPPs functions in wheat.
2. Materials and Methods
2.1. Identification of Putative TPPs in the Wheat Genome
To find putative TPPs in wheat, we utilized TPPs from Arabidopsis and rice. Ensembl Plants database was used to collect TPP protein sequences from Arabidopsis and rice and a BLASTp search was conducted against the most recent wheat assembly from the IWGSC (RefSeq v1.0) (http://plants.ensembl.org/index.html, 10−5 cut-off e-value and bit-score > 100, accessed on 12 March 2021). After eliminating duplicated sequences, SMART (http://smart.embl-heidelberg.de/, accessed on 12 March 2021) or InterPro (https://www.ebi.ac.uk/interpro, accessed on 12 March 2021) and NCBI CDD (https://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi, accessed on 12 March 2021) were used to examine the remaining sequences for the presence of transmembrane domains. TPP-related domain-containing protein sequences were collected and designated consecutively according to their chromosomal locations after the sequences without transmembrane domains were deleted. The ProtParam software (https://web.expasy.org/protparam/, accessed on 13 March 2021) was used to calculate the length, molecular weight, isoelectric point (pI), and grand average of hydropathicity (GRAVY) of TPP proteins.
2.2. Chromosome Localization, Gene Duplication and Synteny Analysis
TPPs genomic locations were acquired from the Ensembl Plants BioMart (http://plants.ensembl.org/biomart/martview, accessed on 14 March 2021) for chromosomal distribution. The TPPs were given a ‘Ta’ prefix and were numbered in ascending order according to their ascending chromosomal location. The TaTPPs on the wheat chromosomes were represented using TBtools. A NCBI BlastP search (https://blast.ncbi.nlm.nih.gov/Blast.cgi?PROGRAM=blastp&PAGE_TYPE=BlastSearch&BLAST_SPEC=&LINK_LOC=blasttab&LAST_PAGE=blastn, query conditions: percent identity between 75 and 100 and query coverage between 80 and 100, accessed on 14 March 2021) based on the proportion of query cover to the identity of the TaTPPs against each other was performed to check for gene duplication [47]. Based on a BLAST search and a phylogenetic tree, duplicate gene pairs were identified. TBtools was used to determine the non-synonymous substitution rate (Ka), synonymous substitution rate (Ks), and Ka/Ks ratio [48]. The synteny relationships of wheat TPP genes with different plant species were analyzed using TBtools.
2.3. Phylogenetic Analysis, Exon-Intron Distribution and 3-D Structure Modeling
ClustalW in MEGA X was used to align full-length protein sequences from various species [49]. Following the alignment, MEGA X was used to create a phylogenetic tree with the Maximum Likelihood method [50] and 1000 bootstrap values [51]. To examine the exon-intron distribution of TaTPPs, the TBtool was used to align the CDSs and genomic sequences. SWISS-MODEL Workspace web tools (https://swissmodel.expasy.org/interactive#sequence, accessed on 16 March 2021), GASS and SOPMA secondary structural method (https://npsa-prabi.ibcp.fr/cgi-bin/npsa_automat.pl?page=npsa%20_sopma.html, accessed on 16 March 2021) and MolProbity server (http://molprobity.biochem.duke.edu/, accessed on 16 March 2021) were used to conduct 3-D structure analyses of TaTPP proteins [52,53,54,55,56,57].
2.4. Subcellular Localization Prediction and Protein Domain Analysis
PredSL (http://aias.biol.uoa.gr/PredSL/index.html, accessed on 17 March 2021) was used to predict subcellular localizations. The TPP domain (trehalose-phosphatase (Trehalose PPase); PF02358) was retrieved from the Pfam database and the structures were created with TBtools [58,59]. We utilized MEME suite 5.1.1 to examine TaTPP motifs and The site distribution was set to any number of repetitions, the maximum number of motifs to locate was set to 9, the minimum width was set to 6, the maximum width was set to 50, and the maximum number of motifs to locate was set to 9 [60].
2.5. Analysis of Publicly Accessible Expression Data and Cis-Regulatory Elements (CREs)
We used the NCBI database (https://www.ncbi.nlm.nih.gov/, accessed on 19 March 2021) to obtain 2 kb upstream from start codon promoter sequences of 11 TaTPPs, which we subsequently submitted to PlantCARE to find the CREs [61]. Netbeans IDE 8.0 (https://netbeans.org., accessed on 25 March 2021) was used to organize data [62] and subsequently TBtools Heatmap was used for data visualization. The Genevestigator RNAseq public anatomy was used to examine gene expression [63] and the MeV tool was then used to visualize expression [64].
2.6. Plant Materials and Treatments
T. aestivum L. cultivar Jinmai39 was used to investigate the transcription of TaTPPs in the presence of salt, drought, and ABA treatments. The seedlings were grown in a growth chamber at 22 °C with 16 h/8 h of light/ darkness and a light intensity of 9000 lux. Wheat plants were treated with either double-distilled water (control) or a 20% PEG-6000 or a 250 mM NaCl solution at the 2–3 leaf stage for drought and salt stress, respectively. For abscisic acid (ABA) treatment, plants at the same stage are sprayed with 100 mM abscisic acid (ABA) or 0.1% (v/v) ethanol (control). To analyze the expression of TaTPPs during leaf senescence, the delayed senescence wheat cultivar Yannong19 was grown in field conditions and collected samples from flag leaf at 0, 7, 10, 16, 19, 22, 24, and 25 days after anthesis. All the leaves after collection are immediately frozen into liquid nitrogen and stored at −80 °C for further RNA extraction.
2.7. RNA Extraction, Quantitative Real-Time Reverse Transcription PCR Analysis and Protein Interaction Network
The Quick RNA isolation Kit (Huayueyang Biotechnology, Beijing, China) was used to extract RNA according to the manufacturer’s instructions and DNase I treatment was used to remove DNA contamination. The RevertAid First Strand cDNA Synthesis Kit (Thermo Scientific, Waltham, MA, USA) was used to synthesize cDNA from a 3-µg aliquot of total RNA from each sample. To measure the expression of TaTPPs qRT-PCR analysis was performed with specific primers (Table S1), as described previously [65]. The ABI PRISM 7500 system (Applied Biosystems, Foster City, CA, USA) was used to generate threshold values (CT) and the transcription level of TaTPPs was measured using the comparative 2−∆∆CT technique that was standardized with the Elongation factor 1α (TaEF-1α) (GenBank accession no. Q03033) [66,67] (Table S1). All of the studies were carried out three times. The TaTPP protein interaction network was examined using the STRING online server (https://string-db.org/, accessed on 27 April 2021).
3. Results
3.1. Identification and Annotation of Wheat TPPs
We identified a total of 31 TPP protein sequences in the wheat genome (Table 1 and Table S2). This number is relatively large when compared to TPPs previously identified in Arabidopsis, rice, and maize (Table S3). Wheat has a greater ploidy level and a larger genome size as it originated from the natural hybridization of three closely related genomes (A, B, and D), which may justify this result [68]. These protein sequences were encoded by 31 genes, three of which were chosen as representatives because they showed splice variants with full domains. A detailed description of TaTPPs is summarized in Table 1. The ORF of TaTPPs ranged from 750 to 1755 bp, with protein lengths ranging from 249 to 584 amino acids (Table 1). The molecular weight of the genes ranged from 28.67 KDa to 65.02 KDa. (Table 1). Fifteen genes were found to be basic (>7) and 16 genes were found to be acidic (<7) based on the predicted pI value (Table 1).

Table 1.
Detailed annotations of the TaTPPs in wheat.
In addition, the Aliphatic Index and Instability Index were computed. The Aliphatic Index measures how much space is taken up by aliphatic side chains in Alanine, Isoleucine, Leucine, and Valine amino acids [69]. The Aliphatic Index ranges observed were 72.28 to 86.42, and the Instability Index ranges were 32.59 to 55.74 (Table 1). The high Aliphatic Index of a protein sequence suggests that it can function at a broad range of temperatures, whereas the Instability Index shows whether the protein is stable or unstable [70]. All the TaTPPs had negative GRAVY values ranging from −0.700 to −0.142 (Table 1). A protein with a negative GRAVY value is non-polar and hydrophilic in nature [69].
3.2. Subcellular Localization Prediction and Chromosomal Distribution of TaTPPs
PredSL (http://aias.biol.uoa.gr/PredSL/index.html accessed on 22 September 2021) was used to predict subcellular localization. Subcellular localization of the TaTPPs was predicted mostly in the chloroplast, whereas, TaTPP1-A, TaTPP7-D, TaTPP10-B appeared to be localized in the mitochondrion (Table 1). Moreover, TaTPP5-B, TaTPP7-A were predicted as secreted proteins (Table 1). However, TaTPP5-A, TaTPP10-A, TaTPP10-D were predicted with unknown localization (Table 1). A schematic diagram was created to explain the chromosomal location of TaTPPs. The TaTPPs are present on 17 wheat chromosomes (Figure 2 and Table 1). On the chromosomes of the A subgenome, the highest number of TaTPP genes (11 genes) were mapped. B and D subgenomes had 10 TaTPP genes in each subgenome. The maximum 14 genes of TaTPPs were located on chromosome 2 (Figure 2). Chromosome 6A, 6B and 6D, had 2 genes on each chromosome and 1A, 1B, 1D, 3A, 3D, 5A, 5B, and 5D had only a single gene. On the other hand, no TaTPPs were found on chromosomes 3B, 4A, 4B, or 4D (Figure 2 and Table 1), suggesting that TPP family genes were unevenly distributed throughout the three subgenomes of wheat.
