Abstract
Abiotic stresses, such as salt and drought, significantly affect plant development and are the major limiting factors for crop quality and productivity. The manipulation of genes involved in plant stress response facilitates plant mitigation of adverse environments. In this study, we characterized CgbZIP1, a differentially expressed gene under normal and salinity conditions in Chrysanthemum grandiflora. CgbZIP1 was significantly upregulated by salt stress and also strongly responsive to drought stress and ABA treatments. Bioinformatics and subcellular localization analyses revealed that CgbZIP1 is a bZIP transcription factor and localized to the nucleus. Transgenic tobacco plants overexpressing CgbZIP1 exhibited significantly enhanced salt and drought stress tolerance associated with characteristic morphological and physiological indexes. The results demonstrate the important role CgbZIP1 plays in plant stress response and suggest its potential use in other crops for improved stress resistance.
1. Introduction
Plants living in nature face a variety of environmental adversities, including biotic (pests, diseases, weeds, among others) and abiotic (low temperature, drought, high salinity among others) stresses. Saline soil accounts for about 10% of the global cultivated area [1]. High salinity can damage plants from the toxic effect of excess sodium (Na+) and chlorine (Cl−) ions on critical biochemical processes. Sodium accumulation also leads to osmotic stress, thus reducing plant water utilization efficiency. Salt stress inhibits plant growth and, affects productivity and ornamental value of crops. For coping with saline environments, plants have developed various molecular, physiological, and biochemical mechanisms for adaptation [2,3,4]. For example, a recent study suggests that night-time transpiration and expansive leaf growth may be a potential acclimation mechanism under salt stress [5]. Changes in different biochemical and physiological processes also contribute to plant adaptation to a saline environment, such as regulated H+-ATPase activity in the plasma membrane, increased γ-aminobutyric acid shunt activity [6], and development of more effective N metabolism regulation [7].
The bZIP TFs are the most widely distributed and most conserved TFs in eukaryotes. At present, a large number of bZIP TFs have been identified in the genomes of different plant species, including Arabidopsis thaliana, tepary beans (Phaseolus acutifolius) and rice (Oryza sativa). They participate in various biological processes, including seed maturation, flower development, light signaling, pathogen defense and response to various stresses [8,9,10,11]. Particularly, accumulating evidence has demonstrated their involvement in regulating plant response to various environmental adversities. Overexpression of ZmbZIP72 TF in A. thaliana enhanced drought tolerance of transgenic plants [12]. Constitutive expression of a bZIP TF in tomato significantly increased plant tolerance to drought and salt stresses [13]. Overexpression of an endogenous bZIP TF gene, OsbZIP23, in O. sativa led to upregulation of hundreds of stress-responsive genes, largely in ABA-dependent stress response pathway, and thereby enhanced plant tolerance to salt and drought stresses [14].
Chrysanthemum includes traditional flower plants in China that are popular and widely cultivated. Chrysanthemum grandiflora is an important ornamental chrysanthemum in Northern China with many advantageous characteristics, including low plant height, concentrated flowering period, multiple flowers, rich colors, easy to manage, and strong resistance to certain stressful conditions. Despite its adaptation to the bitter winter in Northern China, its susceptibility to other environmental adversities, including drought and salinity, significantly impacts its ornamental value and wide application. Efforts have been made to better understand genetic determinants governing plant response to salt stress in Chrysanthemum and for the identification of different candidate genes which provide salt stress resistance. For example, the overexpression of DgWRKY4 and DgNAC1 enhances plant salt tolerance in both Nicotiana tabacum and Chrysanthemum [15,16,17].
At present, a large number of functional genes in many ornamental plants, such as Paeonia lutea [18], Helianthus annuus [19], Rose chinensis Jacq. [20], Rhododendron pulchurum [21], and Phalaenopsisi aphrodite [22] have been annotated by transcriptome sequencing, and explored to understand molecular mechanisms underlying ornamental plant growth and development, flower formation, and stress response. Understanding molecular mechanisms in plant response to various abiotic stresses in Chrysanthemum is the key to the development of novel biotechnology approaches to genetically improve Chrysanthemum species for enhanced performance under adverse environmental conditions. In this study, we analyzed TFs using transcriptome of C. grandiflora in response to salinity [23]. The whole-genome transcriptome profiling led to the identification and cloning of a bZIP TF gene, CgbZIP1 that was significantly induced by salt stress. Transgenic tobacco plants constitutively expressing CgbZIP1 exhibited significantly improved tolerance to salt and drought stresses, indicating the importance of the CgbZIP1 gene for use in genetically engineering new cultivars in Chrysanthemum and other crops with enhanced performance under environmental adversities.
2. Materials and Methods
2.1. Plant Materials and Growth Conditions
The wild-type seedlings of the C. grandiflora ‘9717’ preserved in the tissue culture room of the College of Landscape Architecture of Northeast Forestry University in Harbin were used as the plant material of this experiment, which originally came from Flower Research Center of Northeast Forestry University. The seedlings of the C. grandiflora were grown to a root length of about 2–3 cm in MS solid medium and then transplanted into a sterilized matrix prepared by mixing nutrient soil and vermiculite in a 1:1 ratio. Salt stress was applied 40 days after transplanting. According to the pre-experimental results, the NaCl concentration of 200 mmol·L−1 was selected, and the treatment was carried out every 5 days for 20 days. The untreated plants were used as controls. Each group consisted of three biological replicates, and the leaves were quickly placed in liquid nitrogen and then stored in a −80 °C refrigerator for RNA extraction.
2.2. Gene Expression Analysis
The six-leaf seedlings of the C. grandiflora plants grown in the MS solid medium were transferred to Hoagland nutrient solution, grew hydroponically for one week, and subjected to stress. Salt stress, drought stress and ABA stress were simulated in Hoggar culture medium with 200 mmol·L−1 NaCl, 20% PEG 6000 and 200 μmol·L−1 ABA, respectively. Samples were collected at 0 h, 1 h, 2 h, 4 h, 6 h, 12 h and 24 h with three biological replicates at each time point. The salinity and drought treatment, ABA concentration and treatment time duration were determined according to previous publications with minor adjustment [24,25,26]. RNA was extracted and analyzed by real-time quantitative PCR (RT-qPCR) using the SYBR Green Real-Time PCR Master Mix Kit from Beijing Dixon.
