Abstract
Juvenile and vegetative adult shoot apical meristems (SAM) are actively involved in acquisition of flowering competence, while the embryonic SAM is regarded as “responsible” only for germination. Comparative analyses of imbibed and stratified seeds of garlic Allium sativum show that only stratified seedlings produced bulbs and flower stems at the end of the season. Since the seed morphology of stratified and non-stratified seeds prior to sowing was similar, the differences are attributed to the molecular alterations in the embryonic SAM. Functional annotation analysis of 3000 differentially expressed genes suggests that seed imbibition reactivates the embryonic cell cycle, initiates the metabolism, and primes garlic seed germination. Stratification enhances DNA modifications, biosynthesis, cellular transport, and tissue development. Similar to vernalization of the vegetative buds, stratification of the embryonic SAM resulted in altered expression of meristem-identity and flowering homologs. Phase transitions from seed germination to flowering and bulbing in A. sativum are tightly connected, and possibly associated with downregulation of specific flowering repressor(s). The embryonic SAM plays an important role not only in seed germination, but in the entire plant life cycle, providing the foundation for the genetic regulation of major functional shifts in metabolism and development.
1. Introduction
The regulation of plant developmental transitions and organ growth are carefully orchestrated by internal and environmental signals. The combined effect of multiple cues affects the timing of the progress from seed germination to completion of the life cycle [1]. Post-embryonic development proceeds through the successive juvenile, adult vegetative, and reproductive phases. Although morphologically distinct, these phases are linked by the same regulatory networks. It is generally accepted that plant transition from juvenility to adulthood is accompanied by the acquisition of reproductive competence, followed by morphological makeover [2]. However, the embryonic shoot apical meristem (SAM) lacks the competence to develop reproductive organs, and such a transition is possible only in the post-juvenile phase [3].
In temperate environments, seed dormancy release and germination often require a prolonged exposure to cold, wet conditions, called stratification [4]. Similarly, in order to be competent to flower, many temperate plants must undergo exposure to a prolonged cold period termed vernalization [5]. The co-existence of these processes in the life cycle suggest that natural selection for the adaptation to cold winter should act jointly in seeds and vegetative buds. Traditionally, most of the research on stratification and vernalization mechanisms was carried out separately, but recent reports have established associations between seed germination and flowering in Arabidopsis and other plants [6,7,8]. Since adaptation to long winters evolved several times in the course of plant evolution, various regulatory mechanisms were developed in different plant groups [1]. In general, the accumulation of chilling hours regulates changes in water status, hormonal balance, respiration, and carbohydrate mobilization [9], and consequent increases in plant sensitivity to photoperiodic changes [5].
Such mechanisms are especially important in geophytes—perennial herbs that survive unfavorable periods as underground dormant storage organs—such as bulbs, corms, and rhizomes. Geophytes were metaphorically named “plant computers” because their long-term environment-dependent “memory” strongly affects life cycle and phase transitions to flowering and storage-organ formation [10,11]. During harsh weather in the course of the annual cycle, adult geophytes rest as storage organs, but when the environment becomes favorable for growth, storage organs provide the energy required for re-sprouting and initial growth [12]. The physiology of economically important geophytes (e.g., onion, tulip, lily), has been thoroughly studied [10,13], yet there is only limited understanding of the molecular regulation of their life processes and phase transitions [14,15,16,17].
Many economically important geophytes are clonally propagated (e.g., potatoes, garlic, shallots, ornamental bulbs, and corms), therefore, their seed biology got only a little attention. Most spring-flowering geophytes depend on 6–8 weeks at 4–10 °C germination-inducing stratification, but the seed of geophytes from warm regions do not require low temperatures for germination [18]. In many Allium and Tulipa species that have evolved in Central Asia, seeds become dormant on maturation, and their germination requires cold stratification [19,20,21]. When grown from seeds, most genera undergo an extended period of juvenility of up to seven years before they can produce flowers [22]. In post-juvenile geophytes, vernalization is required for meristem transition, and then for both bulbing and flowering [9,10,23].
Garlic Allium sativum, an important bulbous geophyte, responds to thermoperiodic cues, thus the timing and performance of its annual life cycle depends on cue from the surrounding environment. In many garlic genotypes, vernalization is required for the transition of SAM from vegetative to reproductive state, for the growth of axillary buds and for bulbing [24]. All commercial garlic cultivars do not produce seeds and are propagated only vegetatively. Recent fertility restoration of bolting genotypes opened the way for garlic breeding, with the potential for the production of garlic from true seeds [24,25]. To accomplish this task, an understanding of garlic seed biology, especially the effect of environment on germination and seedling development is of high importance.
We studied the effect of stratification on seed germination and seedling development, as well as on plant morphology and architecture during the entire annual cycle of A. sativum. Under ambient Mediterranean conditions, only seedlings raised from stratified seeds underwent transition to the reproductive stage and formed bulbs. We therefore propose that cold stratification and vernalization share common mechanisms that affect phase transitions and reproductive success during the entire life cycle of the herbaceous perennial plant.
2. Materials and Methods
2.1. Plant Material and Growth Conditions
True hybrid garlic seeds (Allium sativum L. 96 × 87) were harvested in July 2016 and 2018, from plants grown in a living collection at the Agricultural Research Organization (ARO), Volcani Center, Israel. Upon ripening, seeds were threshed, cleaned, and sorted by weight using a Zigzag type 1 Selecta Gravity Seed Separator (The Netherlands), and stored in paper bags in an open-sided shed. Stratification experiments, including both morpho–physiological and transcriptome analyses, were performed in the 2016/17 and 2018/19 growing seasons. Seeds of the same batches were used in the comparative experiments and sown at the same day.
2.2. Pre-Sowing Treatments and Germination Test
The seeds of the same batch were first disinfected by immersion in 6% sodium hypochlorite for 10 min. Thereafter, one group of seeds were dipped in 0.2% Merpan fungicide (Captan) solution (Makhteshim Chemicals Ltd., Beer Sheva, Israel) solution for 10 min followed by 8 h imbibition in sterilized tap water. For stratification, disinfected seeds were mixed with moist vermiculite, and stratified in darkness at 4 °C for eight weeks. Moisture was maintained by controlled watering with 0.2% Merpan.
Treated seeds were sown in Petri dishes on MS medium mixed with 0.2% sucrose and 0.08% agar, in three replicates, 20 per replicate. Each Petri dish was wrapped in polystyrene linear low-density polyethylene (LLDPE) film and then wrapped in aluminum foil, for dark incubation at 20 °C. Germination percentage was recorded every four days for 50 days, and was determined by when the radicle reached a length of 4 mm.
2.3. Growth and Development In Situ
Two hundred treated seeds were sown in 40 L plastic boxes containing 80% ground coconut husk: 10% Styrofoam: 10% compost (Even Ari, Israel) in triplicates. The boxes were placed in an insect-proof 50-mesh screen-house providing 30% shade, at ARO. Regular fertigation consisted of “Shefer” liquid fertilizer (N:P:K = 59:35:94 g L−1, Dshanim, Haifa, Israel). Temperature and photoperiod records during plant growth are presented in Supplementary Figure S1. Emerging seedlings were counted when the cotyledon was visible above ground. Records were taken once every four days, for 60 days. At the end of each growing period, all plants were harvested, and measurements taken on bulb and neck diameter in cm, in order to calculate the bulbing index (BI = bulb diameter/neck diameter) [26].
2.4. Statistical Analysis
A completely randomized one-way analysis of variance (ANOVA) with mean separation by the Tukey–Kramer test was applied using the general linear model (GLM) procedure SAS software package JMP version 12 Pro (SAS Institute Inc., Cary, NC, USA, 1989–2007). Logistic regression was used to distinguish differences in germination and emergence percentages between treatments.