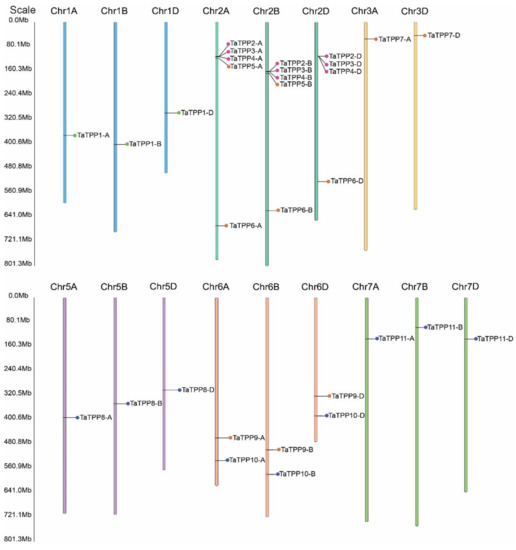
Figure 2.
Graphical presentations of TaTPPs chromosomal distribution of on wheat chromosomes. The name of the gene on the right side and the location of the TaTPPs is indicated by the colored circular circle on the chromosomes. The three subgenomes chromosomal numbers are shown at the top of each bar.
We further investigated the duplication events in the TaTPP gene family in the context of wheat being hexapolyploid and having big genomes. Genes are usually considered duplicated when the query cover and identity value of gene sequences are more than 80% [71]. It has also been reported that genes are considered duplicated when protein sequence similarity and identity are more than 70% and 75%, respectively [72]. By analyzing the sequences, we found 27 pairs of TaTPPs with a sequence identity ranges from 82.14% to 95.25% and 100% query cover within all gene pairs (Tables S4 and S5) and identified in the same phylogenetic tree clade (Figure 3). We further computed the non-synonymous (Ka) and synonymous (Ks) substitutions, as well as the Ka/Ks ratios, for the 27 TaTPP gene pairs to determine the selection pressure on the duplicated TaTPPs (Table S5). These gene pairs Ka/Ks ratios were smaller than one, indicating that they developed under functional restriction with negative or purifying selection. The divergence period ranged from 2.93 to 13.3 million years ago (MYA), showing that these gene pairs were duplicated recently (Table S5).
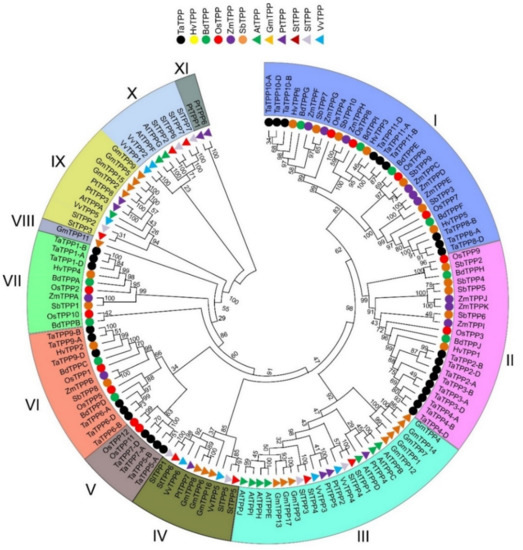
Figure 3.
Phylogenetic analysis of TaTPP proteins. The tree was generated using MEGA X by the maximum likelihood method with 1000 bootstrap values. All the species and protein ID used for constructing tree were presented in Table S6.
3.3. Phylogenetic and Conserved Domain Analyses of TaTPP Proteins
A phylogenetic tree containing full length TPP protein sequences from twelve plant species was constructed by the maximum likelihood method to better understand the evolutionary relations among the TaTPP proteins with other species (Figure 3, Table S6), including five species from monocot: Hordeum vulgare, Brachypodium distachyon, Oryza sativa, Zea maize, Sorghum bicolor; and 6 species from dicot: Arabidopsis thaliana, Glycine max, Populus trichocarpa, Solanum tuberosum, Solanum lycopersicum, Vitis vinifera. The results indicated that TPP proteins were divided into eleven clades, where clade I was the largest with 30 members. Clades II to XI (total 10 clades in order) included 21, 24, 10, 6, 13, 10, 1, 10, 8, and 2 members, respectively (Figure 3).
Plants classified as dicots and monocots were divided into distinct clades. Proteins from monocot plants were grouped into clade I, clade II, clade V, clade VI, and clade VII, whereas proteins from dicot plants were grouped into clade III, clade IV, clade VIII, clade IX, clade X, and clade XI. The highest number of TaTPP proteins were grouped into clade I and clade II, which had nine proteins in each clade. In addition, clades V, VI, VII contained four, six, and three TaTPP proteins, respectively (Figure 3). Most of the wheat TPP proteins were closely related to H. vulgare, B. distachyon, and O. sativa, suggesting their conserved function with those plant species and offering information that can be used to conduct a more in-depth functional analysis. All the TaTPPs were assembled into 11 groups, as sequences from A, B, and D subgenome of 11 groups clustered together in the phylogenetic tree (Figure 3) and protein sequence identity was more the 88% between A, B, and D subgenome of each group (Table S4). Thus, we considered the protein sequences from A, B, and D subgenome of each group are copies of separate TaTPP genes and named them according to the ascending order of the chromosomal location (Table 1).
Further, the Pfam database was utilized to find the important component domains of TaTPP proteins [59]. All the TaTPP proteins contain a specific Trehalose PPase domain (PF02358). In addition, a stress antifungal domain was found in TaTPP-5A, TaTPP7-A and TaTPP7-D (Figure 4a). We used MEME suite 5.1.1 to evaluate motif sequences for 31 TaTPPs and found six significant motifs (motifs 1–6) (Figure 4b). All the motifs were found to be conserved in all TaTPP proteins except for TaTPP5-A, which lacks lacks motif 3 (Figure 4b).
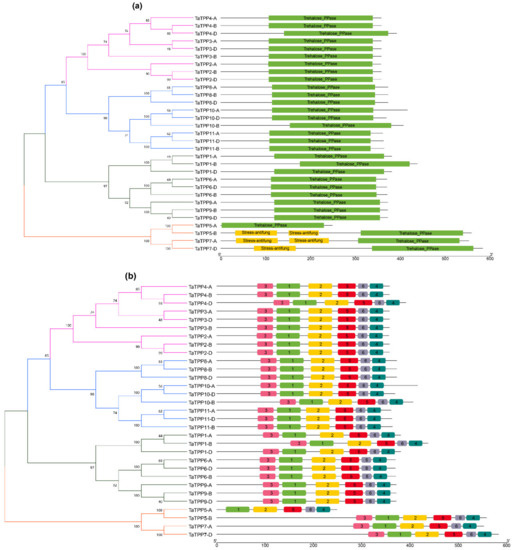
Figure 4.
Conserved domain and motif of TaTPPs. (a) The conserved domain of TaTPP members was identified from Pfam and SMART databases and presented using TBtools. (b) The conserved motifs of TaTPP members. Six motifs were identified using MEME program and presented with different colored boxes.
3.4. Gene Structure and Evolution Analyses of TaTPPs
The exon-intron structures of TaTPPs were studied to better understand their structural features (Figure 5). The TaTPP gene family had a lot of variation in terms of gene structure, according to gene structure analyses as introns ranged from 4 to 13. Most of the TaTPPs contain eight or nine introns. A maximum of 13 introns was found in TaTPP7-D and a minimum of four introns were observed in TaTPP8-B (Figure 5). Moreover, different TaTPPs showed different intron phase patterns. TaTPP1-A, TaTPP1-D, TaTPP5-A, TaTPP6, TaTPP8, TaTPP9 showed phase 0 and TaTPP2, TaTPP3, TaTPP4, TaTPP5-B, TaTPP7-A, TaTPP10, 11 showed phase 2 patterns, whereas TaTPP1-B and TaTPP7-D exhibited all phases (Phase 0,1,2) (Figure 5).
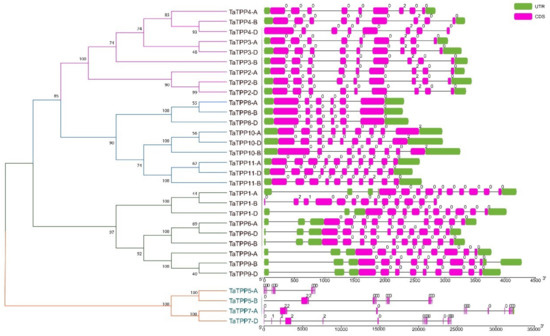
Figure 5.
Structural organizations of TaTPPs. The introns are shown by black lines, whereas the exons are represented by pink boxes and untranslated regions (UTRs) are represented with green boxes. Intron phase, 0: phase 0, 1: phase 1 and 2: phase 2 denotes that a codon is not disrupted by introns, a codon between the first and second bases is disrupted by an intron and a codon between the second and third bases is disrupted by an intron, respectively.
Further, the Multiple Collinearity Scan toolkit was used to investigate the synteny networks between TaTPPs and other wheat relatives and model plants. The results showed that 27, 26, 13, 33, 26, and 22 orthologous gene pairs were identified between TaTPPs and other TPPs in B. distachyon, O. sativa, A. thaliana, H. vulgare, Z. mays, and S. bicolor, respectively (Figure 6 and Table S7). A collinear relation was observed for 19, 18, 9, 22, 17 and 19 TaTPPs with other TPPs in B. distachyon, O. sativa, A. thaliana, H. vulgare, Z. mays, and S. bicolor, respectively. TaTPP6, TaTPP9, TaTPP10 and TaTPP11 were shown to have more than one pair of orthologs. Thus, these TaTPPs might have a crucial role in the evolution of TPPs. These findings imply that TaTPPs in wheat may have evolved from other plant species orthologous genes.