2.3. Cloning of CgbZIP1 Gene and Construction of Expression Vector
Total RNA was extracted from C. grandiflora leaves, and reverse transcription reaction was conducted using the ReverTra Ace® qPCR RT Master Mix with gDNA Remover Reverse Transcription cDNA Synthesis Kit (TOYOBO). The upstream and downstream primers, 5′-GGAAACCTTATGCTTCACAC-3′ and 5′-ACGGAGGGTA TCTAAATGCT-3′, designed based on the EST sequence of CgbZIP1 gene (accession number of MK986728) from the C. grandiflora transcriptome database, were used to amplify CgbZIP1 by PCR. The amplified CgbZIP1 gene was ligated to the pMD-18 T vector. The cloned CgbZIP1 gene was then inserted in a pBI121-GFP binary vector after a cauliflower mosaic virus 35S gene promoter (CaMV35S), linked to a CaMV35S-driven kanamycin resistance selectable marker gene (Kan), generating a plant expression vector, p35S/CgbZIP1-35S/Kan.
To construct a CgbZIP1–GFP fusion gene, we amplified the CgbZIP1 gene without the stop codon using 3′ and 5′ specific primers, 5′- GGTACCATGTCGAGGCAACCT CGACT -3′ and 5′- ACTAGTGTTAATATGAAGCTTGCCCA -3′ (underlined are restriction sites added, KpnI and SpeI). The amplified CgbZIP1 gene without a stop codon was ligated to the 5′ terminal of the GFP coding region in a pBI121-GFP binary vector after a cauliflower mosaic virus 35S gene promoter (CaMV35S), linked to a CaMV35S-driven kanamycin resistance selectable marker gene (Kan), generating a plant expression vector, p35S/CgbZIP1–GFP-35S/Kan.
2.4. Subcellular Localization of CgbZIP1 Protein
The CgbZIP1–GFP recombinant gene construct, p35S/CgbZIP1–GFP-35S/Kan and the control vector, p35S/GFP-p35S/Kan were introduced into the onion epidermis by the gene gun method, and the fluorescence was observed by a confocal laser scanning microscope (LSM 700, Zeiss, Germany).
2.5. Production and Identification of Transgenic Tobacco
Transgenic tobacco plants (Nicotiana tabacum cv K326) harboring p35S/CgbZIP1-35S/Kan were generated using Agrobacterium-mediated transformation of the leaf discs as previously described [27]. Putative transgenics were selected in a rooting screening medium supplemented with kanamycin, and the leaves of different transgenic lines were sampled for DNA and RNA extraction to confirm transgene insertion and expression.
2.6. Assessment of Stress Resistance in Transgenic Tobacco
Plant salt stress response was first evaluated by testing seed germination in culture medium. To this end, the seeds of the WT and transgenic lines T9, T10, and T11 were disinfected in sterile console, and uniformly seeded on the divided four areas in ½ MS solid medium supplemented with 150 mM NaCl. Seed germination and plant growth were observed and photographed 20 d after treatment.
Evaluation of plant response to salt and drought stresses in soil was started by first transplanting the well-grown seedlings of the WT and transgenic plants with 5–6 leaves from MS solid medium to soil and maintaining them for two weeks. Salt stress was then conducted by applying 200 mmol·L−1 NaCl every 5 days, and drought stress was applied by water withholding. Three biological replicates in each line for both salt and drought stresses were taken. Plant phenotypic changes were recorded by photograph.
Samples of stressed plants were taken as described above to measure the physiological parameters of both transgenic and WT tobacco plants. Superoxide dismutase (SOD), peroxidase (POD), and malondialdehyde (MDA) were measured using Superoxide Dismutase Activity Assay Kit, Peroxidase Activity Assay Kit, and Micro Malondialdehyde Assay Kit, respectively, following manufacturer’s instruction (Beijing Solarbio Science & Technology Co., Ltd., Beijing, China).
For stress treatment, there were six replicates for each line in every experiment for salt and drought stress test. Every experiment was repeated twice.
3. Results
3.1. Identification of Differentially Expressed TF Genes in C. grandiflora in Response to Salt Stress
In search for salinity-responsive TFs (TFs) among unigenes in the transcriptome of C. grandiflora previously obtained by RNA-seq [22], we identified a total of 1049 putative TFs. The TF gene families with five or more unigenes identified in the C. grandiflora transcriptome are displayed in Figure 1A, of which the top five are MYB-like (256), AP2/ERF (135), bHLH (120), WRKY (111), and bZIP (61).
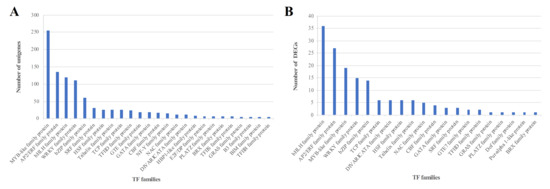
Figure 1.
(A) Distribution of transcription factors (TFs) in C. grandiflora’s transcriptome. (B) Distribution of transcription factors (TFs) differentially expressed in salt-treated and control plants of C. grandiflora.
Further analysis of the TFs identified in salt-treated and control plants revealed a total of 159 differentially expressed genes (DEGs) classified in 20 categories, of which the top five are bHLH (36.23%), AP2/ERF (27.17%), MYB (19.12%), WRKY (15.90%), and bZIP (14.90%) as shown in (Figure 1B). Members of different TF gene families, such as AP2/ERF, bHLH, MYB, WRKY, and bZIP play important regulatory roles in plant growth and development, as well as plant response to biotic and abiotic stress. Some of them, like NAC, WRKY, and MYB TFs, have previously been implicated in plant stress response in Chrysanthemum [28,29,30]. However, the role of bZIP TFs in plant stress response in Chrysanthemum remains largely unknown. The salt responsive DEGs in the bZIP TF gene family identified in Chrysanthemum were listed in Table 1; c26412.graph_c0, one of the bZIP TFs most significantly induced by salt stress, was chosen for further analysis and named CgbZIP1.

Table 1.
Annotation of differentially expressed bZIP TFs in salt-treated and control plants of C. grandiflora.