2.5. Histological Studies
Prior to sowing, 50 seeds per treatment were fixed in FAA solution (formaldehyde: acetic acid: ethanol 70%, 10:5:85, v/v/v). Fixation was followed by an ethanol dilution series (50, 70, 95, 100%; 1 h each and 100% over night) and then by a subsequent stepwise exchange of ethanol with X-tra solvent (Xylene substitute). Samples were embedded in paraffin and sliced by microtome (Leica RM2245, Leica Biosystems, Buffalo Grove, IL, USA) into 12 µm sections, then deparaffinized twice with X-tra solvent for 12 min, rehydrated (ethanol 100, 95, 70 and 50%, 2 min each), stained with Safranin O (1% w/v) for 30 min, washed with tap water and dehydrated in ethanol (50, 70, 2 × 95%, 30 sec each) and fast green (0.3% in 95% EtOH 1:1 clove oil) for 4 min. The stained samples were examined under a light microscope (Leica DMLB, Leica Microsystems GmbH, Germany) at ×20 and ×40 magnifications. Photos were made with a Nikon DS-fi1 camera and analyzed using NIS Elements software (Nikon Corporation, Tokyo, Japan).
2.6. Tissue Sampling for RNA/DNA Analyses and Extraction Procedures
Two replications of 50 seeds each, either dry, stratified, or imbibed, were sampled for nucleic acid analyses prior to sowing. The seeds were dipped in liquid nitrogen and stored at −80 °C.
Total RNA was extracted using the CTAB protocol [27], and extract quality was assessed using an Agilent 2100 Bioanalyzer (Agilent, Santa Clara, CA, USA). Only extracts with a minimum RNA integrated value of seven were used for analysis. This resulted in six libraries. The pairs of replication libraries were compared and the data was averaged for further evaluation.
2.7. Transcriptome Assembly
Library preparation and sequencing were performed at the Genome Center, Life Sciences and Engineering, Technion, Israel Institute of Technology, Haifa, Israel. Six libraries of paired-end RNA sequences of 100 nucleotides were prepared for analysis using Illumina Hiseq2500 and Trueseq protocols. Raw reads were subjected to a cleaning procedure with the FASTX Toolkit (http://hannonlab.cshl.edu/fastx_toolkit/index.html, version 0.0.13.2) as follows:
- (1)
- Trimming read-end nucleotides with quality scores <30 using fastq_quality_trimmer;
- (2)
- Read pairs were discarded if either one had less than 70% base pairs with quality score ≤ 30 as determined using fastq_quality_filter.
Following processing and cleaning, a total of ca. 263 million cleaned paired-end reads were assembled de novo, using Trinity software (Trinityrnaseq version v2.3.2) [28] with default parameters and a Trimmomatic option [29]. The resulting data were compiled using the transcriptome catalog of fertile garlic [26]. The newly assembled catalog contained a total of 120,319 Trinity ‘genes’ with an average length of 909.6 bp and N50 of 1481 bp. The entire catalog has been deposited in the NCBI (National Center for Biotechnology Information) Sequence Read Archive (SRA) database as bioproject PRJNA647152.
2.8. Abundance Estimation and Differential Expression Analysis
The cleaned reads from each library were aligned with the newly assembled transcriptome, using the Bowtie2 aligner [30]. The abundance estimation was calculated using the Trinity protocol [31] and the Expectation–Maximization method (RSEM). The normalized expression was calculated by TMM (trimmed mean of M values) normalization and by FPKM (fragments per kilobase of transcript per million mapped reads).
Comparisons between each pair of samples and differential expression analyses of the sequence count data were performed by the Bioconductor edgeR package in the R environment [32]. Differential expression was defined as a value larger than a two-fold difference in transcription expression with a false discovery-corrected statistical significance below 0.05. The differentially expressed transcripts with a minimum of one pairwise comparison between treatments were examined, using cluster analysis. Based on the means of two replications, hierarchical clustering of transcripts and samples was performed, and clusters were extracted using R scripts.
2.9. Functional Annotations
The assembled transcriptome was used for a search of the NCBI non-redundant (nr) protein database, employing DIAMOND software [33]. Homologous sequences were identified within the Swiss-Prot database using the BLASTX tool [34] at E-value threshold of 10−5. The results were exported to Blast2GO version 4.0 [35] for gene ontology (GO) assignments.
The KAAS tool (KEGG Automatic Annotation Server http://www.genome.jp/tools/kaas) was used for KEGG ontology and KEGG pathway assignments. Gene ontology enrichment analyses were carried out using Blast2GO [35] software based on Fisher’s Exact Test [36] with multiple testing corrections of false discovery rates (FDR) [37]. The threshold was set as FDR with corrected P-value smaller than 0.05. The REViGO web server, which sorts out long lists of gene ontology terms and summarizes them by removing redundant GO terms, was used for visualization of the GO terms in a semantic similarity-based scatterplot (http://revigo.irb.hr) [38].
3. Results
3.1. Effect of Stratification on Seed Germination and Seedling Phenology
On the day of sowing, there were no histological and morphological differences in seed structure between imbibed and stratified seeds. The seeds had normal embryos, with no deformations or degenerated tissues (Figure 1A). The embryonic SAM was visible (Figure 1B), and in some seeds from both groups initial differentiation of the first leaf primordium was evident (Figure 1C).
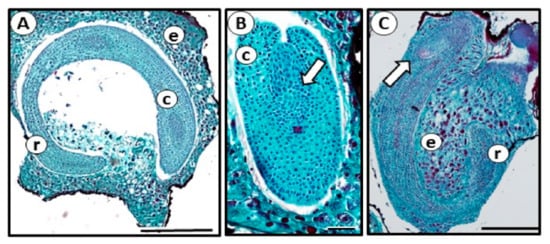
Figure 1.
Representative microphotographs of garlic seed structure. Seeds were either imbibed for eight hours (A,B) or stratified at 4 °C for eight weeks (C). No structural differences were visible. e—endosperm, r—radicle, c—cotyledon. (A) Longitudinal section just before sowing. Note dense meristematic area in the upper part of cotyledon. Bar= 0.5 mm. (B) Cross-section via upper part of cotyledon. Embryonic SAM (arrow) is visible. Bar = 0.1 mm. (C) Longitudinal section of the seed, SAM differentiates the first leaf primordium (arrow). Bar = 0.5 mm.
In comparison with imbibed seeds, stratification clearly enhanced germination and emergence, in both Petri dishes and the planting mixture (Figure 2). During juvenile development, stratification had no effect on leaf number, but it significantly affected leaf elongation. Leaf length in two-month-old seedlings raised from imbibed seeds was 7.7 ± 0.6 cm, compared with 10.9 ± 1.0 cm in stratified seedlings (data not shown).
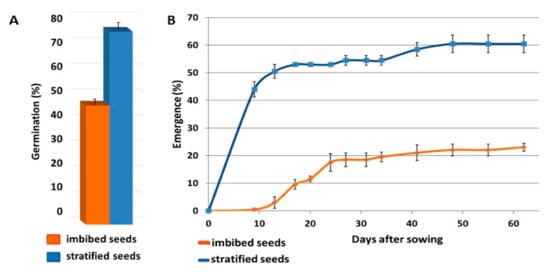
Figure 2.
Germination and emergence of imbibed and stratified A. sativum seeds. Seeds from the July 2016 harvest were either imbibed for eight hours or stratified at 4 °C for eight weeks. Bars represent Standard eror (SE) (A) Germination percentage in Petri dishes after 50 days at 20 °C in dark. n = 2 (20 seeds/replicate). (B) Emergence rate in growing boxes during the first 60 days after sowing. n = 2 (200 seeds/replicate).