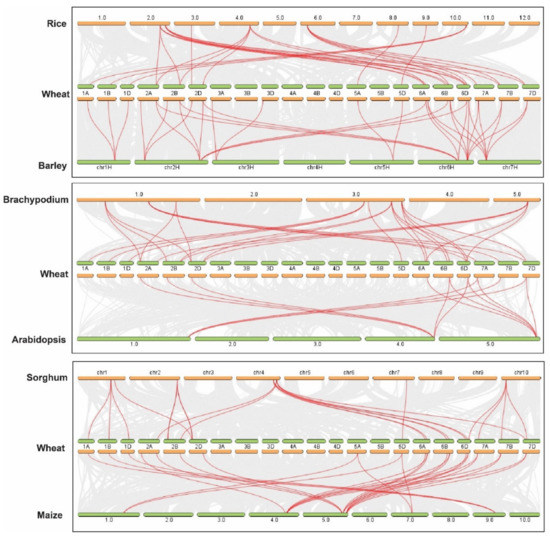
Figure 6.
Syntenic relationship between TaTPPs with rice, Arabidopsis, Brachypodium, sorghum, and maize. The collinear blocks within wheat and other plant genomes are shown by gray lines in the background, while the syntenic TaTPP gene pairs are highlighted by red lines.
3.5. 3-D Protein Structure Analysis
The 3-D structure reveals a few key residues linked to biological processes or intended outcomes [73]. Thus, we used SWISS-MODEL to identify the 3-D model of TaTPP proteins (Figure S1a). For all TaTPP proteins, the 3-D structures were analyzed using template “5gvx.1.A.” and predicted 3-D structures covering the N-terminus and C-terminus regions of 31 TaTPP proteins (Figure S1a). Within 4 A°, three conserved residues that worked as ligands were identified. The interaction of those ligands with chain A and the magnesium ion (Mg2+) indicates that TaTPP proteins have distinct catalytic activities, which have also been reported for AtTPP, ZmTPP, ScTPP, CaTPP, and EcTPP that have a catalytic function and they are all similar to one other by 80% [39,74]. Further, we used SOPMA to calculate the secondary structure elements of protein sequences (Table S8). TaTPP proteins were found to contain a range of 35.70% to 47.99% α helix, 13.41% to 18.18% extended strand, 6.62% to 9.93% β turn and 8.38% to 41.21% random coil (Table S8). All TaTPPs except TaTPP5, TaTPP7, TaTPP9-A, and TaTPP10-B had a coiled coil-like structure in the C-terminus and one Mg2+ ligand each was observed in all the TaTPPs (Figure S1a).
To validate TaTPP protein structures, we employed SWISS-MODEL analysis and the MolProbity server (Figure S1b and Table S9). The produced Ramachandran plot has an average favored region of 94.07%, an average allowed region of 99.08%, and an average outer region of 0.91% (Table S9). The average sequence identity was 34.95%, with a similarity of 37%, covering 68% of the query sequences obtained by the X-ray Method in 2.6 A° (Table S9). The ligand interaction between chain A and Mg2+ was confirmed with the Protein–Ligand Interaction Pipeline (PLIP), and the residue site was noticed to be highly conserved. We investigated these conserved residues further in all TaTPP protein sequence alignments and found that they include aspartic acid (D/Asp), which is conserved in motif 3 and motif 6. (Figure S2, Table S9). For improved visual clarity, the side chains of the catalytic triads were expanded with the TaTPP1-A residues (Figure S3).
3.6. Analysis of Cis-Regulatory Elements
To examine the responses of TaTPPs members to various stimuli, the 2 kb promoter sequences upstream of the start codon of these genes were submitted to the PlantCARE service to predict their Cis-regulatory elements (CREs). A total of 90 CREs with a frequency of 1985 were identified in all TaTPP promoters (Figure 7, Table S10). Among them, 72 CREs were related to phytohormones, stress, growth, and development (Figure 7, Table S10). All of the identified CREs were divided into five groups according to their known functions (Table S10 and Table S11). Group I contained four core Cis-elements, including AT~TATA-box, CAAT-box, TATA, TATA-box. TATA-box (which comprises TATA and AT TATA-box) is a critical promoter element found in approximately 30% of transcription start sites and the CAAT-box is a kind of promoter that may influence the choice of transcription start location [75]. TATA-box and CAAT-box are generally present 25–30 bp and ~75 bp upstream of the transcription start site, respectively, and both of them are found in a wide range across all the promoters.
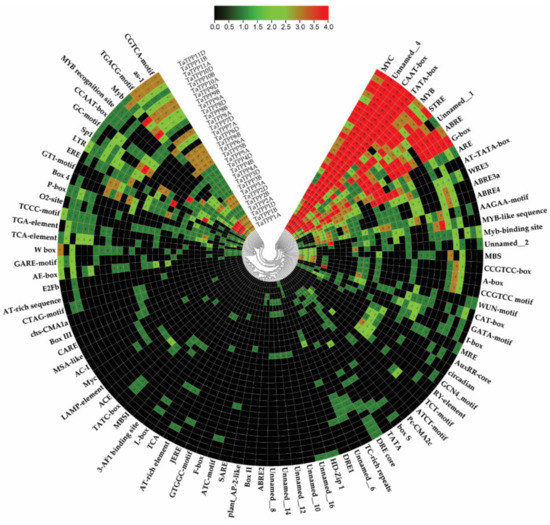
Figure 7.
Putative Cis-acting regulatory elements (CREs) of TaTPPs. The CREs were identified with the 2 kb upstream sequences of the start codon using the PlantCARE online server and presented using TBtools. Red color indicates the CREs with high frequency, while black color indicates CREs with zero frequency.
Group II contained 44 stress-related CREs, among them 20 were light-responsive Cis-elements such as 3-AF1 binding site, 3-AF1 binding site, ABRE4, ACE, ATCT-motif, Box 4, AE-box, Box II, LAMP-element. The stress-responsive CREs consist of one anaerobic-responsive element (ARE), one low-temperature-responsive element (LRT), one drought-responsive element (MBS), two wound-responsive elements (WUN-motif, box S), one cold- and dehydration-responsive (DRE core) and 17 defense- and stress-responsive elements (as-1, TC-rich repeats, W box, CCAAT-box, MYB, MYB recognition site, Myb, Myb-binding site, MYB-like sequence, MYC, Myc, STRE, WRE3, Unnamed_1, Unnamed_8, GC-motif, AT-rich sequence). There were 12 CREs in group III, involved in cell development including seed specific expression (AAGAA-motif, RY-element), cellular development and cell cycle regulation (AC-I, MSA-like), meristem expression (CAT-box, CCGTCC-box, CCGTCC-box), circadian control (circadian), differentiation of palisade mesophyll cells (HD-Zip I), cell cycle regulation (MSA-like), and endosperm expression (GCN4_motif).
Additionally, the hormone-responsive CREs in group IV included 16 CREs such as abscisic-acid-responsive element (ABRE, ABRE2), auxin-responsive elements (AuxRR-core and TGA-element), salicylic-acid-responsive element (TCA-element, TCA, SARE), methyl-jasmonate-responsive elements (TGACG and CGTCA motifs), ethylene-responsive element (ERE) and gibberellin-responsive elements (GARE-motif, P-box, and TATC-box). There were also 14 CREs in group V with unknown functions. CTAG-motif and A-box might act as a CRE, Unnamed_2 might act as an antisense transcript, BOX III might function as a protein binding site and Unnamed_16 was found to be involved in sugar transporter family genes. Most TaTPPs possessed one or more CREs associated with hormone and stress-related activities, suggesting that TaTPPs may be engaged in a variety of physiological processes as a result of diverse environmental adaptations.
3.7. Transcriptional Patterns of TaTPPs in Different Organs and Developmental Stages of Wheat
To investigate the transcription level of TaTPP genes in different wheat organs and development stages, mRNA transcripts data was collected from Genevestigator and visualized with a heatmap in Figure 8 and Figure S4. The transcript data were divided into six groups. Group I included callus, Group II included primary cells (cell culture, spike cell, spikelet cell, floret cell, stamen cell, anther cell, meiocyte, microspore), Group III included seedlings (seedling, coleoptile, root, radicle, radicle tip), Group IV included inflorescence (inflorescence, spike, rachis, spikelet, floret, stamen, anther, pistil, ovary, lemma, awn, glume, caryopsis, embryo, endosperm, aleurone layer, starchy endosperm, endosperm transfer layer, pericarp, outer pericarp), Group V included shoot (shoot, culm (stem), internode, peduncle, leaf, blade (lamina), sheath, flag leaf, blade (lamina), ligule, sheath, crown, shoot apex, shoot apical meristem, axillary bud) and Group VI included rhizome (rhizome, roots, nodal root, unspecified root type, root tip, root, apical meristem). Our results showed that TaTPP1, TaTPP8, TaTPP9-A, TaTPP9-D and TaTPP10-B had the highest transcriptions in most of the organs compared to other TaTPPs (Figure 8). In addition, high expression was observed for TaTPP2-D, TaTPP5-B, TaTPP6-A and TaTPP6-B only in the rhizome group. In contrast, other TaTPPs had no expression in most of the organs (Figure 8).
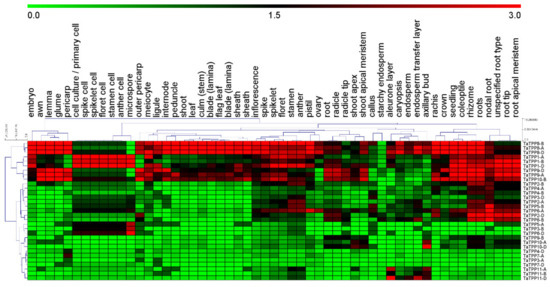
Figure 8.
