Identification and Validation of Candidate Genes Conferring Resistance to Downy Mildew in Maize (Zea mays L.)
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials
2.2. Evaluation of the RILs for DM
2.3. DNA Preparation
2.4. Molecular Marker Assay
2.5. Linkage Map Construction and QTL Analysis for P. sorghi, P. maydis, and S. macrospora Resistance
2.6. RNA Extraction and Candidate Gene Screening with Quantitative Real-Time PCR (qRT-PCR)
3. Results
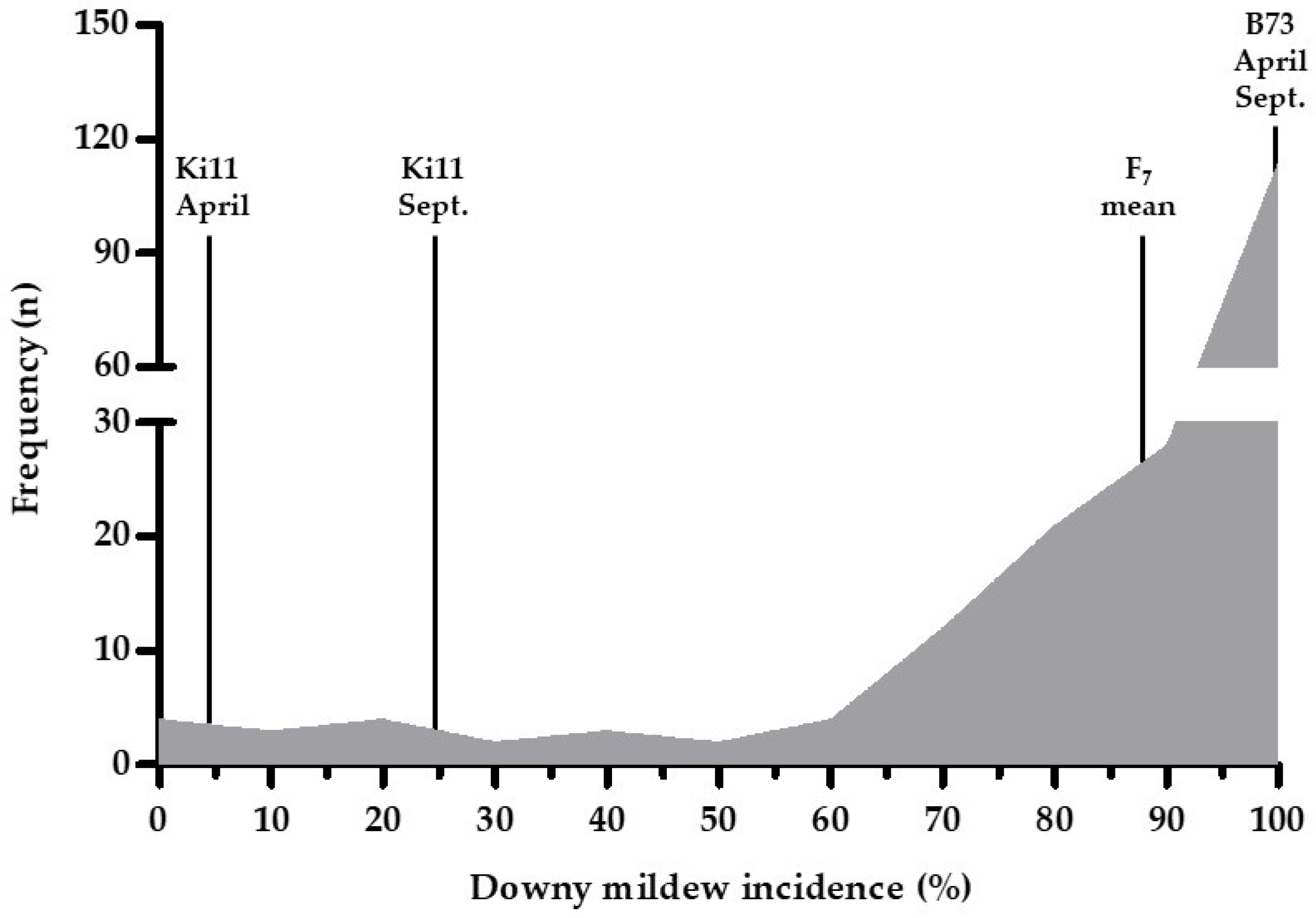
3.1. Phenotypic Data for the DM Analysis
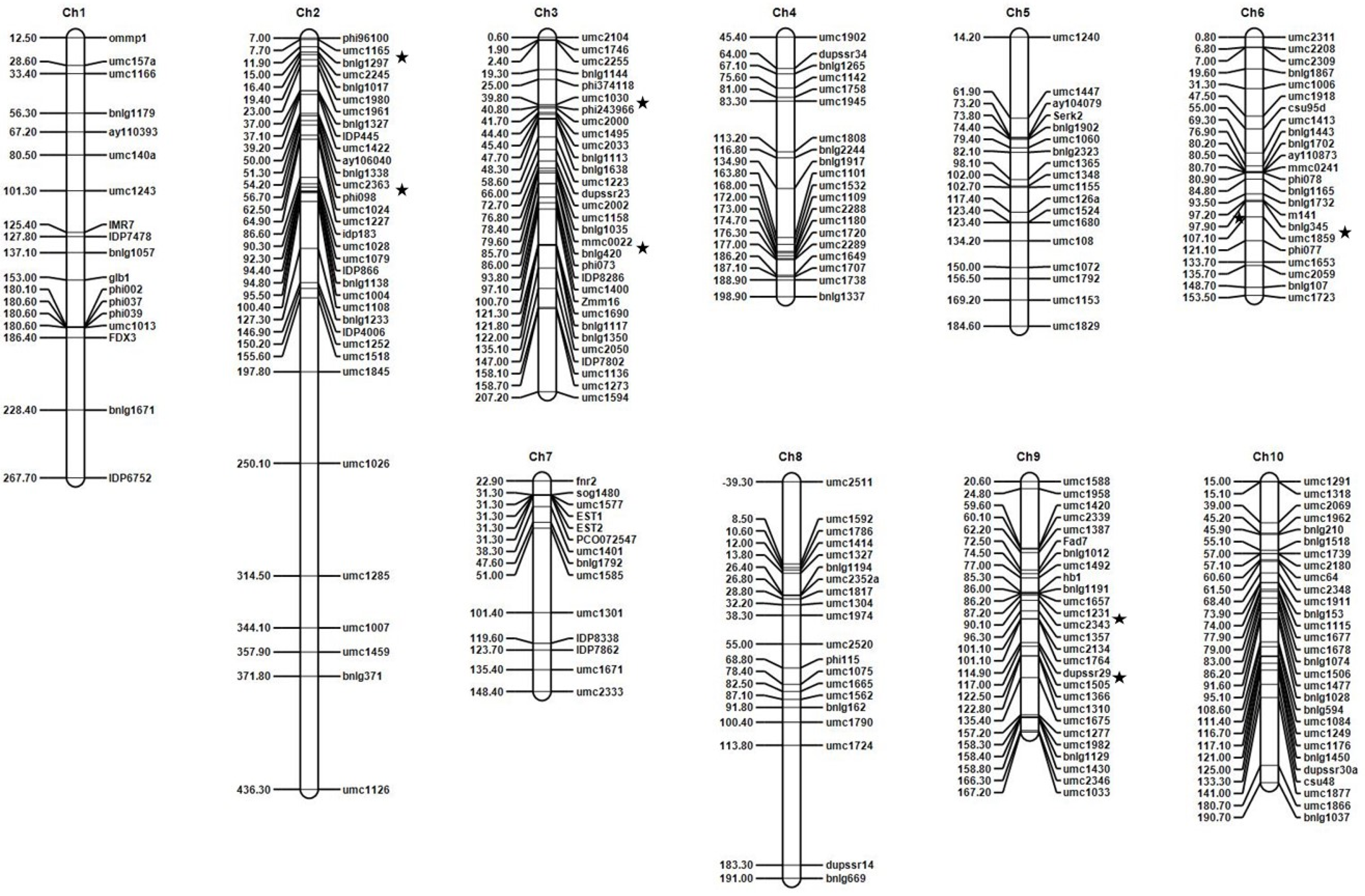
3.2. Marker Data Analysis and Linkage Mapping
3.3. QTL Analysis
3.4. qRT-PCR Validation of the Candidate Genes for P. sorghi, P. maydis, and S. macrospora Resistance
4. Discussion
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Bonde, M. Epidemiology of downy mildew diseases of maize, sorghum and pearl millet. Trop. Pest Manag. 1982, 28, 49–60. [Google Scholar] [CrossRef]
- George, M.; Prasanna, B.; Rathore, R.; Setty, T.; Kasim, F.; Azrai, M.; Vasal, S.; Balla, O.; Hautea, D.; Canama, A. Identification of QTLs conferring resistance to downy mildews of maize in Asia. Theor. Appl. Genet. 2003, 107, 544–551. [Google Scholar] [CrossRef] [PubMed]
- Jeffers, D.; Cordova, H.; Vasal, S.; Srinivasan, G.; Beck, D.; Barandiaran, M. Status in breeding for resistance to maize diseases at CIMMYT. In Proceedings of the 7th Asian Regional Maize Workshop, Los Baños, Phillipines, 23–27 February 1998; Vasal, S.K., Gonzalez, C.F., Fan, X.M., Eds.; CIMMYT: El Batan, Mexico, 2000; pp. 257–266. [Google Scholar]
- Centro Internacional de Mejoramiento de Maíz y Trigo (CIMMYT); Pingali, P.L. CIMMYT 1999/2000 World Maize Facts and Trends-Meeting World Maize Needs: Technological Opportunities and Priorities for the Public Sector; CIMMYT: El Batan, Mexico, 2001. [Google Scholar]
- Rathore, R.; Siradhana, B. Estimation of losses caused by Peronosclerospora heteropogoni on Ganga-5 maize hybrid. Phytophylactica 1988, 19, 119–120. [Google Scholar]
- Rifin, A. Downy mildew resistance of single cross progenies between Indonesia and Philippine corn inbred lines. Penelitian Pertanian 1983, 3, 81–83. [Google Scholar]
- Broyles, J. Observations on time and location of penetration in relation to amount of damage and chemical control of Physoderma maydis. Phytopathology 1956, 46, 8. [Google Scholar]
- Borges, F.; Orangel, L. Diallel analysis of maize resistance to sorghum downy mildew 1. Crop Sci. 1987, 27, 178–180. [Google Scholar] [CrossRef]
- Frederiksen, R.; Renfro, B. Global status of maize downy mildew. Ann. Rev. Phytopathol. 1977, 15, 249–271. [Google Scholar] [CrossRef]
- Payak, M.; Renfro, B. A new downy mildew disease of maize. Phytopathology 1967, 57, 394–397. [Google Scholar]
- Sharma, R.; De Leon, C.; Payak, M. Diseases of maize in South and South-East Asia: Problems and progress. Crop Prot. 1993, 12, 414–422. [Google Scholar] [CrossRef]
- Spencer, M.; Dick, M. Aspects of graminicolous downy mildew biology: Perspectives for tropical plant pathology and Peronosporomycetes phylogeny. Trop. Mycol. 2002, 2, 63–81. [Google Scholar]
- Yen, T.T.O.; Prasanna, B.; Setty, T.; Rathore, R. Genetic variability for resistance to sorghum downy mildew (Peronosclerospora sorghi) and Rajasthan downy mildew (P. heteropogoni) in the tropical/sub-tropical Asian maize germplasm. Euphytica 2004, 138, 23–31. [Google Scholar] [CrossRef]
- Ullstrup, A. Observations on crazy top of Corn. Phytopathology 1952, 42, 675–680. [Google Scholar]
- Putnam, M.L. Brown stripe downy mildew (Sclerophthora rayssiae var. zeae) of maize. Plant Health Prog. 2007, 8, 34. [Google Scholar] [CrossRef]
- Geetha, K.; Jayaraman, N. Inheritance of sorghum downy mildew resistance in maize. Indian J. Agric. Res. 2002, 36, 234–240. [Google Scholar]
- Jinahyon, S. The genetics of resistance and its implications to breeding for resistance in corn. In Proceedings of the Inter-Asian Corn Improvement Workshop, Kuala Lampur, Malyasia, 10–12 December 1973; pp. 30–39. [Google Scholar]