3.2. Molecular Characterization of CgbZIP1
We cloned CgbZIP1 by RT-PCR based on its EST sequence from the C. grandiflora transcriptome database. Its ORF is 867 bp long encoding a protein of 288 amino acids. The deduced polypeptide has a molecular weight (MW) of 32.32 kd and an isoelectric point (pI) of approximately 8.21. Protein sequence analysis revealed that CgbZIP1 has a coiled-coil domain, the primary and leucine zipper (bZIP) conserved region located at positions 162–206 (Figure 2A,B). Phylogenetic analysis based on the amino acid sequences of the CgbZIP1 protein and bZIP proteins from other plant species classifies CgbZIP1 protein to the group E Figure 2C [7], in the same branch with AtbZIP79 from Arabidopsis and close to SlbZIP64 from tomato (Solanum lycopersicum), and IbbZIP1 from (Ipomoea batatas). Of the bZIP proteins belonging to this group, StbZIP61, a TF in potato, involves in plant defense against Phytophthora infestans infection by regulating the biosynthesis of salicylic acid [31], and the sweet potato IbbZIP1 improves plant tolerance to drought and salt stress when overexpressed in Arabidopsis plants [32].
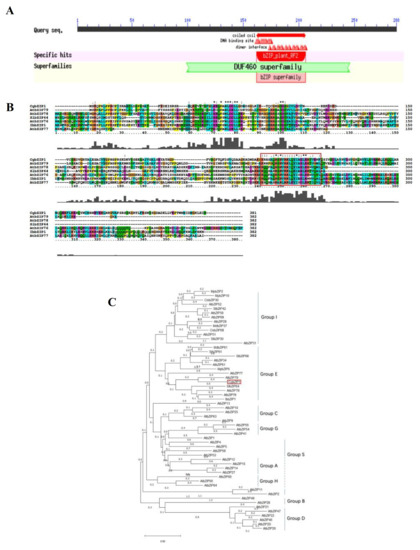
Figure 2.
Sequence analysis of CgbZIP1. (A). Prediction of the conserved domain of CgbZIP1. Conserved Domain Search Service on NCBI was used to identify the conserved domain of CgbZIP1. (B). Amino acids sequence alignment between CgbZIP1 and six homologous bZIP proteins from Arabidopsis, tomato, and sweet potato, including AtbZIP79 (NM120566.6), AtbZIP78 (NM001340558.1), SlbZIP64 (SGN-U572343), AtbZIP76 (NM001036128.2), IbbZIP1 (XP_019169177.1), and AtbZIP77 (NM001333158.1). The red frame shows the conserved basic leucine zipper (bZIP) domain. (C). Phylogenetic relationship of CgbZIP1 with other bZIP proteins. MAGA7 and DNAMAN software were used to compare multiple sequences and construct the phylogenetic tree by the Neighbor-Joining method [33].
3.3. Spatiotemporal Expression Pattern of CgbZIP1 Gene under Abiotic Stresses and ABA Treatment
The CgbZIP1 gene expression in the leaves of the C. grandiflora was significantly induced by salt stress. CgbZIP1 expression increased sharply 1 h after treatment, then declined, and increased again at 4 h and eventually became stabilized, but remained twice as high as that in the untreated control. Interestingly, CgbZIP1 expression in the roots was down-regulated compared with that in untreated control Figure 3A.
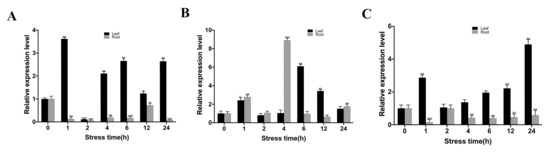
Figure 3.
(A). Expression pattern of CgbZIP1 under salt stress. (B). Expression pattern of CgbZIP1 under PEG drought stress. (C). Expression pattern of CgbZIP1 under ABA stress. Error bars represent SE. Asterisks (*, ** or ***) indicate a significant difference of relative expression level between different stress time points and controls at p < 0.05, 0.01, or 0.001, respectively, by the Student’s t-test.
Under drought stress simulated by PEG treatment, CgbZIP1 was upregulated 1h after treatment in leaves, then declined. It was significantly induced again 6 h after treatment, and then gradually went down but remained higher than the control 24 h after treatment. Similarly, CgbZIP1 was also upregulated in roots 1 h after treatment, then declined. Its expression rose again significantly 4 h after treatment, and then dropped sharply to the levels comparable to that in the controls Figure 3B.
When treated with the hormone ABA, the expression of CgbZIP1 gene was upregulated in the leaves of the C. grandiflora, and the trend was to rise first, then flatten and eventually increase steadily. At 2 h, the expression level was equivalent to that in the control and then continued to grow steadily at the following time points, reaching a peak at 24 h. The expression of CgbZIP1 in the roots was initially down-regulated compared with the control, dropping to the lowest at 1 h, only 1/5 of that in the control. At 2 h, the expression rose to the same level as that in the control, then declined to about half of that in the control for the remaining time points Figure 3C. Intriguingly, there was a discrepancy in gene expression between leaves and roots, induced vs. declined, triggered by various environmental stimuli, especially by salt stress Figure 3A and ABA treatment Figure 3C for the CgbZIP1 gene. This may be associated with different requirements of plant organs in CgbZIP1-mediated target gene expression responding to environmental stimuli. Similar observations have also been reported in other genes and plant species. For example, HKT Na+ transporter genes play an important role in plant salt stress response. The levels of expression of the different rice HKT Na+ transporter genes exhibit differentiated expression patterns in leaves and roots upon salt stress [34].
3.4. Subcellular Localization of CgbZIP1
The CgbZIP1 gene, registered in NCBI with an accession number of MK986728, has an open reading frame (ORF) of 867 bp encoding 288 amino acids. To examine the subcellular localization of the CgbZIP1 protein, the CgbZIP1–GFP fusion gene construct, p35S/CgbZIP1–GFP-p35S/Kan, and the control vector, p35S/GFP-p35/Kan were introduced into the onion epidermis. As shown in Figure 4, the fluorescent signal of the PBI121-CgbZIP1–GFP protein was only found in the nucleus, however, the fluorescent signal of the positive control was present throughout the cell. This confirmed that the CgbZIP1 protein localizes to the nucleus.