Transcription profiles of TaTPPs in different wheat organs. mRNA transcription data of TaTPPs in different wheat organs were retrieved from Genevestigator and presented using MeV software.
Further, we observed the mRNA transcripts level of TaTPPs during different developmental stages of wheat, such as germination, seedling growth, tillering, stem elongation, booting, inflorescence emergence, anthesis, milk development, dough development, and ripening (Figure S4). A number of TaTPPs were expressed differently at various stages of wheat development. For example, TaTPP8 and TaTPP4-A were found to be expressed in all stages, whereas TaTPP1 and TaTPP9 were induced in all except the ripening stage. TaTPP4-D was expressed in all except stem elongation and TaTPP3-D was expressed in all except tillering and ripening stages. TaTPP5 and TaTPP7 showed very low expression in all wheat developmental stages and other TaTPPs were either slightly or highly expressed in one or more developmental stages (Figure S4). These findings suggest that various TaTPPs may have a role in the development of various tissues at different development stages.
3.8. Transcriptions of TaTPPs Were Induced in Response to ABA, Abiotic Stresses and Leaf Senescence
Wheat seedlings treated with ABA or abiotic stress (drought and salinity) were used to analyze the transcriptional pattern of the TaTPPs in wheat. Under ABA treatment, TaTPP1 and TaTPP4 exhibited upregulated transcriptions at most of the time points and significant upregulation was observed from 3 to 12 hpt (hours post treatment). Moreover, three TaTPPs (TaTPP7, TaTPP8 and TaTPP9) were upregulated immediately after ABA treatment and transcriptions decreased with an increase in ABA treatment time points. Transcriptions were significantly downregulated for TaTPP2, TaTPP3 and TaTPP11 at most of the time points after ABA treatments compared to control (0 hpt) (Figure 9a). The transcriptional patterns of TaTPP members were examined following drought stress in wheat to provide insight into the underlying functional roles of wheat TPPs in response to drought stress. During the drought stress treatment, only TaTPP1 and TaTPP4 showed significant upregulations at a later time post treatment. A slight upregulation or significant downregulation was observed for all other TaTPPs after drought stress in wheat (Figure 9b). The transcriptional levels of TaTPP members were examined to elucidate the mechanism of gene responses to leaf senescence in wheat. Most of the TaTPP members were slightly or highly induced during leaf senescence, TaTPP1 showed obvious upregulated transcriptions at 19 and 22 days after anthesis compared to the control (0 days after anthesis) (Figure 9c).
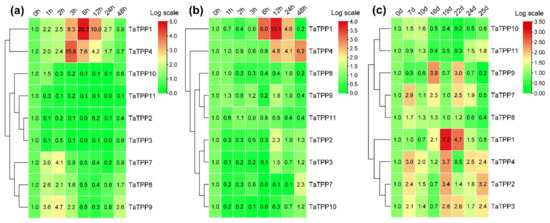
Figure 9.
Relative transcript profiles of TaTPPs in response to (a) abscisic acid (ABA), (b) drought stress and (c) leaf senescence. The relative transcripts of all genes were analyzed using qRT-PCR. The relative transcript levels of TaTPPs were measured using the comparative threshold (2−ΔΔCT) method. Data normalized with the transcripts of wheat elongation factor, TaEF-1α. The 0 h post treatment (a,b) or 0 days after anthesis (c) was used as a control and standardized with 1. Red and green colors denote strong and weak transcription of TaTPPs, respectively. The heat map was generated with TBtools and tree was constructed with the average linkage clustering method.
Further, we analyzed the transcriptions of TaTPP members under salt stress by qRT-PCR to observe the involvement of TaTPPs in wheat salt tolerance (Figure 10). Significant upregulation of the transcripts was observed for TaTPP1, TaTPP2, TaTPP4 and TaTPP9 at an early stage of salt treatment and downregulations were observed at the later stage of salt treatment. Moreover, TaTPP7 showed a significant upregulation only at 12 h post treatment (hpt) compared to the control (0 hpt). In contrast, either no changes or significant downregulations were observed for other TaTPP members compared to the control (Figure 10). However, no expression was observed for TaTPP5 and TaTPP6 by qRT-PCR in all aspects. Overall, these findings suggest that TaTPPs act as an important regulator of wheat abiotic stress and leaf senescence responses.
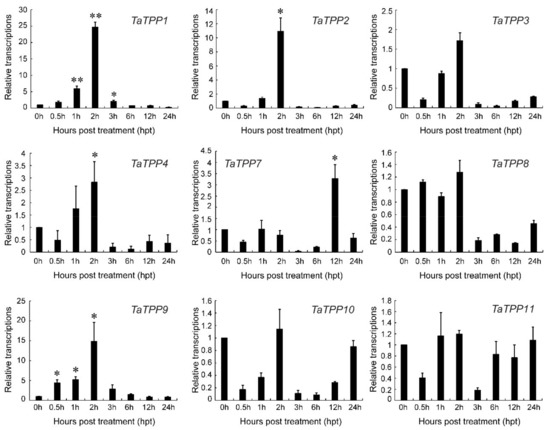
Figure 10.
Relative transcript profiles of TaTPPs in response to salt stress. The relative transcripts of all genes were analyzed using qRT-PCR. The relative transcript levels of TaTPPs were measured using the comparative threshold (2−ΔΔCT) method. Data normalized with the transcripts of wheat elongation factor, TaEF-1α. The 0 h post treatment was used as a control and standardized with 1. Values represent the mean ± SD from three independent biological samples. Asterisks (p < 0.05) or double asterisks (p < 0.01) designate significant differences from 0 hpt by the Student’s t-test.
3.9. Protein–Protein Interaction Analysis of TaTPPs
The STRING database was used to build a network to study protein–protein interactions between TaTPPs and other wheat proteins (Figure S5 and Table S12). From prediction results, it was found that TaTPPs can interact with five other wheat proteins. Traes_1AL_7531AC097.1, Traes_1BL_2AE952A77.1 and Traes_1DL_50B29C62B.2 have encoded an enzyme called TRE, which is hydrolyzed Trehalose to synthesize two molecules of glucose. Moreover, Traes_6DL_33F8A5EF4.1 has encoded TPS enzyme which produces T6P, a phosphorylated intermediate, from UDPG and G6P and Traes_4AS_4B8E78B13.1 was an unknown protein. Thus, our results suggesting that TaTPPs might interact with other enzymes that are involved trehalose biosynthesis pathway to accelerate the trehalose biosynthesis process.
4. Discussion
The TPP gene family has been characterized as catalytic enzymes that mainly function in trehalose biosynthesis [18,76,77]. Despite their catalytic function, a portion of TPP genes has been identified to be involved in growth and development, response in abiotic and biotic stress and senescence [27,28,29,31,32,33,35,42,78,79]. Although wheat is one of the most economically important cereal crops, systemic studies on TPP homologs in wheat have not been reported yet.
In the present study, we analyzed wheat TPPs with other species and identified 31 TPPs in wheat based on the Chinese Spring genome sequence (Table 1). The highest number of TPPs were found in wheat and these genes were distributed over 17 chromosomes (Figure 2). In comparison to previously described TPPs in Arabidopsis, rice, maize, and purple false brome, the wheat TPP gene family has been significantly extended with relatively more TPPs [38,74,80,81,82]. The major driving forces for extending the gene family in various plant species are gene duplication mechanisms, which include segmental, tandem, and whole-genome duplications [83,84]. All the TaTPPs are distributed unevenly on the wheat chromosome and the number ranges from 1 to 5 on each chromosome (Figure 2). Gene duplication analysis revealed that 27 pairs of TaTPPs duplicated within the wheat genome (Tables S4 and S5). The gap between genes on the chromosomal map of common wheat was higher than 200 kb (Figure 2), indicating that these genes were not formed via tandem duplication [85]. In addition, Ka/Ka ratio was less than one for all pairs of duplicated genes, suggesting that TaTPPs were subjected to a rigorous purifying selection (Table S5) and a comparable segmental duplication event was also observed for TPPs in rice [74]. Natural whole-genome duplicating processes might have led to the expansion of the TaTPP gene family. Thus, these findings suggest that whole-genome and segmental duplications might be vital in the expansion and evolution of TaTPPs.
Phylogenetic analysis of 31 TaTPP proteins and 11 other plant species showed that these proteins clustered into 11 groups, where TPPs from monocots and dicots species were grouped into separate clades (Figure 3). TaTPP proteins were grouped into clade I, clade II, clade V, clade VI, and clade VII and closely related to Brachypodium, rice, and barley TPPs, suggesting that TaTPP proteins might originate from a common ancestor. TaTPP5 and TaTPP7 have moved far away from the cluster of all other TPPs in the radiation tree (Figure S6) that was similar to OsTPP11 and OsTPP12 as previously reported [74]. The TaTPP gene structure study demonstrated that the majority of TaTPPs had highly conserved gene structures. The size of an intron has a significant impact on the size of a gene. The number of introns in TaTPPs ranged from 4 to 13 and most of the TaTPPs had 8 or 9 introns (Table 1). The difference in total intron length between the largest gene TaTPP7-A (32 kb) and the shortest gene TaTPP-8B (2.3 kb), resulted in a significant variation in gene size. Further, multiple alignments of TaTPP protein sequences revealed that the Trehalose_PPase domain and conserved motif are conserved within the TaTPPs (Figure S2). Among the identified six motifs, all the motifs were highly conserved in all TaTPPs except TaTPP5-A, which lacks motif 3. All the TaTPPs had a complete Trehalose_PPase domain, suggesting the various proteins’ functional equivalence and evolutionary relationships. In addition, TaTPP5-B and TaTPP7 had a stress-antifungal domain which has been reported to be involved in disulphide bridges and response to salt stress [86,87]. Subcellular localization prediction showed that most of the TaTPPs are localized in the chloroplast, whereas some of them are found in the mitochondrion or secreted protein (Table 1). In Arabidopsis or rice, different localizations were also detected. For instance, AtTPPD and AtTPPE were localized in the chloroplast whereas AtTPPA, AtTPPB, AtTPPC, AtTPPF, and AtTPPH were found in the cytosol and AtTPPG, AtTPPI, and AtTPPJ showed localization in the nucleus [88]. This variation in the localization of TaTPPs might be due to a lack of conserved N-terminus (Figure S2). According to the 3-D structure analysis, all TaTPPs were highly conserved and showed Mg2+ ligand-binding sites in SWISSMODEL (Figure S1a), which are shown to have a role in catalysis by activating or inhibiting a variety of enzymes [89,90]. To investigate the TPP gene synteny relationship in wheat and other plant species, we identified 27, 26, 13, 33, 26 and 22 orthologous gene pairs between TaTPPs and other TPPs in B. distachyon, O. sativa, A. thaliana, H. vulgare, Z. mays, and S. bicolor, respectively (Figure 6 and Table S7). These findings imply that TaTPPs in wheat might have evolved from other plant species orthologous genes.