- Kaneko, K.; Aday, B. Inheritance of resistance to philippine downy mildew of maize, Peronosclerospora philippinensis 1. Crop Sci. 1980, 20, 590–594. [Google Scholar] [CrossRef]
- Leon, C.D.; Ahuja, V.; Capio, E.R.; Mukherjee, B. Genetics of resistance to Philippine downy mildew in three maize populations. Indian J. Gen. Plant Breeding 1993, 53, 406–410. [Google Scholar]
- Nair, S.K.; Prasanna, B.; Rathore, R.; Setty, T.; Kumar, R.; Singh, N. Genetic analysis of resistance to sorghum downy mildew and Rajasthan downy mildew in maize (Zea mays L.). Field Crops Res. 2004, 89, 379–387. [Google Scholar] [CrossRef]
- Nair, S.K.; Prasanna, B.M.; Garg, A.; Rathore, R.; Setty, T.; Singh, N. Identification and validation of QTLs conferring resistance to sorghum downy mildew (Peronosclerospora sorghi) and Rajasthan downy mildew (P. heteropogoni) in maize. Theor. Appl. Genet. 2005, 110, 1384–1392. [Google Scholar] [CrossRef]
- Nallathambi, P.; Sundaram, K.M.; Arumugachamy, S. Inheritance of Resistance to Sorghum Downy mildew (Peronosclerospera sorghi) in Maize (Zea mays L.). Int. J. Agric. Environ. Biotechnol. 2010, 3, 285–293. [Google Scholar]
- Singburaudom, N.; Renfro, B. Heritability of resistance in maize to sorghum downy mildew (Peronosclerospora sorghi (Weston and Uppal) CG Shaw). Crop Prot. 1982, 1, 323–332. [Google Scholar] [CrossRef]
- Young, N. QTL mapping and quantitative disease resistance in plants. Ann. Rev. Phytopathol. 1996, 34, 479–501. [Google Scholar] [CrossRef] [PubMed]
- Austin, D.; Lee, M. Comparative mapping in F 2:3 and F 6:7 generations of quantitative trait loci for grain yield and yield components in maize. Theor. Appl. Genet. 1996, 92, 817–826. [Google Scholar] [CrossRef] [PubMed]
- Burr, B.; Burr, F.A. Recombinant inbreds for molecular mapping in maize: Theoretical and practical considerations. Trends Genet. 1991, 7, 55–60. [Google Scholar] [CrossRef]
- Knapp, S.; Bridges, W. Using molecular markers to estimate quantitative trait locus parameters: Power and genetic variances for unreplicated and replicated progeny. Genetics 1990, 126, 769–777. [Google Scholar] [PubMed]
- Krakowsky, M.; Lee, M.; Woodman-Clikeman, W.; Long, M.; Sharopova, N. QTL mapping of resistance to stalk tunneling by the European corn borer in RILs of maize population B73× De8 1. Crop Sci. 2004, 44, 274–282. [Google Scholar]
- Frova, C.; Gorla, M.S. Quantitative expression of maize HSPs: Genetic dissection and association with thermotolerance. Theor. Appl. Genet. 1993, 86, 213–220. [Google Scholar] [CrossRef]
- Guo, J.; Su, G.; Zhang, J.; Wang, G. Genetic analysis and QTL mapping of maize yield and associate agronomic traits under semi-arid land condition. Afr. J. Biotechnol. 2008, 7, 1829–1838. [Google Scholar]
- Agrama, H.; Moussa, M.; Naser, M.; Tarek, M.; Ibrahim, A. Mapping of QTL for downy mildew resistance in maize. Theor. Appl. Genet. 1999, 99, 519–523. [Google Scholar] [CrossRef]
- Sabry, A.; Jeffers, D.; Vasal, S.; Frederiksen, R.; Magill, C. A region of maize chromosome 2 affects response to downy mildew pathogens. Theor. Appl. Genet. 2006, 113, 321–330. [Google Scholar] [CrossRef]
- Jampatong, C.; Jampatong, S.; Jompuk, C.; Sreewongchai, T.; Grudloyma, P.; Balla, C.; Prodmatee, N. Mapping of QTL affecting resistance against sorghum downy mildew (Peronosclerospora sorghi) in maize (Zea mays L). Maydica 2013, 58, 119–126. [Google Scholar]
- Lohithaswa, H.; Jyothi, K.; Kumar, K.S.; Hittalmani, S. Identification and introgression of QTLs implicated in resistance to sorghum downy mildew (Peronosclerospora sorghi (Weston and Uppal) CG Shaw) in maize through marker-assisted selection. J. Genet. 2015, 94, 741–748. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Ding, Q.; Wang, F.; Li, H.; Zhang, Y.; Liu, L.; Jiao, Z.; Gao, J. Genome-wide gene expression profiles in response to downy mildew in Chinese cabbage (Brassica rapa L. ssp. pekinensis). Eur. J. Plant Pathol. 2018, 151, 861–873. [Google Scholar] [CrossRef]