Five-month-old plants grown from imbibed seeds produced approximately 7–9 prostrate leaves, compared with 7–8 erect leaves in plants from stratified seeds (Figure 3A). The latter formed bulbs and flower stems, while the apical meristem of the plants raised from non-stratified seeds remained vegetative and new leaf production continued (Figure 3B). No bulbing, branching, or flowering were evident in these plants throughout the experiment.
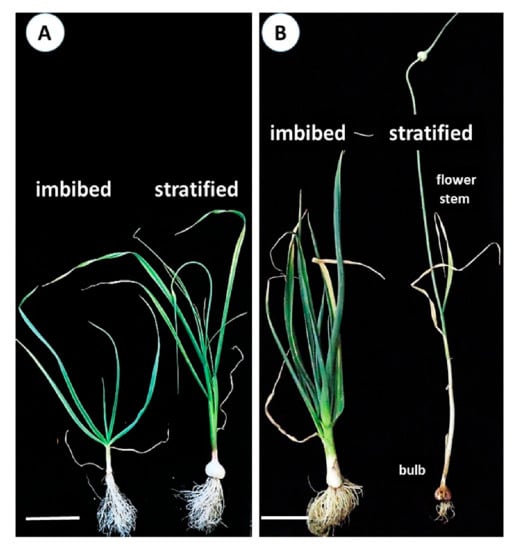
Figure 3.
Effect of seed stratification on the phenotypic expression of garlic seedlings. Seeds were either imbibed for eight hours or stratified for eight weeks at 4 °C prior to sowing in November 2018. (A) Five-month old plants raised from imbibed and stratified seeds, March 2019, bar = 7 cm. (B) Seven-month old plants, May 2019. Plants from imbibed seeds had 14–15 foliage leaves, no flower stem and/or bulb; stratified plants at the end of their annual cycle had dried-out leaves, with a defined bulb, elongated flower stem, and developed inflorescence. Bar = 10 cm.
Early in June, the plants were harvested for morphological evaluation. All plants raised from stratified seeds produced mature dormant bulbs up to 4–6 cm in diameter with closed necks, thus ranked as BI = 3 or 4+ (Figure 4). No bulbs were formed by any plant raised from imbibed seeds.
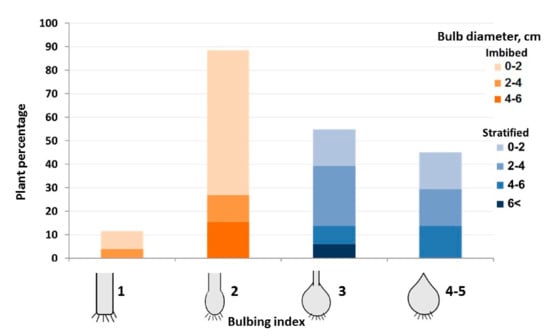
Figure 4.
Effect of seed imbibition and stratification on bulbing index (BI) and bulb size in garlic hybrid 96 × 87. Sown November 2016, harvested June 2017. Orange columns represent plants raised from imbibed seed. Blue columns represent bulbs obtained from stratified seeds. n = 200.
3.2. Global Transcriptome Analysis of Garlic Seeds
The high-throughput parallel RNA-Seq libraries extracted from dry, imbibed, and stratified garlic seeds, yielded 19.8 million 100-bp pair-end reads. Following quality trimming and filtration 16.7 million clean reads were generated and then assembled with the published garlic transcriptome catalog [39] (NCBI bioproject PRJNA243415) using Trinity software. The combined data produced a new comprehensive catalog of about 120,000 Trinity ‘genes’ with a mean length of 909.6 bp and N50 of 1481 bp.
The global gene expression profiles show sharp increases in the number of genes expressed as a result of both imbibition and stratification, compared to dry seeds (Figure 5A).
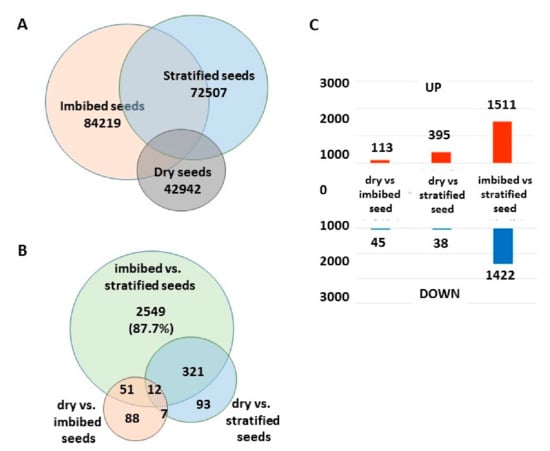
Figure 5.
Comparison between the total transcript distribution and the common differentially expressed genes (DEGs) in dry, imbibed, and stratified seeds of A. sativum. (A) Total number of expressed genes in the three seed samples. (B) Distribution of 2933 differentially expressed genes in three comparisons between samples. (C) Upregulation and downregulation of gene expression in the comparisons between the three seed samples.
The total number of expressed genes was relatively low in dry seeds, about 43,000 transcripts, out of which approximately 40% is shared with the imbibed and stratified seeds. Seed imbibition and stratification led to a doubling of the total number of transcripts to about 84,000 and 72,000, respectively, but only half of the expressed genes were common to both imbibed and stratified seeds (Figure 5A). In the comparison between imbibed and stratified seeds, 2933 differentially expressed genes (DEGs) were found (Figure 5B), of which more than half, 1511, were upregulated in imbibed seeds compared to stratified seeds (Figure 5C).
Functional analysis of the 2933 DEGs revealed enrichment of 24 and 160 biological processes in imbibed and in stratified seeds, respectively (Figure 6). The main enriched terms in imbibed seeds were related to lipid storage, cellular biosynthesis, and to regulation of DNA endoreduplication (Figure 6A). In stratified seeds, the biological processes with significantly enriched terms included DNA replication, cell cycle, glucose metabolism, meristem development, epigentic chromatin silencing, photosynthesis carbon fixation, and more (Figure 6B). These terms are all associated with enhanced plant metabolism and growth.
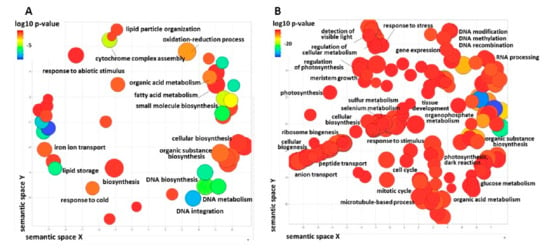
Figure 6.
Biological processes in imbibed and in stratified seeds of A. sativum revealed by GO term distribution analysis using Blast2Go and REViGO algorithms. GO terms are represented by circles and are plotted according to semantic similarities to other GO terms (adjoining circles are most closely related). Circle size is proportional to the abundance of the GO terms within a cluster, while color differences indicate semantic dissimilarities. Only GO terms with higher than 1% frequency are presented. (A) Enriched biological processes in imbibed seeds. (B) Enriched biological processes in stratified seeds.
3.3. Differential Expression of the Genes Associated with Seed Germination
Seed stratification significantly enhanced germination, emergence, and seedling vigor (Figure 2). This physiological activity is associated, on the transcriptome level, with intensive gene expression (Figure 6B). Hence, differential expression of genes associated with breaking dormancy, embryo development, sugar transport, and cell proliferation processes was expected. Data mining for functional annotations narrowed the list of 2933 DEGs between imbibed and stratified seeds to 38 DEGs (Supplementary Table S1). The patterns of expression of the individual gene functions were compared between dry seeds, imbibed seeds, and stratified seeds (Figure 7).