A non-coding DNA sequence found in the promoter region of a gene is known as a CREs. Different CREs distribution in promoter regions may indicate variations in gene regulation and function [91]. To identify the CREs, we used 2kb promoter regions of all TaTPPs and classified into five groups according to their known functions (Figure 7, Table S10). Stress related CREs were identified in high frequency compared to cellular development and hormone related CREs, suggesting the involvement of TaTPPs in response to stress. ABRE, At~ABRE, ABRE3a, and ABRE4 are ABA-responsive CREs that play important roles in seed dormancy, stomatal closure, leaf senescence, and plant biotic and abiotic stress responses. Multiple ABREs or their combinations have been reported to act as CEs (Coupling Elements) in the formation of ABA-responsive complex (ABRC) [92,93,94,95,96,97]. The ABRE CREs were predicted in all TaTPPs promotor with high frequency and ABRE3a and ABRE4 CREs were found in most of the TaTPPs except TaTPP5, TaTPP7, TaTPP9 and TaTPP11. Following that, we also discovered TGA-element, and AuxRR-core (auxin-responsive element), TCA-element (salicylic acid responsiveness), CGTCA-motif (MeJA responsiveness) and p-box and TATC-box (gibberellin responsive element), among other hormone-related CREs [98], that might potentially induce possible signal transduction pathways for wheat TPPs during stress response. Furthermore, other CREs linked to a variety of development and stress were also predicted in TaTPP promoters with high frequency, including MBS (drought inducibility), MYC (drought-responsive CRE) MYB and STRE (stress response element), as-1(Defense response), Unnamed_1 (ABRE-like CRE, responsible for biotic and abiotic stress responses), ARE (anaerobic induction CRE), Unnamed_4 (might responsible for tissue specific expression) and AAGAA-motif (involved in seed specific expression) [91]. These findings suggest that TPP gene family members in wheat may be controlled by a variety of developmental events, hormones, and stress; however, additional experimental investigations will be required to validate this.
Higher transcriptional levels of TaTPPs were observed in different wheat organs and developmental stages. TaTPP1, TaTPP8 and TaTPP9 were expressed in most organs and developmental stages but predominantly expressed in the roots, suggesting that they could be important for root physiology. (Figure 8, Figure S4). Previous evidence showed that AtTPPE modulates ABA-mediated root growth and a rice TPP, OsTPP7, enhanced the anaerobic germination [41,99]. Wang et. al. [27] reported that seed germination was regulated by OsTPP1 via crosstalk with the ABA catabolism pathway. In addition, high expression was also observed for TaTPP1, TaTPP8 and TaTPP9 in all developmental stages except dough development and ripening, suggesting that these genes might have a significant association with wheat developmental processes. Plants have developed sensory and response systems that enable them to adjust physiologically to environmental stress conditions such as drought, excessive salt, and low temperature stress. Previous studies in rice and Arabidopsis demonstrated the involvement of TPP genes in various environmental stresses and ABA signaling [32,37,38,40,41,100]. Under ABA treatment, TaTPP1 and TaTPP4 exhibited upregulated transcriptions at most of the time points and significant upregulations were observed from 3 to 12 hpt. Moreover, three TaTPPs (TaTPP7, TaTPP8 and TaTPP9) were upregulated immediately after ABA treatment and transcriptions were decreased with the duration after ABA treatment and a significant downregulated transcription level was observed for TaTPP2, TaTPP3 and TaTPP11 at most of the time points after ABA treatment (Figure 9a). During the drought stress treatment, only TaTPP1 and TaTPP4 showed significant upregulations at later time post treatment. A slight upregulation or significant downregulation was observed for all other TaTPPs after drought stress in wheat (Figure 9b). An obvious significant upregulation was observed for TaTPP1, TaTPP2, TaTPP4 and TaTPP9 at an early stage of salt stress and downregulated transcriptions were observed at the latter stage of salt treatment. A similar expression pattern was also observed for rice TPP and BdTPPC genes that were upregulated in the first hour under abiotic stress [28,38]. Moreover, TaTPP7 showed a significant upregulation only at 12 dpt. In contrast, either no changes or significant downregulation was observed for other TaTPP members compared to the control (Figure 10). The phenotype of mature otsB-overexpressing Arabidopsis plants included delayed senescence and decreased anthocyanin accumulation, suggesting that the role of TPP may perform a crucial role during leaf senescence in plants [42,43,44,45]. Our results showed that most of the TaTPP members were slightly or highly induced during leaf senescence, especially TaTPP1 showed an obvious upregulated transcription (Figure 9c). Overall, these findings suggest that TaTPPs might act as an important regulator of wheat abiotic stress and leaf senescence responses and could be good candidate genes for wheat improvement under environmental stimuli. Moreover, protein network prediction revealed that TaTPP proteins possible interact with TaTPS or TaTRE protein which involved in trehalose biosynthesis pathway to accelerate the trehalose biosynthesis process (Figure S5).
Furthermore, we suggested a feasible working model based on TaTPPs transcription profiling to illustrate the roles of TaTPPs in a range of biological processes in wheat (Figure 11). TPS produces T6P, a phosphorylated intermediate, from UDPG and G6P and then TPP dephosphorylates T6P to produce trehalose in the second phase. Trehalose is then hydrolyzed by an enzyme called TRE to synthesize two molecules of glucose [16]. The expression of TaTPPs was induced by both endogenous and exogenous stimuli in this model. These signals were detected by multiple Cis-regulatory elements, which then regulated the transcription and functions of TaTPPs involved in numerous plant developmental stages and stress situations, affecting plant growth and tolerance mechanisms (Figure 11).
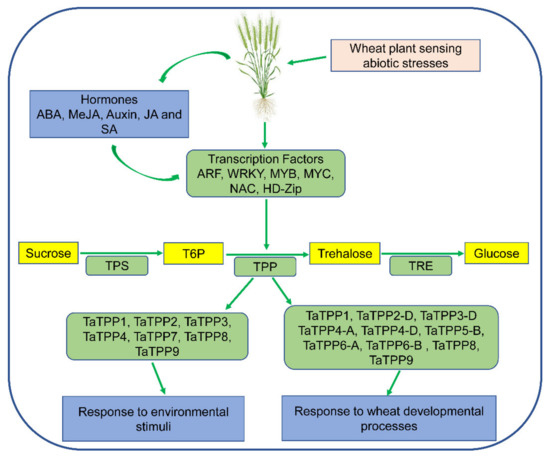
Figure 11.
A proposed model for TaTPP genes functions in various wheat developmental processes and diverse stress conditions. ABA: Abscisic acid; MeJA: Methyl jasmonate; JA: Jasmonic acid; SA: Salicylic acid; ARF: Auxin response factors; MYB: Myeloblastosis; NAC: No apical meristem; TPS: trehalose-6-phosphate synthase; T6P: trehalose-6-phosphate; TPP: trehalose-6-phosphate phosphatase; TRE: trehalase. Yellow boxes indicate carbohydrates and light green boxes indicate proteins.
5. Conclusions
In conclusion, a relatively comprehensive analysis of the TaTPP gene family was performed in this study, which may help to explain the biological activities of TaTPP proteins in developmental processes, stress responses and leaf senescence of wheat. However, our knowledge of their precise biological role is still lacking. Thus, in order to give important insights to help wheat breeders for developing resistant crops cultivars to unfavorable stress conditions, an extensive functional validation study of TaTPPs is necessary.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/genes12111652/s1 Figure S1. Schematic illustration of 3-D protein structures and the Ramachandran plot for TaTPP proteins, Figure S2. Schematic representation of protein alignment of all TaTPPs, Figure S3. Schematic representation of TaTPP1-A side chains catalytic triads, Figure S4. Heat map of TaTPPs transcriptions in wheat development stages, Figure S5. Protein–protein interaction analysis of TaTPP proteins, Figure S6. Radiation tree of TaTPPs along with TPPs from other species, Table S1. List of primers used for TaTPPs qRT-PCR analysis, Table S2. TPP protein sequences identified from wheat genome, Table S3. Number of TPP proteins in different plant species, Table S4. Sequence identity and query cover of TaTPP proteins, Table S5. Pairwise identities and divergence between TaTPP genes and details about the duplication of those genes, Table S6. Phylogenetic tree member with their gene ID, Table S7. The synteny relationships of wheat TPP genes with different plant species, Table S8. Details of the calculated secondary structure elements TaTPPs by SOPMA, Table S9. Validation of TaTPP protein structures, Table S10. Frequency of all identified Cis-Regulatory Elements (CREs) in different TaTPPs promoters, Table S11. Cis-Regulatory Elements (CREs) with sequences and functions, Table S12: The protein-protein interaction network between TaTPPs and other proteins in wheat.