- Burkhardt, A.; Day, B. Transcriptome and small RNAome dynamics during a resistant and susceptible interaction between cucumber and downy mildew. Plant Genome 2016, 9, 1–19. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Wu, J.; Yin, L.; Zhang, Y.; Qu, J.; Lu, J. Comparative transcriptome analysis reveals defense-related genes and pathways against downy mildew in Vitis amurensis grapevine. Plant Physiol. Biochem. 2015, 95, 1–14. [Google Scholar] [CrossRef]
- Kulkarni, K.S.; Zala, H.N.; Bosamia, T.C.; Shukla, Y.M.; Kumar, S.; Fougat, R.S.; Patel, M.S.; Narayanan, S.; Joshi, C.G. De novo transcriptome sequencing to dissect candidate genes associated with pearl millet-downy mildew (Sclerospora graminicola Sacc.) interaction. Front. Plant Sci. 2016, 7, 847. [Google Scholar] [CrossRef]
- Okubara, P.A.; Anderson, P.A.; Ochoa, O.E.; Michelmore, R.W. Mutants of downy mildew resistance in Lactuca sativa (lettuce). Genetics 1994, 137, 867–874. [Google Scholar]
- Zhu, Q.; Gao, P.; Wan, Y.; Cui, H.; Fan, C.; Liu, S.; Luan, F. Comparative transcriptome profiling of genes and pathways related to resistance against powdery mildew in two contrasting melon genotypes. Sci. Hortic. 2018, 227, 169–180. [Google Scholar] [CrossRef]
- Aekatasanawan, C.; Jampatong, S.; Aekatasanawan, C.; Chulchoho, N.; Balla, C. Supporting the hybrid maize breeding research in Thailand. In Proceedings of the 7th Asian Regional Maize Workshop, Los Baños, Phillipines, 23–27 February 1998; Vasal, S.K., Gonzalez, C.F., Fan, X.M., Eds.; CIMMYT: El Batan, Mexico, 2000; pp. 82–91. [Google Scholar]
- Hallauer, A. Specialty Corns, 2nd ed.; CRC Press: Boca Raton, FL, USA, 2001. [Google Scholar]
- Coles, N.D.; McMullen, M.D.; Balint-Kurti, P.J.; Pratt, R.C.; Holland, J.B. Genetic control of photoperiod sensitivity in maize revealed by joint multiple population analysis. Genetics 2010, 184, 799–812. [Google Scholar] [CrossRef]
- Kim, K.; Moon, J.; Kim, J.; Kim, H.; Shin, S.; Song, K.; Baek, S.; Lee, B. Evaluation of maize downy mildew using spreader row technique. Korean J. Crop Sci. 2016, 61, 41–49. [Google Scholar] [CrossRef]
- Liu, K.; Goodman, M.; Muse, S.; Smith, J.S.; Buckler, E.; Doebley, J. Genetic structure and diversity among maize inbred lines as inferred from DNA microsatellites. Genetics 2003, 165, 2117–2128. [Google Scholar]
- Russell, W. Registration of B70 and B73 parental lines of Maize1 (Reg. Nos. PL16 and PL17). Crop Sci. 1972, 12, 721. [Google Scholar] [CrossRef]
- Kim, J.Y.; Moon, J.C.; Kim, H.C.; Shin, S.; Song, K.; Kim, K.H.; Lee, B.M. Identification of downy mildew resistance gene candidates by positional cloning in maize (Zea mays subsp. mays; Poaceae). Appl. Plant Sci. 2017, 5, 1600132. [Google Scholar] [CrossRef] [PubMed]
- Craig, A.; Bockholt, J.; Frederiksen, R.A.; Zuber, M.S. Reaction of important corn inbred lines to Peronosclerospora sorghi. Plant Dis. Rept. 1977, 61, 563–564. [Google Scholar]
- Nagabhushan, T.; Lohithaswa, H.; Sreeramasetty, T.; Puttaramanaik, H.S.; Hittalmani, S. Identification of stable source of resistance to sorghum downy mildew in maize (Zea mays L.). J. Agroecol. Nat. Resour. Manag. 2014, 1, 176–178. [Google Scholar]
- Rao, B.; Prakash, H.; Shetty, H. Relationship of cultivars with sporulation and morphology of asexual propagules of Peronosclerospora sorghi on maize. Int. J. Trop. Plant Dis. 1984, 2, 175–180. [Google Scholar]
- Li, H.; Ye, G.; Wang, J. A modified algorithm for the improvement of composite interval mapping. Genetics 2007, 175, 361–374. [Google Scholar] [CrossRef]
- Wang, J.; Li, H.; Zhang, L.; Meng, L. User’s Manual of QTL IciMapping ver. 4.1; CIMMYT: El Batan, Mexico, 2016. [Google Scholar]
- Kosambi, D.D. The estimation of map distances from recombination values. Ann. Eugen. 1944, 12, 172–175. [Google Scholar] [CrossRef]