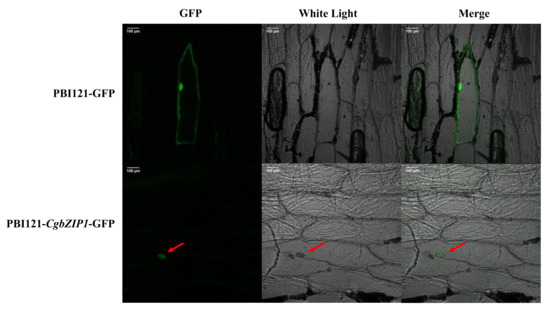
Figure 4.
Subcellular localization of CgbZIP1 in onion epidermal cells. Genes encoding GFP and CgbZIP1–GFP fusion proteins were driven by the CaMV35S promoter. Nuclear localized signals of the CgbZIP1–GFP fusion protein were indicated by arrows. Bars = 100 µm.
3.5. Production of Transgenic Tobacco Overexpressing CgbZIP1
To examine whether or not CgbZIP1 is involved in plant stress response, we generated transgenic tobacco plants harboring the CgbZIP1 expression vector, p35S/CgbZIP1-35S/Kan. PCR analysis of genomic DNA from a total of 9 putative transformation events confirmed the integration of the transgene into the host genome (see example in Figure 5A), and RT-PCR analysis demonstrated the expression of CgbZIP1 in all transgenic lines (see example in Figure 5B). There were no observable differences between transgenics and wild-type controls in plant morphology and development under normal conditions. Four homozygous TG lines, T9, T10, T11 and T17, representative of all the independent TG lines obtained, were chosen for further analysis.
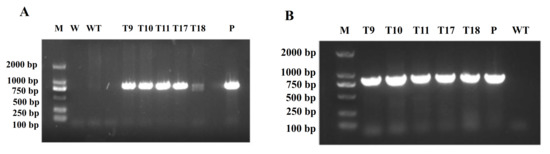
Figure 5.
(A) PCR analysis of plant genomic DNA to confirm the presence of CgbZIP1 in transgenic tobacco genome. M: Molecular-weight standard; W: water; WT: wild-type; P: pBI121-CgbZIP1–GFP plasmid DNA. (B) RT-PCR analysis of CgbZIP1 expression in transgenic lines and wild-type (WT) controls. M: Molecular-weight standard; T9,10,11,17,18: CgbZIP1 transgenic lines; P: pBI121-CgbZIP1–GFP.
3.6. Transgenic Plants Overexpressing CgbZIP1 Exhibit Enhanced Tolerance to Salt Stress
To investigate how CgbZIP1 expression would impact plant response to salt stress, we first examined seed germination in ½ MS solid medium prepared using 150 mM NaCl solution instead of water. The seeds from transgenic tobacco lines 9, 10, and 11 and the wild-type controls were simultaneously sown in the same medium divided into different regions. Although the germination rates and growth of the three transgenic lines were slightly different, they all performed significantly better than wild-type controls (Figure 6A).
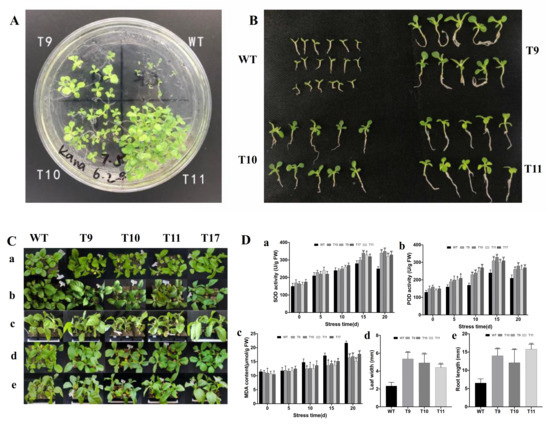
Figure 6.
Comparison of the seed germination and growth between wild-type (WT) and transgenic (TG) plants under salinity stress. (A). Germination of WT and TG seeds on ½ MS medium containing 150 mM NaCl for 20 d. (B). Comparison of the WT and TG tobacco seedlings from (A) 20 d after salt stress. (C). Phenotypic characteristics of the TG and WT control plants under salt stress. (a): 0 d, top view; (b,c): 10 d after salt stress, top view and side view, respectively; (d,e): 20 d after salt stress, top view and side view, respectively. (D). Physiological parameters associated with plant stress response measured at various time points after salt treatment. (a) Effect of salt stress on plant SOD activity. (b) Effect of salt stress on plant POD activity. (c) Effect of the salt stress on plant MDA content. (d) Effect of salt stress on leaf width of the tobacco seedlings. (e) Effect of the salt stress on root length of the tobacco seedlings. Each column represents mean of three biological replicates. Error bars represent SE. A significant difference between WT and each TG line at p < 0.05, 0.01 or 0.001 by Student’s t-test was represented as “*”, “**” or “***”, respectively.
Under salt stress, the growth of wild-type plants was significantly inhibited, the root length was significantly shorter than that of the transgenic plants, and the roots had no branches. Wild-type controls had only two true leaves, whereas the transgenic plants had four true leaves. The wild-type leaves were yellow, whereas the leaves in transgenic plants were green and significantly larger than the wild-type controls Figure 6B.
The involvement of CgbZIP1 gene in plant salt tolerance was further studied by assessing the performance of both the CgbZIP1 transgenic plants and wild-type controls grown in soil and subjected to 200 mmol·L−1 NaCl treatment every 5 d. As shown in Figure 6C, the CgbZIP1 transgenic plants were largely similar to wild-type under normal conditions, but significantly superior to wild-type controls in both plant height and growth status under salt-stressed conditions.
In order to study the role CgbZIP1 plays in plant response to salt stress, several physiological parameters including SOD, POD, MDA were analyzed as shown in Figure 6D. The three physiological parameters showed similar trends in change between normal and salt stress conditions. In the absence of stress, the SOD activity was similar between wild-type and transgenic plants. When subjected to stressful conditions, however, with the prolonged stress time, the SOD activity in wild-type controls increased first and then rapidly declined, whereas that of the transgenic plants increased steadily. Similar patterns of the change in the POD activity between wild-type and transgenic plants were also observed. The MDA content in wild-type controls gradually increased with the prolonged stress time and was always higher than that in transgenic plants. The leaf width and root length in transgenic lines were both significantly higher than those in wild-type controls.
3.7. Transgenic Plants Overexpressing CgbZIP1 Exhibit Enhanced Tolerance to Drought Stress
To study how overexpression of CgbZIP1 would impact plant response to water deficiency, drought stress was applied to both wild-type and CgbZIP1 transgenic plants by water withholding for 20 days. As shown in Figure 7A, although both impacted by the stressful condition, transgenic plants performed much better than wild-type controls. Most transgenic plants grew normally, whereas the wild-type controls all wilted.