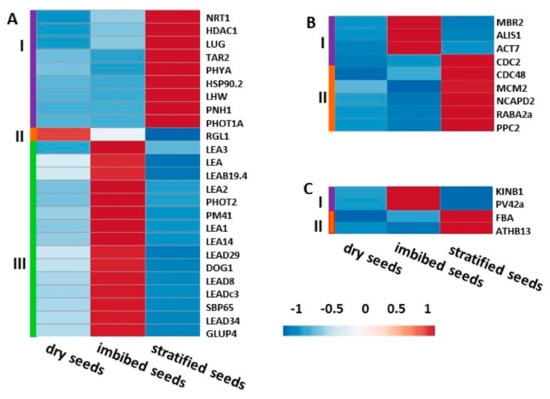
Figure 7.
Hierarchical cluster analysis of gene expression patterns in dry, imbibed, and stratified garlic seeds. Unit variance scaling is applied to FPKM expression values. Genes are clustered using Pearson correlation distance and average linkage. Main clusters are shown by colors and Roman numerals. (A) Genes associated with seed dormancy and germination. (B) Genes associated with cell proliferation and division. (C) Key genes in glycolysis and glucose transport.
The homologs of genes associated with seed dormancy and with germination are grouped in three clusters (Figure 7A). With a single exception, the lowest expression of all DEGs was recorded in dry seeds. Genes in cluster I (Figure 7A, violet) have relatively low expression in both dry and imbibed seeds and are over-expressed in stratified seeds. This cluster includes homologs of TRYPTOPHAN AMINOTRANSFERASE-RELATED PROTEIN 2 (TAR2), which is involved in the auxin signaling pathway that regulates embryo and seedling growth, PHOTOTROPIN 1A (PHOT1A), a blue light photoreceptor responsible for phototropism during germination and PHYTOCROME A (PHYA), which enhances germination even under low light. HEAT SHOCK PROTEIN 90.2 (HSP90.2) is also presented in this cluster.
Cluster II (Figure 7A, orange) has only the single gene RGA-like1 (RGL1), which is involved in repression of gibberellin-induced germination. This gene exhibits a unique pattern in that it is highly expressed in dry seeds, poorly expressed in imbibed seeds, and barely expressed in stratified seeds, corresponding inversely to the germinability of the respective seed lots.
The largest cluster III (Figure 7A, green) consists of genes upregulated only in imbibed seeds. Many members of the family of LATE EMBYOGENESIS ABUNDANT PROTEINS (LEA), associated with seed development and stress tolerance, share this cluster. Notably, DELAY OF GERMINATION1 (DOG1), known to regulate dormancy and juvenility, and GLUTELIN PRECURSOR 4 (GLUP4) involved in storage protein trafficking in seeds, are also overrepresented only in imbibed seeds.
Genes associated with cell proliferation and division, are clustered into two groups. Cluster I (Figure 7B, violet) is highly expressed only in imbibed seeds, and includes E3 UBIQUITIN- LIGASE MBR2-LIKE (MBR2) involved in cell proliferation and cell division. Cluster II (Figure 7B, orange) is overrepresented in stratified seeds and includes, among others, CELL DIVISION CONTROL 2 HOMOLOG A- LIKE (CDC2) and MINICHROMOSOME MAINTENANCE PROTEIN 2 (MCM2), associated with cell proliferation processes.
DEGs associated with glycolysis, SNF1-RELATED KINASE REGULATORY SUBUNITS (KINB1) and SNF1-RELATED PROTEIN KINASE REGULATORY SUBUINT GAMMA-LIKE (PV42A), are involved in the signal transduction cascade and carbohydrate metabolism. These DEGs are upregulated only in imbibed seeds. HOMEOBOX-LEUCINE ZIPPER (ATHB13), a transcription factor in the sucrose-signaling pathway, and FRUCTOSE-BIPHOSPHATE ADOLASE (FBA), which plays a key role in glycolysis and gluconeogenesis, are overrepresented only in stratified seeds (Figure 7C). Enrichment in carbon metabolism and glycolysis/gluconeogenesis in stratified seeds was confirmed by KEGG analysis (Supplementary Figure S2).
3.4. Differential Expression of Meristem Identity Genes in Germinating Seeds
Pre-sowing seed treatments greatly affected plant phenotype throughout the growth period. From seeding to maturation, plants from both treatments experienced the same environment, hence the differences in meristem transition are attributed to the pre-sowing embryonic meristematic response to seed treatment. Data mining for meristem identity genes, including those associated with meristem transition, vernalization, photoperiod, circadian clock and flowering, resulted in 26 DEGs between imbibed and stratified seeds (Supplementary Table S1). The two pre-sowing treatments had a significant effect on their activity (Figure 8).
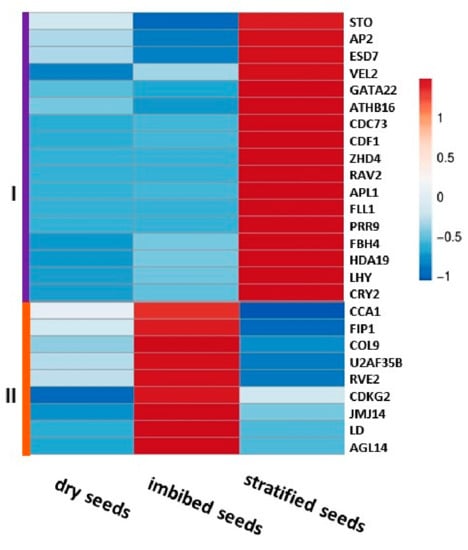
Figure 8.
Hierarchical cluster analysis of the meristem identity genes’ expression patterns in dry, imbibed and stratified garlic seeds. Unit variance scaling is applied to the FPKM (fragments per kilobase of transcript per million mapped reads) expression values. Genes are clustered using Pearson correlation distance and average linkage; main clusters are shown by colors and Roman numerals.
Cluster analysis revealed two DEGs modules. Cluster I (Figure 8, violet) is highly expressed only in stratified seeds. Some genes of this cluster are directly associated with flowering and photoperiodic control. The cluster includes, among others, a homolog of SALT TOLERANCE –LIKE (STO), which in Arabidopsis downregulates FLC and upregulates the key flowering-time genes FT and SOC1. In addition, RELATED TO ABI3/VP1 2 (RAV2) and VERNALIZATION5/VIN3-like PROTEIN 2 (VEL2), involved in both the vernalization and photoperiod pathways, and HOMEOTIC APETALA 2-LIKE (AP2) that plays a role in floral transition are present in this group. CRYPTOCHROME (CRY2) promotes the expression of FT in vascular bundles and is negatively regulated by FLC in Arabidopsis. TWO-COMPONENT RESPONSE REGULATOR 9 (PRR9) controls photoperiodic flowering response and temperature compensation, and FLOWERING BHLH 4 (FBH4) positively regulates CO transcription for photoperiodic flowering.
Cluster II (Figure 8, orange) is over-represented only in imbibed seeds. Two of these genes act as repressors in the photoperiod pathway of Arabidopsis, e.g., CONSTANS-LIKE 9-like (COL9) and JUMONJI 14 (JMJ14). AGAMOUS-LIKE14 (AGL14), a key player in at the complex gene regulatory network underlying SAM transitions promoting flowering in response to multiple signals, is also included in this module.