Author Contributions
The present study was conceptualized by D.S. and M.A.I.; bioinformatics analysis and visualization were conducted by M.M.R. (Md Mustafzur Rahman), M.A.I. and M.M.R. (Md Mizanor Rahman); experiments were investigated by M.A.I., X.J., L.S., K.Z., S.W., and H.N.; writing—original draft was prepared by M.A.I. and M.M.R. (Md Mustafzur Rahman); writing—reviewed and edited by J.-S.J., M.M.R. (Md Mizanor Rahman), D.S., A.S. and W.Z.; funding was acquired by D.S. All authors have read and agreed to the published version of the manuscript.
Funding
This research was sponsored by the State Key Laboratory of Sustainable Dryland Agriculture, Shanxi Agricultural University (No. 202002-2).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
References
- USDA. World Agricultural Production. 2019. Available online: https://www.fas.usda.gov/data/world-agricultural-production (accessed on 20 January 2020).
- FAO. World Food Situation. 2021. Available online: http://www.fao.org/worldfoodsituation/csdb/en/ (accessed on 10 July 2021).
- Yin, J.-L.; Fang, Z.-W.; Sun, C.; Zhang, P.; Zhang, X.; Lu, C.; Wang, S.-P.; Ma, D.-F.; Zhu, Y.-X. Rapid identification of a stripe rust resistant gene in a space-induced wheat mutant using specific locus amplified fragment (SLAF) sequencing. Sci. Rep. 2018, 8, 1–9. [Google Scholar] [CrossRef]
- Tester, M.; Bacic, A. Abiotic Stress Tolerance in Grasses. From Model Plants to Crop Plants. Plant Physiol. 2005, 137, 791–793. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shafi, M.; Zhang, G.; Bakht, J.; Khan, M.A.; Islam, U.; Khan, M.D.; Raziuddin, G. Effect of cadmium and salinity stresses on root morphology of wheat. Pak. J. Bot. 2010, 42, 2747–2754. [Google Scholar]
- Foolad, M.R. Recent Advances in Genetics of Salt Tolerance in Tomato. Plant Cell Tissue Organ Cult. 2004, 76, 101–119. [Google Scholar] [CrossRef]
- Genc, Y.; Taylor, J.; Lyons, G.; Li, Y.; Cheong, J.; Appelbee, M.; Oldach, K.; Sutton, T. Bread Wheat with High Salinity and Sodicity Tolerance. Front. Plant Sci. 2019, 10, 1280. [Google Scholar] [CrossRef] [Green Version]
- Rahneshan, Z.; Nasibi, F.; Moghadam, A.A. Effects of salinity stress on some growth, physiological, biochemical parameters and nutrients in two pistachio (Pistacia vera L.) rootstocks. J. Plant Interact. 2018, 13, 73–82. [Google Scholar] [CrossRef] [Green Version]
- Wingler, A. The function of trehalose biosynthesis in plants. Phytochemistry 2002, 60, 437–440. [Google Scholar] [CrossRef]
- Mostafa, M.R.; Mervat, S.S.; Safaa, R.E.-L.; Ebtihal, M.A.E.; Magdi, T.A. Exogenous α-tocopherol has a beneficial effect on Glycine max(L.) plants irrigated with diluted sea water. J. Hortic. Sci. Biotechnol. 2015, 90, 195–202. [Google Scholar] [CrossRef]
- Patist, A.; Zoerb, H. Preservation mechanisms of trehalose in food and biosystems. Colloids Surf. B Biointerfaces 2005, 40, 107–113. [Google Scholar] [CrossRef]
- Iturriaga, G.; Suárez, R.; Nova-Franco, B. Trehalose Metabolism: From Osmoprotection to Signaling. Int. J. Mol. Sci. 2009, 10, 3793–3810. [Google Scholar] [CrossRef] [Green Version]
- Lunn, J.E.; Delorge, I.; Figueroa, C.M.; Van Dijck, P.; Stitt, M. Trehalose metabolism in plants. Plant J. 2014, 79, 544–567. [Google Scholar] [CrossRef]
- Klähn, S.; Hagemann, M. Compatible solute biosynthesis in cyanobacteria. Environ. Microbiol. 2010, 13, 551–562. [Google Scholar] [CrossRef]
- Chang, S.-W.; Chang, W.-H.; Lee, M.-R.; Yang, T.-J.; Yu, N.-Y.; Chen, C.-S.; Shaw, J.-F. Simultaneous Production of Trehalose, Bioethanol, and High-Protein Product from Rice by an Enzymatic Process. J. Agric. Food Chem. 2010, 58, 2908–2914. [Google Scholar] [CrossRef]
- Elbein, A.D.; Pan, Y.T.; Pastuszak, I.; Carroll, D. New insights on trehalose: A multifunctional molecule. Glycobiology 2003, 13, 17R–27R. [Google Scholar] [CrossRef]
- Lunn, J.E. Gene families and evolution of trehalose metabolism in plants. Funct. Plant Biol. 2007, 34, 550–563. [Google Scholar] [CrossRef]
- Avonce, N.; Mendoza-Vargas, A.; Morett, E.; Iturriaga, G. Insights on the evolution of trehalose biosynthesis. BMC Evol. Biol. 2006, 6, 109. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ponnu, J.; Wahl, V.; Schmid, M. Trehalose-6-Phosphate: Connecting Plant Metabolism and Development. Front. Plant Sci. 2011, 2, 70. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lunn, J.E.; Feil, R.; Hendriks, J.H.M.; Gibon, Y.; Morcuende, R.; Osuna, D.; Scheible, W.-R.; Carillo, P.; Hajirezaei, M.-R.; Stitt, M. Sugar-induced increases in trehalose 6-phosphate are correlated with redox activation of ADPglucose pyrophosphorylase and higher rates of starch synthesis in Arabidopsis thaliana. Biochem. J. 2006, 397, 139–148. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meitzel, T.; Radchuk, R.; McAdam, E.L.; Thormählen, I.; Feil, R.; Munz, E.; Hilo, A.; Geigenberger, P.; Ross, J.J.; Lunn, J.E.; et al. Trehalose-6-phosphate promotes seed filling by activating auxin biosynthesis. New Phytol. 2021, 229, 1553–1565. [Google Scholar] [CrossRef] [PubMed]
- Yadav, U.P.; Ivakov, A.; Feil, R.; Duan, G.Y.; Walther, D.; Giavalisco, P.; Piques, M.; Carillo, P.; Hubberten, H.-M.; Stitt, M.; et al. The sucrose–trehalose 6-phosphate (Tre6P) nexus: Specificity and mechanisms of sucrose signalling by Tre6P. J. Exp. Bot. 2014, 65, 1051–1068. [Google Scholar] [CrossRef] [Green Version]
- Acevedo Hernández, G.J.; León, P.; Herrera-Estrella, L.R. Sugar and ABA responsiveness of a minimal RBCS light-responsive unit is mediated by direct binding of ABI4. Plant J. 2005, 43, 506–519. [Google Scholar] [CrossRef]
- Gómez, L.D.; Gilday, A.; Feil, R.; Lunn, J.E.; Graham, I.A. AtTPS1-mediated trehalose 6-phosphate synthesis is essential for embryogenic and vegetative growth and responsiveness to ABA in germinating seeds and stomatal guard cells. Plant J. 2010, 64, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Fichtner, F.; Olas, J.J.; Feil, R.; Watanabe, M.; Krause, U.; Hoefgen, R.; Stitt, M.; Lunn, J.E. Functional Features of TREHALOSE-6-PHOSPHATE SYNTHASE1, an Essential Enzyme in Arabidopsis [OPEN]. Plant Cell 2020, 32, 1949–1972. [Google Scholar] [CrossRef] [Green Version]
- Fichtner, F.; Barbier, F.F.; Annunziata, M.G.; Feil, R.; Olas, J.J.; Mueller-Roeber, B.; Stitt, M.; Beveridge, C.A.; Lunn, J.E. Regulation of shoot branching in Arabidopsis by trehalose-6-phosphate. New Phytol. 2021, 229, 2135–2151. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Li, X.; Ye, N.; Huang, M.; Feng, L.; Li, H.; Zhang, J. OsTPP1 regulates seed germination through the crosstalk with abscisic acid in rice. New Phytol. 2021, 230, 1925–1939. [Google Scholar] [CrossRef]
- Wang, S.; Ouyang, K.; Wang, K. Genome-Wide Identification, Evolution, and Expression Analysis of TPS and TPP Gene Families in Brachypodium distachyon. Plants 2019, 8, 362. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Satoh-Nagasawa, N.; Nagasawa, N.; Malcomber, S.; Sakai, H.; Jackson, D. A trehalose metabolic enzyme controls inflorescence architecture in maize. Nature 2006, 441, 227–230. [Google Scholar] [CrossRef]