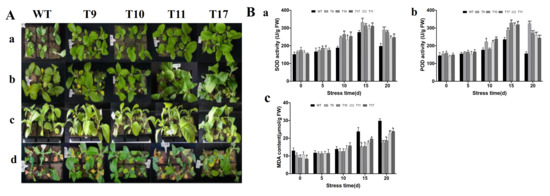
Figure 7.
(A). Performance of transgenic (TG) and wild-type (WT) control plants under drought stress. (a): 0 d, top view; (b,c): 10 d after water withholding, top view and side view; (d): 20 d after water withholding, top view. (B). Physiological parameters associated with plant stress response measured at various time points after drought stress. (a) Effect of the drought stress on plant SOD activity. (b) Effect of the drought stress on plant POD activity. (c) Effect of drought stress on plant MDA content. Each column represents the mean of three biological replicates. Error bars represent SE. A significant difference between WT and each TG line at p < 0.05, 0.01 or 0.001 by the Student’s t-test was represented as “*”, “**” or “***”, respectively.
We further compared several physiological parameters, including SOD, POD, and MDA, between wild-type and transgenic plants. As shown in Figure 7B. The three physiological parameters displayed similar trends in change between normal and drought stress conditions. In normal conditions, the SOD activity, POD activity and MDA content were all similar in both wild-type and transgenic lines. After 5, 10 and 15 d of drought treatment, the activities of SOD, POD in transgenic lines were significantly higher than those in wild-type controls, while the content of MDA was lower in transgenic lines than in wild-type control plants.
4. Discussion
4.1. TF Analysis Based on Transcriptome in Plants under Salt Stress
Based on high-throughput transcriptome sequencing data analyzing global gene expression in salt-stressed C. grandiflora plants and their controls, we have compared differentially expressed regulatory genes in the control group and the salt-stressed group, revealing the possible involvement of several families of TFs in plant response to salt stress in C. grandiflora. We found 159 DEGs can be classified in 20 TF families. While genes from most of these families, including MYB, bHLH, and AP2/ERF have been demonstrated to be involved in plant response to salinity [35,36,37,38,39], less study has been conducted on genes from the TF families like BRX, Dof and GRAS. In our transcriptome data, a high percentage of DEGs enriched in the plant hormone signaling pathway was found by comparing the DEGs between the control and salt-stressed groups. This indicates that the plant hormone signal transduction pathway plays an important role in the plant stress response, and is worth further investigation into related genes. In the plant hormone signal transduction pathway, the largest number of differentially expressed genes are related to abscisic acid (ABA) signaling. In the ABA signal recognition and transduction pathway, seven PYR/PYL genes and three genes in SnRK2 family were down-regulated, while seven PP2C genes and two ABF genes were upregulated. PYR/PYL proteins are a type of ABA receptors and signal regulators at the most upstream of the ABA signaling pathway [40]. Type 2C protein phosphatases (PP2C) are negative regulators of ABA signal transduction, and SnRK2 protein kinases are positive regulators of ABA signal transduction. These three core components constitute a double negative regulator in the ABA signal transduction system. By regulating these three signal factors in the ABA signaling pathway, plants can adjust hormone levels to respond when they are under stress [41]. Type 2C protein phosphatases negatively regulate the ABA response. Almost all PP2C members in Arabidopsis and Oryza sativa are upregulated in response to abiotic stresses such as cold, heat, PEG and salt treatments [42], consistent with the observation in our research. ABFs, the ABRE-binding proteins, are TFs in the Arabidopsis alkaline region leucine zipper (bZIP) TF family that recognize and bind to the abscisic acid response element in the promoter and regulate the expression of downstream stress response genes in ABA signaling. They demonstrated a key role in salinity stress response in various plant species, such as cotton [43], Oryza sativa [13] and rapeseed [44]. TFs that regulate the expression of different target genes thus play an important role in plant abiotic stress control.
4.2. Molecular Mechanism of the bZIP TF-Mediated Plant Response to Abiotic Stress
Abiotic stress impairs plant growth by causing various damages at the cellular level [45]. TFs play an important role in various plant biological processes. TFs such as MYC/MYB, basic leucine zipper (bZIP), and WRKY interact with specific and conserved DNA sequences (corresponding regulatory elements) in the promoter of abiotic stress-responsive genes, regulating at the transcriptional level their expression. Plant-specific bZIP TFs are among the transcriptional regulators associated with abiotic stress conditions. A number of bZIP genes have previously been shown to be transcriptionally responsive to drought and osmotic stress [46,47,48]. Some of them have been implicated in abiotic stress response and ABA signaling in various species [49,50,51]. Consistent with this, in our study, real-time quantitative PCR was used to examine the expression of CgbZIP1 gene in roots and leaves of C. grandiflora under three different abiotic stresses revealed that the CgbZIP1 gene responded significantly to salt, drought and ABA stresses.
In this study, drought and salt treatments were applied to transgenic tobacco overexpressing the CgbZIP1 gene of C. grandiflora. Transgenic tobacco exhibited higher antioxidant enzyme activity, lower MDA content, and better growth response under salt and drought stress conditions, indicating an improved tolerance to salt and drought stress compared to wild-type tobacco. Similar results that bZIP TFs positively regulate plant abiotic stress response have also been reported in other species. For example, the transgenic Arabidopsis overexpressing a wheat bZIP TF gene, TabZIP showed healthier plant growth than wild-type controls under both salt and drought stress conditions [52]. Heterologous overexpression of the TF, ThbZIP1 from Tamarix hispida in transgenic tobacco led to changes in stress-related parameters such as malondialdehyde, superoxide dismutase, peroxidase and chlorophyll content and enhanced plant tolerance to salt and drought stress, but increased ABA sensitivity [49]. Ying et al. [11] overexpressed the maize ZmbZIP72 TF in Arabidopsis and observed enhanced drought and salt stress resistance associated with changes in various physiological parameters, including electrolyte leakage, proline content, and water retention capacity in transgenic lines. Our study in C. grandiflora provides new evidence substantiating the role some bZIP TFs play in positively regulating plant response to salt and drought stress. The fact that CgbZIP1 expression is spatiotemporally regulated by ABA Figure 3C suggests that the CgbZIP1-mediated plant stress response likely involves an ABA-dependent pathway.