Several major components of the circadian clock are differentially expressed in response to the pre-sowing treatments. PHYTOCHROME A (PHYA) and CRYPTOCHROME2 (CRY), as well as oscillator output CYCLIC DOF FACTOR1 (CDF1) and the morning component of the circadian clock LATE ELONGATED HYPOCOTYL (LHY) were upregulated in stratified seeds. On the other hand, CIRCADIAN CLOCK ASSOCIATED1 (CCA1), usually co-expressed with LHY, showed the opposite pattern—it was highly expressed in imbibed seeds.
4. Discussion
Following germination, seedlings undergo several developmental phases, and the apical meristem gradually acquires competence for flowering. It is generally accepted that this process begins in the juvenile stage and is regulated by the miR156-SPL-miR172 cascade [2,3]. The plant’s life cycle, however, is not a simple sequence of separate developmental events, but a chain of interactive linked processes, regulated by short- and long-term mechanisms. In this context, the role of the embryonic meristem in phase transitions and further development of the adult plant is still not clear.
Our results show that germination, flowering, and bulbing of A. sativum are tightly connected and are affected by similar environmental cues. Seed stratification promoted not only germination and emergence (Figure 1 and Figure 2), but also had a long-term effect on meristem transition in adult plants (Figure 9).
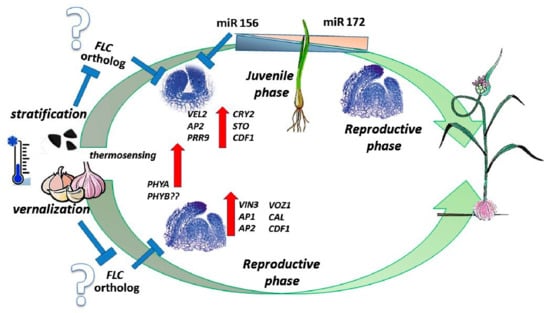
Figure 9.
Effect of cold period on seeds (stratification) or bulbs (vernalization) of Allium sativum. Putative flower repressor(s) (FLC orthologs) are downregulated by cold in both embryonic and adult meristems. In seedling development, acquisition of flowering competence depends on the miR156-miR172 cascade [2] and meristem enlargement, while in bud development the meristem is ready for phase transition to flowering [63].
4.1. Seed Germination is Enhanced by Low Temperatures
Similar to other Allium species [40], when seed moisture content decreases upon maturation, garlic seeds become dormant [20]. Gene expression in dry seeds is rather low, but one specific gene, a homolog of RGL1, was highly expressed. RGL1 belongs to the DELLA group (GAI, RGA, RGL1, and RGL2), and participates in coding for large multiprotein complexes that repress transcription of GA-inducible genes. In Arabidopsis it enhances RGL2 repression of germination and is downregulated by imbibition [41]. We have now provided evidence of a significant decrease in RGL1 expression by cold stratification, in garlic (Figure 7). We therefore propose RGL1 as a possible marker for seed dormancy and embryo meristem activation, but its role in the genetic control of seed germination has to be further investigated.
Imbibition of the mature seed reactivates a variety of metabolic systems, such as mitochondrial biogenesis and the synthesis of new components, required for renewed cell expansion and cell division [42]. In onion, imbibition prompted the swelling of storage tissues, elongation of embryo cells, and initiation of sugar metabolism [43]. Our histological observations showed structural similarities between imbibed and stratified garlic seeds, including their embryonic meristems producing a single leaf primordium prior to germination (Figure 1). However, this morphological similarity did not correlate with germination and emergence. As in many species from temperate regions [44], cold treatment induced uniform and rapid germination (Figure 2). Water uptake doubled the number of total expressed genes, compared with dry seeds (Figure 5). Similar to other plant species [45], imbibition facilitates the initiation of carbohydrate and lipid metabolism. Dry seeds harbor most of their cells’ activities at the G1-phase, and imbibition reactivates meristematic cells into the cell cycle in preparation for seedling establishment [46]. We found that DNA endoreduplication and DNA metabolism processes are enriched in imbibed garlic seeds, and that active metabolism restarts prior to germination sensu stricto (Figure 6A).
The expression of DOG1, a homolog of the major dormancy gene, especially drew our attention due to its dual role in seed germination [47] and in flowering control [48]. It is also involved in juvenility regulation governed by the miR156-SPL-miR172 cascade and the age-dependent flowering pathway [49]. In garlic, however, seed imbibition led to DOG1 over-expression, while after stratification its expression decreased. This DOG1 expression may precede the initiation of epigenetic silencing [50], or act to delay the germination of imbibed seeds, enabling pre-germination processes to complete.
Following an eight-week cold period, metabolic and developmental activities increased considerably (Figure 6B). These were associated with cell cycle, sugar metabolism (e.g., FBA), cell division, and proliferation (e.g., CDC2, MCM2), as expected when storage substances gradually break down to support metabolic processes during embryo development and growth [51,52].
We conclude that initially seed imbibition reactivates the embryonic cell cycle, initiates metabolism (KINB1 and PV42A) and stress tolerance (as evidence by the large group of LEA genes, Figure 7A), and primes garlic seed in support of germination. Further, wet cold stratification serves two functions: it provides an ample supply of water to the dry seeds and the chilling initiates a number of biological pathways. These include enhanced carbon metabolism, desired DNA modifications, biosynthesis, cellular transport, and tissue development (Figure 6, Figure 7 and Figure 8). On a physiological level, this dynamic is translated into rapid and synchronized radicle protrusion, followed by seedling emergence.
4.2. Embryonic Meristem Stratification Affects Phase Transitions in Adult Plant
Information on the long-term effect of seed treatments on the entire annual cycle and especially on phase transition is scarce. Most research is focused on the acquisition of flowering competence by juvenile and vegetative adult meristems [2,3,53], while embryonic SAM is regarded as “responsible” only for germination. We found that a long period of moist chilling of garlic seeds contributes to meristem transition from vegetative to reproductive state, even without vernalization of the adult plant. Phenotypic differences between seedlings produced from imbibed and stratified seeds were obvious after three months of growth (Figure 3), and only plants raised from stratified seeds formed flower stems and mature bulbs six to seven months after sowing (Figure 4). Bulb formation was independent of plant size (Figure 4), and the dramatic differences between plants raised from imbibed vs. stratified seed should thus be attributed to meristematic changes on the molecular level, initiated by the stratification of the embryonic SAM.
In adult plants, meristem transition from the vegetative to the reproductive state is coded by a multifaceted gene network involving photoperiod, gibberellins, vernalization, and autonomous signaling modules [54,55]. In the vernalization pathway, core transcription factors such as FLC in Brassicaceae, and VRN1 and VRN2 in cereals act as main repressors of flower transition [56,57,58]. However, since climate dependence evolved several times in the history of plants, the occurrence of different regulatory mechanisms in various plant groups is not surprising [1,5]. Thus, in Beta vulgaris the FLC homolog does not play a major role in the control of vernalization response [59]. The discovery of alternative mechanisms of flowering in wheat, maize, and rice, suggest that monocots may also be FLC-independent for flower initiation [58]. Bulbous monocots probably evolved different FLC-dependent pathways, which modulate the activity of other genes that might act as cold-regulated flowering repressors [60]. It is possible, therefore, that the effect of cold on garlic embryonic and adult meristems is associated with downregulation of a specific repressor(s) of meristem transition to flowering. We found that two homologs of the known flowering repressors COL9, a repressor of CO and FT in Arabidopsis [61], and JMJ14, whichrepresses FT and TSF [62], were expressed in imbibed seeds, but downregulated by cold stratification. The same patterns were found in a few other meristem-identity genes (e.g., LD, FIP, and AGL14). Perhaps one or more of these genes are putative FLC ortholog(s) that act as repressors of meristem phase transition in garlic (Figure 9). Further research, employing more sensitive tools (e.g., transformation systems or genome sequencing), is required to find specific flower repressors in geophyte species.