- Doerge, R.W.; Churchill, G.A. Permutation tests for multiple loci affecting a quantitative character. Genetics 1996, 142, 285–294. [Google Scholar]
- Cui, F.; Fan, X.; Chen, M.; Zhang, N.; Zhao, C.; Zhang, W.; Han, J.; Ji, J.; Zhao, X.; Yang, L. QTL detection for wheat kernel size and quality and the responses of these traits to low nitrogen stress. Theor. Appl. Genet. 2016, 129, 469–484. [Google Scholar] [CrossRef]
- Lander, E.; Kruglyak, L. Genetic dissection of complex traits: Guidelines for interpreting and reporting linkage results. Nat. Genet. 1995, 11, 241. [Google Scholar] [CrossRef]
- Rozen, S.; Skaletsky, H. Primer3 on the WWW for general users and for biologist programmers. In Bioinformatics Methods and Protocols; Springer: Berlin/Heidelberg, Germany, 2000; pp. 365–386. [Google Scholar]
- Manoli, A.; Sturaro, A.; Trevisan, S.; Quaggiotti, S.; Nonis, A. Evaluation of candidate reference genes for qPCR in maize. J. Plant Physiol. 2012, 169, 807–815. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2− ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Shekhar, M.; Kumar, S. Inoculation Methods and Disease Rating Scales for Maize Diseases; Directorate of Maize Research: New Delhi, India, 2012. [Google Scholar]
- Rashid, Z.; Singh, P.K.; Vemuri, H.; Zaidi, P.H.; Prasanna, B.M.; Nair, S.K. Genome-wide association study in Asia-adapted tropical maize reveals novel and explored genomic regions for sorghum downy mildew resistance. Sci. Rep. 2018, 8, 366. [Google Scholar] [CrossRef] [PubMed]
- Nagabhushan; Lohithaswa, H.C.; Pandravada, A.S. Construction of high-density linkage map and identification of QTLs for resistance to sorghum downy mildew in maize (Zea mays L.). Mol. Breeding 2017, 37, 2. [Google Scholar] [CrossRef]
- Jeuken, M.J.W.; Pelgrom, K.; Stam, P.; Lindhout, P. Efficient QTL detection for nonhost resistance in wild lettuce. Theor. Appl. Genet. 2008, 116, 845–857. [Google Scholar] [CrossRef] [PubMed]
- Magalhaes, J.V. Aluminum tolerance genes are conserved between monocots and dicots. Proc. Natl. Acad. Sci. USA 2006, 103, 9749–9750. [Google Scholar] [CrossRef]
- Miller, R.N.; Bertioli, D.J.; Baurens, F.C.; Santos, C.M.; Alves, P.C.; Martins, N.F.; Togawa, R.C.; Júnior, M.T.S.; Júnior, G.J.P. Analysis of non-TIR NBS-LRR resistance gene analogs in Musa acuminata Colla: Isolation, RFLP marker development, and physical mapping. BMC Plant Biol. 2008, 8, 15. [Google Scholar] [CrossRef]
- Körösi, K.; Bán, R.; Barna, B.; Virányi, F. Biochemical and molecular changes in downy mildew-infected sunflower triggered by resistance inducers. J. Phytopathol. 2011, 159, 471–478. [Google Scholar] [CrossRef]
- Toffolatti, S.L.; De Lorenzis, G.; Costa, A.; Maddalena, G.; Passera, A.; Bonca, M.C.; Pindo, M.; Stefani, E.; Cestaro, A.; Casati, P.; et al. Uique resistance traits against downy mildew from the center of origin of grapevine (Vitis vinifera). Sci. Rep. 2018, 8, 1–11. [Google Scholar] [CrossRef]
- Jiang, M.; Jiang, J.J.; He, C.M.; Guan, M. Broccoli plants over-expressiong a cytosolic ascorbate peroxidase gene increase resistance to downy mildew and heat stress. J. Plant Pathol. 2016, 98, 413–420. [Google Scholar]
- Lin, H.; Leng, H.; Guo, Y.; Kondo, S.; Zhao, Y.; Guo, X. QTLs and candidate genes for downy mildew resistance conferred by interspecific grape (V. vinifera L. x V. amurensis Rupr.) crossing. Sci. Hortic. 2019, 244, 200–207. [Google Scholar] [CrossRef]
- She, H.; Quan, W.; Zhang, H.; Liu, Z.; Wang, X.; Wu, J.; Feng, C.; Correll, J.C.; Xu, Z. Fine mapping and candidate gene screening of the downy mildew resistance gene RPF1 in spinach. Theor. Appl. Genet. 2018, 131, 2529–2541. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Ding, F.; Peng, H.; Huang, Y.; Lu, J. Molecular cloning of a CC-NBS-LRR gene from Vitis quinquangularis and its expression pattern in response to downy mildew pathogen infection. Mol. Genet. Genomics 2017, 293, 61–68. [Google Scholar] [CrossRef] [PubMed]