5. Conclusions
In conclusion, we cloned and studied a bZIP TF gene, CgbZIP1, from C. grandiflora. Heterologous expression of CgbZIP1 in tobacco led to enhanced plant tolerance to salt and drought stress, suggesting its potential use as a candidate in other crop species for genetically engineering new cultivars better adapting to adverse environments. To our knowledge, this is the first bZIP TF from C. grandiflora implicated in plant stress response. Its further characterization in unravelling the molecular mechanisms underlying CgbZIP1-mediated plant response to abiotic stress would allow additional new strategies to facilitate the molecular breeding of crops for improved plant stress resistance and enhanced agriculture productivity.
Author Contributions
Y.Z. and M.H. designed the experiments. L.W. cloned the bZIP gene; L.W., C.L., H.L. (He Liu) and H.Y. participated in sample collection and physiological studies; Y.L. analyzed the data and wrote the manuscript, H.L. (Hong Luo) reviewed and edited the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This research was supported by the National Natural Science Foundation of China (31870687), and the National Key Research and Development Program of China (grant number 2019YFD1001500 and 2018YFD1000406).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
All data generated or analysed during this study are included in this published article. The accession number for CgbZIP1 is MK986728 on the NCBI database; the link is https://www.ncbi.nlm.nih.gov/nuccore/MK986728.1/ (accessed on 1 January 2022). All methods were carried out in accordance with relevant guidelines and regulations.
Acknowledgments
The support provided by China Scholarship Council (CSC) during a visit of Yu Liu to Clemson is acknowledged.
Conflicts of Interest
The authors declare that they have no conflict of interests.
References
- Sun, J.; Yang, R.; Li, W.; Pan, Y.; Zheng, M.; Zhang, Z. Effect of biochar amendment on water infiltration in a coastal saline soil. J. Soils Sediments 2018, 18, 3271–3279. [Google Scholar] [CrossRef]
- Zelm, E.V.; Zhang, Y.; Testerink, C. Salt Tolerance Mechanisms of Plants. Annu. Rev. Plant Biol. 2020, 71, 403–433. [Google Scholar] [CrossRef] [Green Version]
- Ayaz, A.; Huang, H.; Zheng, M.; Zaman, W.; Li, D.; Saqib, S.; Zhao, H.; Lü, S. Molecular Cloning and Functional Analysis of GmLACS2-3 Reveals Its Involvement in Cutin and Suberin Biosynthesis along with Abiotic Stress Tolerance. Int. J. Mol. Sci. 2021, 22, 9175. [Google Scholar] [CrossRef] [PubMed]
- Manghwar, H.; Hussain, A.; Ali, Q.; Liu, F. Brassinosteroids (BRs) Role in Plant Development and Coping with Different Stresses. Int. J. Mol. Sci. 2022, 23, 1012. [Google Scholar] [CrossRef] [PubMed]
- Fricke, W. Energy costs of salinity tolerance in crop plants: Night-time transpiration and growth. New Phytol. 2020, 225, 1152–1165. [Google Scholar] [CrossRef]
- Che-Othman, M.H.; Jacoby, R.P.; Millar, A.H.; Taylor, N.L. Wheat mitochondrial respiration shifts from the tricarboxylic acid cycle to the GABA shunt under salt stress. New Phytol. 2020, 225, 1166–1180. [Google Scholar] [CrossRef]
- de la Torre-González, A.; Navarro-León, E.; Blasco, B.; Ruiz, J.M. Nitrogen and photorespiration pathways, salt stress genotypic tolerance effects in tomato plants (Solanum lycopersicum L.). Acta Physiol Plant. 2020, 42, 1–8. [Google Scholar] [CrossRef]
- Jakoby, M.; Weisshaar, B.; Dröge-Laser, W.; Vicente-Carbajosa, J.; Tiedemann, J.; Kroj, T.; Parcy, F. bZIP transcription factors in Arabidopsis. Trends Plant Sci. 2002, 7, 106–111. [Google Scholar] [CrossRef]
- Rodriguez-Uribe, L.; O’Connell, M.A. A root-specific bZIP transcription factor is responsive to water deficit stress in tepary bean (Phaseolus acutifolius) and common bean (P. vulgaris). J. Exp. Bot. 2006, 57, 1391–1398. [Google Scholar] [CrossRef]
- Schütze, K.; Harter, K.; Chaban, C. Post-translational regulation of plant bZIP factors. Trends Plant Sci. 2008, 13, 247–255. [Google Scholar] [CrossRef]
- Amir Hossain, M.; Lee, Y.; Cho, J.I.; Ahn, C.H.; Lee, S.K.; Jeon, J.S.; Kang, H.; Lee, C.H.; An, G.; Park, P.B. The bZIP transcription factor OsABF1 is an ABA responsive element binding factor that enhances abiotic stress signaling in rice. Plant Mol. Biol. 2010, 72, 557–566. [Google Scholar] [CrossRef] [PubMed]
- Ying, S.; Zhang, D.F.; Fu, J.; Shi, Y.S.; Song, Y.C.; Wang, T.Y.; Li, Y. Cloning and characterization of a maize bZIP transcription factor, ZmbZIP72, confers drought and salt tolerance in transgenic Arabidopsis. Planta 2012, 235, 253–266. [Google Scholar] [CrossRef] [PubMed]
- Hsieh, T.H.; Li, C.W.; Su, R.C.; Cheng, C.P.; Sanjaya Tsai, Y.C.; Chan, M.T. A tomato bZIP transcription factor, SlAREB, is involved in water deficit and salt stress response. Planta 2010, 231, 1459–1473. [Google Scholar] [CrossRef]
- Xiang, Y.; Tang, N.; Du, H.; Ye, H.; Xiong, L. Characterization of OsbZIP23 as a key player of the basic leucine zipper transcription factor family for conferring abscisic acid sensitivity and salinity and drought tolerance in rice. Plant Physiol. 2008, 148, 1938–1952. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, K.; Wu, Y.H.; Tian, X.Q.; Bai, Z.Y.; Liang, Q.Y.; Liu, Q.L.; Pan, Y.Z.; Zhang, L.; Jiang, B.B. Overexpression of DgWRKY4 Enhances Salt Tolerance in Chrysanthemum Seedlings. Front Plant Sci. 2017, 8, 1592. [Google Scholar] [CrossRef] [Green Version]
- Wang, K.; Zhong, M.; Wu, Y.H.; Bai, Z.Y.; Liang, Q.Y.; Liu, Q.L.; Pan, Y.Z.; Zhang, L.; Jiang, B.B.; Jia, Y.; et al. Overexpression of a chrysanthemum transcription factor gene DgNAC1 improves the salinity tolerance in chrysanthemum. Plant Cell Rep. 2017, 36, 571–581. [Google Scholar] [CrossRef]
- Liu, Q.L.; Xu, K.D.; Zhao, L.J.; Pan, Y.Z.; Jiang, B.B.; Zhang, H.Q.; Liu, G.L. Overexpression of a novel chrysanthemum NAC transcription factor gene enhances salt tolerance in tobacco. Biotechnol. Lett. 2011, 33, 2073–2082. [Google Scholar] [CrossRef]
- Zhou, L.; Shi, Q.; Qi, Y.; Miao, K.; Wang, Y. Transcriptome Analysis of Paeonia lutea and Its Differentially Expressed Genes Screening at Different Flower Opening Stages. Molecular Plant Breeding. 2018. Available online: http://kns.cnki.net/kcms/detail/46.1068.s.20181008.0928.016.html (accessed on 1 January 2022).