In adult garlic plants, the molecular cascade of vernalization includes, to some extent, genes coding for the photoperiodic pathway [63]. The latter are homologs of Arabidopsis STO (which has a major role in response to light and photoperiodism), LHY (circadian rhythm and photoperiodism), FKF1 (regulation of circadian clock-dependent transition to flowering time), VOZ1 (promotion flowering downstream of phytochrome B (phyB), and CKA1 and CKA2 (circadian rhythm). In lily, homologs of flowering genes FT, VRN1, VRN2, SOC1, and genes of circadian rhythm were differentially expressed in vernalized bulbs, in comparison with non-vernalized [64], while during floral induction in tulip bulbs 57 homologs of Arabidopsis flowering genes were generated, including those associated with photoperiodism [65]. Similar to the results found in adult geophyte plants, our results for stratified garlic seeds show abundant processes related to meristem development, photosynthesis, light reaction, and over-representation of genes involved in photoperiodic flowering response (e.g., CRY2, PRR9, FBH4), as well as downregulation of photoperiodic repressors (e.g., JMJ14, COL9) (Figure 6 and Figure 9).
We conclude that in garlic the processes associated with chilling-dependent meristem transition, such as increased sugar metabolism, molecular SAM activation, and an upsurge in photoperiod-associated genes, are similar for both seeds and buds/bulbs [63]. Similar phenomena were reported for peach [6], almonds [66], Persian walnut [67], and other fruit trees. Although initial genetic work on model plants identified independent molecular mechanisms for stratification [68,69] and vernalization [70,71], recent studies suggest that overlapping molecular pathways may indeed coordinate seed dormancy and vernalization-induced flowering [3,49,72,73,74].
Embryonic and adult SAMs differ in function, size, and genetic regulation. In garlic clove, prior to vernalization, the adult SAM has already produced several leaf primordia, and is morphologically ready for the flower transition [63]. In contrast, the embryonic SAM is very small (Figure 1) and has to reach maturity (formation of 7–8 leaves and more [75]) prior to achieving flowering competence (Figure 9). It is obvious that a juvenile garlic plant has to reach a critical mass and physiological age prior to acquisition of flower competence, and the pivotal role of miR156-miR172 interplay in this process has to be further investigated.
We propose that the embryonic SAM functions not only in seed germination and emergence, but plays an important role in the entire plant life cycle, providing the foundation for the molecular regulation of major functional transitions. The similarities we have highlighted between seed and bud chilling requirements and responses shed light on the evolution of cold-mediated plant development. Future research should address shared genetic pathways involved in the response to cold at the two separate yet critical life stages. The study of molecular mechanisms that regulate the embryonic meristem, including the epigenetic memory of the cold winter experienced by seeds, will help to elucidate successful flowering and bulb formation in adult geophytes.
Supplementary Materials
The following are available online at https://www.mdpi.com/2073-4395/10/11/1651/s1, Figure S1: Daylength and average air temperatures (based on 8 measures/day) in Bet Dagan, Israel during growth season 2015–2016; Figure S2: Pathways of glycolysis and gluconeogenesis in imbibed and stratified garlic seeds as revealed by KEGG algorithm osa00010. Over-representation in stratified seeds is marked in red, over-representation in imbibed seeds is marked in blue; Table S1: List of 61 homologous genes associated with seed dormancy and germination, proliferation, glycolysis and glucose transport and meristem identity in seeds of Allium sativum. Short description according to UniProt database and literature survey.
Author Contributions
Conceptualization, T.E.B.M., R.K.G. and H.D.R.; methodology, investigation, T.E.B.M., L.R., A.F., E.S.-M., I.F.; writing—original draft preparation, T.E.B.M.; writing—review and editing, H.D.R., R.P., J.D.K. and R.K.G. All authors have read and agreed to the published version of the manuscript.
Funding
This research was partly supported by Classeed LTD.
Acknowledgments
The authors thank Hanita Zemah (ARO, Israel) for sharing her expertise in histology.
Conflicts of Interest
The authors declare no conflict of interest.
Data Availability:
The data deposited in the NCBI Sequence Read Archive (SRA) database, bioproject RJNA647152 https://www.ncbi.nlm.nih.gov/sra/?term=PRJNA647152.
References
- Bouché, F.; Woods, D.P.; Amasino, R.M. Winter memory throughout the plant kingdom: Different paths to flowering. Plant Physiol. 2017, 173, 27–35. [Google Scholar] [CrossRef]
- Trindade, I.; Schubert, D.; Gaudin, V. Epigenetic Regulation of Phase Transitions in Arabidopsis thaliana. In Plant Epigenetics; Rajewsky, N., Jurga, S., Barciszewski, J., Eds.; Springer: Cham, Germany, 2017; pp. 359–383. [Google Scholar]
- Huijser, P.; Schmid, M. The control of developmental phase transitions in plants. Development 2011, 138, 4117–4129. [Google Scholar] [CrossRef]
- Finch-Savage, W.E.; Leubner-Metzger, G. Seed dormancy and the control of germination. New Phytol. 2006, 171, 501–523. [Google Scholar] [CrossRef]
- Andrés, F.; Coupland, G. The genetic basis of flowering responses to seasonal cues. Nat. Rev. Genet. 2012, 13, 627–639. [Google Scholar] [CrossRef]
- Leida, C.; Conejero, A.; Arbona, V.; Gomez-Cadenas, A.; Llácer, G.; Badenes, M.L.; Ríos, G. Chilling-dependent release of seed and bud dormancy in peach associates to common changes in gene expression. PLoS ONE 2012, 7, e35777. [Google Scholar] [CrossRef] [PubMed]
- Vidigal, D.S.; Marques, A.C.; Willems, L.A.; Buijs, G.; Méndez-Vigo, B.; Hilhorst, H.W.; Bentsink, L.; Picó, F.X.; Alonso-Blanco, C. Altitudinal and climatic associations of seed dormancy and flowering traits evidence adaptation of annual life cycle timing in Arabidopsis thaliana. Plant Cell Environ. 2016, 39, 1737–1748. [Google Scholar] [CrossRef] [PubMed]
- Rubin, M.J.; Friedman, J. The role of cold cues at different life stages on germination and flowering phenology. Am. J. Bot. 2018, 105, 749–759. [Google Scholar] [CrossRef] [PubMed]
- Khodorova, N.V.; Boitel-Conti, M. The role of temperature in the growth and flowering of geophytes. Plants 2013, 2, 699–711. [Google Scholar] [CrossRef]
- De Hertogh, A.; Le Nard, M. The Physiology of Flower Bulbs; Elsevier: Amsterdam, The Netherlands, 1993; p. 795. [Google Scholar]