- Prabhu, S.A.; Kini, K.R.; Raj, S.N.; Moerschbacher, B.M.; Shetty, H.S. Polygalacturonase-inhibitor proteins in pearl millet: Possible involvement in resistance against downy mildew. Acta Biochim. Biophys. Sin. 2012, 44, 415–423. [Google Scholar] [CrossRef]
- Nandeeshkumar, P.; Sudisha, J.; Ramachandra, K.K.; Prakash, H.; Niranjana, S.; Shekar, S.H. Chitosan induced resistance to downy mildew in sunflower caused by Plasmopara halstedii. Physiol. Mol. Plant Pathol. 2008, 72, 188–194. [Google Scholar] [CrossRef]
- Diez-Navajas, A.; Wiedemann-Merdinoglu, S.; Greif, C.; Merdinoglu, D. Nonhost versus host resistance to the grapevine downy mildew, Plasmopara viticola, studied at the tissue level. Phytopathology 2008, 98, 776–780. [Google Scholar] [CrossRef]
- Jürges, G.; Kassemeyer, H.H.; Dürrenberger, M.; Düggelin, M.; Nick, P. The mode of interaction between Vitis and Plasmopara viticola Berk. & Curt. Ex de Bary depends on the host species. Plant Biol. 2009, 11, 886–898. [Google Scholar]
- Kortekamp, A. Expression analysis of defence-related genes in grapevine leaves after inoculation with a host and a non-host pathogen. Plant Physiol. Biochem. 2006, 44, 58–67. [Google Scholar] [CrossRef]
- Pezet, R.; Gindro, K.; Viret, O.; Spring, J.-L. Glycosylation and oxidative dimerization of resveratrol are respectively associated to sensitivity and resistance of grapevine cultivars to downy mildew. Physiol. Mol. Plant Pathol. 2004, 65, 297–303. [Google Scholar] [CrossRef]
- Reuveni, R.; Bothma, G. The relationship between peroxidase activity and resistance to Sphaerotheca fuliginea in melons. J. Phytopathol. 1985, 114, 260–267. [Google Scholar] [CrossRef]
- Reuveni, R.; Shimoni, M.; Crute, I. An association between high peroxidase activity in lettuce (Lactuca sativa) and field resistance to downy mildew (Bremia lactucae). J. Phytopathol. 1991, 132, 312–318. [Google Scholar] [CrossRef]
- Reuveni, R.; Karchi, Z. Peroxidase activity-a possible marker for resistance of melon against downy mildew. Phytopathology 1987, 77, 1724. [Google Scholar]
- Reuveni, R.; Ferreira, J. The relationship between peroxidase activity and the resistance of tomatoes (Lycopersicum esculentum) to Verticillium dahliae. J. Phytopathol. 1985, 112, 193–197. [Google Scholar] [CrossRef]
- Bartsch, M.; Gobbato, E.; Bednarek, P.; Debey, S.; Schultze, J.L.; Bautor, J.; Parker, J.E. Salicylic acid–independent enhanced disease susceptibility1 signaling in Arabidopsis immunity and cell death is regulated by the monooxygenase FMO1 and the nudix hydrolase NUDT7. Plant Cell 2006, 18, 1038–1051. [Google Scholar] [CrossRef]
- Van Der Biezen, E.A.; Freddie, C.T.; Kahn, K.; Parker, J.E.; Jones, J.D. Arabidopsis RPP4 is a member of the RPP5 multigene family of TIR-NB-LRR genes and confers downy mildew resistance through multiple signalling components. Plant J. 2002, 29, 439–451. [Google Scholar] [CrossRef]
- Babitha, M.; Prakash, H.; Shetty, H. Purification and properties of lipoxygenase induced in downy mildew resistant pearl millet seedlings due to infection with Sclerospora graminicola. Plant Sci. 2004, 166, 31–39. [Google Scholar] [CrossRef]
- Nagatathna, K.C.; Shetty, S.A.; Bhat, S.G.; Shetty, H.S. The possible involvement of lipoxygenase in downy mildew resistance in pearl millet. J. Exp. Bot. 1992, 43, 1283–1287. [Google Scholar] [CrossRef]
- Pushpalatha, H.; Sudisha, J.; Geetha, N.; Amruthesh, K.; Shetty, H.S. Thiamine seed treatment enhances LOX expression, promotes growth and induces downy mildew disease resistance in pearl millet. Biol. Plantarum 2011, 55, 522–527. [Google Scholar] [CrossRef]
- Shivakumar, P.; Geetha, H.; Shetty, H. Peroxidase activity and isozyme analysis of pearl millet seedlings and their implications in downy mildew disease resistance. Plant Sci. 2003, 164, 85–93. [Google Scholar] [CrossRef]