- Guo, S.C. Tanscriptome Sequencing Analysis of Sunflower Responsive to Verticillium Dahliae Infection and Mining of Resistance-related Genes. Ph.D. Thesis, Inner Mongolia University, Inner Mongolia, Hohhot, China, March 2017. [Google Scholar]
- Liu, C. Transcriptome Dynamics and Functions of RhIAA16 Genes during Rose Petal Abscission. Ph.D. Thesis, China Agricultural University, Beijing, China, September 2016. [Google Scholar]
- Wang, H.; Wang, W.; Wang, D.; Zhang, S.; Hu, X.; Lu, S.; Gong, X. De novo assembly and functional annotation of transcriptome data of Rhododendron pulchurum cv. Baifeng 4 leaf. Acta Agric. Zhejiangensis 2018, 30, 1149–1159. [Google Scholar]
- Zhang, J.; Shi, X.; Liu, H.; Ma, G.; Zou, Q.; Zhu, K.; Zhou, J.; Mao, J. Study on the Differential Accumulation of Anthocyanin in Different-colored Phalaenopsis Based on Transcriptomics. Mol. Plant Breed. 2018, 16, 4530–4542. [Google Scholar]
- Wang, L.; Yang, Y.; Liu, Y.; Yang, L.; Liu, Y.; Zhou, Y. Transcriptome analysis of Chrysanthemum × grandiflora in salt stress based on high-through-put sequencing. Mol. Plant Breed. 2020, 18, 1419–1427. [Google Scholar]
- Gao, W.J.; He, M.; Liu, J.; Ma, X.; Zhang, Y.; Dai, S.; Zhou, Y. Overexpression of Chrysanthemum lavandulifolium ClCBF1 in Chrysanthemum morifolium White Snow improves the level of salinity and drought tolerance. Plant Physiol. Biochem. 2018, 124, 50–58. [Google Scholar] [CrossRef] [PubMed]
- Fan, Q.; Song, A.; Jiang, J.; Zhang, T.; Sun, H.; Wang, Y.; Chen, S.; Chen, F. CmWRKY1 enhances the dehydration tolerance of chrysanthemum through the regulation of ABA-associated genes. PLoS ONE 2016, 11, e0150572. [Google Scholar] [CrossRef] [Green Version]
- Hong, S.; Chen, S.; Jiang, J.; Chen, F.; Chen, Y.; Gu, C.; Li, P.; Song, A.; Zhu, X.; Gao, H.; et al. Heterologous expression of the chrysanthemum r2r3-myb transcription factor cmmyb2 enhances drought and salinity tolerance, increases hypersensitivity to aba and delays flowering in arabidopsis thaliana. Mol. Biotechnol. 2012, 51, 160–173. [Google Scholar]
- Gallois, P.; Marinho, P. Leaf disk transformation using Agrobacterium tumefaciens-expression of heterologous genes in tobacco. Methods Mol. Biol. 1995, 49, 39–48. [Google Scholar] [CrossRef]
- Liu, Y.; He, M.; Dong, F.; Cai, Y.; Gao, W.; Zhou, Y.; Dai, S. The Chrysanthemum lavandulifolium ClNAC9 gene positively regulates saline, alkaline and drought stress in transgenic Chrysanthemum grandiflorum. J. Am. Soc. Hortic. Sci. 2019, 144, 280–288. [Google Scholar] [CrossRef] [Green Version]
- Liang, Y.; Li, X.; Zhang, D.; Gao, B.; Yang, H.; Wang, Y.; Guan, K.; Wood, A.J. ScDREB8, a novel A-5 type of DREB gene in the desert moss Syntrichia caninervis, confers salt tolerance to Arabidopsis. Plant Physiol. Biochem. 2017, 120, 242–251. [Google Scholar] [CrossRef]
- Liu, X.F.; Xiang, L.L.; Yin, X.R.; Grierson, D.; Li, F.; Chen, K.S. The identification of a MYB transcription factor controlling anthocyanin biosynthesis regulation in Chrysanthemum flowers. Sci. Hortic. 2015, 194, 278–285. [Google Scholar] [CrossRef]
- Zhou, X.T.; Jia, L.J.; Wang, H.Y.; Zhao, P.; Wang, W.Y.; Liu, N.; Song, S.W.; Wu, Y.; Su, L.; Zhang, J.; et al. The potato transcription factor StbZIP61 regulates dynamic biosynthesis of salicylic acid in defense against Phytophthora infestans infection. Plant J. 2018, 95, 1055–1068. [Google Scholar] [CrossRef] [Green Version]
- Kang, C.; Zhai, H.; He, S.; Zhao, N.; Liu, Q. A novel sweetpotato bZIP transcription factor gene, IbbZIP1, is involved in salt and drought tolerance in transgenic Arabidopsis. Plant Cell Rep. 2019, 38, 1373–1382. [Google Scholar] [CrossRef] [Green Version]
- Yuan, C.; Shi, J.; Zhao, L. The CmbZIP1 transcription factor of chrysanthemum negatively regulates shoot branching. Plant Physiol. Biochem. 2020, 151, 69–76. [Google Scholar] [CrossRef] [PubMed]
- Khan, I.; Mohamed, S.; Regnault, T.; Mieulet, D.; Guiderdoni, E.; Sentenac, H.; Véry, A.-A. Constitutive Contribution by the Rice OsHKT1;4 Na+ Transporter to Xylem Sap Desalinization and Low Na+ Accumulation in Young Leaves Under Low as High External Na+ Conditions. Front. Plant Sci. 2020, 11, 1130. [Google Scholar] [CrossRef] [PubMed]