- Duran-Nebreda, S.; Bassel, G.W. Plant behaviour in response to the environment: Information processing in the solid state. Philos. Tran. R. Soc. B. 2019, 374, 20180370. [Google Scholar] [CrossRef]
- Kamenetsky, R. Biodiversity of Geophytes: Phytogeography, Morphology, and Survival Strategies. In Ornamental Geophytes: From Basic Science to Sustainable Production; Kamenetsky, R., Okubo, O., Eds.; CRC Press: Boca Raton, FL, USA, 2012; pp. 57–75. [Google Scholar]
- Rabinowitch, H.D.; Kamenetsky Goldstein, R. Allium crops. In The Physiology of Vegetable Crops; Wien, C., Ed.; CABI: Wallingford, UK, 2020; pp. 581–619. [Google Scholar]
- Karjee, S.; Mahapatra, S. Physiological Studies of Ornamental Bulb Dormancy. Int. J. Curr. Microbiol. Appl. Sci. 2019, 8, 2305–2314. [Google Scholar] [CrossRef]
- Lee, R.; Baldwin, S.; Kenel, F.; McCallum, J.; Macknight, R. FLOWERING LOCUS T genes control onion bulb formation and flowering. Nat. Commun. 2013, 4, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Leeggangers, H.A.; Moreno-Pachon, N.; Gude, H.; Immink, R.G. Transfer of knowledge about flowering and vegetative propagation from model species to bulbous plants. Int. J. Dev. Biol. 2013, 57, 611–620. [Google Scholar] [CrossRef] [PubMed]
- Leeggangers, H.A.; Rosilio-Brami, T.; Bigas-Nadal, J.; Rubin, N.; van Dijk, A.D.; Nunez de Caceres Gonzalez, F.F.; Saadon-Shitrit, S.; Nijveen, H.; Hilhorst, H.W.; Immink, R.G.; et al. Tulipa gesneriana and Lilium longiflorum PEBP genes and their putative roles in flowering time control. Plant Cell Physiol. 2018, 59, 90–106. [Google Scholar] [CrossRef] [PubMed]
- Rees, A.R. Ornamental Bulbs, Corms and Tubers; CAB International: Wallingford, UK, 1992. [Google Scholar]
- Rouhi, H.R.; Shakarami, K.; Afshari, R. Seed treatments to overcome dormancy of waterlily tulip (Tulipa kaufmanniana Regel.). Aust. J. Crop Sci. 2010, 4, 718. [Google Scholar]
- Rozenblat, L. Seed Development and Germination Mechanisms in Allium Species. Master’s Thesis, The Hebrew University of Jerusalem, Jerusalem, Israel, 2017. [Google Scholar]
- Kirmizi, S.; Guleryuz, G.; Arslan, H. Effects of environmental and storage conditions on the germination of Allium species. Fresen. Environ. Bull. 2017, 26, 3470–3478. [Google Scholar]
- Bach, A.; Sochacki, D. Propagation of ornamental geophytes: Physiology and management systems. In Ornamental Geophytes: From Basic Science to Sustainable Production; Kamenetsky, R., Okubo, O., Eds.; CRC Press: Boca Raton, FL, USA, 2012; pp. 280–305. [Google Scholar]
- Ben Michael, T.; Shemesh-Mayer, E.; Kimhi, S.; Gershberg, C.; Forer, I.; de Ávila, V.T.; Rabinowitch, H.D.; Kamenetsky Goldstein, R. Temporal and spatial effect of low pre-planting temperatures on plant architecture and flowering in bolting garlic. Sci. Hortic. 2018, 242, 69–75. [Google Scholar] [CrossRef]
- Kamenetsky, R. Garlic: Botany and Horticulture. Hortic. Rev. 2007, 33, 123–172. [Google Scholar]
- Etoh, T.; Simon, P.W. Diversity, fertility and seed production of garlic. In Allium Crop Science—Recent Advances; Rabinowitch, H.D., Currah, L., Eds.; CABI Publishing: Wallingford, UK, 2002; pp. 101–117. [Google Scholar]
- Mann, L. Anatomy of the garlic bulb and factors affecting bulb development. Hilgardia 1952, 21, 195–251. [Google Scholar] [CrossRef][Green Version]
- Chang, S.; Puryear, J.; Cairney, J. A simple and efficient method for isolating RNA from pine trees. Plant Mol. Biol. Report 1993, 11, 113–116. [Google Scholar] [CrossRef]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q.; et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011, 29, 644–652. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Langmead, B.; Trapnell, C.; Pop, M.; Salzberg, S.L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009, 10, R25. [Google Scholar] [CrossRef] [PubMed]
- Haas, B.J.; Papanicolaou, A.; Yassour, M.; Grabherr, M.; Blood, P.D.; Bowden, J.; Couger, M.B.; Eccles, D.; Li, B.; Lieber, M.; et al. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat. Protoc. 2013, 8, 1494–1512. [Google Scholar] [CrossRef] [PubMed]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef] [PubMed]
- Buchfink, B.; Xie, C.; Huson, D.H. Fast and sensitive protein alignment using DIAMOND. Nat. Methods 2015, 12, 59–60. [Google Scholar] [CrossRef] [PubMed]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Conesa, A.; Götz, S.; García-Gómez, J.M.; Terol, J.; Talón, M.; Robles, M. Blast2GO: A universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 2005, 21, 3674–3676. [Google Scholar] [CrossRef]
- Upton, G.J. Fisher’s exact test. J. R. Stat. Soc. Ser. A 1992, 155, 395–402. [Google Scholar] [CrossRef]
- Benjamini, Y.; Hochberg, Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Supek, F.; Bošnjak, M.; Škunca, N.; Šmuc, T. REVIGO summarizes and visualizes long lists of gene ontology terms. PLoS ONE 2011, 6, e21800. [Google Scholar] [CrossRef]
- Kamenetsky, R.; Faigenboim, A.; Shemesh-Mayer, E.; Ben-Michael, T.; Gershberg, C.; Kimhi, S.; Esquira, I.; Rohkin Shalom, S.; Eshel, D.; Rabinowitch, H.D.; et al. Integrated transcriptome catalogue and organ-specific profiling of gene expression in fertile garlic (Allium sativum L.). BMC Genomics 2015, 16, 12. [Google Scholar] [CrossRef] [PubMed]
- Baskin, C.C.; Baskin, J.M. Seeds: Ecology, Biogeography and Evolution of Dormancy and Germination, 2nd ed.; Academic Press: San Diego, CA, USA, 2014. [Google Scholar]
- Cao, D.; Hussain, A.; Cheng, H.; Peng, J. Loss of function of four DELLA genes leads to light-and gibberellin-independent seed germination in Arabidopsis. Planta 2005, 223, 105–113. [Google Scholar] [CrossRef] [PubMed]
- Kermode, A.R. Regulatory Mechanisms in the Transition from Seed Development to Germination: Interactions between the Embryo and the. Seed Dev. Germination 1995, 41, 273. [Google Scholar]
- Brewster, J.L. Onions and Other Vegetable Alliums; CABI: Wallingford, UK, 2008; Volume 15. [Google Scholar]
- Baskin, J.M.; Baskin, C.C. A classification system for seed dormancy. Seed Sci. Res. 2004, 14, 1–16. [Google Scholar] [CrossRef]
- Tuan, P.A.; Sun, M.; Nguyen, T.N.; Park, S.; Ayele, B.T. Molecular mechanisms of seed germination. In Sprouted Grains; Feng, H., Nemzer, B., DeVries, J.W., Eds.; AACC International Press: Washington, DC, USA, 2019; pp. 1–24. [Google Scholar]
- Vázquez-Ramos, J.M.; de la Paz Sánchez, M. The cell cycle and seed germination. Seed Sci. Res. 2003, 13, 113. [Google Scholar] [CrossRef]
- Nonogaki, H. Seed biology updates-highlights and new discoveries in seed dormancy and germination research. Front Plant Sci. 2017, 8, 524. [Google Scholar] [CrossRef]