- Mustafa, G.; Khong, N.G.; Tisserant, B.; Randoux, B.; Fontaine, J.; Magnin-Robert, M.; Reignault, P.; Sahraoui, A.L.H. Defence mechanisms associated with mycorrhiza-induced resistance in wheat against powdery mildew. Funct. Plant Biol. 2017, 44, 443–454. [Google Scholar] [CrossRef]
- Dangl, J.L.; Jones, J.D. Plant pathogens and integrated defence responses to infection. Nature 2001, 411, 826. [Google Scholar] [CrossRef] [PubMed]
- Hammond-Kosack, K.E.; Parker, J.E. Deciphering plant–pathogen communication: Fresh perspectives for molecular resistance breeding. Curr. Opin. Biotechnol. 2003, 14, 177–193. [Google Scholar] [CrossRef]
- Warren, R.F.; Henk, A.; Mowery, P.; Holub, E.; Innes, R.W. A mutation within the leucine-rich repeat domain of the Arabidopsis disease resistance gene RPS5 partially suppresses multiple bacterial and downy mildew resistance genes. Plant Cell 1998, 10, 1439–1452. [Google Scholar] [CrossRef] [PubMed]
- Heath, M.C. Nonhost resistance and nonspecific plant defenses. Curr. Opin. Plant Biol. 2000, 3, 315–319. [Google Scholar] [CrossRef]
- King, S.R.; McLellan, H.; Boevink, P.C.; Armstrong, M.R.; Bukharova, T.; Sukarta, O.; Win, J.; Kamoun, S.; Birch, P.R.; Banfield, M.J. Phytophthora infestans RXLR effector PexRD2 interacts with host MAPKKKε to suppress plant immune signaling. Plant Cell 2014, 26, 1345–1359. [Google Scholar] [CrossRef] [PubMed]
- White, P.; Broadley, M. Calcium in plants. Ann. Bot. 2003, 92, 487–511. [Google Scholar] [CrossRef] [PubMed]
- Melvin, P.; Prabhu, S.A.; Anup, C.P.; Shailasree, S.; Shetty, H.S.; Kini, K.R. Involvement of mitogen-activated protein kinase signalling in pearl millet–downy mildew interaction. Plant Sci. 2014, 214, 29–37. [Google Scholar] [CrossRef]
- Di Matteo, A.; Bonivento, D.; Tsernoglou, D.; Federici, L.; Cervone, F. Polygalacturonase-inhibiting protein (PGIP) in plant defence: A structural view. Phytochemistry 2006, 67, 528–533. [Google Scholar] [CrossRef]
- Wu, J.; Zhang, Y.; Zhang, H.; Huang, H.; Folta, K.M.; Lu, J. Whole genome wide expression profiles of Vitis amurensis grape responding to downy mildew by using Solexa sequencing technology. BMC Plant Biol. 2010, 10, 234. [Google Scholar] [CrossRef]
- Van Damme, M.; Zeilmaker, T.; Elberse, J.; Andel, A.; de Sain-van der Velden, M.; Van den Ackerveken, G. Downy mildew resistance in Arabidopsis by mutation of homoserine kinase. Plant Cell 2009, 21, 2179–2189. [Google Scholar] [CrossRef]
- Marsh, E.; Alvarez, S.; Hicks, L.M.; Barbazuk, W.B.; Qiu, W.; Kovacs, L.; Schachtman, D. Changes in protein abundance during powdery mildew infection of leaf tissues of Cabernet Sauvignon grapevine (Vitis vinifera L.). Proteomics 2010, 10, 2057–2064. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Liu, H.; Zhang, H.; Wang, X.; Song, F. OsBIRH1, a DEAD-box RNA helicase with functions in modulating defence responses against pathogen infection and oxidative stress. J. Exp. Bot. 2008, 59, 2133–2146. [Google Scholar] [CrossRef] [PubMed]
- Asakura, Y.; Galarneau, E.; Watkins, K.P.; Barkan, A.; Van Wijk, K.J. Chloroplast RH3 DEAD box RNA helicases in maize and Arabidopsis function in splicing of specific group II introns and affect chloroplast ribosome biogenesis. Plant Physiol. 2012, 159, 961–974. [Google Scholar] [CrossRef] [PubMed]
- Hok, S.; Danchin, E.G.; Allasia, V.; Panabieres, F.; Attard, A.; Keller, H. An Arabidopsis (malectin-like) leucine-rich repeat receptor-like kinase contributes to downy mildew disease. Plant Cell Environ. 2011, 34, 1944–1957. [Google Scholar] [CrossRef] [PubMed]
- Bruggmann, R.; Abderhalden, O.; Reymond, P.; Dudler, R. Analysis of epidermis-and mesophyll-specific transcript accumulation in powdery mildew-inoculated wheat leaves. Plant Mol. Biol. 2005, 58, 247–267. [Google Scholar] [CrossRef] [PubMed]
- Zheng, L.; Huang, J.; Yu, D. Isolation of genes expressed during compatible interactions between powdery mildew (Blumeria graminis) and wheat. Physiol. Mol. Plant Pathol. 2008, 73, 61–66. [Google Scholar] [CrossRef]