- Tang, Y.; Bao, X.; Zhi, Y.; Wu, Q.; Guo, Y.; Yin, X.; Zeng, L.; Li, J.; Zhang, J.; He, W.; et al. Overexpression of a MYB Family Gene, OsMYB6, Increases Drought and Salinity Stress Tolerance in Transgenic Rice. Front. Plant Sci. 2019, 10, 168. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Y.B.; Wei, T.; Wang, L.H.; Hu, Y.W.; Liu, X.W.; Liu, Y.S. Kiwifruit (Actinidia chinensis) R1R2R3-MYB transcription factor AcMYB3R enhances drought and salinity tolerance in Arabidopsis thaliana. J. Integr. Agric. 2019, 18, 417–427. [Google Scholar] [CrossRef]
- Sun, X.; Wang, Y.; Sui, N. Transcriptional regulation of bHLH during plant response to stress. Biochem. Biophys. Res. Commun. 2018, 503, 397–401. [Google Scholar] [CrossRef]
- Bouaziz, D.; Pirrello, J.; Ben Amor, H.; Hammami, A.; Charfeddine, M.; Dhieb, A.; Bouzayen, M.; Gargouri-Bouzid, R. Ectopic expression of dehydration responsive element binding proteins (StDREB2) confers higher tolerance to salt stress in potato. Plant Physiol. Biochem. 2012, 60, 98–108. [Google Scholar] [CrossRef] [Green Version]
- Li, H.; Zhang, D.; Li, X.; Guan, K.; Yang, H. Novel DREB A-5 subgroup transcription factors from desert moss (Syntrichia caninervis) confers multiple abiotic stress tolerance to yeast. J. Plant Physiol. 2016, 194, 45–53. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D. An Abscisic Acid Signaling Pathway Starting from Plastid/Chloroplast. Chin. Bull. Bot. 2011, 46, 361–369. [Google Scholar]
- Cutler, S.R.; Rodriguez, P.L.; Finkelstein, R.R.; Abrams, S.R. Abscisic acid: Emergence of a core signaling network. Annu. Rev. Plant Biol. 2010, 61, 651–679. [Google Scholar] [CrossRef] [Green Version]
- Cao, J.; Jiang, M.; Li, P.; Chu, Z. Genome-wide identification and evolutionary analyses of the PP2C gene family with their expression profiling in response to multiple stresses in Brachypodium distachyon. BMC Genom. 2016, 17, 175. [Google Scholar] [CrossRef] [Green Version]
- Liang, C.; Meng, Z.; Meng, Z.; Malik, W.; Yan, R.; Lwin, K.M.; Lin, F.; Wang, Y.; Sun, G.; Zhou, T.; et al. GhABF2, a bZIP transcription factor, confers drought and salinity tolerance in cotton (Gossypium hirsutum L.). Sci. Rep. 2016, 6, 35040. [Google Scholar] [CrossRef] [Green Version]
- Zhao, B.Y.; Hu, Y.F.; Li, J.J.; Yao, X.; Liu, K.D. BnaABF2, a bZIP transcription factor from rapeseed (Brassica napus L.), enhances drought and salt tolerance in transgenic Arabidopsis. Bot. Stud. 2016, 57, 12. [Google Scholar] [CrossRef] [Green Version]
- Llorca, C.M.; Potschin, M.; Zentgraf, U. bZIPs and WRKYs: Two large transcription factor families executing two different functional strategies. Front. Plant Sci. 2014, 5, 169. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, X.; Chu, Z. Genome-wide evolutionary characterization and analysis of bZIP transcription factors and their expression profiles in response to multiple abiotic stresses in Brachypodium distachyon. BMC Genom. 2015, 16, 227. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, J.; Chen, N.; Chen, F.; Cai, B.; Dal Santo, S.; Tornielli, G.B.; Pezzotti, M.; Cheng, Z.M. Genome-wide analysis and expression profile of the bZIP transcription factor gene family in grapevine (Vitis vinifera). BMC Genom. 2014, 15, 281. [Google Scholar] [CrossRef] [Green Version]
- Baloglu, M.C.; Eldem, V.; Hajyzadeh, M.; Unver, T. Genome-wide analysis of the bZIP transcription factors in cucumber. PLoS ONE 2014, 9, e96014. [Google Scholar] [CrossRef] [Green Version]
- Ji, X.; Liu, G.; Liu, Y.; Zheng, L.; Nie, X.; Wang, Y. The bZIP protein from Tamarix hispida, ThbZIP1, is ACGT elements binding factor that enhances abiotic stress signaling in transgenic Arabidopsis. BMC Plant Biol. 2013, 13, 151. [Google Scholar] [CrossRef] [Green Version]
- Yoshida, T.; Fujita, Y.; Maruyama, K.; Mogami, J.; Todaka, D.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Four Arabidopsis AREB/ABF transcription factors function predominantly in gene expression downstream of SnRK2 kinases in abscisic acid signalling in response to osmotic stress. Plant Cell Environ. 2015, 38, 35–49. [Google Scholar] [CrossRef] [Green Version]
- Zhang, L.; Zhang, L.; Xia, C.; Zhao, G.; Liu, J.; Jia, J.; Kong, X. A novel wheat bZIP transcription factor, TabZIP60, confers multiple abiotic stress tolerances in transgenic Arabidopsis. Physiol. Plant. 2015, 153, 538–554. [Google Scholar] [CrossRef]
- Agarwal, P.; Baranwal, V.K.; Khurana, P. Genome-wide Analysis of bZIP Transcription Factors in wheat and Functional Characterization of a TabZIP under Abiotic Stress. Sci. Rep. 2019, 9, 4608. [Google Scholar] [CrossRef] [PubMed] [Green Version]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).