- Graeber, K.; Linkies, A.; Steinbrecher, T.; Mummenhoff, K.; Tarkowská, D.; Turečková, V.; Ignatz, M.; Sperber, K.; Voegele, A.; De Jong, H.; et al. DELAY OF GERMINATION 1 mediates a conserved coat-dormancy mechanism for the temperature-and gibberellin-dependent control of seed germination. Proc. Natl. Acad. Sci. USA 2014, 111, E3571–E3580. [Google Scholar] [CrossRef]
- Huo, H.; Wei, S.; Bradford, K.J. DELAY OF GERMINATION1 (DOG1) regulates both seed dormancy and flowering time through microRNA pathways. Proc. Natl. Acad. Sci. USA 2016, 113, E2199–E2206. [Google Scholar] [CrossRef]
- Chen, N.; Wang, H.; Abdelmageed, H.; Veerappan, V.; Tadege, M.; Allen, R.D. HSI2/VAL1 and HSL1/VAL2 function redundantly to repress DOG1 expression in Arabidopsis seeds and seedlings. New Phytol. 2020, 227, 840–856. [Google Scholar] [CrossRef]
- Ali, A.S.; Elozeiri, A.A. Metabolic processes during seed germination. Adv. Seed Biol. 2017, 141–166. [Google Scholar]
- Li, F.H.; Yu, P.; Song, C.H.; Wu, J.J.; Tian, Y.; Wu, X.F.; Zhang, X.W.; Liu, Y.M. Differential protein analysis of Heracleum moellendorffii Hance seeds during stratification. Plant Physiol. Biochem. 2019, 145, 10–20. [Google Scholar] [CrossRef] [PubMed]
- Hyun, Y.; Richter, R.; Coupland, G. Competence to flower: Age-controlled sensitivity to environmental cues. Plant Physiol. 2017, 173, 36–46. [Google Scholar] [CrossRef] [PubMed]
- Albani, M.C.; Coupland, G. Comparative Analysis of Flowering in Annual and Perennial Plants. In Current Topics in Developmental Biology; Timmermans, M.C.P., Ed.; Academic Press: Cold Spring Harbor, NY, USA, 2010; Volume 91, pp. 323–348. [Google Scholar]
- Fornara, F.; de Montaigu, A.; Coupland, G. SnapShot: Control of flowering in Arabidopsis. Cell 2010, 141, 550. [Google Scholar] [CrossRef]
- Song, J.; Irwin, J.; Dean, C. Remembering the prolonged cold of winter. Curr. Biol. 2013, 23, R807–R811. [Google Scholar] [CrossRef]
- Mateos, J.L.; Tilmes, V.; Madrigal, P.; Severing, E.; Richter, R.; Rijkenberg, C.W.; Krajewski, P.; Coupland, G. Divergence of regulatory networks governed by the orthologous transcription factors FLC and PEP1 in Brassicaceae species. Proc. Natl. Acad. Sci. USA 2017, 114, E11037–E11046. [Google Scholar] [CrossRef]
- Sharma, N.; Ruelens, P.; D’hauw, M.; Maggen, T.; Dochy, N.; Torfs, S.; Kaufmann, K.; Rohde, A.; Geuten, K. A flowering locus C homolog is a vernalization-regulated repressor in Brachypodium and is cold regulated in wheat. Plant Physiol. 2017, 173, 1301–1315. [Google Scholar] [CrossRef] [PubMed]
- Vogt, S.H.; Weyens, G.; Lefèbvre, M.; Bork, B.; Schechert, A.; Müller, A.E. The FLC-like gene BvFL1 is not a major regulator of vernalization response in biennial beets. Front Plant Sci. 2014, 5, 146. [Google Scholar] [CrossRef]
- Aljaser, J. Gladiolus Breeding for Rapid Generation Cycling for Potted Plant Production and the Discovery of Gladiolus Genes, UFC and FLX; University of Minnesota: Saint Paul, MN, USA, 2020. [Google Scholar]
- Cheng, X.F.; Wang, Z.Y. Overexpression of COL9, a CONSTANS-LIKE gene, delays flowering by reducing expression of CO and FT in Arabidopsis thaliana. Plant J. 2005, 43, 758–768. [Google Scholar] [CrossRef]
- Yang, W.; Jiang, D.; Jiang, J.; He, Y. A plant-specific histone H3 lysine 4 demethylase represses the floral transition in Arabidopsis. Plant J. 2010, 62, 663–673. [Google Scholar] [CrossRef]
- Michael, T.E.B.; Faigenboim, A.; Shemesh-Mayer, E.; Forer, I.; Gershberg, C.; Shafran, H.; Rabinowitch, H.D.; Kamenetsky-Goldstein, R. Crosstalk in the darkness: Bulb vernalization activates meristem transition via circadian rhythm and photoperiodic pathway. BMC Plant Biol. 2020, 20, 1–16. [Google Scholar] [CrossRef]
- Villacorta-Martin, C.; de Cáceres González, F.F.N.; de Haan, J.; Huijben, K.; Passarinho, P.; Hamo, M.L.B.; Zaccai, M. Whole transcriptome profiling of the vernalization process in Lilium longiflorum (cultivar White Heaven) bulbs. BMC Genomics 2015, 16, 550. [Google Scholar] [CrossRef] [PubMed]
- Leeggangers, H.A.; Nijveen, H.; Bigas, J.N.; Hilhorst, H.W.; Immink, R.G. Molecular regulation of temperature-dependent floral induction in Tulipa gesneriana. Plant Physiol. 2017, 173, 1904–1919. [Google Scholar] [CrossRef] [PubMed]
- Kester, D.E.; Raddi, P.; Asay, R. Correlations of chilling requirements for germination, blooming and leafing within and among seedling populations of almond. J. Am. Soc. Hortic. Sci. 1977, 102, 145–148. [Google Scholar]
- Aslamarz, A.A.; Vahdati, K.; Rahemi, M.; Hassani, D. Relationship between Seed and Bud Chilling Requirement of Persian Walnut. Acta Hortic. 2010, 861, 279–282. [Google Scholar] [CrossRef]
- Bentsink, L.; Jowett, J.; Hanhart, C.J.; Koornneef, M. Cloning of DOG1, a quantitative trait locus controlling seed dormancy in Arabidopsis. Proc. Natl. Acad. Sci. USA 2006, 103, 17042–17047. [Google Scholar] [CrossRef] [PubMed]
- Huang, D.; Xu, Y.; Hu, H. The changes of seed structure in Osmanthus americanus seed during cold stratification. J. Cent. South Univ. Technol. 2010, 4. Available online: https://en.cnki.com.cn/Article_en/CJFDTOTAL-ZNLB201004022.htm (accessed on 22 October 2020). (In Chinese).
- Simpson, G.G.; Dean, C. Arabidopsis, the Rosetta stone of flowering time? Science 2002, 296, 285–289. [Google Scholar] [CrossRef]
- Caicedo, A.L.; Stinchcombe, J.R.; Olsen, K.M.; Schmitt, J.; Purugganan, M.D. Epistatic interaction between Arabidopsis FRI and FLC flowering time genes generates a latitudinal cline in a life history trait. Proc. Natl. Acad. Sci. USA 2004, 101, 15670–15675. [Google Scholar] [CrossRef]
- Chiang, G.C.K.; Barua, D.; Kramer, E.M.; Amasino, R.M.; Donohue, K. Major flowering time gene, FLOWERING LOCUS C, regulates seed germination in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 2009, 106, 11661–11666. [Google Scholar] [CrossRef]
- Auge, G.A.; Blair, L.K.; Neville, H.; Donohue, K. Maternal vernalization and vernalization-pathway genes influence progeny seed germination. New Phytol. 2017, 216, 388–400. [Google Scholar] [CrossRef]
- Blair, L.; Auge, G.; Donohue, K. Effect of FLOWERING LOCUS C on seed germination depends on dormancy. Funct. Plant Biol. 2017, 44, 493–506. [Google Scholar] [CrossRef] [PubMed]
- Shemesh-Mayer, E.; Winiarczyk, K.; Błaszczyk, L.; Kosmala, A.; Rabinowitch, H.D.; Kamenetsky, R. Male gametogenesis and sterility in garlic (Allium sativum L.): Barriers on the way to fertilization and seed production. Planta 2013, 237, 103–120. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).