| Variety | Mid-April to June | Early September to October | ||
|---|---|---|---|---|
| 4 Weeks | 6 Weeks | 4 Weeks | 6 Weeks | |
| Resistant Genotype | ||||
| CML228 | 0% | 0% | 20% | 25% |
| Ki3 | 0% | 0% | 16.6% | 22.2% |
| Ki11 | 0% | 5% | 10% | 25% |
| Susceptible Genotype | ||||
| B73 | 50% | 100% | 100% | 100% |
| CML270 | 100% | 100% | 75% | 100% |
| Trait | Sample Size | Mean of DM Incidence | Standard Deviation | Minimum | Maximum | Range | Skewness | Kurtosis |
|---|---|---|---|---|---|---|---|---|
| DM | 192 | 86.49 | 23.87 | 0 | 100 | 0–100 | −2.35 | 5.14 |
| QTLs | Chr 1 | Bin Location | Position | Left CI | Right CI | Left Marker | Right Marker | LOD 2 | PVE (%) 3 | Add 4 | Donor of DM-Resistant Genotype |
|---|---|---|---|---|---|---|---|---|---|---|---|
| qDM1 | 2 | 2.01 | 11.03 | 10.53 | 11.53 | umc1165 | bnlg1297 | 14.12 | 12.95 | −32.87 | Ki11 |
| qDM2 | 2.02 | 55.03 | 54.53 | 55.53 | umc2363 | phi098 | 3.60 | 0.75 | −13.93 | Ki11 | |
| qDM3 | 3 | 3.04 | 40.60 | 40.10 | 41.10 | umc1030 | phi243966 | 3.21 | 0.48 | −8.39 | Ki11 |
| qDM4 | 3.05 | 80.60 | 80.10 | 81.10 | mmc0022 | bnlg420 | 18.16 | 2.90 | −31.68 | Ki11 | |
| qDM5 | 6 | 6.05/6.06 | 98.80 | 98.30 | 99.30 | umc1859 | bnlg345 | 10.77 | 2.85 | −33.18 | Ki11 |
| qDM6 | 9 | 9.05 | 88.58 | 88.08 | 90.08 | umc1231 | umc2343 | 3.84 | 0.77 | −11.09 | Ki11 |
| qDM7 | 9.07 | 116.58 | 115.08 | 117.08 | dupssr29 | umc1505 | 3.17 | 0.47 | −8.27 | Ki11 |
| Gene ID | Cultivar | Chromosome | Location | Description |
|---|---|---|---|---|
| EDM2 | A. thaliana | 5 | 22,447,937–22,454,805 | Enhanced downy mildew 2 |
| SGT1B | A. thaliana | 4 | 6,851,184–6,853,912 | SGT1b |
| HSK | A. thaliana | 2 | 7,508,473–7,509,887 | Homoserine kinase; downy mildew resistance 1 |
| DMR6 | A. thaliana | 5 | 8,378,759–8,383,401 | Putative 2OG-Fe(II) oxygenase |
| LOC4324025 | O. sativa | 1 | 17,932,375–17,947,080 | Enhanced downy mildew 2 |
| LOC107275878 | O. sativa | 3 | 8,811,797–8,818,014 | Enhanced downy mildew 2 |
| LOC107275863 | O. sativa | 8 | 15,057,975–15,070,064 | Enhanced downy mildew 2-like |
| LOC4345309 | O. sativa | 8 | 15,114,902–15,130,493 | Enhanced downy mildew 2 |
| LOC4345959 | O. sativa | 8 | 24,800,574–24,812,015 | Enhanced downy mildew 2 |
| LOC4351808 | O. sativa | 12 | 6,760,146–6,763,038 | Enhanced downy mildew 2 |
| LOC103647182 | Z. mays | 2 | 160,527,731–160,580,975 | Enhanced downy mildew 2 |
| LOC103648264 | Z. mays | 2 | 241,693,442–241,703,192 | Enhanced downy mildew 2 |
| LOC100191339 | Z. mays | 3 | 135,359,547–135,361,124 | Downy mildew resistance 6 |
| LOC103650325 | Z. mays | 3 | 102,996,061–102,998,690 | Enhanced downy mildew 2 |
| LOC100382073 | Z. mays | 4 | 173,897,007–173,895,527 | Downy mildew resistance 6 |
| LOC103654479 | Z. mays | 4 | 202,734,383–202,764,701 | Enhanced downy mildew 2 |
| IDC1 | Z. mays | 6 | 20,483,352–20,484,802 | Iron deficiency candidate 1; downy mildew resistance 6 |
| LOC103632498 | Z. mays | 7 | 79,090,176–79,133,359 | Enhanced downy mildew 2 |
| LOC103642860 | Z. mays | 10 | 135,816,580–135,824,835 | Flavanone 3-dioxygenase 2 |
| Transcript ID | Chr 1 | Bin Location | Type | Length (bp) | Predicted Protein Size (aa) | Description | Protein Information |
|---|---|---|---|---|---|---|---|
| AC210003.2_FG004 | 2 | 2.01/2.02 | T01 | 999 | 332 | Uncharacterized LOC100274427 | Peroxidase 16 |
| GRMZM2G020043 | T01 | 1938 | 260 | - | - | ||
| GRMZM2G039345 | T01 | 697 | 206 | Ribulose bisphosphate carboxylase/oxygenase activase 2, chloroplastic | P-loop containing nucleoside triphosphate hydrolase superfamily protein | ||
| GRMZM2G045049 | T01 | 2111 | 521 | Probable phosphoribosylformylglycinamidine synthase, chloroplastic/mitochondrial | Probable phosphoribosylformylglycinamidine synthase, chloroplastic/mitochondrial | ||
| GRMZM2G314171 | T01 | 1395 | 464 | - | - | ||