- Pellny, T.K.; Ghannoum, O.; Conroy, J.P.; Schluepmann, H.; Smeekens, S.; Andralojc, J.; Krause, K.P.; Goddijn, O.; Paul, M. Genetic modification of photosynthesis with E. coli genes for trehalose synthesis. Plant Biotechnol. J. 2004, 2, 71–82. [Google Scholar] [CrossRef]
- Miranda, J.A.; Avonce, N.; Suárez, R.; Thevelein, J.M.; Van Dijck, P.; Iturriaga, G. A bifunctional TPS–TPP enzyme from yeast confers tolerance to multiple and extreme abiotic-stress conditions in transgenic Arabidopsis. Planta 2007, 226, 1411–1421. [Google Scholar] [CrossRef] [Green Version]
- Lin, Q.; Yang, J.; Wang, Q.; Zhu, H.; Chen, Z.; Dao, Y.; Wang, K. Overexpression of the trehalose-6-phosphate phosphatase family gene AtTPPF improves the drought tolerance of Arabidopsis thaliana. BMC Plant Biol. 2019, 19, 381. [Google Scholar] [CrossRef] [Green Version]
- Pilon-Smits, E.A.; Terry, N.; Sears, T.; Kim, H.; Zayed, A.; Hwang, S.; van Dun, K.; Voogd, E.; Verwoerd, T.C.; Krutwagen, R.W.; et al. Trehalose-producing transgenic tobacco plants show improved growth performance under drought stress. J. Plant Physiol. 1998, 152, 525–532. [Google Scholar] [CrossRef]
- Wu, B.; Su, X. Identification of drought response genes in Zygophyllum xanthoxylum by suppression subtractive hybridization. J. Plant Biol. 2016, 59, 377–385. [Google Scholar] [CrossRef]
- Garg, A.K.; Kim, J.-K.; Owens, T.G.; Ranwala, A.P.; Choi, Y.D.; Kochian, L.; Wu, R.J. Trehalose accumulation in rice plants confers high tolerance levels to different abiotic stresses. Proc. Natl. Acad. Sci. USA 2002, 99, 15898–15903. [Google Scholar] [CrossRef] [Green Version]
- Shi, Y.; Sun, H.; Wang, X.; Jin, W.; Chen, Q.; Yuan, Z.; Yu, H. Physiological and transcriptomic analyses reveal the molecular networks of responses induced by exogenous trehalose in plant. PLoS ONE 2019, 14, e0217204. [Google Scholar] [CrossRef] [Green Version]
- Jiang, D.; Chen, W.; Gao, J.; Yang, F.; Zhuang, C. Overexpression of the trehalose-6-phosphate phosphatase OsTPP3 increases drought tolerance in rice. Plant Biotechnol. Rep. 2019, 13, 285–292. [Google Scholar] [CrossRef]
- Ge, L.; Chao, D.-Y.; Shi, M.; Zhu, M.-Z.; Gao, J.-P.; Lin, H.-X. Overexpression of the trehalose-6-phosphate phosphatase gene OsTPP1 confers stress tolerance in rice and results in the activation of stress responsive genes. Planta 2008, 228, 191–201. [Google Scholar] [CrossRef] [PubMed]
- Acosta-Pérez, P.; Camacho-Zamora, B.D.; Espinoza-Sánchez, E.A.; Gutiérrez-Soto, G.; Zavala-García, F.; Abraham-Juárez, M.J.; Sinagawa-García, S.R. Characterization of Trehalose-6-phosphate Synthase and Trehalose-6-phosphate Phosphatase Genes and Analysis of its Differential Expression in Maize (Zea mays) Seedlings under Drought Stress. Plants 2020, 9, 315. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lin, Q.; Wang, S.; Dao, Y.; Wang, J.; Wang, K.; Wang, S. Arabidopsis thaliana trehalose-6-phosphate phosphatase gene TPPI enhances drought tolerance by regulating stomatal apertures. J. Exp. Bot. 2020, 71, 4285–4297. [Google Scholar] [CrossRef] [Green Version]
- Wang, W.; Chen, Q.; Xu, S.; Liu, W.C.; Zhu, X.; Song, C.P. Trehalose-6-phosphate phosphatase E modulates ABA-controlled root growth and stomatal movement in Arabidopsis. J. Integr. Plant Biol. 2020, 62, 1518–1534. [Google Scholar] [CrossRef] [Green Version]
- Wingler, A.; Delatte, T.L.; O’Hara, L.; Primavesi, L.; Jhurreea, D.; Paul, M.; Schluepmann, H. Trehalose 6-Phosphate Is Required for the Onset of Leaf Senescence Associated with High Carbon Availability. Plant Physiol. 2012, 158, 1241–1251. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Noodén, L.D.; Guiamét, J.J.; John, I. Senescence mechanisms. Physiol. Plant. 1997, 101, 746–753. [Google Scholar] [CrossRef]
- Pourtau, N.; Jennings, R.; Pelzer, E.; Pallas, J.; Wingler, A. Effect of sugar-induced senescence on gene expression and implications for the regulation of senescence in Arabidopsis. Planta 2006, 224, 556–568. [Google Scholar] [CrossRef] [PubMed]
- Wingler, A.; Purdy, S.; MacLean, J.A.; Pourtau, N. The role of sugars in integrating environmental signals during the regulation of leaf senescence. J. Exp. Bot. 2006, 57, 391–399. [Google Scholar] [CrossRef] [Green Version]
- Zhang, P.; He, Z.; Tian, X.; Gao, F.; Xu, D.; Liu, J.; Wen, W.; Fu, L.; Li, G.; Sui, X.; et al. Cloning of TaTPP-6AL1 associated with grain weight in bread wheat and development of functional marker. Mol. Breed. 2017, 37, 78. [Google Scholar] [CrossRef]
- Johnson, M.; Zaretskaya, I.; Raytselis, Y.; Merezhuk, Y.; McGinnis, S.; Madden, T.L. NCBI BLAST: A better web interface. Nucleic Acids Res. 2008, 36, W5–W9. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Guindon, S.; Gascuel, O. A Simple, Fast, and Accurate Algorithm to Estimate Large Phylogenies by Maximum Likelihood. Syst. Biol. 2003, 52, 696–704. [Google Scholar] [CrossRef] [Green Version]
- Felsenstein, J. Confidence limits on phylogenies: An approach using the bootstrap. Evolution 1985, 39, 783–791. [Google Scholar] [CrossRef]
- Combet, C.; Blanchet, C.; Geourjon, C.; Deléage, G. NPS@: Network Protein Sequence Analysis. Trends Biochem. Sci. 2000, 25, 147–150. [Google Scholar] [CrossRef]
- Izidoro, S.; De Melo-Minardi, R.C.; Pappa, G.L. GASS: Identifying enzyme active sites with genetic algorithms. Bioinformatics 2015, 31, 864–870. [Google Scholar] [CrossRef] [Green Version]
- Lovell, S.C.; Davis, I.W.; Arendall III, W.B.; De Bakker, P.I.; Word, J.M.; Prisant, M.G.; Richardson, J.S.; Richardson, D.C. Structure validation by Cα geometry: ϕ, ψ and Cβ deviation. Proteins 2003, 50, 437–450. [Google Scholar] [CrossRef]
- Moraes, J.P.A.; Pappa, G.L.; Pires, D.E.V.; Izidoro, S.C. GASS-WEB: A web server for identifying enzyme active sites based on genetic algorithms. Nucleic Acids Res. 2017, 45, W315–W319. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Waterhouse, A.; Bertoni, M.; Bienert, S.; Studer, G.; Tauriello, G.; Gumienny, R.; Heer, F.T.; De Beer, T.A.P.; Rempfer, C.; Bordoli, L.; et al. SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Res. 2018, 46, W296–W303. [Google Scholar] [CrossRef] [Green Version]
- Williams, C.J.; Headd, J.J.; Moriarty, N.W.; Prisant, M.G.; Videau, L.L.; Deis, L.N.; Verma, V.; Keedy, D.A.; Hintze, B.J.; Chen, V.B.; et al. MolProbity: More and better reference data for improved all-atom structure validation. Protein Sci. 2017, 27, 293–315. [Google Scholar] [CrossRef] [PubMed]
- Ren, J.; Wen, L.; Gao, X.; Jin, C.; Xue, Y.; Yao, X. DOG 1.0: Illustrator of protein domain structures. Cell Res. 2009, 19, 271–273. [Google Scholar] [CrossRef]
- Finn, R.D.; Bateman, A.; Clements, J.; Coggill, P.; Eberhardt, R.Y.; Eddy, S.R.; Heger, A.; Hetherington, K.; Holm, L.; Mistry, J.; et al. Pfam: The protein families database. Nucleic Acids Res. 2014, 42, D222–D230. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.; Li, W.W.; Noble, W.S. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 2009, 37, w202–w208. [Google Scholar] [CrossRef]
- Lescot, M.; Déhais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouzé, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef]
- NetBeans, IDE 8.0. Oracle Co., Redwood City, CA, 2015. Available online: https://netbeans.org (accessed on 25 March 2021).