| GRMZM2G363066 | T01 | 1450 | 422 | G-type lectin S-receptor-like serine/threonine-protein kinase At1g34300 | G-type lectin S-receptor-like serine/threonine-protein kinase At1g34300 | ||
| GRMZM2G133707 | 3 | 3.04 | T01 | 217 | - | - | - |
| GRMZM2G047677 | 6 | 6.05/6.06 | T01 | 1199 | 271 | Uncharacterized LOC100216590 | Abscisic acid receptor PYL5; abscisic acid receptor PYL3; pyrabactin resistance-like protein |
| GRMZM2G062031 | T01 | 1809 | 382 | Uncharacterized LOC100276496 | Uncharacterized LOC100276496 | ||
| GRMZM2G128315 | T01 | 3343 | 964 | Probable LRR receptor-like serine/threonine-protein kinase IRK | Putative leucine-rich repeat receptor-like protein kinase family protein | ||
| GRMZM2G005984 | 9 | 9.05 | T01 | 951 | 222 | Photosystem II 11 kd protein | Photosystem II repair protein PSB27-H1 chloroplastic; photosystem II protein |
| AC191071.3_FG001 | 9.07 | T01 | 1605 | 534 | Probable flavin-containing monooxygenase 1 | Probable flavin-containing monooxygenase 1 | |
| GRMZM2G028643 | T01 | 1954 | 523 | Putative leucine-rich repeat receptor-like serine/threonine-protein kinase At2g14440 | Putative leucine-rich repeat receptor-like serine/threonine-protein kinase At2g14440; Leucine-rich repeat (LRR) family protein | ||
| GRMZM2G178880 | T01 | 1894 | 574 | Uncharacterized LOC100191890 | Putative mannan synthase 7 | ||
| GRMZM2G330907 | T01 | 3005 | 759 | Uncharacterized LOC541659 | Leucine-rich repeat transmembrane protein kinase 3; leucine-rich transmembrane protein kinase2; putative STRUBBELIG family receptor protein kinase |
| Transcript ID | Family | Description |
|---|---|---|
| AC210003.2_FG004 | Peroxidase | Peroxidase (PF00141) |
| GRMZM2G020043 | Hydrolase | Haloacid dehalogenase-like hydrolase (PF00702) |
| GRMZM2G039345 | - | - |
| GRMZM2G045049 | AIRS_C, GATase_5 | AIR synthase related protein, C-terminal domain (PF13507); CobB/CobQ-like glutamine amidotransferase domain (PF13507) |
| GRMZM2G314171 | DEAD, Metalloenzyme, Metalloenzyme | DEAD/DEAH box helicase (PF00270); Metalloenzyme superfamily (PF01676); Metalloenzyme superfamily (PF01676) |
| GRMZM2G363066 | Pkinase_Tyr | Protein tyrosine kinase (PF07714) |
| GRMZM2G133707 | - | - |
| GRMZM2G047677 | Polyketide_cyc2 | Polyketide cyclase/dehydrase and lipid transport (PF10604) |
| GRMZM2G062031 | Microtub_bd | Microtubule binding (PF16796) |
| GRMZM2G128315 | LRRNT_2 | Leucine rich repeat N-terminal domain (PF08263) |
| GRMZM2G005984 | PSII_Pbs27 | Photosystem II Pbs27 (PF13326) |
| AC191071.3_FG001 | FMO-like | Flavin-binding monooxygenase-like (PF00743) |
| GRMZM2G028643 | Malectin_like, LRR_1 | Malectin-like domain (PF00560); Leucine rich repeat (PF00560) |
| GRMZM2G178880 | Glyco_trans_2_3 | Glycosyl transferase family group 2 (PF13632) |
| GRMZM2G330907 | LRRNT_2, LRR_8, LRR_1, LRR_8, Pkinase | Leucine rich repeat N-terminal domain (PF08263); Leucine rich repeat (PF13855); Leucine rich repeat (PF00560); Leucine rich repeat (PF13855); Protein kinase domain (PF00069) |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, H.C.; Kim, K.-H.; Song, K.; Kim, J.Y.; Lee, B.-M. Identification and Validation of Candidate Genes Conferring Resistance to Downy Mildew in Maize (Zea mays L.). Genes 2020, 11, 191. https://doi.org/10.3390/genes11020191
Kim HC, Kim K-H, Song K, Kim JY, Lee B-M. Identification and Validation of Candidate Genes Conferring Resistance to Downy Mildew in Maize (Zea mays L.). Genes. 2020; 11(2):191. https://doi.org/10.3390/genes11020191
Chicago/Turabian StyleKim, Hyo Chul, Kyung-Hee Kim, Kitae Song, Jae Yoon Kim, and Byung-Moo Lee. 2020. "Identification and Validation of Candidate Genes Conferring Resistance to Downy Mildew in Maize (Zea mays L.)" Genes 11, no. 2: 191. https://doi.org/10.3390/genes11020191
APA StyleKim, H. C., Kim, K.-H., Song, K., Kim, J. Y., & Lee, B.-M. (2020). Identification and Validation of Candidate Genes Conferring Resistance to Downy Mildew in Maize (Zea mays L.). Genes, 11(2), 191. https://doi.org/10.3390/genes11020191