- Hruz, T.; Laule, O.; Szabo, G.; Wessendorp, F.; Bleuler, S.; Oertle, L.; Widmayer, P.; Gruissem, W.; Zimmermann, P. Genevestigator V3: A Reference Expression Database for the Meta-Analysis of Transcriptomes. Adv. Bioinform. 2008, 2008, 1–5. [Google Scholar] [CrossRef]
- Howe, E.; Holton, K.; Nair, S.; Schlauch, D.; Sinha, R.; Quackenbush, J. MeV: Multi Experiment Viewer. In Biomedical Informatics for Cancer Research; Springer: Berlin/Heidelberg, Germany, 2010; pp. 267–277. [Google Scholar]
- Duan, Y.-H.; Guo, J.; Ding, K.; Wang, S.-J.; Zhang, H.; Dai, X.-W.; Chen, Y.-Y.; Govers, F.; Huang, L.-L.; Kang, Z.-S. Characterization of a wheat HSP70 gene and its expression in response to stripe rust infection and abiotic stresses. Mol. Biol. Rep. 2010, 38, 301–307. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Paolacci, A.R.; Tanzarella, O.A.; Porceddu, E.; Ciaffi, M. Identification and validation of reference genes for quantitative RT-PCR normalization in wheat. BMC Mol. Biol. 2009, 10, 11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fleury, D.; Jefferies, S.; Kuchel, H.; Langridge, P. Genetic and genomic tools to improve drought tolerance in wheat. J. Exp. Bot. 2010, 61, 3211–3222. [Google Scholar] [CrossRef] [Green Version]
- Bhattacharya, M.; Hota, A.; Kar, A.; Chini, D.S.; Malick, R.C.; Patra, B.C.; Das, B.K. In silico structural and functional modelling of Antifreeze protein (AFP) sequences of Ocean pout (Zoarces americanus, Bloch & Schneider 1801). J. Genet. Eng. Biotechnol. 2018, 16, 721–730. [Google Scholar] [CrossRef] [PubMed]
- Yaqoob, U.; Kaul, T.; Nawchoo, I.A. In-Silico Analysis, Structural Modelling and Phylogenetic Analysis of Acetohydroxyacid Synthase Gene of Oryza sativa. Med. Aromat. Plants 2016, 5, 2167-0412. [Google Scholar] [CrossRef] [Green Version]
- Kong, X.; Lv, W.; Jiang, S.; Zhang, D.; Cai, G.; Pan, J.; Li, D. Genome-wide identification and expression analysis of calcium-dependent protein kinase in maize. BMC Genom. 2013, 14, 433. [Google Scholar] [CrossRef] [Green Version]
- Gu, Z.; Cavalcanti, A.; Chen, F.-C.; Bouman, P.; Li, W.-H. Extent of Gene Duplication in the Genomes of Drosophila, Nematode, and Yeast. Mol. Biol. Evol. 2002, 19, 256–262. [Google Scholar] [CrossRef] [Green Version]
- Bordoli, L.; Schwede, T. Automated Protein Structure Modeling with SWISS-MODEL Workspace and the Protein Model Portal. In Homology Modeling; Springer: Berlin/Heidelberg, Germany, 2011; Volume 857, pp. 107–136. [Google Scholar] [CrossRef] [Green Version]
- Rahman, M.M.; Rahman, M.M.; Eom, J.S.; Jeon, J.S. Genome-wide identification, expression profiling and promoter analysis of trehalose-6-phosphate phosphatase gene family in rice. J. Plant Biol. 2021, 64, 55–71. [Google Scholar] [CrossRef]
- Grace, M.L.; Chandrasekharan, M.; Hall, T.C.; Crowe, A.J. Sequence and Spacing of TATA Box Elements Are Critical for Accurate Initiation from the β-Phaseolin Promoter. J. Biol. Chem. 2004, 279, 8102–8110. [Google Scholar] [CrossRef] [Green Version]
- Vogel, G.; Aeschbacher, R.A.; Müller, J.; Boller, T.; Wiemken, A. Trehalose-6-phosphate phosphatases from Arabidopsis thaliana: Identification by functional complementation of the yeast tps2 mutant. Plant J. 1998, 13, 673–683. [Google Scholar] [CrossRef]
- Svanström, Å.; van Leeuwen, M.R.; Dijksterhuis, J.; Melin, P. Trehalose synthesis in Aspergillus niger: Characterization of six homologous genes, all with conserved orthologs in related species. BMC Microbiol. 2014, 14, 90. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.-J.; Hao, Y.-J.; Zhang, Z.-G.; Chen, T.; Zhang, J.-S.; Chen, S.-Y. Isolation of trehalose-6-phosphate phosphatase gene from tobacco and its functional analysis in yeast cells. J. Plant Physiol. 2005, 162, 215–223. [Google Scholar] [CrossRef]
- Streeter, J.; Gomez, M. Three enzymes for trehalose synthesis in Bradyrhizobium cultured bacteria and in bacteroids from soybean nodules. Appl. Environ. Microbiol. 2006, 72, 4250–4255. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Henry, C.; Bledsoe, S.W.; Siekman, A.; Kollman, A.; Waters, B.; Feil, R.; Stitt, M.; Lagrimini, L.M. The trehalose pathway in maize: Conservation and gene regulation in response to the diurnal cycle and extended darkness. J. Exp. Bot. 2014, 65, 5959–5973. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schluepmann, H.; van Dijken, A.; Aghdasi, M.; Wobbes, B.; Paul, M.; Smeekens, S. Trehalose Mediated Growth Inhibition of Arabidopsis Seedlings Is Due to Trehalose-6-Phosphate Accumulation. Plant Physiol. 2004, 135, 879–890. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Leyman, B.; Van Dijck, P.; Thevelein, J. An unexpected plethora of trehalose biosynthesis genes in Arabidopsis thaliana. Trends Plant Sci. 2001, 6, 510–513. [Google Scholar] [CrossRef]
- Lawton-Rauh, A. Evolutionary dynamics of duplicated genes in plants. Mol. Phylogenetics Evol. 2003, 29, 396–409. [Google Scholar] [CrossRef]
- Moore, R.C.; Purugganan, M.D. The early stages of duplicate gene evolution. Proc. Natl. Acad. Sci. USA 2003, 100, 15682–15687. [Google Scholar] [CrossRef] [Green Version]
- Holub, E.B. The arms race is ancient history in Arabidopsis, the wildflower. Nat. Rev. Genet. 2001, 2, 516–527. [Google Scholar] [CrossRef]
- Miyakawa, T.; Miyazono, K.-I.; Sawano, Y.; Hatano, K.-I.; Tanokura, M. Crystal structure of ginkbilobin-2 with homology to the extracellular domain of plant cysteine-rich receptor-like kinases. Proteins Struct. Funct. Bioinform. 2009, 77, 247–251. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Tian, L.-H.; Zhao, J.-F.; Song, Y.; Zhang, C.-J.; Guo, Y. Identification of an Apoplastic Protein Involved in the Initial Phase of Salt Stress Response in Rice Root by Two-Dimensional Electrophoresis. Plant Physiol. 2009, 149, 916–928. [Google Scholar] [CrossRef] [Green Version]
- Krasensky-Wrzaczek, J.; Broyart, C.; Rabanal, F.; Jonak, C. The Redox-Sensitive Chloroplast Trehalose-6-Phosphate Phosphatase AtTPPD Regulates Salt Stress Tolerance. Antioxid. Redox Signal. 2014, 21, 1289–1304. [Google Scholar] [CrossRef] [Green Version]
- Cowan, J.A. Metal Activation of Enzymes in Nucleic Acid Biochemistry. Chem. Rev. 1998, 98, 1067–1088. [Google Scholar] [CrossRef] [PubMed]
- Bertini, G.; Gray, H.B.; Gray, H.; Valentine, J.S.; Stiefel, E.I.; Stiefel, E. Biological Inorganic Chemistry: Structure and Reactivity; University Science Books: Sausalito, CA, USA, 2007. [Google Scholar]
- Hernandez-Garcia, C.M.; Finer, J.J. Identification and validation of promoters and cis-acting regulatory elements. Plant Sci. 2014, 217, 109–119. [Google Scholar] [CrossRef] [Green Version]
- Choi, H.-I.; Hong, J.-H.; Ha, J.-O.; Kang, J.-Y.; Kim, S.Y. ABFs, a Family of ABA-responsive Element Binding Factors. J. Biol. Chem. 2000, 275, 1723–1730. [Google Scholar] [CrossRef] [Green Version]
- Hobo, T.; Asada, M.; Kowyama, Y.; Hattori, T. ACGT-containing abscisic acid response element (ABRE) and coupling element 3 (CE3) are functionally equivalent. Plant J. 1999, 19, 679–689. [Google Scholar] [CrossRef]
- Guiltinan, M.J.; Marcotte, W.R.; Quatrano, R.S. A Plant Leucine Zipper Protein That Recognizes an Abscisic Acid Response Element. Science 1990, 250, 267–271. [Google Scholar] [CrossRef] [Green Version]
- Marcotte, W.R., Jr.; Russell, S.H.; Quatrano, R.S. Abscisic acid-responsive sequences from the em gene of wheat. Plant Cell 1989, 1, 969–976. [Google Scholar]
- Narusaka, Y.; Nakashima, K.; Shinwari, Z.K.; Sakuma, Y.; Furihata, T.; Abe, H.; Narusaka, M.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Interaction between two cis-acting elements, ABRE and DRE, in ABA-dependent expression of Arabidopsis rd29A gene in response to dehydration and high-salinity stresses. Plant J. 2003, 34, 137–148. [Google Scholar] [CrossRef]
- Shen, Q.; Zhang, P.; Ho, T. Modular nature of abscisic acid (ABA) response complexes: Composite promoter units that are necessary and sufficient for ABA induction of gene expression in barley. Plant Cell 1996, 8, 1107–1119. [Google Scholar] [PubMed] [Green Version]
- Banerjee, J.; Sahoo, D.K.; Dey, N.; Houtz, R.L.; Maiti, I.B. An Intergenic Region Shared by At4g35985 and At4g35987 in Arabidopsis thaliana Is a Tissue Specific and Stress Inducible Bidirectional Promoter Analyzed in Transgenic Arabidopsis and Tobacco Plants. PLoS ONE 2013, 8, e79622. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kretzschmar, T.; Pelayo, M.A.F.; Trijatmiko, K.; Gabunada, L.F.M.; Alam, R.; Jimenez, R.; Mendioro, M.S.; Slamet-Loedin, I.; Sreenivasulu, N.; Bailey-Serres, J.; et al. A trehalose-6-phosphate phosphatase enhances anaerobic germination tolerance in rice. Nat. Plants 2015, 1, 15124. [Google Scholar] [CrossRef]
- Shima, S.; Matsui, H.; Tahara, S.; Imai, R. Biochemical characterization of rice trehalose-6-phosphate phosphatases supports distinctive functions of these plant enzymes. FEBS J. 2007, 274, 1192–1201. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).